Abstract
Relatively high oxidative stress levels in the prostate are postulated to be a major factor for prostate carcinogenesis and prostate cancer (CaP) progression. We focused on elucidating metabolic pathways of oxidative stress generation in CaP cells. Previously, we showed that transcription factor JunD is essential for androgen-induced reactive oxygen species (ROS) production in androgen-dependent human prostate cancer cells. We also recently demonstrated that androgen induces the first and regulatory enzyme spermidine/spermine N1-acetyl transferase (SSAT) in a polyamine catabolic pathway that produces copious amounts of metabolic ROS. Here, we present co-immunoprecipitation and Gaussia luciferase reconstitution assay data that show JunD forms a complex with androgen-activated AR in situ. Our chromatin immunoprecipitation assay data demonstrate that JunD binds directly to a specific SSAT promoter sequence only in androgen-treated LNCaP cells. Using a vector containing a luciferase reporter gene connected to the SSAT promoter and a JunD-silenced LNCaP cell line, we show that JunD is essential for androgen-induced SSAT gene expression. The elucidation of JunD-AR complex inducing SSAT expression leading to polyamine oxidation establishes the mechanistic basis of androgen-induced ROS production in CaP cells and opens up a new prostate specific target for CaP chemopreventive/chemotherapeutic drug development.
Keywords: androgen receptor, AP1, protein-protein interaction, SSAT, reactive oxygen species
Introduction
Approximately 1–5% of the oxygen that we breathe in is converted to reactive oxygen species (ROS) such as hydrogen peroxide, superoxide, hydroxyl radical, etc (reviewed in 1). When cellular ROS production exceeds detoxification capacity, oxidative stress occurs (2,3). Damage to cellular proteins, DNA, RNA and phospholipids due to oxidative stress (4–9) as well as ROS molecules acting as signals for promoting cell proliferation (10) are believed to contribute to cancer development. The strongest evidence thus-far is ROS-induced oxidative damage products in human and transgenic mice prostatectomy specimens. Both mouse and human prostate tumor cells are reported to have higher ROS-induced macromolecular damages as compared to normal prostatic epithelium (11,12). Accumulating evidence shows that ROS play a key role in occurrence and recurrence of prostate cancer (CaP) as well as its progression from androgen-dependence to androgen-independence (11–15).
Androgen signaling is one source of ROS generation in prostatic epithelial cells (16,17). Androgen binding to androgen receptor (AR) initiates a cascade of events leading to ROS generation in prostate cells (17–19). We established that one pathway of androgen-induced oxidative stress involves activation of AP-1 transcription factor JunD (19, 20), followed by induction of enzyme spermidine/spermine N1-acetyl transferase (SSAT) that initiates a major polyamine oxidation pathway (21). As prostatic epithelia produce a large excess of polyamines, induction of polyamine oxidation could result in high ROS levels in the prostate. We have demonstrated that inhibiting androgen-induced ROS production using different small molecule inhibitors of the androgen signaling pathway or polyamine oxidation inhibits cell growth and androgen-induced ROS generation in cultured human CaP cells as well as tumor growth in TRansgenic Adenocarcinoma in the Mouse Prostate (TRAMP) model (21,22, and unpublished data). Since the SSAT gene promoter sequence contains no AR binding Element (ARE), the exact mechanism of how AR may induce SSAT gene expression remains unknown. To develop more potent CaP chemopreventive agents that can specifically block this pathway, we focused on elucidating the mechanism of androgen-induced SSAT gene expression.
We previously demonstrated that androgen activation of AR in LNCaP human CaP cells induces AP-1 transcription factors Fra-2 and JunD (19). However, only JunD levels and its functional activity remained elevated for 96h after androgen treatment, when androgen-induced oxidative stress was observed (17–20). JunD may either inhibit (23–25) or help (26) cellular ROS production, depending on cell type, presence of ROS-generating proteins, growth conditions, etc. Since androgen-induced ROS generation was abrogated by either blocking androgen-induced JunD overexpression with anti-androgen bicalutamide or silencing JunD protein expression using siRNA (19,20), we concluded that JunD activity is necessary for androgen-induced oxidative stress in LNCaP cells.
Here, we present data clearly demonstrating that 1) JunD is required for androgen–induced SSAT gene expression, 2) activated AR interacts with JunD in situ, and 3) JunD binds directly to the SSAT promoter sequence only in androgen-treated human CaP cells. Based on these results, we hypothesize that AR and JunD form a complex that binds to the SSAT promoter, resulting in SSAT gene expression and consequent high levels of ROS in androgen-treated prostate cells. Induction of SSAT and polyamine oxidation as a main source of ROS production in prostatic epithelia was first reported from our laboratory (27), and further confirmed in our subsequent publication (21). Although other studies implicating the effect of AR-induced CaP cell growth stimulation via ROS production through changes in mitochondrial function and gene expression were published within the last couple years (28,29), none of those publications probed deep into the actual biochemical pathway(s) of ROS production in CaP cells. To the best of our knowledge, this is the first demonstration of a possible molecular mechanism of androgen-induced activation of an enzymatic pathway that can be directly related to ROS generation in prostate cells. A clear understanding of this mechanism may open a new avenue of research in the field of therapy and/or prevention of CaP occurrence and progression.
Materials and Methods
Antibodies
Primary antibodies: monoclonal antibody against AR (AR (441); sc-7305; Santa Cruz Biotechnology, Santa Cruz, CA); polyclonal antibody against JunD (sc-74X, Santa Cruz); polyclonal antibody against Gaussia luciferase (Nanolight Technology, Pinetop, AZ); monoclonal antibody against β-actin (A5441; Sigma, St. Louis, MO). Secondary antibodies for immunohistochemistry: Alexa Fluor 594 goat anti-rabbit IgG (Invitrogen, Carlsbad, CA); Alexa Fluor 488 donkey anti-mouse IgG (Invitrogen).
Cell culture
Androgen sensitive LNCaP human prostate carcinoma cells were obtained from ATCC and maintained in DMEM supplemented with 10% FBS (F10 medium) as described before (17). Hep3B human hepatoma cells were obtained from the Small Molecule Screening Facility (SMSF) at the UW Carbone Cancer Center, Madison, WI, and were maintained in RPMI supplemented with 10% FBS and antibiotics. Cell lines are tested annually for mycoplasma.
Culture conditions for LNCaP androgen response studies included use of cells passage 40–90, hormone depleted media containing 4% charcoal-stripped FBS plus 1% non-stripped FBS (F1C4), and synthetic androgen R1881 (methyltrienolone; NEN, Boston, MA) at 1nM for maximal induction of JunD and ROS as described before (17,19,20). For AR-JunD interaction studies in AR-transfected Hep3B cells, R1881 was used at 2nM in DMEM to maximally induce AR (data not shown).
Vector construction
cDNA for human androgen receptor (AR) was obtained from Open Biosystems (Huntsville, AL). The whole human junD gene (20) was subcloned in a pCI-based vector (Promega, Madison, WI). Two sections of the humanized Gaussia luciferase gene, N-terminus hGluc1 and C-terminus hGluc2, in two separate vectors (30) were kind gifts from Prof. Stephen Michnick (University of Montreal, Canada). hGluc1 was cloned in frame with the N-terminal end of AR in a pcDNA3.1-based vector (Invitrogen) to create vector Gluc1-AR.
The pCI-junD vector was used to fuse hGluc2 in frame at the end of the junD gene after removing the junD stop codon to construct vector JunD-Gluc2. The authenticity of each construct was verified by using Big Dye terminator and sequencing via the Biotechnology Center of UW-Madison. The in-frame fusion of each construct was also verified by transfecting each into AR-negative Hep3B cells and analyzing cell lysate by western blot with AR antibody for Gluc1-AR or antibody for Gaussia luciferase at the C-terminal end of the fusion protein for JunD-Gluc2. β-actin was used to control for protein loading in all western blot analyses.
Transfection of constructs into Hep3B cells
5×105 Hep3B cells were seeded, then one day later co-transfected with 3µg each of Gluc1-AR and JunD-Gluc2 constructs, or transfected with Gluc1-AR or JunD-Gluc2 alone as negative controls using Lipofectamine 2000 reagent (Invitrogen), following the manufacturer-supplied protocol. Two to three hours after transfection, cells were washed and refed DMEM without serum and treated with 2nM R1881 for 48h prior to collecting cell lysates. Corresponding untreated cells were used as negative controls.
Bioluminescence activity of Gaussia luciferase in lysates from Hep3B cells transfected with Gluc1-AR and JunD-Gluc2
Gaussia luciferase activity was measured in 25µl of Hep3B cell lysates from R1881-treated or untreated control cells using a Gaussia luciferase assay kit from New England Biolabs (Ipswich, MA), following the manufacturer-supplied protocol. Bioluminescence activity of the lysate-substrate mixture was read on a single tube Monolight 2010 luminometer (Analytical Luminescence Laboratory, Spaarks, MD) at 480nm.
Immunocytochemistry
LNCap cells were grown in F1C4 medium on coverslips for 2 days followed by 3 days of treatment with 1nM R1881. Immunofluorescent staining was carried out following a published procedure (31), using primary-secondary antibody pairs JunD-AlexaFlour594 or AR-AlexaFluor488.
Immunoprecipitation
For whole cell lysates, LNCaP cells were lysed using modified RIPA buffer containing complete protease inhibitors (Roche Applied Sciences, Indianapolis, IN). Nuclear and cytoplasmic fractions were prepared and checked for purity using NE-PER Nuclear and Cytoplasmic extraction reagents (Thermo scientific, Pierce Biotechnology, Rockford, IL) following manufacturer-supplied protocol. For immunoprecipitation, lysates were pre-cleared by incubation with 50% Protein A-agarose slurry (Pierce, Rockford, IL). Six µg AR antibody and 500µl pre-cleared lysate (500µg total protein) were mixed and rocked overnight at 4°C. The immunocomplex was captured with 100µl of 50% protein A-agarose slurry, then analyzed by western blotting using JunD antibody. The same immunoprecipitation procedure was repeated for capturing the immunocomplex using JunD antibody and western blotting using AR antibody. Proper controls with IgG and protein A-agarose were run in parallel.
Transcriptional activity of full length SSAT-promoter in siJunD and vector control cell lines
The full length (FL) SSAT promoter sequence, kindly provided by Dr. Robert Casero (Johns Hopkins University, Baltimore, MD), was amplified and cloned into pGL4-basic vector (Promega) with a firefly luciferase reporter gene. This vector, pGL4-SSAT-luc, was transiently transfected into our published (20) JunD-silenced (siJunD) and vector control LNCaP cell lines. Briefly, 5×105 siJunD or vector control LNCaP cells were seeded, then transfected one day later with 1µg pGL4-SSAT-luc DNA construct using Lipofectamine2000 (Invitrogen). After transfection, cells were treated with 1nM R1881 or left untreated for 72h, then lysed. Luciferase activity was measured in cell lysates by a luciferase assay system kit (Promega) following the manufacturer-supplied protocol.
Chromatin Immunoprecipitation (ChIP) assay
ChIP assay was performed using a commercially available ChIP assay kit (Millipore, Billerica, MA), essentially following the manufacturer supplied protocol. Briefly, 2×106 LNCaP cells were treated with 1nM R1881 for 24h, protein-DNA were cross-linked by addition of formaldehyde (1% final concentration), cells were lysed, and lysates were sonicated for twenty 10sec pulses with 30sec intervals to shear the chromatin into approximately 500bp fragments.
Cross-linked protein-DNA were separated into four parts and immunoprecipitated with either 6µg JunD antibody, 6µg AR antibody, non-specific rabbit IgG or no antibody. Chromatin-antibody complexes were isolated by incubation with 50% salmon sperm DNA/protein-agarose slurry. Pelleted agarose was eluted and DNA was recovered by phenol/chloroform extraction followed by ethanol precipitation. Two microliters of this DNA was used for each PCR reaction to determine the presence of SSAT promoter fragment bound to either JunD or AR in the immunoprecipitates. Primers were designed based on the SSAT gene promoter sequence to cover the complete SSAT promoter sequence (GenBank accession#1103903), as follows:
F1:5’ggaggctgaagcaggagaatc; R1:5’ctcactctattgcccaggctggag
F2:5’cagcctgggcaatagagtgag; R2:5’gagatggcgccattgcactcc
F3:5’gagtgcaatggcgccatctcg; R3:5’ctcaccatcttgcccaggctg
F4:5’cagcctgggcaagatggtgaggcc; R4:5’ggagaccctgcagatcccaag
F5:5’tctgagggtctcccggatcacac; R5:5’acctcggcgagtgacggatagg
PCR products were run on a 1% agarose gel, purified, cloned into pCR2.1 TOPO vector (Invitrogen) and transformed into TOP10F’ competent cells (Invitrogen). Ten colonies were selected and their plasmids were extracted and sequenced using M13 primer (5’ caggaaacagctatgac).
qRT-PCR
Quantitative RT-PCR analysis of SSAT mRNA levels was performed as previously described (21).
Results
AR and JunD co-immunoprecipitate from LNCaP cell lysates
The co-immunoprecipitation of AR and JunD was first shown in whole cell lysates from LNCaP cells grown under normal F10 medium conditions (Figures 1A,B).
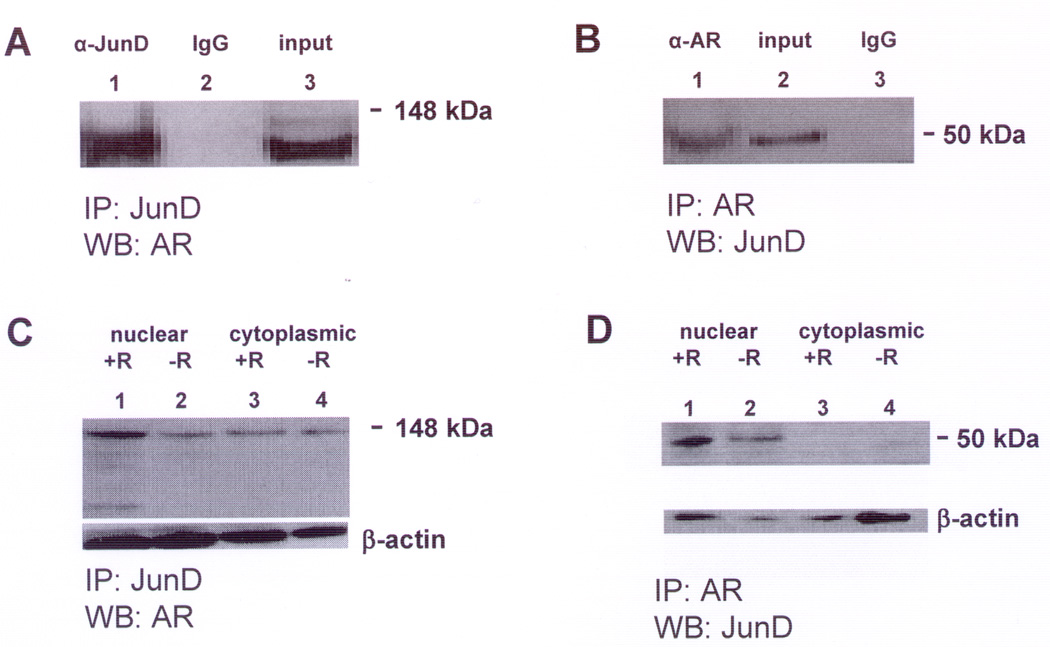
Figure 1. Representative western blots showing JunD and Androgen Receptor (AR) in an immunocomplex in LNCaP cell lysates.
A. Immunoprecipitation (IP) of LNCaP whole cell lysate was performed using a rabbit polyclonal antibody against JunD (sc-74X) (IP:JunD). Immunoprecipitates were analyzed by western blot (WB) using monoclonal antibody against human AR (sc-7305) (WB:AR). Lane (1): IP with JunD antibody, Lane (2): IP with non-specific IgG, Lane (3): total LNCaP cell lysate.
B. IP of LNCaP whole cell lysate was performed using AR antibody (sc-7305) (IP:AR) and WB analysis using JunD antibody (sc-74X) (WB:JunD). Lane (1): IP with AR antibody, Lane (2): total LNCaP cell lysate, Lane (3): IP with non-specific IgG.
Positions of molecular size markers 148 kDa (A) and 50 kDa (B) are shown on the right. The experiment was repeated 3 times with similar results.
C. IP:JunD/WB:AR of LNCaP nuclear and cytoplasmic extracts was performed as described in (A) above. Lane (1): nuclei from cells treated with R1881 (+R), Lane (2): nuclei from untreated (−R) cells, Lane (3): cytoplasm from cells treated with R1881 and Lane (4): cytoplasm from untreated cells.
D. IP:AR/WB:JunD of LNCaP nuclear and cytoplasmic extracts was performed as described in (B) above. Lane (1): nuclei from cells treated with R1881 (+R), Lane (2): nuclei from untreated (−R) cells, Lane (3): cytoplasm from cells treated with R1881 and Lane (4): cytoplasm from untreated cells.
The immunoprecipitation of AR by rabbit polyclonal antibody against JunD (IP:JunD) was visualized by western blot analysis using monoclonal antibody against human androgen receptor (WB:AR) as shown in Figure 1A. The immunoprecipitation of JunD by monoclonal antibody against AR (IP:AR) was visualized by western blotting using antibody against JunD (WB:JunD) as shown in Figure 1B.
To specifically investigate the effect of androgen, LNCaP cell lysates were prepared after incubation with 1nM R1881 for 72h and analyzed for co-immunoprecipitation of AR and JunD. Our published time kinetic studies established that SSAT enzyme activity and cellular ROS production maximizes under these treatment conditions (21). The corresponding results for co-IP of AR and JunD in nuclear and cytoplasmic fractions of untreated versus androgen-treated LNCaP cells are shown in Figures 1C,D. Immunopreciptate using JunD antibody that was probed in a western blot using AR antibody (Fig.1C) showed AR-JunD immunocomplex in the nuclear fraction was increased by approximately 3-fold (normalized to β-actin) in 1nM R1881-treated cells compared to low androgen untreated cells growing in F1C4. Only a small increase was observed in nuclear fractions by IP:AR,WB:JunD (Fig.1D). This may be due to a difference between the nature of interaction between JunD with its antibody as compared to that between AR and its antibody. No difference in AR-JunD immunocomplex was observed in cytoplasmic fractions of R1881-treated versus untreated cells.
Androgen induces nuclear translocation of JunD in LNCaP cells
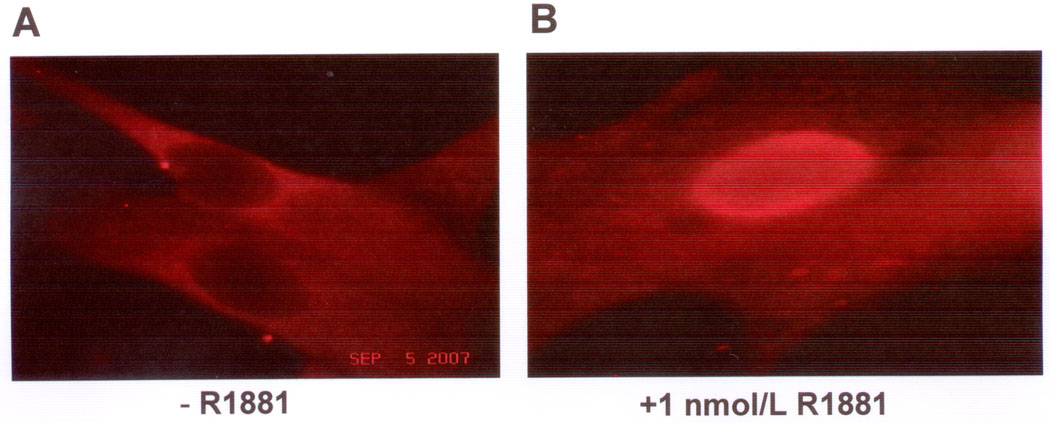
To further investigate the effect of androgen treatment on JunD activity in LNCaP cells, immunofluorescence staining was performed to determine JunD localization in untreated versus androgen (1nM R1881) treated LNCaP cells. Representative pictures for each condition are shown in Figure 2. In untreated LNCaP cells, JunD is mostly dispersed in the cytoplasm with negative staining for the nuclei (Fig.2A). After R1881 treatment, a substantial amount of JunD translocates into the nucleus as shown in Figure 2B. Using AR antibody and its related fluorescence-tagged secondary antibody, the translocation of AR into the nucleus in R1881-treated LNCaP cells also was observed under the same condition (data not shown), which is consistent with observation reported elsewhere (31). These data suggest androgen induces simultaneous translocation of JunD and AR into cell nuclei.
Figure 2. Representative immunocytochemistry showing JunD translocated into LNCaP cell nuclei after androgen treatment.
LNCaP cells were untreated (− R1881) (A) or treated with androgen (1 nM R1881) (B) for 72h, then fixed, stained with JunD primary-AlexaFluor594 secondary antibody pair, and observed using an Olympus microscope model Mercury 100 fluorescence. (100x-magnification). Nuclei were identified by DAPI staining (data not shown). The experiment was repeated 2 times, including 6 slides in 2 replicates per condition with similar results.
Expression of Gluc1-AR and JunD-Gluc2 in Hep3B cells
Because Hep3B cells do not have endogenous AR, this cell line was chosen as a model for AR and JunD interaction studies using a Protein Complementation Assay (PCA) developed by Remy and Michnick (30). This technique is based on reconstitution of the reporter enzyme Gaussia luciferase in live cells. The gene coding for the enzyme was split into two sections: N-terminal section (Gluc1) and C-terminal section (Gluc2). Gluc1 and Gluc2 sequences were separately fused to the N-terminus of AR (Gluc1-AR) and the C-terminus of JunD (JunD-Gluc2), respectively, as shown schematically in Figure 3A.
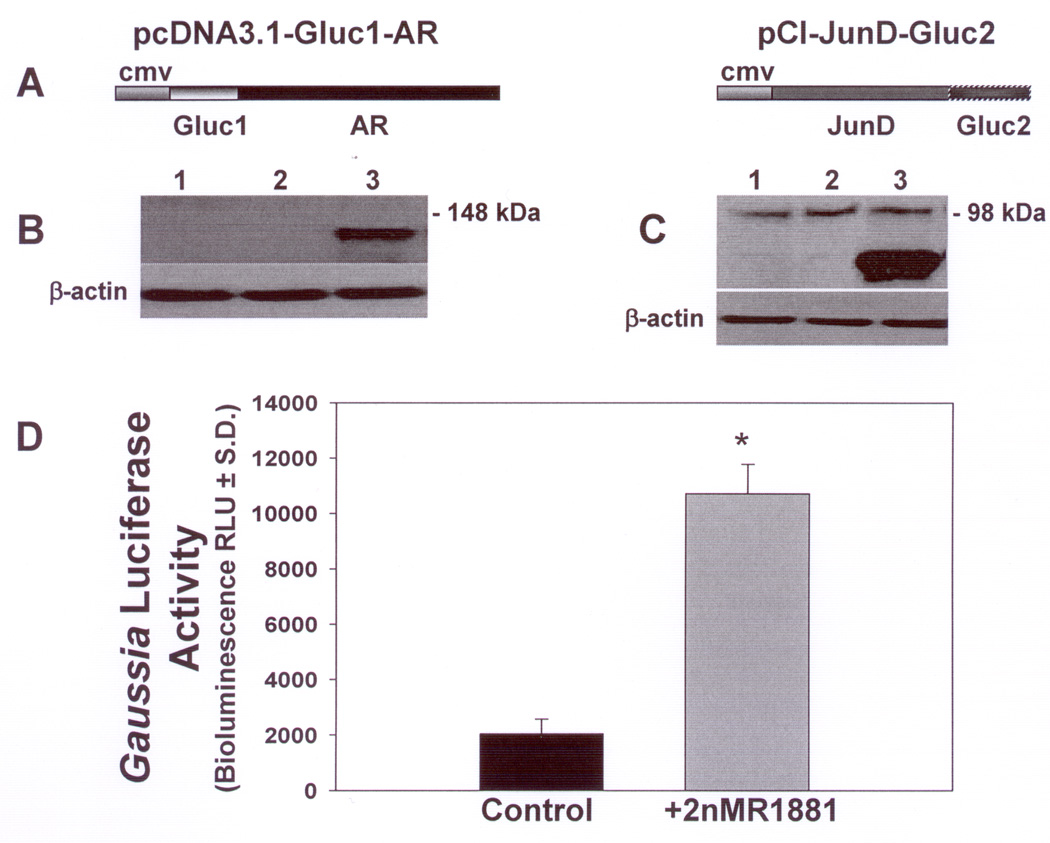
Figure 3. Western blot and bioluminescence analysis demonstrating expression of Gluc1-AR and JunD-Gluc2 and reconstitution of Gaussialuciferase activity following androgen stimulation in transfected Hep3B cells.
A Schematic diagrams for Gluc1-AR and JunD-Gluc2 fusion constructs.
B. Representative Western blot of Hep3B cell lysates analyzed using AR antibody. Lane (1): Hep3B cells transfected with control vector (pcDNA3.1); Lane (2): control untransfected Hep3B cells; Lane (3): Hep3B cells transfected with Gluc1-AR.
C. Representative Western blot of Hep3B cell lysates analyzed using Gaussia luciferase antibody. Lane (1): Hep3B cells transfected with control vector (pCI); Lane (2): control untransfected Hep3B cells; Lane (3): Hep3B cells transfected with JunD-Gluc2. Membranes were stripped and probed with monoclonal antibody against β-actin to control for protein loading (B,C). Positions of molecular size markers 148 kDa (B) and 98 kDa (C) are shown on the right. Cell lysates were obtained and analyzed by western blot from six independent transfection experiments for each construct with similar results.
D. Gaussia luciferase activity in co-transfected cells: Hep3B cells were co-transfected with Gluc1-AR and JunD-Gluc2, then treated with androgen (2nM R1881) (gray bar) or left untreated (−R1881) (black bar). Cell lysates were collected after 48h and bioluminescence activity of Gaussia luciferase was assayed by measuring light emitted from reconstituted Gluc1-Gluc2 at 480nm. Reconstitution of Gluc1-Gluc2 and resulting Gaussia luciferase activity was significantly increased >5-fold in androgen treated cells compared to untreated control cells. Lysates used for these studies were collected from six independent transfections each run in triplicate. * P < 10−8.
To verify in-frame fusion of Gluc1-AR and JunD-Gluc2, cell lysates from Hep3B cells transfected with Gluc1-AR or JunD-Gluc2 were analyzed by western blotting. Figure 3B shows western blot analysis using monoclonal antibody against AR. Hep3B cells alone (Fig.3B,lane 1) or Hep3B cells transfected with control vector pcDNA3.1 (Fig.3B,lane 2) do not show any protein band related to AR. However, expression of the fusion protein (Gluc1-AR) is observed in cells transfected with Gluc1-AR (Fig.3B,lane 3). As AR is fused at the C-terminal end of Gluc1, presence of the AR band in the western blot confirms the in-frame fusion of AR with Gluc1. Similarly, presence of the C-terminal portion of Gaussia luciferase in the western blot of lysate from Hep3B cells transfected with JunD-Gluc2 using polyclonal antibody against Gaussia luciferase confirms the in-frame fusion of JunD with Gluc2 (Fig.3C,lane 3). Hep3B cells alone or Hep3B cells transfected with control vector pCI (Fig.3C,lanes 1 and 2 respectively) neither expected nor show any band related to the Gaussia luciferase. β-actin was used for protein loading control (Fig.3B,C). In-frame gene fusions (Gluc1-AR and JunD-Gluc2) were also confirmed by sequencing using specific primers across the fusion junctions in both vectors (data not shown).
Bioluminescence activity of reconstituted Gaussia luciferase in Hep3B cells co-transfected with Gluc1-AR and JunD-Gluc2 is markedly enhanced by androgen treatment
Cell lysates from Hep3B cells that were co-transfected with both Gluc1-AR and JunD-Gluc2 with or without treatment with androgen (R1881) were collected 48h after transfection and analyzed for Gaussia luciferase bioluminescence activity. Results are shown in Figure 3D. Lysates from co-transfected cells that were not treated with androgen (−R1881) showed very low Gaussia luciferase activity. Lysates from co-transfected cells that were treated with 2nM R1881 (+R1881) showed significantly higher Gaussia luciferase activity (>5 fold, P < 10−8) than the untreated co-transfected cells. Cells transfected with either of the fusion constructs Gluc1-AR and JunD-Gluc2 individually did not show any measurable Gaussia luciferase activity (data not shown) confirming that the enzyme activity is only observed after both fragments Gluc1 and Gluc2 associate with each other (30). Minor baseline reporter enzyme activity in cell lysates from androgen-untreated co-transfected cells (−R1881) might be due to interaction of residual activated-AR remaining after the transfection process, which was performed in medium containing serum that was not stripped of androgen. These data clearly establish an interaction of JunD and androgen-activated AR in situ that brings their corresponding fusion proteins Gluc1 and Gluc2 together to reconstitute Gaussia luciferase activity.
Activated androgen receptor requires JunD to induce transcriptional activity of the SSAT promoter
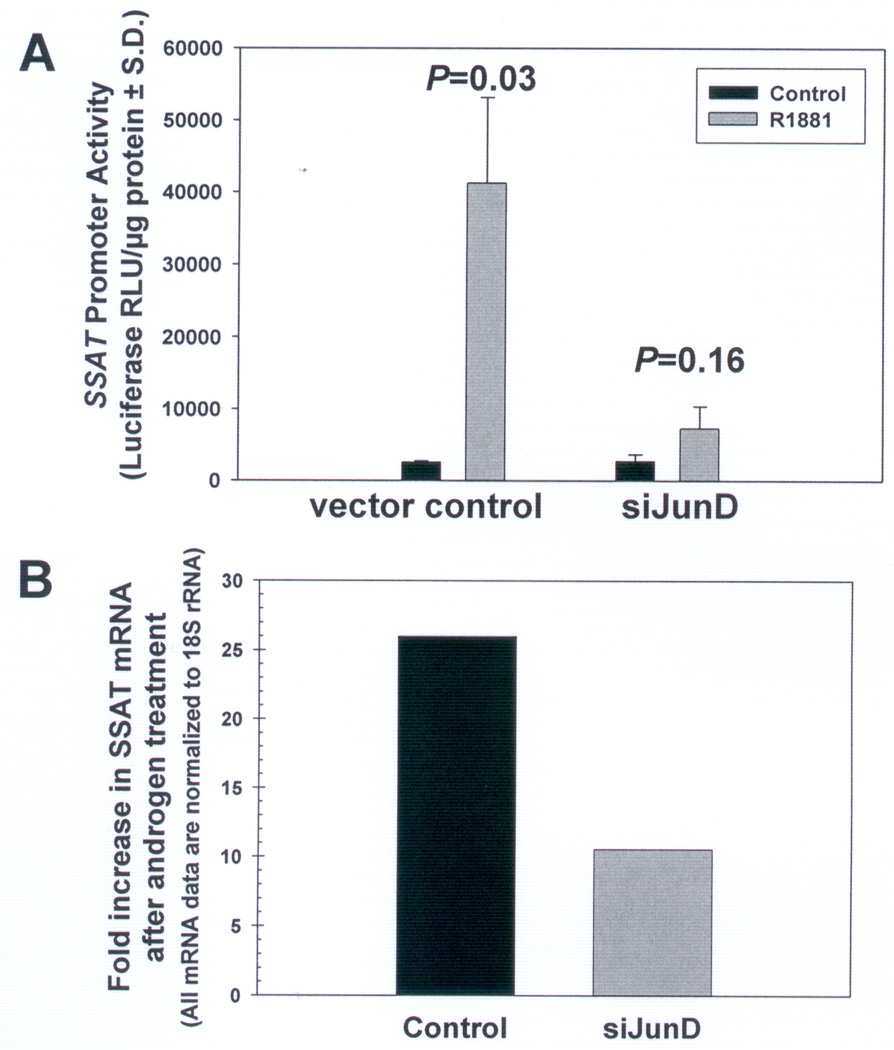
The effect of AR activation by androgen on transcriptional activity of full length SSAT promoter and the necessity of JunD for such effect was studied in LNCaP cells stably transfected with either junD silenced (siJunD) or control vector that were then transiently transfected with a full length SSAT promoter-firefly luciferase reporter vector (FLSSAT-luc) followed by androgen treatment (Figures 4A,B). Androgen treatment (1nM R1881) caused a significant >16-fold increase in SSAT promoter activity in vector control LNCaP cells (Control) compared to corresponding untreated cells (Fig.4A,P=0.03). Although androgen also caused a small increase in SSAT promoter activity in siJunD cells, where JunD expression is 70% suppressed, the extent of induction was not statistically significant (Fig.4A,P=0.16). SSAT mRNA levels in LNCaP cells determined by qRT-PCR assay are shown in Figure 4B. There is an androgen-induced increase in SSAT mRNA (~10-fold) in siJunD cells, but this increase is much less than that observed in vector control cells (~25-fold). Thus, even though some increase in SSAT mRNA expression in siJunD cells was observed, the increase is not enough to significantly enhance cellular SSAT protein expression as evident from the insignificant increase in luciferase reporter expression.
Figure 4. Androgen-induced increase in SSAT promoter activity is abrogated when JunD is silenced in LNCaP cells.
A. LNCaP cells in which JunD is silenced (siJunD) and respective vector control LNCaP cells were transiently transfected with an SSAT promoter luciferase reporter vector, then treated with androgen (1nM R1881) (gray bars) or left untreated (Control) (black bars). Cell lysates were collected after 72h and firefly luciferase activity was measured. Data and error bars are respectively the mean and standard deviation from 18 data points of measured relative light units (RLU) normalized to protein concentration from 6 independent repeat transfection/treatment experiments (n=3 samples per condition for each experiment). P-values were calculated using a two-tailed Student’s t-test assuming unequal variance. B. SSAT mRNA levels as determined by qRT-PCR showing a >25-fold increase in SSAT mRNA in vector control cells and only 10-fold increase in SSAT mRNA in JunD silenced cells (siJunD). Results are presented as a ratio of mRNA in androgen-treated/androgen-untreated cells after normalizing for corresponding 18rRNA. Data are mean of three independent observations.
JunD binds to the SSAT promoter in situ by Chromatin Immunoprecipitation (ChIP) assay
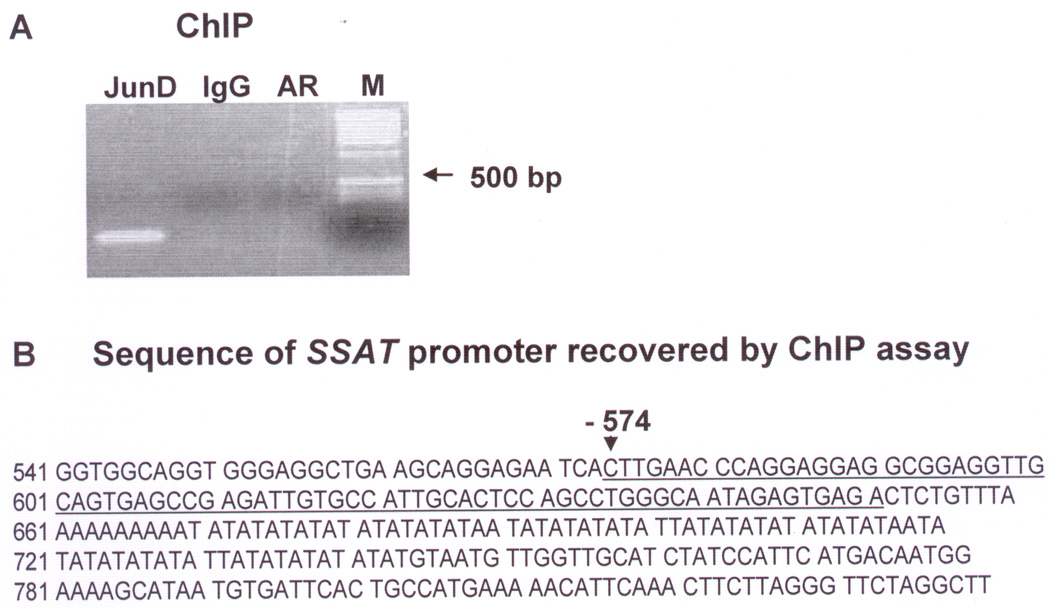
ChIP assay was performed in LNCaP cells with R1881 treatment to ascertain if activated AR and/or JunD bind to the SSAT promoter site using primer sets targeted to identify the SSAT promoter sequence in the immunoprecipitates (see Materials and Methods). ChIP assay was performed under conditions where only protein-DNA and not protein-protein interactions can be detected (reviewed in 32). Under these conditions, the only PCR product obtained using those primers was from chromatin fragment(s) immunoprecipitated by rabbit polyclonal antibody against JunD using the F1R1 primer pair as shown in Figure 5A. Using the same F1R1 primer pair, no PCR product was obtained from immunoprecipitates obtained by monoclonal antibody against AR, nor from the non-specific IgG and no antibody controls. None of the other primer pairs yielded any PCR product from either JunD-, AR- or IgG-immunoprecipitated chromatin fragments (data not shown).
Figure 5. ChIP assay identifying a binding site for JunD but not AR within the SSAT promoter sequence.
ChIP assay studies were carried out in LNCaP cells treated with androgen (1nM R1881) using primer pairs targeted to identify the SSAT promoter sequence (see text). A. Agarose gel electrophoresis of PCR products showing the only PCR product obtained, which was from DNA fragments immunoprecipitated by JunD antibody (Lane:JunD) using the F1R1 primer pair. Using the same F1R1 primer pair, no PCR product was obtained from immunoprecipitation of chromatin fragments by AR antibody (Lane:AR), nor from the non-specific IgG (Lane:IgG) controls. M: DNA Ladder size marker. B. Sequence of the PCR product, which was cloned into PCR2.1TOPO and sequenced using M13 primer, matches −574 to −651bp of the SSAT gene promoter (NCBI accession#1103903).
The sequence data obtained from the PCR product that was cloned in pCR2.1TOPO indicates the existence of JunD binding sites at −574 to −651bp upstream in the SSAT promoter sequence that contains multiple half sites (TGA) of the AP1 consensus sequence (TGAG/CTCA) (Fig.5B).
Discussion
Although it is well established that androgens produce oxidative stress in prostate cells that plays a key role in the occurrence and progression of CaP, the exact molecular mechanism of androgen-induced oxidative stress generation in prostatic epithelia and prostate cancer cells is only recently being elucidated. We previously reported that AP-1 transcription factor JunD plays a key role in androgen induction of ROS (20). Recently we reported that androgen significantly induces the expression and enzymatic activity of “spermidine/spermine acetyl transferase” (SSAT) (21), a regulatory enzyme in the polyamine catabolic pathway that produces excess amount of ROS in polyamine-rich prostate cells. In this report, we show a relationship at the molecular level between these two components that establishes the mechanism of androgen-induced ROS generation in prostate cells.
Discovering the mechanism that regulates expression of SSAT is the focus of many studies (33–35). Thus-far, binding sites of many important transcription factors in the SSAT gene promoter have been identified (33–35). Because the SSAT gene promoter sequence lacks an AR binding site (ARE), the mechanism of androgen-induced SSAT expression is unclear. Here we show a direct binding of androgen-activated AR with JunD, and that an induction of SSAT by androgen occurs following an interaction of JunD with a specific sequence in the SSAT promoter only in androgen treated LNCaP cells, probably due to the formation of an activated AR-JunD complex.
We previously showed that androgen-activated AR induces overexpression of transcription factor JunD as well as activates JunD binding to the AP-1 DNA-binding sequence in LNCaP cells (19). Here we demonstrate that androgen treatment causes AR and JunD to co-precipitate as an immunocomplex from LNCaP cell lysate. Relatively more complex precipitates from the nuclear than from the cytoplasmic fraction (Fig.1). Androgen treatment induces translocation of JunD into the nucleus in LNCaP cells (Fig.2) at the same time as AR translocates into the nucleus as shown by immunoprecipitation/western blot of nuclear extract (Fig.1C) and also reported by other laboratories (31). These observations suggest an interaction of activated AR with JunD in androgen-treated CaP cells that also causes functional activation of JunD.
More direct evidence of AR and JunD interaction was demonstrated using the Gaussia luciferase reconstitution assay recently developed to study in situ protein-protein interactions (30). The significant reconstitution of Gaussia luciferase activity only in androgen-treated Hep3B cells transfected with vectors expressing N-terminal and C-terminal fragments of Gaussia luciferase enzyme linked to AR and JunD, respectively, provides clear and direct evidence of JunD interaction with androgen-activated AR in situ (Fig.3D). While immunoprecipitation and co-localization of AR with another AP-1 family member, c-Jun, has been reported (31), to the best of our knowledge this is the first direct demonstration of androgen-activated AR and JunD complex formation.
Since overexpression of JunD is necessary for the induction of ROS following androgen exposure (19, 20), presumably the AR-JunD complex regulates expression of genes involved in ROS production in LNCaP cells. The complex may bind via JunD to sequences containing binding sites for members of the AP1 family of transcription factors (TGAG/CTCA) (36). These sequences may or may not contain any ARE sequence. Thus, many genes such as SSAT that are not directly regulated by AR might be regulated by an AR-JunD complex.
By scanning the SSAT gene promoter sequence in silico, we identified six putative AP1 binding sites. Using our siJunD clone of the LNCaP cell line (20), we demonstrated that in the absence of JunD, androgen-activated AR does not induce SSAT expression (Fig.4). Thus we conclude that androgen activated AR requires JunD for SSAT expression.
A direct binding of JunD to the SSAT promoter sequence was demonstrated by ChIP assay (Fig.5). By PCR analysis with primers designed to identify the SSAT promoter, we obtained a PCR product that corresponds to a DNA fragment of the SSAT promoter only in the chromatin fragment precipitated by JunD antibody and not in the chromatin fragment precipitated by AR antibody (Fig.5A). This suggests that under these conditions, where JunD directly binds to the −574bp to −651bp of the SSAT promoter, there may not be a direct binding of AR to the SSAT promoter. Elucidation of this mechanism also explains the delay in SSAT expression (72h) and ROS generation after androgen treatment as previously reported (21).
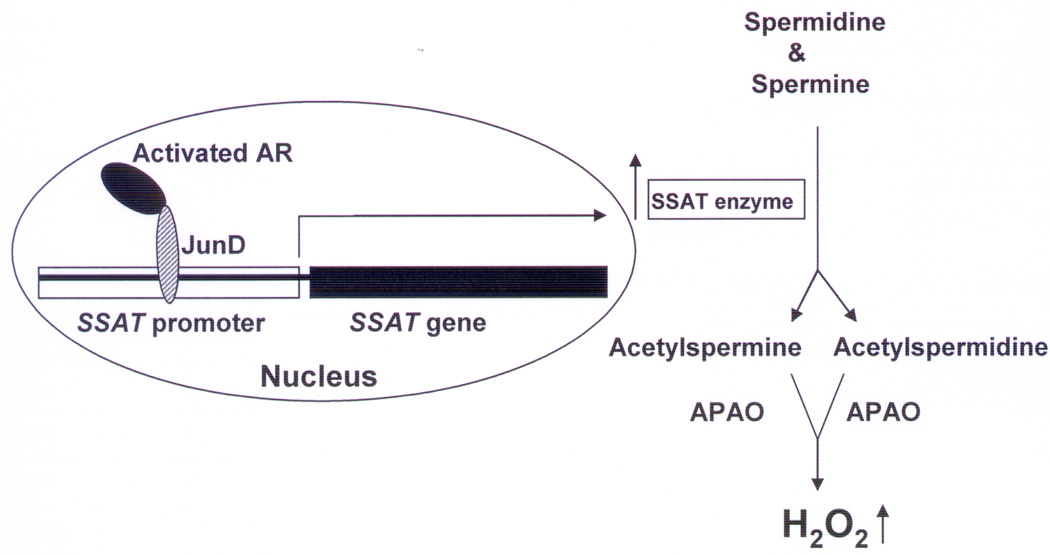
Collectively, our data suggest that activated AR forms a complex with JunD that binds to an AP-1 DNA-binding sequence in the SSAT promoter to activate SSAT gene transcription resulting in overproduction of H2O2 in CaP cells, as shown schematically in Figure 6. To the best of our knowledge, the data presented above provides for the first time a molecular mechanism of androgen-induced increase in SSAT activity and consequent ROS overproduction in CaP cells.
Figure 6. Schematic diagram showing a possible mechanism of androgen-induced increase in cellular ROS production in CaP cells through an AR-JunD complex.
APAO:N’-acetylpolyamine oxidase; H2O2:hydrogen peroxide
The demonstration of a mechanistic pathway of androgen-induced ROS production opens up a new avenue for development of drugs that specifically target steps in this ROS generating pathway in CaP cells and thus can be effective in therapy and prevention of CaP without major systemic toxicity.
Acknowledgments
Supported by Department of Defense, National Institutes of Health and Prostate Cancer Foundation
References
- 1.Karihtala P, Soini Y. Reactive oxygen species and antioxidant mechanisms in human tissues and their relation to malignancies. Acta Pathol Microbiol et Immunol Scandinavica. 2007;115:81–103. doi: 10.1111/j.1600-0463.2007.apm_514.x. [DOI] [PubMed] [Google Scholar]
- 2.Newmeyer DD, Ferguson-Miller S. Mitochondria: Releasing power for life and unleashing the machineries of death. Cell. 2003;112:481–490. doi: 10.1016/s0092-8674(03)00116-8. [DOI] [PubMed] [Google Scholar]
- 3.Klaunig JE, Kamendulis LM. The role of oxidative stress in carcinogenesis. Ann. Rev Pharmacol Toxicol. 2004;44:239–267. doi: 10.1146/annurev.pharmtox.44.101802.121851. [DOI] [PubMed] [Google Scholar]
- 4.Bokov A, Chaudhuri A, Richardson A. The role of oxidative damage and stress in aging. Mech Ageing Dev. 2004;125:811–826. doi: 10.1016/j.mad.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 5.Cerutti PA. Prooxidant states and tumor promotion. Science. 1985;227:375–381. doi: 10.1126/science.2981433. [DOI] [PubMed] [Google Scholar]
- 6.Dargel R. Lipid peroxidation- a common pathogenic mechanism? Exp Toxic Pathol. 1992;44:169–181. doi: 10.1016/S0940-2993(11)80202-2. [DOI] [PubMed] [Google Scholar]
- 7.Oberley TD, Oberley LW. Oxygen radicals and cancer. In: Yu BP, editor. Free Radicals in Aging. Ann Arbor: CRC Press; 1993. pp. 247–267. [Google Scholar]
- 8.Nelson WG, De Marzo AM, DeWeese TL, Issacs WB. The role of inflammation in the pathogenesis of prostate cancer. J Urol. 2004;172:S6–S12. doi: 10.1097/01.ju.0000142058.99614.ff. discussion S11–S12. [DOI] [PubMed] [Google Scholar]
- 9.Bostwick DG, Alexander EE, Singh R, et al. Antioxidant enzyme expression and reactive oxygen species damage in prostatic intraepithelial neoplasia and cancer. Cancer. 2000;89:123–134. [PubMed] [Google Scholar]
- 10.Schulze-Osthoff K, Bauer M, Vogt M, Wesselborg S, Baeuerle PA. Reactive oxygen species as primary and second messengers in the activation of transcription factors. In: Forman HJ, Cadenas E, editors. Oxidative Stress and Signal Transduction. New York: Chapman and Hall; 1997. pp. 239–259. [Google Scholar]
- 11.Oberley TD, Zhong W, Szweda LI, Oberley LW. Localization of antioxidant enzymes and oxidative damage products in normal and malignant prostate epithelium. Prostate. 2000;44:144–155. doi: 10.1002/1097-0045(20000701)44:2<144::aid-pros7>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 12.Tam NN, Nyska A, Maronpot RR, et al. Differential attenuation of oxidative/nitrosative injuries in early prostatic neoplastic lesion in TRAMP mice by dietary antioxidants. Prostate. 2006;66:57–69. doi: 10.1002/pros.20313. [DOI] [PubMed] [Google Scholar]
- 13.Sun XY, Donald SP, Phang JM. Testosterone and prostate specific antigen stimulate generation of reactive oxygen species in prostate cancer cells. Carcinogenesis. 2001;22:1775–1780. doi: 10.1093/carcin/22.11.1775. [DOI] [PubMed] [Google Scholar]
- 14.Feig DI, Reid TM, Loeb LA. Reactive oxygen species in tumorigenesis. Cancer Res. 1994;54:1890s–1894s. [PubMed] [Google Scholar]
- 15.Hatziapostolou M, Polytarchou C, Katsoris P, Courty J, Papadimitriou E. Heparin affin regulatory peptide/pleiotrophin mediates fibroblast growth factor 2 stimulatory effects on human prostate cancer cells. J Biol Chem. 2006;281:32217–32226. doi: 10.1074/jbc.M607104200. [DOI] [PubMed] [Google Scholar]
- 16.Wilding G. Endocrine control of prostate cancer. Cancer Surveys. 1995;23:43–62. [PubMed] [Google Scholar]
- 17.Ripple MO, Henry WF, Rago RP, Wilding G. Prooxidant-antioxidant shift induced by androgen treatment of human prostate carcinoma cells. J Natl Cancer Inst. 1997;89:40–48. doi: 10.1093/jnci/89.1.40. [DOI] [PubMed] [Google Scholar]
- 18.Ripple MO, Hagopian K, Oberley TD, Schatten H, Weindruch R. Androgen-induced oxidative stress in human LNCaP prostate cancer cells is associated with multiple mitochondrial modifications. Antiox Redox Signal. 1999;1:71–81. doi: 10.1089/ars.1999.1.1-71. [DOI] [PubMed] [Google Scholar]
- 19.Church DR, Lee E, Thompson TA, et al. Induction of AP-1 activity by androgen activation of the androgen receptor in LNCaP human prostate carcinoma cells. Prostate. 2005;63:155–168. doi: 10.1002/pros.20172. [DOI] [PubMed] [Google Scholar]
- 20.Mehraein-Gomi F, Lee E, Church DR, Thompson TA, Basu HS, Wilding G. JunD mediates androgen-induced oxidative stress in androgen dependent LNCaP human prostate cancer cells. Prostate. 2008;68:924–934. doi: 10.1002/pros.20737. [DOI] [PubMed] [Google Scholar]
- 21.Basu HS, Thompson TA, Church DR, et al. A small molecule polyamine oxidase inhibitor blocks androgen-induced oxidative stress and delays prostate cancer progression in the Transgenic Adenocarcinoma of the Mouse Prostate model. Cancer Res. 2009;69:7689–7695. doi: 10.1158/0008-5472.CAN-08-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson TA, Wilding G. Androgen antagonist activity by the antioxidant moiety of vitamin E, 2,2,5,7,8-pentamethyl-6-chromanol in human prostate carcinoma cells. Mol Cancer Therapeutics. 2003;2:797–803. [PubMed] [Google Scholar]
- 23.Gerald D, Berra E, Frapart YM, et al. JunD reduces tumor angiogenesis by protecting cells from oxidative stress. Cell. 2004;118:781–794. doi: 10.1016/j.cell.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 24.Tsuji Y. JunD activates transcription of the human ferretin H gene through an antioxidant response element during oxidative stress. Oncogene. 2005;24:7567–7578. doi: 10.1038/sj.onc.1208901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou H, Gao J, Lu ZY, Lu L, Dai W, Xu M. Role of c-Fos/JunD in protecting stress-induced cell death. Cell Prolif. 2007;40:431–444. doi: 10.1111/j.1365-2184.2007.00444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hock TD, Liby K, Wright MM, et al. JunB and JunD regulate human heme oxygenase-1 gene expression in renal epithelial cells. J Biol Chem. 2007;282:6875–6886. doi: 10.1074/jbc.M608456200. [DOI] [PubMed] [Google Scholar]
- 27. http://www.medpagetoday.com/HematologyOncology/ProstateCancer/4465.
- 28.Veeramani S, Yuan TC, Lin FF, Lin MF. Mitochondrial redox signaling by p66Shc is involved in regulating androgenic growth stimulation of human prostate cancer cells. Oncogene. 2008;27:5057–5068. doi: 10.1038/onc.2008.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharifi N, Hurt EM, Thomas SB, Farrar WL. Effects of manganese superoxide dismutase silencing on androgen receptor function and gene regulation: implications for castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6073–6080. doi: 10.1158/1078-0432.CCR-08-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Remy I, Michnick SW. A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat Methods. 2006;3:977–979. doi: 10.1038/nmeth979. [DOI] [PubMed] [Google Scholar]
- 31.Cai C, Hsieh CL, Shemshedini L. c-Jun has multiple enhancing activities in the novel cross talk between the androgen and Ets variant gene 1 in prostate cancer. Mol Cancer Res. 2007;5:725–735. doi: 10.1158/1541-7786.MCR-06-0430. [DOI] [PubMed] [Google Scholar]
- 32.Gingras AC, Gstaiger M, Raught B, Aebersold R. Analysis of protein complexes using mass spectrometry. Nat Rev Mol Cell Biol. 2007 Aug;8(8):645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- 33.Babbar N, Hacker A, Huang Y, Casero RA., Jr Tumor necrosis factor alpha induces spermidine/spermine N1- acetyltransferase through nuclear factor kappa B in non-small cell lung cancer cells. J Biol Chem. 2006;281:24182–24192. doi: 10.1074/jbc.M601871200. [DOI] [PubMed] [Google Scholar]
- 34.Ignatenko NA, Babbar N, Mehta D, Casero RA, Jr, Gerner EW. Suppression of polyamine catabolism by activated ki-ras in human colon cancer cells. Mol Carcinog. 2004;39:91–102. doi: 10.1002/mc.10166. [DOI] [PubMed] [Google Scholar]
- 35.Tomitori H, Nenoi M, Mita K, Daino K, Igrashi K, Ichimura S. Functional characterization of the human spermidine/spermine N(1)-acetyltransferase gene promoter. Biochim Biophys Acta. 2002;1579:180–184. doi: 10.1016/s0167-4781(02)00545-6. [DOI] [PubMed] [Google Scholar]
- 36.Angel P, Karin M. The role of Jun, Fos, and the AP-1 complex in cell proliferation and transformation. Biochimica Biophys Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]