Abstract
Developing, targeting, and evaluating genomic strategies for population-based disease prevention require population-based data. In response to this urgent need, genotyping has been conducted within the Third National Health and Nutrition Examination (NHANES III), the nationally-representative household-interview health survey in the U.S. However, before these genetic analyses can occur, family relationships within households must be accurately ascertained. Unfortunately, reported family relationships within NHANES III households based on questionnaire data are incomplete and inconclusive with regards to actual biological relatedness of family members. We inferred family relationships within households using DNA fingerprints (Identifiler®) that contain the DNA loci used by law enforcement agencies for forensic identification of individuals. However, performance of these loci for relationship inference is not well understood. We evaluated two competing statistical methods for relationship inference on pairs of household members: an exact likelihood ratio relying on allele frequencies to an Identical By State (IBS) likelihood ratio that only requires matching alleles. We modified these methods to account for genotyping errors and population substructure. The two methods usually agree on the rankings of the most likely relationships. However, the IBS method underestimates the likelihood ratio by not accounting for the informativeness of matching rare alleles. The likelihood ratio is sensitive to estimates of population substructure, and parent-child relationships are sensitive to the specified genotyping error rate. These loci were unable to distinguish second-degree relationships and cousins from being unrelated. The genetic data is also useful for verifying reported relationships and identifying data quality issues. An important by-product is the first explicitly nationally-representative estimates of allele frequencies at these ubiquitous forensic loci.
Keywords: Forensics, allele sharing, population structure, CODIS, IBS, IBD
1 Introduction
The recent revolution in genetics promises enormous gains for understanding and improving health. In all genome-wide association studies since 2007, genetic variants at nearly 100 regions of the genome have been associated with an increased risk for diseases with complex genetic causes, such as diabetes, inflammatory bowel disease, heart disease, and cancer (Chanock and Hunter, 2008). Twenty-eight specific genetic variants have been linked to cancers of the breast, prostate, colon, lung, and skin (Easton and Eeles, 2008). Research is progressing rapidly (Lin et al., 2006) to determine risks conferred by newly-discovered types of genetic variation such as copy-number variants (Feuk et al., 2006), and to elucidate the joint effects of multiple genetic variants in concert with non-genetic factors.
However, the hotly-debated question remains about how to use genetic information to better develop, target, and evaluate policies for population-level disease prevention (Pharoah et al., 2008; Gail, 2008). Although the found genetic variants are common, each has small effect on disease risks, and so modify disease risks only slightly for most individuals. However, reliable identification of population subgroups at high disease risk has major implications for population health (Pharoah et al., 2008).
As genetic findings accrue, evaluating their potential impact on population health requires population-representative data. In response to this pressing need, the Centers for Disease Control and Prevention and the National Cancer Institute have collaborated to conduct genotyping on a subset of the Third National Health and Nutrition Examination Survey (NHANES III). NHANES III is the nationally-representative household-interview and medical examination survey of the U.S. non-institutionalized civilian population conducted from 1988-1994 by the National Center for Health Statistics (NCHS) (NCHS, 1994). The nationally representative sample is obtained from a complex, stratified, multistage probability sample design with unequal selection probabilities.
These NHANES genetic data are the first U.S.-population-based genetic data. The continuing NHANES survey is the first major periodic official health survey in the world to collect genetic data. These data are a unique and paramount resource for analyzing the distribution of genetic variation in the U.S. and for estimating the potential population impact of genomic strategies for disease prevention. In addition, NHANES III oversamples non-Hispanic blacks and Mexican-Americans, important yet genetically understudied populations who also suffer from health disparities. These NHANES III data will integrate existing social, environmental, behavioral, and biologic data with genetic data to understand the determinants of health and health disparities in the U.S. (Chang et al., 2009).
However, before these impending analyses can be conducted, accurate information about familial relationships within households must be available. Related individuals in a household cannot be treated as an independent sample for genetic analyses. NHANES III collected no self-reported family relationship information. Instead, family relationships were reported with respect to a single person in the household who is often not in the sample (U.S. Department of Health and Human Services (DHHS). National Center for Health Statistics., 1996, see HFRELR). As a result, it is impossible to determine exactly the reported relationship between two sample members. For example, one cannot presume that the adult female sample persons in the household are the mothers of the children/youth sample people in the household. Thus the data on reported family relationships within NHANES III households are incomplete and inconclusive with regards to actual biological relatedness of family members.
We use the NHANES III genetic data to infer familial relationships within NHANES III households. DNA labs usually track biosamples using what is colloquially called a 'DNA fingerprint' (more properly, a DNA profile), a system of DNA loci useful for forensic identification. One popular system is AmpFlSTR® Identifiler® PCR Amplification Kit (Applied Biosystems, Foster City, CA, USA). Identifiler® contains the DNA loci used by the Combined DNA Index System (CODIS; http://www.fbi.gov/hq/lab/html/codis1.htm) that is commonly used by law enforcement agencies for forensic identification. While these loci have a track-record for addressing if two DNA profiles are from the same person (or, equivalently, identical twins), the performance of these loci for inferring family relationships more distant than identical twin is less understood (Bieber et al., 2006).
We assess the use of the Identifiler® DNA loci for inferring family relationships with nationally-representative survey data. We compared two methods that estimate the likelihood ratio that a pair of household members have a hypothesized relationship versus being unrelated. The first method (”exact method” (Evett and Weir, 1998, Ch. 5-8)) uses allele frequencies and the second (”IBS (Identical By State) method” (Presciuttini et al., 2002)) uses only the fact that alleles match between individuals. The exact method extracts information out of matches on rare alleles, as matching rare alleles are more indicative of a familial relationship than matching common alleles. However, the IBS method does not require allele frequencies and is thus robust to inaccurate or inappropriate allele frequencies. Since the genotyped DNA samples were cell lysates with widely varying DNA concentrations, we modified both methods to account for genotyping errors. Finally, we used a modification of the exact method to account for “cryptic relatedness” (Devlin and Roeder, 1999) (also called population substructure): the fact that all ostensibly unrelated humans still share small amounts of DNA from distant common ancestors. Cryptic relatedness implies that ostensibly unrelated individuals have a residual relatedness, which can violate the independence assumptions of standard methods for relationship inference. We assess how much cryptic relatedness reduces the evidence in favor of familial relationships. We also hope that this work will introduce survey statisticians to the swiftly-arriving era of genetic data from surveys.
A by-product of our work are the first explicitly nationally-representative and ethnicallyspecific estimates of these important allele frequecies. Our allele frequency estimates could be relevant to forensic calculations requiring U.S. population-based allele frequencies.
1.1 Data Description
During the second phase of NHANES III (1991-1994), lymphocytes were frozen and cell lines were immortalized to create a DNA bank. Genetic variation data were collected from 7,159 participants aged 12 years and older. DNA was extracted by cell lysis and the genotyping used in this paper was conducted by the Core Genotyping Facility at the National Cancer Institute (http://cgf.nci.nih.gov). See (Chang et al., 2009) for all details.
We use genetic data from Identifiler® for each participant. Identifiler® tests for genetic variants at 15 DNA loci called Short Tandem Repeats (STRs). STRs are multiple copies of an identical DNA sequence arranged in direct succession in a particular region of a chromosome (Butler, 2006). For example, the DNA locus D7S820 is in Figure 1. This locus is on chromosome 7 (hence the D7). In the middle of this locus, the tetranucleotide sequence gata is repeated 13 times. The number of repeats names the genetic variant (called an allele), and a person has two alleles (one on each chromosome 7 inherited from the mother and father). D7S820 typically has 6-14 gata repeats. However, there can be variants in the repeated sequence motif as well; for example, the allele named 13.1 has an extra DNA base inserted in the sequence of 13 ”gata” repeats in D7S820. See (Butler, 2006) for details on each possible allele.
Figure 1.
DNA locus D7S820 with the tetranucleotide motif repeat gata upcased. This version has 13 gata repeats, so is named allele 13. The locus is broken into chunks of length 10 for ease of counting the position of each nucleotide (the numbers give the position of the nucleotide at the far left). The DNA bases are a for adenine, c for cytosine, g for guanine, and t for thymine.
Identifiler® contains the 13 CODIS loci commonly used by law enforcement agencies for forensic identification: TPOX, CSF1PO, D5S818, D13S317, D16S539, TH01, D18S51, D7S280, VWA, FGA, D3S1358, D8S1179, D21S11; Identifiler® also includes D19S433 and D2S1338. Both CODIS and Identifiler® also have the STR AMEL, but AMEL provides information only on sex. For all details on these loci, see (Butler, 2006).
A fictitious example of a participant's DNA profile is in Figure 1. Each allele at each locus is shown, e.g. 13/10 means alleles 13 and 10 are observed. The pair of alleles is called the genotype. We also have the demographic variables of race/ethnicity, sex, and age. Sex and age for each pair of household members can help narrow down the possible familial relationships, and ethnicity is needed to select the proper allele frequencies to use in relationship inference. Given a feasible region of familial relationships, we use the genetic information to infer family relationships.
From the 7159 participants, we excluded 346 due to poor DNA quality or low DNA concentration (samples with less than 250 relative flourescence units; these samples had data at fewer than 12 of the 16 Identifiler® loci). Furthermore, 72 participants who had a mismatch between the reported sex and the (AMEL) genetically-determined sex (indicative of lack of data quality) were excluded, yielding 6741 participants. The distribution of genotyped household size is 1:2781, 2:1070, 3:329, 4:137, 5:27, 6:13, 7:3, 8:4, 9:1, and 11:1. The genotyped household size does not count individuals who were not genotyped. Thus 3960 were in multiple-person households, yielding 3610 possible pairs of genotyped relatives within households. The 2781 participants who are the only genotyped member of their household are included to estimate allele frequencies. To estimate nationally-representative allele frequencies, NCHS statisticians provided a sample weight for each participant to weight our dataset up the U.S. population. We categorized the race/ethnicity of participants as ’non-Hispanic White’, ’non-Hispanic Black’, and ’Mexican-American’. Participants who self-identified as Mexican-American in NHANES III represent a heterogeneous race-ethnic population of primarily Hispanic American Indian and Hispanic White. Because specific information on which current Office of Management and Budget categorization each of these participants represents is not available, we will use the term Mexican-American for the purposes of this publication.
2 Methods
Denote the genotype (the pair of alleles) for participant k at locus j as lj,k. The full DNA profile of the 15 Identifiler® loci is Pk = (l1,k, l2,k, …, l15,k). Statistical evidence in favor of a hypothesized familial relationship R (such as parent-child, full-siblings, etc.) between the two participants providing DNA profiles (P1, P2) is measured by the likelihood ratio (LR)
| (1) |
We use the exact method and the IBS method to compute the LR as well as maximum likelihood estimates of relationships.
2.1 Exact Method
The likelihood for two profiles P1, P2 within the same household given a relationship is
| (2) |
because the loci are on different chromosomes and are thus independent.
To make further progress, relationships can be parameterized in terms of Identical-By-Descent (IBD) probabilities. Two people can share 0, 1, or 2 alleles IBD. For the pair in Table 1, they share 0 alleles IBD at D13S317, at most 1 allele IBD at D16S539 (at most 1 because their matching allele 11 could be from two different ancestors, so they could share 0 alleles IBD), and at most 2 alleles IBD at CSF1PO. The probability of sharing i alleles IBD is denoted by ki and Σki = 1.
Table 1.
Example Identifiler® genetic test results and demographic variables for two fictitious household members.
| AMEL | CSF1PO | D13S317 | D16S539 | D18S51 | D19S433 | D21S11 | D2S1338 | D3S1358 | |
|---|---|---|---|---|---|---|---|---|---|
| X/Y | 7/7 | 13/10 | 11/13 | 16/21 | 30/31 | 21/24 | 22/16 | 17/15 | |
| X/X | 7/7 | 11/12 | 11/14 | 16/20 | 26/31 | 21/22 | 22/15 | 17/17 | |
|
| |||||||||
| D5S818 | D7S820 | D8S1179 | FGA | TH01 | TPOX | vWA | Sex | Race | Age |
|
| |||||||||
| 10/11 | 10/14 | 14/14 | 24/28 | 9/9 | 8/8 | 17/20 | Male | non-Hispanic white | 17 |
| 9/12 | 10/14 | 14/13 | 23/27 | 9/9 | 8/8 | 17/20 | Female | non-Hispanic white | 14 |
All familial relationships are defined by their IBD probabilities (Thompson, 1991) (Table 2). For example, a person must share both alleles IBD within himself or his monozygotic (identical) twin. Two unrelated people cannot share any alleles IBD (they can merely appear to share to due to chance; their shared alleles would be from different ancestors and so cannot be IBD). Since each parent contributes 1 allele at each locus for their child, they must share exactly 1 allele IBD. For example, the pair in Table 1 cannot be the same person or monozygotic twins, and they cannot be parent-child because they share no alleles at D13S317. As Table 2 shows, the 2nd degree relationships (grandparent-grandchild, uncle-nephew, half-sibling) have the exact same IBD probabilities and so cannot be distinguished based on IBD alone. However, IBD plus age information usually suffices. We note that 2nd degree relationships can be distinguished from each other by using correlated genetic loci (McPeek and Sun, 2000).
Table 2.
IBD probabilities k0, k1, and k2 for defined relationships. 2nd degree relationships include half-siblings, uncle-nephew, and grandparent-grandchild.
| Relationship | k0 | k1 | k2 |
|---|---|---|---|
| Self or identical twin | 0 | 0 | 1 |
| Unrelated | 1 | 0 | 0 |
| Parent-Child | 0 | 1 | 0 |
| Full Sibilings | 0.25 | 0.5 | 0.25 |
| 2nd Degree | 0.5 | 0.5 | 0 |
| Cousin | 0.75 | 0.25 | 0 |
Using IBD, the likelihood for two profiles is
| (3) |
The second term is the k0, k1 and k2 probabilities for each relationship in Table 2. The genotype probabilities P(lj,1, lj,2) are independent of R given the IBD sharing because IBD defines which alleles are fixed (from a common ancestor) and the others are random.
The genotype probability calculations P(lj,1, lj,2|IBD = i) are in Table 3 (Thompson, 1991). Although the Identifiler® alleles are labeled by numbers, we label the four alleles in a pair of genotypes generically by A, B, C, D. The constant factors of two and four in Table 3 reflect the fact that alleles within genotype are unordered (i.e. AB is equivalent to BA). When IBD=0, the two genotypes are independent, so P(lj,1, lj,2|IBD = 0) = P(lj,1)P(lj,2). When IBD=2, the two genotypes are completely dependent, so P(lj,1,lj,2|IBD = 2) = P(lj,1) = P(lj,2). When IBD=1, the calculations are more complex, e.g.
because the conditional probability involves the probabilities of having an A, the probability the IBD allele is indeed A (1), and the probability that the IBD allele is in the second position (0.5) (first term) or in the first position (0.5) (second term). See (Wagner et al., 2006; Evett and Weir, 1998) for more details. In Table 3, we also represent each probability as the probability over the alleles not IBD, so that, e.g. P(lj,1 = AA, lj,2 = AA|IBD = 1) = P(AAA). This notation will be convenient for section 2.1.1 that relaxes the assumption of independent non-IBD alleles.
Table 3.
Probability of observing genotypes given IBD sharing.
| Genotypes lj,1, lj,2 | IBD=0 | IBD=1 | IBD=2 | |||
|---|---|---|---|---|---|---|
| AA,AA |
|
|
|
|||
| AA,AB |
|
|
0 | |||
| AA,BB |
|
0 | 0 | |||
| AA,BC |
|
0 | 0 | |||
| AB,AB |
|
P(AAB) + (ABB) = pApB(pA + pB) | 2P(AB) = 2pApB | |||
| AB,AC |
|
P(ABC) = pApBpC | 0 | |||
| AB,CD | 4P(ABCD) = 4pApBpCpD | 0 | 0 |
We note that throughout this paper, we assume that no participants within households are inbred and that all particiants between households are unrelated. Furthermore, we assume that no loci are missing data in any way informative of relatedness; in this way, the product (2) can be safely done over the observed loci alone.
2.1.1 Accounting for Cryptic Relatedness
Identifiler® allele frequencies vary between ethnicities, and even within ethnicity, allele frequencies can vary between subpopulations within ethnicities (Budowle et al., 2001). For example, the particular European/African ancestry of the non-Hispanic whites/blacks in NHANES III is not collected, and allele frequencies can vary within these groups. The effect of unknown subpopulations means that heterogeneous allele frequences between the unknown subpopulations will cause intraclass correlation of alleles within ethnicity (Devlin and Roeder, 1999), violating the independence assumption required to calculate the genotype probabilities of Table 3. Furthermore, all unrelated humans still share small amounts of DNA IBD from distant common ancestors, and this cryptic relatedness (Devlin and Roeder, 1999) results in nebulous subpopulations that further increase the intraclass correlation.
Genotype probability calculations can be extended to account for the intraclass correlation of alleles within ethnicity, called FST (Wright, 1969) (or the coancestry coefficient (Evett and Weir, 1998)). FST is positive and can be interpreted as the probability that two alleles are IBD from an unknown common ancestor. FST is accounted for by using a Dirichlet-Multinomial distribution (Evett and Weir, 1998, pg. 123-5). The genotype probability calculation for a single participant is altered as (Balding and Nichols, 1994)
since with probability FST the two A's are IBD from a distant common ancestor. Similarly,
since different alleles cannot be IBD. The genotype probability calculations for pairs of genotypes (for IBD=0) can be calculated using the Dirichlet-Multinomial recursion relation (Balding and Nichols, 1995)
The recursion relation is used to calculate the probability of observing the alleles that are not IBD; these alleles are no longer independent but have correlation FST . For concreteness, the genotype probabilty calculations for these IBD=0 alleles are
For example, P(AAAA) = P(A|AAA)P(AAA). Under independent alleles P(A|AAA) = P(A), but with positive intraclass correlation FST , the probability of observing another A given that 3 A's have been observed is higher and is specified by the recursion relation. Similarly, P(ABCD) = P(D|ABC)P(ABC) and P(D|ABC) conditions on not having observed D before, so the probability of observing D decreases.
To calculate the final genotype probability, plug in the above expressions into Table 3. Expressions for the LR accounting for FST exist (Ayres, 2000), but the above likelihood contributions are needed for maximum likelihood estimation of relationships via estimating the IBD probabilities k0, k1, k2.
2.2 IBS Method
The IBS (Identical By State) method (Chakraborty and Jin, 1993; Presciuttini et al., 2002) estimates the LR using only the fact that alleles match at each locus. For the example pair of Table 1, CSF1PO is considered a match on two alleles, D13S317 a match on zero alleles, and D16S539 a match on one allele. The IBS method relies on heterozygosity Hj, the probability that the two alleles at locus j are different. The probability that i = 0, 1, 2 alleles match at locus j in profiles P1 and P2 for a given relationship R is denoted z(i|Hj, R). The z(i|Hj, R) as a function of heterozygosity at each locus, familial relationship, and for each i = 0, 1, 2 are empirically by cubic functions with little residual variation (Presciuttini et al., 2002, Fig. 1). The empirical estimates of the cubic functions ẑ(i|Hj, R) are available (Presciuttini et al., 2002, Table 2), and the IBS method estimates the LR in (1) as
| (4) |
The IBS method does not distinguish between types of alleles and has no need for allele frequencies, and thus loses information versus the exact method by ignoring the rarity or commonality of matches. But the IBS method is robust when allele frequencies are unavailable or inappropriate. For example, in a mass disaster (such as a plane crash), allele frequencies are unavailable for use to match DNA from the remains with DNA samples provided by relatives. For another example, it is unclear how allele frequencies from nonU.S.-population-based databases of DNA profiles are for use in the general population.
2.3 Accounting for Genotyping Errors
As noted in section 1.1, the DNA was extracted by cell lysis, a sub-optimal method of DNA extraction that could introduce more genotyping errors than ordinarily expected. We adopt a simple model that the true genotype is observed with probability 1−∈, but with probability ∈, the observed genotype is drawn randomly from the population (Broman and Weber, 1998; Epstein et al., 2000). The genotype probability calculations of section 2.1 and 2.1.1 in Table 3 are altered as
| (5) |
because a randomly drawn genotype from the population has no IBD sharing. The exact LR under errors takes a simple form. By (3), the contribution each locus j makes to the usual LR is
| (6) |
With errors (denoting e = (1 − (1 − ∈)2)), the contribution is now
Since the IBS LR is meant to estimate the exact LR, we can use the above functional form to modify the contributions to the IBS LR in (4).
We note that STRs can, on rare occasions, spontaneously mutate. Thus the overall genotyping error rate combines both measurement error and mutation. Since the cells were crudely lysed to extract the DNA, we believe that measurement error dominates the error rate parameter.
2.4 Maximum-Likelihood Estimation of Relationships
Instead of hypothesis testing for relationships, the best relationship can be directly estimated with maximum-likelihood estimates of the IBD probabilities k0, k1 and k2 (Milligan, 2003). We maximize the exact likelihood (3), modifying the genotype probability calculation P(lj,1, lj,2|IBD = i) to account for errors as in (5) and for cryptic relatedness as in section 2.1.1. Within the simplex formed by k0, k1 and k2, the feasible region of maximization for non-inbred families is (Thompson, 1991).
3 Inferring Family Relationships in NHANES III
3.1 Allele Frequencies
Allele frequencies for the U.S. and for each ethnicity (non-Hispanic white, non-Hispanic black, Mexican American) were estimated in the standard way of estimating a proportion using sample weights in a Horvitz-Thompson estimator (ignoring finite population corrections) (Raj, 1968, pg. 42).
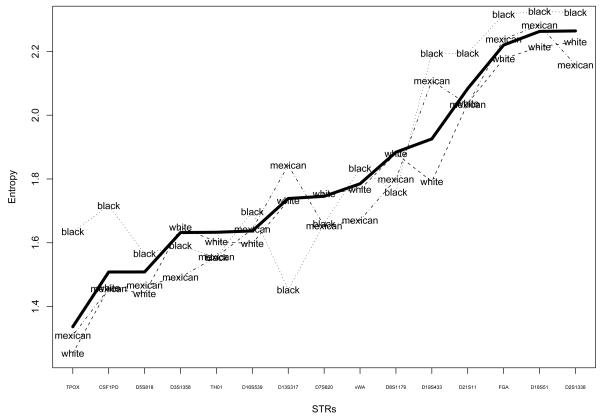
To assess the informativeness of a locus, we calculate the entropy of its allele frequency distribution. The entropy is the sum over each allele i of −piln(pi) where pi is the frequency of allele i. A locus with high entropy will have many alleles and low allele frequencies, and so can better distinguish people than a low entropy locus. Figure 2 plots the entropies for the U.S., each ethnicity, and for each locus, ordered by the entropy of the loci for the U.S.. The least informative locus is TPOX, for which only two alleles account for over 75% of its alleles; at D2S1338 the top two alleles account for only 35% of its alleles. The allele distributions for Non-Hispanic blacks generally have more entropy than those for other ethnicities, especially for the least- and most- informative loci. Thus the Non-Hispanic blacks appear to have more genetic diversity than the other ethnicities in NHANES III.
Figure 2.
Entropy of allele distributions for each locus, for overall U.S. population (thickest line) and for each ethnicity.
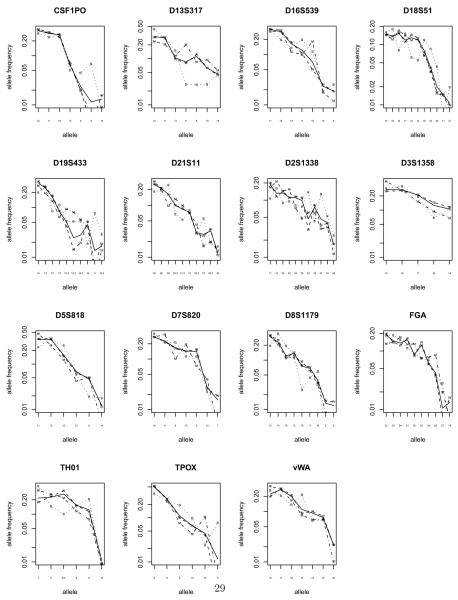
Figure 3 shows the actual allele frequecies. Alleles are ordered by frequency in the U.S. population and alleles with frequency < 1% are not shown. Some loci have only 5 alleles with frequency >= 1%, some have as many as 11. Most loci have many alleles with frequency 1 – 5%, and the presence of such alleles can be very informative for inferring family relationships. D2S1338 has the highest entropy, due to having 11 alleles, many with frequency 1 – 5%. D3S1358 has a flat allele distribution, but only 5 alleles, so has low entropy. TPOX has a sharply dropping distribution, emphasizing its low entropy. There are clear ethnic differences in allele frequencies at many loci, especially for non-Hispanic blacks (e.g. D13S317, CSF1PO, D18S51 ).
Figure 3.
NHANES III Identifiler® allele frequencies (> 1%) for the U.S. (US) [thickest line] and by ethnicity: (W) non-Hispanic white, (B) non-Hispanic black, (M) Mexican-American.
3.2 Exact method vs. IBS method
We classified the most likely relationship for a pair of household members by the highest exact or IBS LR in favor of that relationship. If each LR for each relationship for a pair is less than one, we classify the pair as most likely unrelated. When the pair reported the same ethnicity, the LR used the allele frequencies for that ethnicity. The overall U.S. allele frequencies were used for the LRs for the 54 pairs reporting different ethnicities.
Table 4 classifies the most likely relationship for a pair of household members (highest LR in favor of that relationship) by the exact (FST = 0 and ∈ = 0) and IBS methods. The two methods strongly agree on which pairs are most likely parent-child or siblings. No IBS LR for cousin was presented in Presciuttini et al. (2002), so for the cousin pairs by the exact method, the IBS LR parcels them out to 2nd degree and unrelated. The Spearman correlations of the exact and IBS LR for parent-child, sibling, and 2nd-degree are 0.97, 0.98, 0.94 respectively, underscoring that the two methods rank relationships equally. This strong agreement changes negligibly with different FST or ∈.
Table 4.
Counts of most likely familial relationship, by exact and IBS methods, for each pair.
| most likely by IBS | |||||||
|---|---|---|---|---|---|---|---|
| unrelated | parent-child | Sibling | 2nd Degree | cousin | Sum | ||
| unrelated | 1173 | 0 | 0 | 35 | 0 | 1208 | |
| parent-child | 0 | 1158 | 16 | 0 | 0 | 1174 | |
| most likely by exact | Sibling | 0 | 8 | 436 | 28 | 0 | 472 |
| 2nd Degree | 10 | 0 | 10 | 308 | 0 | 328 | |
| 1st-cousin | 259 | 0 | 3 | 166 | 0 | 428 | |
| Sum | 1442 | 1166 | 465 | 537 | 0 | 3610 | |
We considered whether the most likely relationship is consistent with the reported ages. Only 5% (59) of the parent-child pairs had an age difference of under 16 years and 9% (42) of the sibling pairs had an age difference over 25 years. A more refined analysis might attribute these pairs to another likely relationship consistent with the ages of the pair. Furthermore, seven pairs had identical observed DNA profiles, implying that they are either identical twins or they are the same individual (some of these pairs have differing ages or ethnicities).
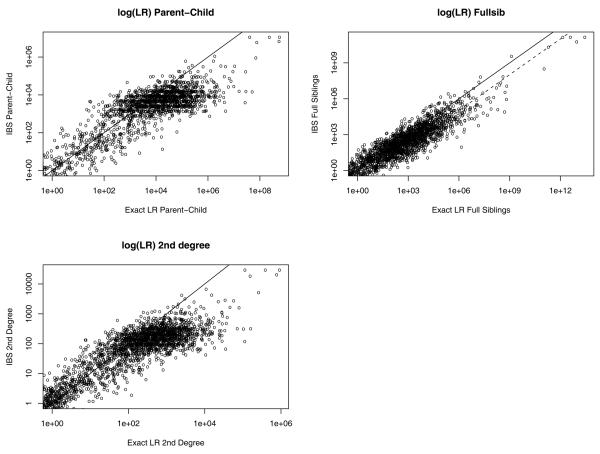
Figure 4 plots the exact and IBS LRs for three relationships, limiting to pairs where either the exact or IBS LR is greater than one. For siblings, the IBS LR underestimates the exact LR (a smooth loess curve is added to make this clear). For parent-child and 2nd-degree, the underestimation is pronouced at higher exact LRs, where the true parent-child or 2nd-degree pairs are likely to be. So while the two methods agree on the ranking of relationships, they can disagree on the quantification of the LR.
Figure 4.
Exact LR vs. IBS LR for 3 familial relationships, with line where the LRs are equal.
3.3 Distribution of exact LRs by ethnicity, error rate, and FST
We did not estimate error rates or FST from our complex survey data, but instead assessed sensitivity to plausible values. We observed a 1% sex mismatch rate (section 1.1), suggesting that perhaps a 2% error rate overall is reasonable; we also considered 0% and 4%. A National Research Council report recommends using FSTs of 1 – 3% (National Research Council II Report, 1996). We considered FSTs of 0%, 1%, and 3%.
Table 5 shows the distribution of the exact LR for the most likely relationship by FST. The most striking observation are the rather low LRs for 2nd-degree, cousin, and unrelated, suggesting that the Identifiler® loci are not informative enough to conclusively determine these three relationships. Second, the parent-child and sibling LRs are sensitive to FST , with median LRs changing by factors of 3-7 as FST increases, and Q3 LRs changing by factors of 10. Although these intraclass correlations (FST) are small, the exact LR changes a lot because the exact method derives powerful information from matching on rare alleles (Ayres, 2000). Any non-zero FST implies that a match on rare alleles could well be a result of sharing unknown distant relatives rather than sharing a close familial relationship. This result is analogous to the inflation of the variance under cluster sampling with a small intraclass correlation but large clusters (Korn and Graubard, 1999).
Table 5.
Distribution of exact LRs by best relationship.
| Relationship | FST | Min | Q1 | Med | Mean | Q3 | Max |
|---|---|---|---|---|---|---|---|
| Parent-child | 0% | 1.54 | 10100 | 66000 | 31400000 | 514000 | 1.45e+10 |
| 1% | 1.49 | 3070 | 14700 | 239000 | 73100 | 5.04e+07 | |
| 3% | 1.41 | 429 | 1750 | 10100 | 6550 | 8.65e+05 | |
|
| |||||||
| Full-sibling | 0% | 2.23 | 1200 | 36700 | 1.32e+12 | 1160000 | 4.20e+14 |
| 1% | 2.15 | 421 | 11400 | 9.48e+10 | 213000 | 2.77e+13 | |
| 3% | 2.02 | 135 | 2440 | 2.99e+09 | 32400 | 6.53e+11 | |
|
| |||||||
| 2nd-Degree | 0% | 1.59 | 5.20 | 13.20 | 121 | 40 | 8370 |
| 1% | 1.44 | 4.42 | 9.45 | 27 | 23 | 730 | |
| 3% | 1.36 | 3.28 | 6.26 | 9 | 10 | 80 | |
|
| |||||||
| Cousin | 0% | 1 | 1.24 | 1.59 | 2.11 | 2.20 | 68.40 |
| 1% | 1 | 1.21 | 1.49 | 1.68 | 1.94 | 4.93 | |
| 3% | 1 | 1.12 | 1.34 | 1.46 | 1.68 | 4.29 | |
|
| |||||||
| unrelated | 0% | 0.0339 | 0.234 | 0.385 | 0.434 | 0.616 | 1.000 |
| 1% | 0.0329 | 0.220 | 0.367 | 0.416 | 0.587 | 0.999 | |
| 3% | 0.0314 | 0.189 | 0.314 | 0.373 | 0.523 | 0.998 | |
Table 6 shows the counts of the best relationship (by exact LR) by error rates and FST . Increasing the error rate increases the number of parent-child relationships because a single genotyping error causing a perfect mismatch at a locus eliminates the possibility of a parent-child relationship. Allowing for an error rate removes this possibility, allowing the other loci to contribute meaningfully to the parent-child LR. Sibling relationships are not sensitive to error rates. 2nd-degree and cousin relationships are sensitive, mostly because the LRs in favor of these relationships are very small and are vulnerable to small changes. Increasing FST decreases the counts of parent-child and sibling relationships on the order of 5%. Thus FST has little effect on the determination of the most likely relationship, but strongly affects the quantification of the LR in its favor.
Table 6.
Counts of best relationship by FST and error rates.
| error | FST | unrelated | parent-child | sibling | 2nd degree | cousin |
|---|---|---|---|---|---|---|
| 0% | 0% | 1208 | 1174 | 472 | 328 | 428 |
| 0% | 1% | 1343 | 1173 | 456 | 293 | 345 |
| 0% | 3% | 1531 | 1174 | 419 | 255 | 231 |
|
| ||||||
| 2% | 0% | 1197 | 1236 | 460 | 297 | 420 |
| 2% | 1% | 1336 | 1223 | 448 | 267 | 336 |
| 2% | 3% | 1522 | 1207 | 430 | 221 | 230 |
|
| ||||||
| 4% | 0% | 1186 | 1262 | 470 | 275 | 417 |
| 4% | 1% | 1325 | 1254 | 456 | 242 | 333 |
| 4% | 3% | 1517 | 1223 | 435 | 212 | 223 |
Table 7 shows the distribution of the exact LR by most likely relationship, by ethnicity. We fixed ∈ = 2%, FST = 1% as our most plausible values. When parent-child is most likely, the LR for non-Hispanic blacks tends to be the highest, possibly due to greater entropy in the non-Hispanic black allele frequencies. However, when sibling is most likely, the LRs seem somewhat more comparable, although somewhat lower for Mexican Americans.
Table 7.
Distribution of exact LRs by ethnicity when parent-child or full-sibling are the most likely relationship, ∊ = 2%, FST = 1%.
| Parent-Child most likely | ||||||
|---|---|---|---|---|---|---|
| Ethnicity | Min | Q1 | Med | Mean | Q3 | Max |
| non-Hispanic white | 2.22 | 2610 | 10600 | 92400 | 55600 | 2760000 |
| non-Hispanic black | 1.49 | 4810 | 17800 | 293000 | 88100 | 29000000 |
| Mexican American | 1.52 | 2140 | 13300 | 130000 | 64800 | 6610000 |
|
| ||||||
| Full-sibling most likely | ||||||
| Ethnicity | Min | Q1 | Med | Mean | Q3 | Max |
| non-Hispanic white | 4.30 | 1050 | 14900 | 1.35e+11 | 175000 | 9.19e+12 |
| non-Hispanic black | 7.06 | 599 | 17500 | 2.63e+11 | 617000 | 2.77e+13 |
| Mexican American | 2.21 | 278 | 6730 | 4.83e+08 | 151000 | 1.09e+11 |
3.3.1 Ability to infer unrelated individuals
Without complete and conclusive reporting of family history, we cannot formally verify how close the inferred familial relationships are to the truth. But as an approximation to truly unrelated individuals, we consider to be unrelated the 500 household pairs where either member is over the age of 40, have ages within 12 years of each other, and are of opposite sexes. Most likely, these are married or unmarried couples, i.e., pairs who are highly likely to be unrelated. We use the LR assuming ∈ = 2% and FST = 1%. To make decisions about relationships, we have set LR cutoffs. To be conservative, we consider an LR > 104 to be strong evidence for the relationship. Since we expect far more unrelated individuals than second-degree relationships, we consider an LR < 103 to be evidence for being unrelated. We are equivocal for LRs between 103 and 104.
Of these 500 pairs, the LR maximizes at unrelated for 382. Another 17 and 86 LRs maximize at halfsib or cousin, respectively. The maximum LR for halfsib is only 16 and for cousin is merely 3, indicating that each of these 103 pairs are most likely unrelated. Four pairs had maximum LR at parent-child (with maximum LR of 457), but these pairs all had age differences under 7 years, so they are not parent-child relationships, and their small LRs indicate that they are most likely unrelated. The remaining 11 pairs have maximum LR for full siblings; 6 have LR under 1000 (most likley unrelated), one has LR of 6000 (equivocal), the remaining four have LRs of 106, 107, 108, and 1013 (overwhelmingly full sibling). Thus we believe the genetic data naturally infers the 495 unrelated pairs in this group of 500 pairs, identifies another 4 who are most likely full siblings, and only one pair is unresolved.
3.4 Inferring Household Family Structure
The LRs can be used to help infer the most likely family structure within each household. We can infer family structure only amongst household members with genotyping results. We use only the exact LR assuming ∈ = 2% and FST = 1%. We restrict our presentation to households with two- or three-persons with genotyping results (88% of the households) as larger households are more likely to contain half-siblings and cousins, non-immediate relationships that our LR has little ability to detect. We assume that all pairs with LRs maximizing at half-sibling or cousin are truly unrelated. A thorough analysis that infers family structure by carefully considering age, race, ethnicity, and other demographic variables, especially to account for household members without genotyping results (who are not in our dataset), is beyond the scope of this article.
For the 1070 households with two individuals with genotyping results, the LR maximizes at: 576 unrelated, 350 parent-child, and 144 full-sibling relationships. Purely full-sibling relationships in a two-person household may imply the presence of other household members for whom we do not have genotyping results. For the 329 households with three individuals with genotyping results, the LR maximixes at: 95 2-parent 1-child trios, 81 parent-child plus an unrelated, 51 single parent raising two full-siblings, 19 unrelated person raising 2 full-siblings, 53 completely unrelated, and 30 maximized at inbred or impossible family structures.
3.5 IBD probability estimates
We estimated maximum-likelihood estimates of the IBD probabilities k0, k1 and k2 numerically using the Nelder-Mead simplex algorithm as implemented by the R function optim(). We fixed ∈ = 2% and FST = 1% as our most plausible values. It took 5 hours on a Pentium 4 3Ghz computer to compute IBD probabilities for all 3610 pairs.
Table 8 shows the distribution of the estimated IBD probabilites by ethnicity. For parent-child, there is not much difference in the distributions of k̂1, k̂2 by ethnicity. For siblings, non-Hispanic whites in our NHANES III sample tend to have the highest k̂1, k̂2, followed by non-Hispanic blacks for k̂2. In particular, both of their median k̂2 are elevated over 0.25 and the non-Hispanic white median k̂1 = 0.524 is also elevated over 0.5. These slight elevations suggest that non-Hispanic white siblings in our NHANES III sample may be more closely related than expected, and suggest the presence of cryptic relatedness.
Table 8.
Distribution of IBD probabilities k̂1, k̂2 for most likely parent-child and sibling relationships (∊ = 2%, FST = 1%).
| most likely parent-child | |||||
|---|---|---|---|---|---|
| Ethnicity | Q1 | Med | Mean | Q3 | |
| non-Hispanic white | 0.910 | 1.000 | 0.953 | 1 | |
| k̂ 1 | non-Hispanic black | 0.939 | 1.000 | 0.959 | 1 |
| Mexican American | 0.901 | 1.000 | 0.949 | 1 | |
|
| |||||
| non-Hispanic white | 0 | 0.0000 | 0.0426 | 0.0754 | |
| k̂ 2 | non-Hispanic black | 0 | 0.0000 | 0.0354 | 0.0485 |
| Mexican American | 0 | 0.0000 | 0.0446 | 0.0763 | |
|
| |||||
| most likely siblings | |||||
| Ethnicity | Q1 | Med | Mean | Q3 | |
| non-Hispanic white | 0.500 | 0.524 | 0.548 | 0.578 | |
| k̂ 1 | non-Hispanic black | 0.498 | 0.500 | 0.512 | 0.550 |
| Mexican American | 0.500 | 0.500 | 0.540 | 0.569 | |
|
| |||||
| non-Hispanic white | 0.246 | 0.299 | 0.281 | 0.302 | |
| k̂ 2 | non-Hispanic black | 0.231 | 0.291 | 0.302 | 0.300 |
| Mexican American | 0.198 | 0.250 | 0.249 | 0.300 | |
An advantage of estimating IBD probabilities is flagging potentially non-standard relationships. Twenty pairs had 0.4 ≤ k̂2 < 1; these are non-standard (possibly inbred) familial relationships for which we do not compute an LR.
4 Discussion
The NHANES III genetics data will be an unparalleled resource for incorporating genetics into a comprehensive understanding of the determinants of health in the U.S. and for developing, targeting, and evaluating policies for disease prevention that use genetic information. However, these analyses are handicapped until family relationships amongst household members are inferred. We evaluated two methods for relationship inference, the exact and IBS methods, and find that while they often agree on the most likely relationship, the IBS method generally underestimates the LR in favor of a relationship. This underestimation occurs because the exact method can take advantage of the informativeness of matches on rare alleles. The IBS method is robust to inadequate or inappropriate allele frequencies, but the NHANES III allele frequencies are a large population-based sample, so the exact method seems appropriate (notwithstanding possible concern about the stability of the less common (1 – 5%) allele frequency estimates). Accounting for genotyping errors and FST is critical for quantifying the LR, but has little effect on which relationship is judged most likely. The genotyping error rate has most effect on parent-child relationships. LRs for non-Hispanic blacks tend to be most informative because their allele frequencies have the most entropy.
Other health surveys worldwide have begun collecting genetic information, such as the Health 2000 survey (Samani et al., 2008) and the Canadian Health Measures Survey. We expect that future health surveys will routinely collect genetic information. Our results are likely relevant to other surveys since Identifiler® is usually conducted for specimen tracking by most DNA labs.
Furthermore, even if family relationships are believed to be accurately reported, genetic data are critical to verify reported relationships and identify data quality issues. We noted an observed gender/chromosomal sex mismatch rate of 1% (so overall would be about 2%). In our experience with other datasets, we have observed non-trival sex mismatch rates of 1-4%. In section 3.2, we found that 5% of the parent-child pairs and 9% of the full-sibling pairs had implausible reported ages, seven participants were either identical twins with household members or else duplicates in the data, and twenty pairs may have non-standard (inbred) relationships. Furthermore, our methods could also be used to detect unsuspected familial relationships across households. Discrepancies could reflect either on misreporting by household members (perhaps lack of knowledge of true paternity) or on specimen handling/analysis problems in the survey. Regardless of the source of the discrepancy, the genetic analysis helps identify such problems.
Both the exact and IBS methods can also be viewed from the perspective of probabilistic record linkage (Herzog et al., 2007) because they compare two data vectors for matches. For relationship inference, there are ”record linkages” of different types based on the different possible relationships. The difference between the exact and IBS methods is whether to extract information from the commonality or rareness of matches, akin to a similar debate in the record linkage literature (Herzog et al., 2007, Ch. 9).
While the LR can be used to infer the most likely familial relationship, estimating IBD probabilities has two advantages. IBD probabilities provide a continuous measure of the amount of DNA shared by two household members. All relatives have, only on average, the IBD sharing in Table 2, and the estimated IBD probabilities estimate the true IBD sharing. Another advantage of IBD probability estimates is their ability to improve regression modeling of survey data, regardless of whether the model uses genetic information, via specifying the correlation structure of a continuous outcome measured on household members. For example, a simplified model for the correlation of two household members' outcomes y1, y2 using the average IBD sharing I = 0k0 + 1k1 + 2k2 is
(Lange, 2002, pg 101). Specified within-household correlation of outcomes can be exploited in regression modeling to improve efficiency of parameter estimates (Korn and Graubard, 1999). This correlation matrix applies regardless of whether the model involves genetic information.
Our ethnically-specific allele frequecy estimates are unique because they are explicitly nationally-representative. Comparing our estimates to other established estimates (Budowle et al., 2001; Einum and Scarpetta, 2004) shows agreement on common allele frequencies, but disagreements on rarer (1 – 5%) allele frequencies that are the most informative yet most vulnerable to small uncertainties. Our estimates may be helpful for calculating the probability that a given DNA profile matches by chance with a random individual from the U.S. population, or to infer whether individuals in a genetic database may be relatives of an individual with a given DNA profile (Bieber et al., 2006).
Our STR loci are not informative enough to conclusively determine that pairs are 2nd-degree relatives, cousins, or unrelated. Relationship inference could be improved by using large numbers of Single Nucleotide Polymorphisms (SNPs) instead of STRs. The available SNPs in NHANES III were chosen as candidate polymorphisms from a priori hypotheses for association with diseases of potential public health significance (Chang et al., 2009). These SNPs are unlikely to be included as a group in SNP panels used for sample tracking, quality control and assessment of cyptic relatedness. However, future NHANES genotyping may involve dense genotyping panels including over one million SNPs, and these data will be important for resolving distant relationships.
Supplementary Material
Acknowledgments
We thank the National Center for Health Statistics for use of their Research Data Center to conduct this research. This research was supported in part by the Intramural Research Program of the NIH/National Cancer Institute.
Footnotes
Conflict of Interest: None declared.
References
- Ayres KL. Relatedness testing in subdivided populations. Forensic Sci Int. 2000;114(2):107–115. doi: 10.1016/s0379-0738(00)00289-9. [DOI] [PubMed] [Google Scholar]
- Balding DJ, Nichols RA. DNA profile match probability calculation: how to allow for population stratification, relatedness, database selection and single bands. Forensic Sci Int. 1994;64(2-3):125–140. doi: 10.1016/0379-0738(94)90222-4. [DOI] [PubMed] [Google Scholar]
- Balding DJ, Nichols RA. A method for quantifying differentiation between populations at multi-allelic loci and its implications for investigating identity and paternity. Genetica. 1995;96(1-2):3–12. doi: 10.1007/BF01441146. [DOI] [PubMed] [Google Scholar]
- Bieber FR, Brenner CH, Lazer D. Human genetics: Finding criminals through DNA of their relatives. Science. 2006;312(5778):1315–1316. doi: 10.1126/science.1122655. [DOI] [PubMed] [Google Scholar]
- Broman KW, Weber JL. Estimation of pairwise relationships in the presence of genotyping errors. Am J Hum Genet. 1998;63(5):1563–1564. doi: 10.1086/302112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budowle B, Shea B, Niezgoda S, Chakraborty R. CODIS STR loci data from 41 sample populations. J Forensic Sci. 2001;46(3):453–489. [PubMed] [Google Scholar]
- Butler JM. Genetics and genomics of core short tandem repeat loci used in human identity testing. J Forensic Sci. 2006;51(2):253–265. doi: 10.1111/j.1556-4029.2006.00046.x. [DOI] [PubMed] [Google Scholar]
- Chakraborty R, Jin L. Determination of relatedness between individuals using DNA fingerprinting. Hum Biol. 1993;65(6):875–895. [PubMed] [Google Scholar]
- Chang M-H, Lindegren ML, Butler MA, Chanock SJ, Dowling NF, Gallagher M, Moonesinghe R, Moore CA, Ned RM, Reichler MR, Sanders CL, Welch R, Yesupriya A, Khoury MJ, CDC/NCI NHANES III Genomics Working Group Prevalence in the united states of selected candidate gene variants: Third National Health and Nutrition Examination Survey, 1991–1994. Am J Epidemiol. 2009;169(1):54–66. doi: 10.1093/aje/kwn286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanock SJ, Hunter DJ. Genomics: when the smoke clears …. Nature. 2008;452(7187):537–538. doi: 10.1038/452537a. [DOI] [PubMed] [Google Scholar]
- Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Easton DF, Eeles RA. Genome-wide association studies in cancer. Hum Mol Genet. 2008;17(R2):R109–R115. doi: 10.1093/hmg/ddn287. [DOI] [PubMed] [Google Scholar]
- Einum DD, Scarpetta MA. Genetic analysis of large data sets of North American Black, Caucasian, and Hispanic populations at 13 CODIS STR loci. J Forensic Sci. 2004;49(6):1381–1385. [PubMed] [Google Scholar]
- Epstein MP, Duren WL, Boehnke M. Improved inference of relationship for pairs of individuals. Am J Hum Genet. 2000;67(5):1219–1231. doi: 10.1016/s0002-9297(07)62952-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evett I, Weir B. Interpreting DNA Evidence: Statistical Genetics for Forensic Scientists. Sinauer Associates; Sunderland, MA: 1998. [Google Scholar]
- Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7(2):85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- Gail MH. Discriminatory accuracy from single-nucleotide polymorphisms in models to predict breast cancer risk. J Natl Cancer Inst. 2008;100(14):1037–1041. doi: 10.1093/jnci/djn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog TN, Scheuren FJ, Winkler WE. Data Quality and Record Linkage Techniques. Springer; 2007. [Google Scholar]
- Korn EL, Graubard BI. Analysis of Health Surveys. John Wiley & Sons; 1999. [Google Scholar]
- Lange K. Mathematical and Statistical Methods for Genetic Analysis. Springer-Verlag Inc; 2002. [Google Scholar]
- Lin BK, Clyne M, Walsh M, Gomez O, Yu W, Gwinn M, Khoury MJ. Tracking the epidemiology of human genes in the literature: the huge published literature database. Am J Epidemiol. 2006;164(1):1–4. doi: 10.1093/aje/kwj175. [DOI] [PubMed] [Google Scholar]
- McPeek MS, Sun L. Statistical tests for detection of misspecified relationships by use of genome-screen data. Am J Hum Genet. 2000;66(3):1076–1094. doi: 10.1086/302800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan BG. Maximum-likelihood estimation of relatedness. Genetics. 2003;163(3):1153–1167. doi: 10.1093/genetics/163.3.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council II Report . The evaluation of forensic evidence. National Academy Press; Washington D.C.: 1996. Technical report. [Google Scholar]
- NCHS National center for health statistics. plan and operation of the third national health and nutrition examination survey, 1988-94. Vital and Health Statistics. 1994;1(32) [PubMed] [Google Scholar]
- Pharoah PDP, Antoniou AC, Easton DF, Ponder BAJ. Polygenes, risk prediction, and targeted prevention of breast cancer. N Engl J Med. 2008;358(26):2796–2803. doi: 10.1056/NEJMsa0708739. [DOI] [PubMed] [Google Scholar]
- Presciuttini S, Toni C, Tempestini E, Verdiani S, Casarino L, Spinetti I, Stefano FD, Domenici R, Bailey-Wilson JE. Inferring relationships between pairs of individuals from locus heterozygosities. BMC Genet. 2002;3:23. doi: 10.1186/1471-2156-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj D. Sampling Theory. McGraw-HIll; 1968. [Google Scholar]
- Samani NJ, Raitakari OT, Sipil K, Tobin MD, Schunkert H, Juonala M, Braund PS, Erdmann J, Viikari J, Moilanen L, Taittonen L, Jula A, Jokinen E, Laitinen T, Hutri-Khnen N, Nieminen MS, Kesniemi YA, Hall AS, Hulkkonen J, Khnen M, Lehtimki T. Coronary artery disease-associated locus on chromosome 9p21 and early markers of atherosclerosis. Arterioscler Thromb Vasc Biol. 2008;28(9):1679–1683. doi: 10.1161/ATVBAHA.108.170332. [DOI] [PubMed] [Google Scholar]
- Thompson EA. Estimation of relationships from genetic data. In: Rao CR, Chakraborty R, editors. Handbook of Statistics Volume 8: Statistical Methods in Biological and Medical Sciences. Elsevier; North-Holland: 1991. pp. 255–269. [Google Scholar]
- U.S. Department of Health and Human Services (DHHS) National Center for Health Statistics . Third National Health and Nutrition Examination Survey, 1988-1994, NHANES III Household Adult Data File. Centers for Disease Control and Prevention; Hyattsville, MD: 1996. Technical report. [Google Scholar]
- Wagner AP, Creel S, Kalinowski ST. Estimating relatedness and relationships using microsatellite loci with null alleles. Heredity. 2006;97(5):336–345. doi: 10.1038/sj.hdy.6800865. [DOI] [PubMed] [Google Scholar]
- Wright S. Evolution and the genetics of populations. Vol II: The theory of gene frequencies. University of Chicago Press; 1969. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.