Summary
Understanding the mechanisms that underlie the organization of bacterial cells has become a significant challenge in the field of bacterial cytology. Of specific interest are early macromolecular sorting events that establish cellular non-uniformity and provide chemical landmarks for later localization events. In this review, we will examine specific examples of lipids and proteins that appear to exploit differences in membrane curvature to drive their localization to particular regions of a bacterial cell. We will also discuss the physical limits of curvature-mediated localization within bacteria, and the use of modelling to infer biophysical properties of curvature-sensing macromolecules.
Introduction
The non-uniform distribution of molecules within cells is a hallmark of life. For many decades now, it has been self-evident that eukaryotic cells, which are clearly partitioned into many subcellular compartments, must be highly organized at a molecular level. Bacterial cells also possess obvious large structures that are capable of assuming specific residences, such as polar flagella, pili, and partitioned chromosomes (Brown et al., 2009; Craig et al., 2004). In addition, bacteria have long been known to sort proteins faithfully into specific chemically distinct subcellular compartments, such as the inner and outer membranes, the periplasm, and the cell wall (Bos et al., 2007; Hegde and Bernstein, 2006; Marraffini et al., 2006; Narita et al., 2004). However, only with recent advances in fluorescence live-cell imaging has it been revealed that, much like eukaryotic cells, many individual macromolecules like proteins and lipids also reside in distinct subcellular locations.
Several newly discovered classes of sorted proteins reside in locations that were not previously thought to be chemically distinct, such as the midcell plane or the poles of rod-shaped bacteria. As the list of proteins with specific localization patterns has grown, understanding the mechanisms by which bacteria sort proteins to these regions of the cell, without the aid of the eukaryotic vesicle-mediated sorting machinery, has become a fundamental challenge in the re-emerging field of bacterial cell biology (Shapiro et al., 2009). Coincident with mounting evidence of the existence of lipid rafts in eukaryotic cells (Lingwood and Simons, 2010), it has also become clear that the bacterial plasma membrane is not simply a homogeneously distributed mix of phospholipids. On the contrary, specific lipids such as cardiolipin tend to cluster at architecturally interesting locations like the poles of a cell (Mileykovskaya and Dowhan, 2000; Nishibori et al., 2005).
The application of traditional genetic and biochemical methods to discern the mechanisms by which proteins are recruited to these locations has typically revealed a similar solution: targeting to another protein that is already localized. However, relying on another localized protein implies a strong dependence on initial conditions for localization to take place. Moreover, this solution simply displaces responsibility onto explaining how the pre-localized protein came to localize in the first place. Cells are inherently polarized by the difference in ages between the two poles, with division leaving behind landmark proteins that specify the new pole (Lam et al., 2006; Perez et al., 2000). Nonetheless, the prevalence and diversity of localization in bacteria suggests that there are several mechanisms that contribute to the non-uniform distribution of macromolecules inside the cell using self-contained components of the cell that are capable of self-organization, highlighting the need for a broad perspective of cellular physiology that includes physical and mechanical forces.
Several recent reports have suggested that some lipids and proteins localize to specific regions of a bacterial cell not by recognizing a chemical cue at all (like a pre-localized protein), but by recognizing a geometric cue. Specifically, the cell architecture itself, which defines the shape of its periphery and, therefore, the membrane that delineates it, can serve as a geometric beacon that recruits macromolecules to their final destination. Accordingly, certain proteins and lipids can localize to specific regions of the cell by “sensing” the curvature of their target membrane. Curvature sensing can be direct detection of the membrane geometry by a single molecule, or the accumulation of a population of a particular molecule to particular regions of a cell that are slightly curved. Moreover, macromolecules can display a preference towards positive (i.e., convex) membrane curvature as found on the surface of cellular organelles, or towards negative (i.e., concave) membrane curvature as found on the cytosolic face of the plasma membrane. In this review, we address the physical limits of curvature sensing within bacterial cells, and the evidence for curvature-mediated localization by intracellular lipids and proteins.
Direct perception of highly curved membranes
Two protein motifs implicated in directly sensing very acute positive membrane curvature have been described extensively in the literature: amphipathic alpha helices (exemplified in the discussion below by the ArfGap1 lipid packing sensor, or “ALPS”) (Bigay et al., 2005; Carlton et al., 2004) and the Bin-Amphiphysin-RVS (BAR) domain (Peter et al., 2004). Both motifs, which have been identified in eukaryotic systems, are structurally distinct, and different mechanisms have traditionally been invoked to explain how they detect curved membranes (reviewed in (Bhatia et al., 2009a)).
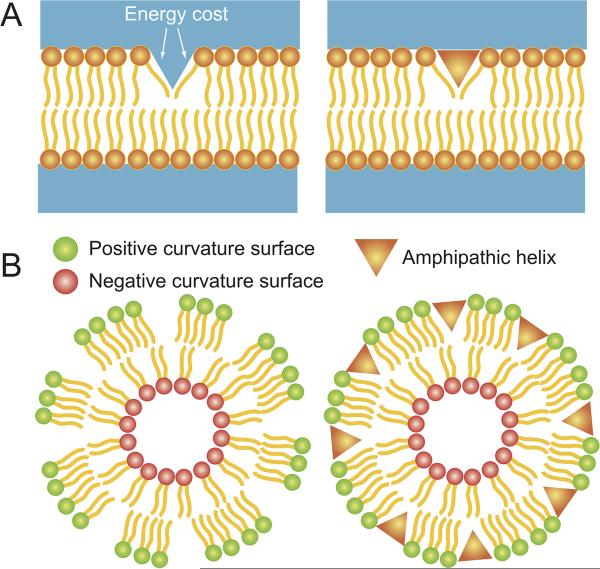
The ALPS domain is an amphipathic alpha helix, one lateral face of which is largely hydrophobic and buried in the phospholipid bilayer, while the opposite face is polar and faces the cytosol (Bigay et al., 2005). One model suggests that a polar face largely devoid of positively charged residues distinguishes the curvature sensing ability of an ALPS domain from a standard amphipathic helix (Drin et al., 2007). ALPS domain proteins cluster on the outside of highly positively curved membranes such as vesicles with a radius on the order of 50 nm, and are thought to do so mechanistically because such surfaces exhibit lipid packing defects (Bigay et al., 2003). Specifically, in order to decrease the membrane radius of curvature, phospholipid head groups must move apart, and this strain comes with an associated energy cost (Fig. 1B). ALPS domains are thought to insert like a wedge into the exposed space created by the curving membrane, thereby alleviating the packing stress. Consistent with this model, increasing packing stress by introducing lipids with small head groups into small vesicles resulted in the increased binding of ArfGAP1 (Drin et al., 2007). Conversely, alleviating this lipid packing stress by incubating highly curved vesicles with molecules that insert into this space abrogated the preferential adsorption of another alpha helix-bearing curvature sensing protein (Davies et al., 2001). Thus, lipid-packing defects on the binding surface provide one mechanism for the preferential insertion of amphipathic alpha helices into highly convex membranes.
Figure 1.
Lipid packing defects at the nanometer scale in (A) flat membranes caused by lipids with intrinsic curvature, or (B) highly curved membranes composed of cylindrical lipids. The positive curvature of the outer leaflet is shown in green, while the negative curvature of the inner leaflet is shown in red. The packing stress at this scale may be alleviated by insertion of an amphipathic helix (depicted as an orange wedge) at the location of the defect. In this manner, an amphipathic helix could also favour the formation of highly curved vesicles.
Historically, an entirely different mechanism has been invoked to explain the mechanism by which BAR domains recognize highly curved membranes (Zimmerberg and McLaughlin, 2004). The crystal structure of the amphiphysin BAR domain revealed a banana-shaped homodimer (Peter et al., 2004). Remarkably, the concave face of this “banana” has clusters of positively charged residues. The structure immediately suggests an elegant model: convex membrane surfaces, which display negatively charged phospholipid head groups, could interact with the positively charged residues that line the concave surface of BAR domains. Such interactions would be especially favourable when the curvature of the membrane matches the curvature of the crescent-shaped “banana”, resulting in the recruitment of BAR domain proteins to these sites. A recent report suggested, however, that the crescent shape of BAR domains is not in fact responsible for its ability to sense curvature (Bhatia et al., 2009b). Varying neither the shape nor the charges of various BAR domains affected the ability of these motifs to detect highly curved surfaces in vitro. Instead, amphipathic alpha helices that are sometimes associated with BAR domains (so-called “N-BAR” domains) were reported to mediate not only membrane binding, but also the sensing of positive membrane curvature. Consistent with this model, alleviating lipid-packing defects on target vesicles abrogated the preferential adsorption of N-BAR domains onto highly curved vesicles. Thus, although a highly intuitive model (one in which a curved protein motif recognizes curved surfaces) may be in doubt, a more unified model of membrane curvature detection centred on amphipathic helices might be emerging.
Quantifying curvature sensation
The direct sensing of highly curved surfaces of very small vesicles (~50nm radius) by proteins harbouring either ALPS or BAR domains might be thought of as akin to wrapping one's arms around a large tree to sense that its surface is curved. Can a single molecule also directly perceive the relatively almost “imperceptible” curvature that exists at the surface of larger entities like organelles, analogous to the challenge that an ancient mariner would have faced in perceiving that the world is round while floating in the middle of the seemingly flat ocean?
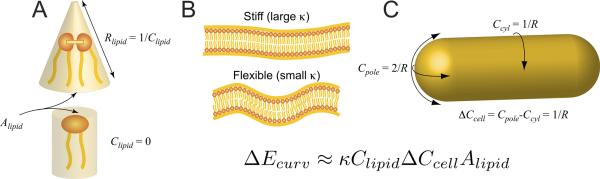
Consider a molecule with a preference for curved surfaces in solution with either a small vesicle surface or the surface of a large organelle. This intrinsic curvature preference might be positive or negative, and is specified by the sum of the reciprocals of the preferred radii of curvature. If presented with two surfaces, one hemispherical (e.g., the poles of a rod-shaped bacterium) and one cylindrical with curvatures 2/R and 1/R, respectively, a molecule inside the cell with a negative curvature preference has a preference for the poles (and vice versa for a molecule with a positive curvature preference). However, the preference of the molecule for a particular curvature need not result in an ability to sense the difference in curvature of two membrane geometries. The strength of this preference can be measured as the elastic energetic cost of bending a patch of membrane containing the molecule into a geometry dictated by the cell shape. This energy is proportional to the preferred curvature Clipid of the molecule, the difference in the curvature of the two surfaces ΔCcell (for a rod-shaped cell ΔCcell ~1/R), and the lipid area A. The scale of this energy is set by a bending modulus k. We note that ΔCcell involves a difference between two curvatures that might have opposite signs; for the purposes of our qualitative analysis, the distinction between a convex and a concave surface with curvatures 1/R and −1/R, respectively, is equivalent to a convex and a flat surface with curvatures 2/R and 0.
For nanometer-sized molecules within a micron-sized cell, the difference in length scales dictates that this energy difference is small compared with the thermal energy; hence the curvature-mediated localization is likely to be washed away by Brownian motion. This establishes a physical limit to the ability of individual molecules to sense differences in curvature at the cellular scale. To illustrate this, we consider the phospholipid cardiolipin that, as discussed in greater detail below, is found preferentially at the negatively curved membranes of cell poles in the inner leaflet of the inner membrane. Estimates based on the crystal structure of cardiolipin suggest that its intrinsic curvature (Clipid) is approximately (1nm)−1–(5nm)−1 (Kooijman et al., 2005). A typical bending modulus for phospholipid bilayers is approximately 25 times the thermal energy kBT (Rawicz et al., 2000; Wiggins and Phillips, 2005). Typical lipid dimensions are 0.5–1nm, yielding an area (A) of <1nm2 (Edholm and Nagle, 2005). Therefore, an estimate of the curvature-mediated energy preference of a single cardiolipin molecule for the poles of a micron-sized bacterium is
| (1) |
and the differential affinity of cardiolipin for the poles versus the lateral walls is then at most a negligible 1 − exp(−ΔEcurv/kBT) ≈ 5%, suggesting that a single molecule of cardiolipin can not reliably detect the curvature difference between the cell poles and the lateral edge of the cell in the face of thermal fluctuations. Each component of the energy calculation in Eq. 1 is depicted schematically in Fig. 2. Moreover, this analysis argues against the possibility that cells become organized by tuning the curvature preferences of a wide range of molecules: molecules with a preference for weakly curved surfaces will face a relatively flat energy landscape that leads to promiscuous localization rather than specific targeting to regions of the membrane matching their preferred geometry.
Figure 2.
Key physical characteristics for quantifying curvature sensitivity of single cardiolipin molecules. (A) Cardiolipin's unique dimeric structure, with two phosphatidyl head groups and four acyl chains, is indicative of a conical shape with a small radius of curvature Rlipid and hence a preference for highly curved geometries. (B) The energy scale κ is set by the stiffness of the membrane for bending. (C) The enhanced curvature of the pole is the difference between the curvature at the pole and at the cylindrical midcell, ΔCcell = 1/R. The combination of κ, Clipid,, ΔCcell, and the surface area Alipid then determines the extent to which the molecule will preferentially localize at the pole.
Similar calculations suggest that strength of the curvature preference of any single nanometer-scale molecule in bacteria is negligible compared with thermal fluctuations. Does this mean that curvature-mediated localization is impossible? An examination of Eq. (1) reveals that the length scale inequality can be resolved if the molecules with high intrinsic curvature are subject to cooperative interactions that cause the formation of stable aggregates. These aggregates would then have a larger effective area (A) that can mediate an increase in ΔEcurv to above kBT, thereby allowing stable localization. The aggregation argument above can be applied similarly to proteins, and applies equally to instances where aggregation of a macromolecule occurs before, during, or after adsorption onto a membrane. Thus, in contrast to the direct curvature sensing of highly curved vesicles by BAR and ALPS domains, curvature sensing within micron-sized bacteria by individual macromolecules is unlikely, but might be achieved if lipids, proteins, and/or lipid-protein form complexes that display a collective preference for curved membranes.
Curvature-mediated partitioning of phospholipids
Although bacterial membranes have a wide variety of membrane components, a natural candidate for geometric sensing is the anionic phospholipid cardiolipin. Cardiolipin is found throughout the prokaryotes and eukaryotes, has a unique structure and subcellular localization, and has been connected with several diverse features of potential biological significance, including osmoregulation and sporulation in bacteria (Kawai et al., 2004; Romantsov et al., 2009b), longevity in yeast (Zhou et al., 2009), and Barth's syndrome and diabetes in humans (Ferreira et al., 2003; Schlame et al., 2002; Vreken et al., 2000). In bacteria, cardiolipin is generally formed through the condensation of two molecules of phosphatidylglycerol, suggesting a small ratio of head-to-tail surface areas and a preference for the concave inner leaflet (Hirschberg and Kennedy, 1972; Fig. 2A). A high-resolution X-ray crystal structure of cardiolipin adjacent to a transmembrane protein from Rhodobacter sphaeroides demonstrates that cardiolipin can have a large negative curvature with a head group oriented towards the cytoplasm (McAuley et al., 1999). In mitochondria from bovine hearts, 75% of the cardiolipin in the inner membrane of bovine heart mitochondria is located in the matrix leaflet, equivalent to the inner leaflet of a bacterial membrane (de Kroon et al., 1997; Krebs et al., 1979). Taken together, the existing evidence currently points towards an asymmetric distribution of cardiolipin across the leaflets of the inner membrane of bacteria, with enhanced accumulation on the cytosolic face of the membrane.
That cardiolipin accumulates at the poles was demonstrated by two approaches (Mileykovskaya and Dowhan, 2009). First, the cardiolipin-specific dye 10-N-nonyl acridine orange (NAO) was used to demonstrate that cardiolipin is enriched at the cell poles and near potential division sites in the Gram-negative bacteria Escherichia coli (Mileykovskaya and Dowhan, 2000) and Pseudomonas putida (Bernal et al., 2007), as well as the Gram-positive bacterium Bacillus subtilis (Kawai et al., 2004). All three of these organisms are rod-shaped, with a width of ~1μm. Second, lipid analysis of E. coli minicells (small round cells formed from polar division) affirmed the high polar concentration of cardiolipin in a dye-independent fashion (Koppelman et al., 2001). Given the structure of cardiolipin, it is enticing to invoke the enhanced curvature of the poles relative to the cylindrical midcell as a possible mechanism for localization. However, as we have discussed above, the relative affinity of a single nanometer-sized cardiolipin molecule for the very slightly curved poles is likely insufficient for stable polar localization in a micron-sized bacterium.
Although aggregation into domains can enhance the curvature preference of cardiolipin, it is also important to consider how the difference in curvature is determined in the first place. For the vast majority of bacteria, the rigid peptidoglycan cell wall dictates cell shape, independent of the membrane composition, and is the primary structure that bears the stress of turgor pressures on the order of atmospheres (Holtje, 1998; Scheffers and Pinho, 2005). In the absence of turgor pressure, the membrane will undergo plasmolysis and separate from the cell wall into highly invaginated structures (Ou and Marquis, 1970). How does one consider the physical effects of the competing geometries of the cell wall and the membrane components?
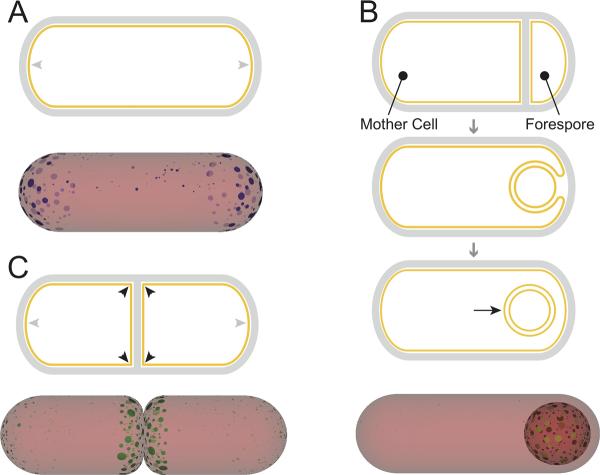
We have recently proposed a biophysical model demonstrating that the osmotic forces coupling the membrane and cell wall geometries create a repulsive interaction that counteracts the tendency of high-curvature lipids such as cardiolipin to aggregate (Huang et al., 2006; Mukhopadhyay et al., 2008). This interaction is long-ranged and, hence, destabilizes very large aggregates, resulting in finite-sized domains of high-curvature lipids. These domains are large enough to overcome thermal fluctuations and entropy to localize stably at the two poles and septal regions, as shown schematically in Fig. 3A.
Figure 3.
Curvature sensing of geometric differences in a rod-shaped bacterium. (A) Polar localization: above, a rod shaped cell is depicted in which the hemispherical cell poles (grey arrowheads) are the preferred localization sites for the lipid cardiolipin. Below, a schematic of results obtained from a simulation in which molecules with a preference for curved surfaces localize via cooperative interactions. By forming clusters, molecules with a preference for negative curvature (shown in blue, representing cardiolipin) can distinguish cell poles (grey arrowheads) from the cylindrical region of the cell membrane (red). (B) Stages of sporulation in B. subtilis: in the top panel, a cell divides asymmetrically to produce the larger mother cell and the smaller forespore. In the middle panel, the mother cell begins to engulf the forespore, which causes the septum to curve. Finally, in the bottom panel, the forespore pinches off as a double-membrane bound organelle, the cytosolic face of which is the only positively curved surface (black arrow) in the mother cell cytosol. Below, by forming clusters, molecules with a preference for positive curvature (shown in green, representing SpoVM) might be able to distinguish the outer spore surface from the rest of the cytosolic surface. (C) Hierarchical curvature localization: above, depiction of an actively dividing B. subtilis cell in which sharp membrane inflections created by a nascent division septum (black arrowheads) are the primary site of DivIVA localization, whereas the hemispherical cell poles (grey arrowheads) are the secondary site of DivIVA localization. Below, by forming clusters, molecules with a preference for negatively curved surfaces (representing DivIVA) might be able to distinguish between the highly curved division septum (black arrowheads), the less curved hemispherical poles (grey arrowheads), and the least curved lateral edge of the cell.
Protein localization via detection of curvature-sensing lipids
Because aggregation is critical for curvature sensing of any molecule, at sufficiently low concentrations such molecules should fail to localize since domains will be entropically unfavourable. For cardiolipin, our biophysical model predicts a sharp phase transition around a critical concentration of 1–2%, below which cardiolipin molecules will be relegated to small clusters of a few lipids that localize uniformly around the membrane (Mukhopadhyay et al., 2008). Fortunately, a simple experiment probes this regime of concentration. By deleting the cardiolipin synthase gene (cls) in E. coli, cardiolipin levels are reduced ~10-fold to 0.1–0.6% and the residual levels of cardiolipin indeed fail to localize at the poles (Romantsov et al., 2009b; Romantsov et al., 2007; Romantsov et al., 2008). Does the delocalization of cardiolipin result in the mislocalization of any proteins? Recent reports demonstrated that the polar localization of ProP (a proline transporter) (Romantsov et al., 2007; Romantsov et al., 2008) and MscS (the mechanosensitive channel of small conductance) (Romantsov et al., 2009a) were diminished coincident with the delocalization of cardiolipin. The identification of these correlations highlights the need for quantitative metrics of localization. Additionally, the amphipathic helix responsible for targeting the cell division protein MinD to the membrane in E. coli and B. subtilis has a preference for anionic phospholipids such as cardiolipin (Kawai et al., 2004; Mileykovskaya and Dowhan, 2000; Mileykovskaya et al., 2003; Szeto et al., 2003; Szeto et al., 2002). These results suggest that curvature-mediated localization of cardiolipin domains may serve as a target for several proteins, providing a raft-like mechanism for concentrating proteins at the poles.
Direct perception of cellular curvature by proteins
Can very slight differences in membrane curvature formed by the architecture of bacterial cells directly drive the localization of a protein, analogous to the manner in which cardiolipin partitions to particular cellular locations? For at least two bacterial proteins, SpoVM and DivIVA, the ability to localize to either the surface of a subcellular organelle or to division septa is mediated directly by the shape of the membrane.
While bacterial proteins do not usually encounter convex membrane surfaces, a rare example is a bacterial organelle called the forespore, which is elaborated during endospore formation in some Gram-positive bacteria. When Bacillus subtilis senses the depletion of nutrients in the surrounding medium, the rod-shaped bacterium divides asymmetrically, producing two unevenly sized compartments (the larger “mother cell” and the smaller “forespore”), instead of two equally sized daughter cells (Stragier and Losick, 1996). The asymmetrically placed polar division septum then bends as the mother cell engulfs the forespore (Fig. 3B). When engulfment of the forespore is complete, it resides as a double membrane-bound organelle inside the mother cell as it develops further into a mature spore. The bending polar septum (later the outer surface of the engulfed forespore) represents the only positively curved membrane in the mother cell cytosol, and over the last several years, a combination of electron and fluorescent microscopy approaches have revealed that many proteins specifically localize to this surface (Aung et al., 2007; Ben-Yehuda et al., 2003; Blaylock et al., 2004; Doan et al., 2005; Driks et al., 1994; Ozin et al., 2001; Rudner and Losick, 2002; van Ooij et al., 2004; van Ooij and Losick, 2003). The proper recruitment of many of these proteins is usually attributed to a pre-localized protein at the forespore surface, but for at least one protein, SpoVM, biochemical and genetic approaches failed to reveal an upstream factor that recruited it to this location.
SpoVM is a short peptide produced exclusively in the mother cell, is encoded by a 26-codon open reading frame, and is not known to undergo any post-translational modifications (Levin et al., 1993). SpoVM directly tethers a protein (SpoIVA) that makes up the innermost layer of a proteinaceous shell, called the spore coat, which assembles around the forespore (Ramamurthi et al., 2006). Although unordered in aqueous solution, the peptide assumes an alpha-helical conformation in the presence of lipid vesicles (Prajapati et al., 2000). Analysis of its primary amino acid sequence suggested a striking amphipathic profile for SpoVM: all six of its positively charged residues cluster along one longitudinal face of the helix, whereas the opposite face is largely hydrophobic. SpoVM inserts into membranes such that its long axis is parallel to the phospholipid bilayer (Ramamurthi et al., 2006). As a result, its hydrophobic face is buried and its charged face is exposed to the mother cell cytosol. Curiously, a proline at position 9 is critical for the proper localization of SpoVM onto the forespore surface. Substitution of P9 with alanine resulted in the indiscriminate mislocalization of SpoVM onto all available membrane surfaces in the mother cell cytosol (van Ooij and Losick, 2003).
The structure of SpoVM is at least superficially reminiscent of the historically cited geometric mechanism of ALPS domain localization to the outside of small vesicles. Instead of a chemical cue such as a pre-localized protein or lipid, could the primary factor driving the localization of SpoVM to the forespore surface be a geometric cue like convex membrane curvature? It is important to note that the ALPS motif differs from the SpoVM helix in several critical ways. First, the consensus motif for an ALPS domain predicts that the hydrophilic face of the helix is largely devoid of charged residues, whereas SpoVM contains six positively charged residues. Second, the SpoVM helix harbours a well-conserved proline residue that is indispensable for its localization, whereas the ALPS domain does not require such a proline. Third, while ALPS domains recognize highly convex surfaces (tiny vesicles that are perhaps 50 nm in diameter), SpoVM localizes in vivo to the more gently curved forespore surface that is approximately 1000 nm in diameter. However, several tantalizing similarities are evident between SpoVM and membrane curvature-sensing ALPS domains. First, both are amphipathic alpha helices. Furthermore, both helices insert similarly into membranes, with the hydrophobic face buried in the lipid bilayer. Most importantly, ALPS domain-containing proteins like ArfGAP1 bind preferentially to highly positively curved membranes, such as the outside of budding vesicles (Bigay et al., 2003), and SpoVM preferentially binds to the convex cytosolic surface of the forespore (van Ooij and Losick, 2003).
Ultimately, three lines of evidence suggested that SpoVM localizes to the surface of the forespore by sensing its convex membrane curvature. First, in a B. subtilis mutant (ΔspoIID/M/P) in which sporulation is arrested at a stage before the start of engulfment (thereby blocking the formation of a convex surface; Fig. 3B, top panel), SpoVM was mislocalized. While the peptide still adsorbed onto membranes, it no longer discriminated between sites of different membrane curvature and, instead, localized promiscuously (Ramamurthi et al., 2009). Upon elaboration of an artificial convex surface in the same genetic background, however, SpoVM resumed its localization to the newly created positively curved surface. Second, production of SpoVM in heterologous cells that harboured internal structures unrelated to forespores resulted in the accumulation of SpoVM on the convex membrane surfaces of these structures, whereas a SpoVM variant harbouring alanine instead of proline at position 9 localized indiscriminately in these cells. Finally, purified SpoVM, when incubated in buffered solution with heterogeneously-sized large membrane vesicles, preferentially adsorbed onto smaller, forespore-sized vesicles and was largely excluded from binding very large vesicles. In contrast, purified SpoVMP9A no longer specifically adsorbed onto forespore-sized vesicles (Ramamurthi et al., 2009). Taken together, these in vitro experiments suggested that positive curvature is at least sufficient to localize wild-type SpoVM to the surface of the forespore.
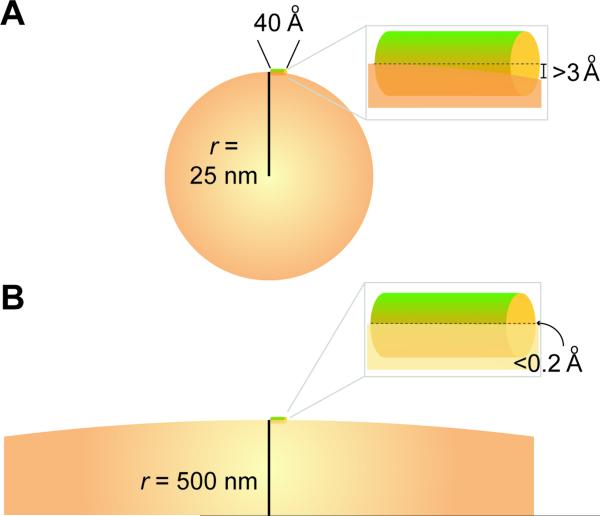
Mechanistically, how do SpoVM molecules detect the positive curvature of the forespore surface? A single SpoVM peptide, if perfectly alpha helical, would form an approximately 40 Å-long rod (7.2 turns of a 26 amino acid-long peptide at 5.4 Å per turn). Laying tangential to the forespore surface with a radius of curvature of ~5000 Å, the deflection of the surface along the length of a 40 Å-long peptide would be less than 0.2 Å (Fig. 4)! Since a typical covalent bond is about 1 Å, the direct detection of such slight curvature by a single SpoVM peptide appears unlikely. This qualitative argument is analogous to the physical limitation on curvature-mediated localization due to a disparity in length scales discussed above. Similar to our analysis of cardiolipin localization to negatively curved surfaces, would multiple molecules of SpoVM display a collective sensitivity for positive curvature? Indeed, at very low concentrations of SpoVM (for which entropy is likely to limit SpoVM distribution to individual peptides), SpoVM adsorbed more promiscuously onto large and small vesicles, independent of curvature. However, increasing the concentration of SpoVM resulted in the aforementioned preferential adsorption onto smaller vesicles (Ramamurthi et al., 2009), suggesting that many molecules of SpoVM are necessary to detect positive curvature collectively. In Fig. 3B, we illustrate a possible model for the localization of clusters of molecules such as SpoVM with a positive curvature preference to the forespore surface.
Figure 4.
Comparison of macromolecule and surface dimensions on very small vesicles and bacterial membranes. A 26 amino acid-long perfectly alpha helical peptide is depicted as a 40 angstrom-long rod lying tangentially on the surface of either a small vesicle of 50 nm diameter (A) or a large organelle of 1 micron diameter (B). The calculated distance between the central axis of the rod and the surface of the vesicle is shown on the right in the magnified inset. The length of the rod, circumference of the spheres, and the inflection of the membranes are all drawn to scale.
A recent report demonstrated that the preferential adsorption of proteins harbouring BAR domains onto very small vesicles is not achieved by increased affinity of the protein for small vesicles (decreased kd) or by a cooperative mechanism (increasing the Hill coefficient, h). Instead, the authors suggested that positively curved membranes simply increase the number of available binding sites (Bmax) for a BAR domain-containing protein by displaying a more hydrophobic surface, presumably by decreasing the packing of lipid head groups (Hatzakis et al., 2009). While the direct sensing of increased hydrophobicity on the surface of small vesicles by a single protein provides a reasonable mechanism by which BAR domain-harbouring proteins localize, it is difficult to imagine that the slightly curved forespore could display an appreciably more hydrophobic surface with which to attract SpoVM. Nonetheless, additional in vitro experiments should explicitly demonstrate which one (or combination) of the sigmoidal parameters SpoVM molecules exploit in order to adsorb preferentially onto convex surfaces, and whether or not this adsorption is strictly cooperative.
An enormous variety of proteins localize to the poles of rod-shaped bacteria (for a review, see (Shapiro et al., 2009)). Can degrees of negative membrane curvature also drive protein localization? Although the entire inner surface of a rod-shaped bacterial cell is negatively curved, some regions are more curved than others. At sites where cell division septa meet the lateral wall of the cell in B. subtilis cells, the membrane is sharply curved inwards and represents the most concave surface inside the cell (Fig. 3C). The hemispherical poles, by comparison, are also curved in two dimensions, but less so than division sites. Finally, the lateral edges of the cell represent the least concave surface, with negative curvature in only one dimension (the circumferential direction). Two recent reports suggested that the cell division protein DivIVA in B. subtilis exploits these differences in membrane curvature to drive its proper localization (Lenarcic et al., 2009; Ramamurthi and Losick, 2009). During normal growth, DivIVA localizes to division septa and cell poles (the two regions with the highest negative curvature) and recruits proteins that prevent the aberrant formation of septa close to these sites (Gregory et al., 2008; Marston et al., 1998). Intriguingly, DivIVA also localizes to the poles and division septa of E. coli and even Schizosaccharomyces pombe cells, two organisms in which there are no known DivIVA homologues (Edwards et al., 2000). Like SpoVM, DivIVA is a peripheral membrane protein and also has an amphipathic alpha helix implicated as a membrane anchor (although, unlike SpoVM, this helical region alone was insufficient for proper localization; (Lenarcic et al., 2009). Accordingly, several lines of evidence, all from in vivo studies, are consistent with the hypothesis that DivIVA localizes properly by sensing membrane geometry. First, DivIVA displayed a hierarchical preference for localization sites according to degree of negative membrane curvature, i.e., division septa, and was largely excluded from the hemispherical poles when both sites were present. However, dismantling division septa by producing an inhibitor of cytokinesis resulted in the redistribution of DivIVA to its second site of preference, cell poles. Treating the rod-shaped B. subtilis cells with a cell wall-degrading enzyme produced spherical cells called protoplasts. In the presence of uniform negative membrane curvature, DivIVA localized indiscriminately (Ramamurthi and Losick, 2009). Furthermore, in mutant strains of B. subtilis that were shaped aberrantly, DivIVA localized preferentially to artificially produced regions of high negative curvature (Lenarcic et al., 2009). In Fig. 3C, we illustrate the strong tendency of clusters of molecules with a preference for negatively curved surfaces to localize first at the septum, and then at the poles.
Could other mechanisms drive the localization of DivIVA? The phenomenon called nucleoid occlusion, which prevents the recruitment and assembly of the cytokinetic protein FtsZ at areas of the cells that contain the nucleoid, was eliminated because in anucleate mutant cells that maintained their rod-shape, DivIVA remained at division septa. These observations suggested that the presence of the nucleoid did not occlude DivIVA localization and, hence, relegate it to the extremities of the cell (Ramamurthi and Losick, 2009). The contribution of specific lipids (phosphatidylethanolamine, phosphatidylglycerol, and cardiolipin) to the localization of DivIVA was also discounted, as DivIVA remained largely localized to cell poles in mutant strains that did not produce the synthases for these phospholipids (Lenarcic et al., 2009). These experiments may be difficult to interpret, given that such lipid synthase mutants in E. coli have been shown to synthesize new negative-curvature phospholipids (Mileykovskaya et al., 2009). Nevertheless, taken together with the observation that localization of DivIVA is dependent on the activity of FtsZ and PBP 2B (proteins that are required for the onset of cell constriction and therefore generation of membrane curvature; Daniel et al., 2000), but not direct contact with these proteins (Hamoen and Errington, 2003), these results are consistent with a model in which concave membrane surfaces recruit DivIVA.
Nevertheless, the localization of DivIVA to curved membranes, like that of SpoVM, poses a problem of scale: how does a tiny protein recognize the very slight membrane curvature that delineates the boundary of a bacterial cell? DivIVA forms higher order structures (Stahlberg et al., 2004), and so may have an aggregation-enhanced capacity to sense membrane geometry similar to cardiolipin. As we have argued above, even if DivIVA has a higher affinity for negatively curved surfaces, the assembly of these higher order structures is likely necessary to facilitate a cooperative mechanism whereby multiple DivIVA molecules are required to localize properly. However, it is unclear whether DivIVA has the requisite intrinsic curvature in two dimensions to generate enhanced affinity for the poles, or whether it relies on the increased availability of curved orientations at the poles relative to the lateral surfaces due to the freedom to rotate. Alternatively, like BAR domains, does membrane curvature somehow provide more binding sites for DivIVA, which results in an apparent preference for concave surfaces, which would be revealed by an increased Bmax? The establishment of a robust in vitro assay that measures the adsorption of DivIVA onto negatively curved membranes might clarify the mechanism by which DivIVA localizes properly.
The impact of curvature
Amongst the many possible mechanisms for establishing spatial heterogeneity (Rafelski and Theriot, 2006; Raskin and de Boer, 1999), the exploitation of membrane curvature as a tool to recruit macromolecules to specific subcellular locations appears to be widespread. This strategy might be organized into two broad categories: sensing of highly curved surfaces (vesicles on the order of 50 nm in diameter) and sensing of cellular curvature (surfaces that are about 1 micron in diameter). Proteins that preferentially detect highly curved surfaces have only been described in eukaryotic systems, where ALPS or BAR domain-harbouring proteins are recruited to the surface of budding vesicles. On the other hand, examples of macromolecules that preferentially localize to areas of very slight curvature have been described exclusively in bacteria. The lipid cardiolipin as well as the cell division protein DivIVA localize to concave cellular membrane surfaces (cell poles and division septa), whereas the peptide SpoVM clusters preferentially onto the convex surface of a developing spore. Although in vitro techniques using optical tweezers (Sorre et al., 2009) and supported membranes (Parthasarathy et al., 2006) have proven useful for demonstrating the curvature sensing properties of several macromolecules, they must ultimately be supplemented by in vivo assays to determine the role of geometric cues within the cellular milieu. Recent cytological studies have helped to elucidate the roles of the bacterial cell wall, membrane, and cytoskeletal elements in establishing cellular morphologies with regions of differing curvature (Cabeen and Jacobs-Wagner, 2005). The ability to alter cell shape through chemical (Gitai et al., 2005), genetic (Young, 2006), and/or mechanical perturbations (Takeuchi et al., 2005), thereby creating artificial surface geometries to which the curvature-dependent localization of lipids and proteins can be measured in vivo, makes bacteria an especially powerful system in which to study curvature-mediated phenomena.
There is growing appreciation that cell shape might serve a regulatory role for a many biological processes. For example, the curvature-mediated microdomain formation of cardiolipin organizes the membrane in response to cell shape and suggests a raft-like mechanism for concentrating proteins at the poles. Accordingly, large increases in the cellular concentration of cardiolipin have been shown to disrupt critical processes such as cell division (Mileykovskaya et al., 2003). In B. subtilis cells lacking the cls gene, there is nevertheless a striking increase in cardiolipin levels during sporulation, and a drop in the heat resistance of spores, suggesting that cells specifically upregulate alternative cardiolipin production pathways (Kawai et al., 2004). Curiously, an E. coli strain completely lacking the major anionic phospholipids phosphatidylglycerol and cardiolipin synthesizes novel anionic phospholipids with small head-to-tail ratios that also localize to the poles and septal regions (Mileykovskaya et al., 2009). These results suggest many general biological processes rely on the localization of phospholipids with similar biophysical properties to cardiolipin, and can serve as motivation to meet the considerable challenge of labelling a wide variety of phospholipids with high specificity to determine their localization and dynamics over time frames as long as the cell cycle.
The assembly of large proteinaceous structures such as the spore coat, flagella, and the divisome often occurs at architecturally specific regions of the cell, and the site of assembly is usually marked by one or a few proteins on which the assembly of the entire structure depends. It appears now that some of these structures owe their proper localization to proteins that directly sense shape of a specific region of a cell. While there is surely a broad spectrum of geometric sensitivities among the intracellular components, perhaps geometry will become a routine addition to chemical and physical considerations when studying the subcellular organization of macromolecules.
Acknowledgements
We thank members of our labs and Sigolene Lecuyer, Kit Pogliano, and Ned Wingreen for helpful comments. KH acknowledges funding from National Institutes of Health Grants K25 GM075000 and DP2 OD006466. KR acknowledges funding from the Intramural Research Program of the NIH National Cancer Institute Center for Cancer Research.
References
- Aung S, Shum J, Abanes-De Mello A, Broder DH, Fredlund-Gutierrez J, Chiba S, Pogliano K. Dual localization pathways for the engulfment proteins during Bacillus subtilis sporulation. Mol Microbiol. 2007;65:1534–1546. doi: 10.1111/j.1365-2958.2007.05887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yehuda S, Rudner DZ, Losick R. Assembly of the SpoIIIE DNA translocase depends on chromosome trapping in Bacillus subtilis. Curr Biol. 2003;13:2196–2200. [PubMed] [Google Scholar]
- Bernal P, Munoz-Rojas J, Hurtado A, Ramos JL, Segura A. A Pseudomonas putida cardiolipin synthesis mutant exhibits increased sensitivity to drugs related to transport functionality. Environ Microbiol. 2007;9:1135–1145. doi: 10.1111/j.1462-2920.2006.01236.x. [DOI] [PubMed] [Google Scholar]
- Bhatia VK, Hatzakis NS, Stamou D. A unifying mechanism accounts for sensing of membrane curvature by BAR domains, amphipathic helices and membrane-anchored proteins. Seminars in cell & developmental biology. 2009a doi: 10.1016/j.semcdb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Bhatia VK, Madsen KL, Bolinger PY, Kunding A, Hedegard P, Gether U, Stamou D. Amphipathic motifs in BAR domains are essential for membrane curvature sensing. Embo J. 2009b;28:3303–3314. doi: 10.1038/emboj.2009.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigay J, Casella JF, Drin G, Mesmin B, Antonny B. ArfGAP1 responds to membrane curvature through the folding of a lipid packing sensor motif. Embo J. 2005;24:2244–2253. doi: 10.1038/sj.emboj.7600714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigay J, Gounon P, Robineau S, Antonny B. Lipid packing sensed by ArfGAP1 couples COPI coat disassembly to membrane bilayer curvature. Nature. 2003;426:563–566. doi: 10.1038/nature02108. [DOI] [PubMed] [Google Scholar]
- Blaylock B, Jiang X, Rubio A, Moran CP, Jr., Pogliano K. Zipper-like interaction between proteins in adjacent daughter cells mediates protein localization. Genes Dev. 2004;18:2916–2928. doi: 10.1101/gad.1252704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos MP, Robert V, Tommassen J. Biogenesis of the gram-negative bacterial outer membrane. Annual review of microbiology. 2007;61:191–214. doi: 10.1146/annurev.micro.61.080706.093245. [DOI] [PubMed] [Google Scholar]
- Brown PJ, Hardy GG, Trimble MJ, Brun YV. Complex regulatory pathways coordinate cell-cycle progression and development in Caulobacter crescentus. Advances in microbial physiology. 2009;54:1–101. doi: 10.1016/S0065-2911(08)00001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabeen MT, Jacobs-Wagner C. Bacterial cell shape. Nat Rev Microbiol. 2005;3:601–610. doi: 10.1038/nrmicro1205. [DOI] [PubMed] [Google Scholar]
- Carlton J, Bujny M, Peter BJ, Oorschot VM, Rutherford A, Mellor H, Klumperman J, McMahon HT, Cullen PJ. Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high- curvature membranes and 3-phosphoinositides. Curr Biol. 2004;14:1791–1800. doi: 10.1016/j.cub.2004.09.077. [DOI] [PubMed] [Google Scholar]
- Craig L, Pique ME, Tainer JA. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol. 2004;2:363–378. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- Daniel RA, Harry EJ, Errington J. Role of penicillin-binding protein PBP 2B in assembly and functioning of the division machinery of Bacillus subtilis. Mol Microbiol. 2000;35:299–311. doi: 10.1046/j.1365-2958.2000.01724.x. [DOI] [PubMed] [Google Scholar]
- Davies SM, Epand RM, Kraayenhof R, Cornell RB. Regulation of CTP: phosphocholine cytidylyltransferase activity by the physical properties of lipid membranes: an important role for stored curvature strain energy. Biochemistry. 2001;40:10522–10531. doi: 10.1021/bi010904c. [DOI] [PubMed] [Google Scholar]
- de Kroon AI, Dolis D, Mayer A, Lill R, de Kruijff B. Phospholipid composition of highly purified mitochondrial outer membranes of rat liver and Neurospora crassa. Is cardiolipin present in the mitochondrial outer membrane? Biochim Biophys Acta. 1997;1325:108–116. doi: 10.1016/s0005-2736(96)00240-4. [DOI] [PubMed] [Google Scholar]
- Doan T, Marquis KA, Rudner DZ. Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum. Mol Microbiol. 2005;55:1767–1781. doi: 10.1111/j.1365-2958.2005.04501.x. [DOI] [PubMed] [Google Scholar]
- Driks A, Roels S, Beall B, Moran CP, Jr., Losick R. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes & development. 1994;8:234–244. doi: 10.1101/gad.8.2.234. [DOI] [PubMed] [Google Scholar]
- Drin G, Casella JF, Gautier R, Boehmer T, Schwartz TU, Antonny B. A general amphipathic alpha-helical motif for sensing membrane curvature. Nature structural & molecular biology. 2007;14:138–146. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- Edholm O, Nagle JF. Areas of molecules in membranes consisting of mixtures. Biophys J. 2005;89:1827–1832. doi: 10.1529/biophysj.105.064329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards DH, Thomaides HB, Errington J. Promiscuous targeting of Bacillus subtilis cell division protein DivIVA to division sites in Escherichia coli and fission yeast. EMBO J. 2000;19:2719–2727. doi: 10.1093/emboj/19.11.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira FM, Seica R, Oliveira PJ, Coxito PM, Moreno AJ, Palmeira CM, Santos MS. Diabetes induces metabolic adaptations in rat liver mitochondria: role of coenzyme Q and cardiolipin contents. Biochim Biophys Acta. 2003;1639:113–120. doi: 10.1016/j.bbadis.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Gitai Z, Dye NA, Reisenauer A, Wachi M, Shapiro L. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell. 2005;120:329–341. doi: 10.1016/j.cell.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Gregory JA, Becker EC, Pogliano K. Bacillus subtilis MinC destabilizes FtsZ-rings at new cell poles and contributes to the timing of cell division. Genes Dev. 2008;22:3475–3488. doi: 10.1101/gad.1732408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamoen LW, Errington J. Polar targeting of DivIVA in Bacillus subtilis is not directly dependent on FtsZ or PBP 2B. J Bacteriol. 2003;185:693–697. doi: 10.1128/JB.185.2.693-697.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzakis NS, Bhatia VK, Larsen J, Madsen KL, Bolinger PY, Kunding AH, Castillo J, Gether U, Hedegard P, Stamou D. How curved membranes recruit amphipathic helices and protein anchoring motifs. Nature chemical biology. 2009;5:835–841. doi: 10.1038/nchembio.213. [DOI] [PubMed] [Google Scholar]
- Hegde RS, Bernstein HD. The surprising complexity of signal sequences. Trends in biochemical sciences. 2006;31:563–571. doi: 10.1016/j.tibs.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Hirschberg CB, Kennedy EP. Mechanism of the enzymatic synthesis of cardiolipin in Escherichia coli. Proc Natl Acad Sci U S A. 1972;69:648–651. doi: 10.1073/pnas.69.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KC, Mukhopadhyay R, Wingreen NS. A curvature-mediated mechanism for localization of lipids to bacterial poles. PLoS Comput Biol. 2006;2:e151. doi: 10.1371/journal.pcbi.0020151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai F, Shoda M, Harashima R, Sadaie Y, Hara H, Matsumoto K. Cardiolipin domains in Bacillus subtilis marburg membranes. J Bacteriol. 2004;186:1475–1483. doi: 10.1128/JB.186.5.1475-1483.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooijman EE, Chupin V, Fuller NL, Kozlov MM, de Kruijff B, Burger KN, Rand PR. Spontaneous curvature of phosphatidic acid and lysophosphatidic acid. Biochemistry. 2005;44:2097–2102. doi: 10.1021/bi0478502. [DOI] [PubMed] [Google Scholar]
- Koppelman CM, Den Blaauwen T, Duursma MC, Heeren RM, Nanninga N. Escherichia coli minicell membranes are enriched in cardiolipin. J Bacteriol. 2001;183:6144–6147. doi: 10.1128/JB.183.20.6144-6147.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs JJ, Hauser H, Carafoli E. Asymmetric distribution of phospholipids in the inner membrane of beef heart mitochondria. J Biol Chem. 1979;254:5308–5316. [PubMed] [Google Scholar]
- Lam H, Schofield WB, Jacobs-Wagner C. A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell. 2006;124:1011–1023. doi: 10.1016/j.cell.2005.12.040. [DOI] [PubMed] [Google Scholar]
- Lenarcic R, Halbedel S, Visser L, Shaw M, Wu LJ, Errington J, Marenduzzo D, Hamoen LW. Localisation of DivIVA by targeting to negatively curved membranes. EMBO J. 2009;28:2272–2282. doi: 10.1038/emboj.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin PA, Fan N, Ricca E, Driks A, Losick R, Cutting S. An unusually small gene required for sporulation by Bacillus subtilis. Mol Microbiol. 1993;9:761–771. doi: 10.1111/j.1365-2958.1993.tb01736.x. [DOI] [PubMed] [Google Scholar]
- Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- Marraffini LA, Dedent AC, Schneewind O. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol Mol Biol Rev. 2006;70:192–221. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston AL, Thomaides HB, Edwards DH, Sharpe ME, Errington J. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev. 1998;12:3419–3430. doi: 10.1101/gad.12.21.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuley KE, Fyfe PK, Ridge JP, Isaacs NW, Cogdell RJ, Jones MR. Structural details of an interaction between cardiolipin and an integral membrane protein. Proc Natl Acad Sci U S A. 1999;96:14706–14711. doi: 10.1073/pnas.96.26.14706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mileykovskaya E, Dowhan W. Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J Bacteriol. 2000;182:1172–1175. doi: 10.1128/jb.182.4.1172-1175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mileykovskaya E, Dowhan W. Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochim Biophys Acta. 2009;1788:2084–2091. doi: 10.1016/j.bbamem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mileykovskaya E, Fishov I, Fu X, Corbin BD, Margolin W, Dowhan W. Effects of phospholipid composition on MinD-membrane interactions in vitro and in vivo. J Biol Chem. 2003;278:22193–22198. doi: 10.1074/jbc.M302603200. [DOI] [PubMed] [Google Scholar]
- Mileykovskaya E, Ryan AC, Mo X, Lin CC, Khalaf KI, Dowhan W, Garrett TA. Phosphatidic acid and N-acylphosphatidylethanolamine form membrane domains in Escherichia coli mutant lacking cardiolipin and phosphatidylglycerol. J Biol Chem. 2009;284:2990–3000. doi: 10.1074/jbc.M805189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R, Huang KC, Wingreen NS. Lipid localization in bacterial cells through curvature-mediated microphase separation. Biophys J. 2008;95:1034–1049. doi: 10.1529/biophysj.107.126920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita S, Matsuyama S, Tokuda H. Lipoprotein trafficking in Escherichia coli. Archives of microbiology. 2004;182:1–6. doi: 10.1007/s00203-004-0682-4. [DOI] [PubMed] [Google Scholar]
- Nishibori A, Kusaka J, Hara H, Umeda M, Matsumoto K. Phosphatidylethanolamine domains and localization of phospholipid synthases in Bacillus subtilis membranes. J Bacteriol. 2005;187:2163–2174. doi: 10.1128/JB.187.6.2163-2174.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou LT, Marquis RE. Electromechanical interactions in cell walls of gram-positive cocci. Journal of bacteriology. 1970;101:92–101. doi: 10.1128/jb.101.1.92-101.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozin AJ, Samford CS, Henriques AO, Moran CP., Jr. SpoVID guides SafA to the spore coat in Bacillus subtilis. J Bacteriol. 2001;183:3041–3049. doi: 10.1128/JB.183.10.3041-3049.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthasarathy R, Yu CH, Groves JT. Curvature-modulated phase separation in lipid bilayer membranes. Langmuir. 2006;22:5095–5099. doi: 10.1021/la060390o. [DOI] [PubMed] [Google Scholar]
- Perez AR, Abanes-De Mello A, Pogliano K. SpoIIB localizes to active sites of septal biogenesis and spatially regulates septal thinning during engulfment in bacillus subtilis. J Bacteriol. 2000;182:1096–1108. doi: 10.1128/jb.182.4.1096-1108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- Prajapati RS, Ogura T, Cutting SM. Structural and functional studies on an FtsH inhibitor from Bacillus subtilis. Biochim Biophys Acta. 2000;1475:353–359. doi: 10.1016/s0304-4165(00)00089-1. [DOI] [PubMed] [Google Scholar]
- Rafelski SM, Theriot JA. Mechanism of polarization of Listeria monocytogenes surface protein ActA. Mol Microbiol. 2006;59:1262–1279. doi: 10.1111/j.1365-2958.2006.05025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi KS, Clapham KR, Losick R. Peptide anchoring spore coat assembly to the outer forespore membrane in Bacillus subtilis. Mol Microbiol. 2006;62:1547–1557. doi: 10.1111/j.1365-2958.2006.05468.x. [DOI] [PubMed] [Google Scholar]
- Ramamurthi KS, Lecuyer S, Stone HA, Losick R. Geometric cue for protein localization in a bacterium. Science. 2009;323:1354–1357. doi: 10.1126/science.1169218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi KS, Losick R. Negative membrane curvature as a cue for subcellular localization of a bacterial protein. Proc Natl Acad Sci U S A. 2009;106:13541–13545. doi: 10.1073/pnas.0906851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin DM, de Boer PA. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci U S A. 1999;96:4971–4976. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawicz W, Olbrich KC, McIntosh T, Needham D, Evans E. Effect of chain length and unsaturation on elasticity of lipid bilayers. Biophys J. 2000;79:328–339. doi: 10.1016/S0006-3495(00)76295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romantsov T, Battle AR, Hendel JL, Martinac B, Wood JM. Protein localization in Escherichia coli cells: comparison of cytoplasmic membrane proteins ProP, LacY, ProW, AqpZ, MscS, and MscL. J Bacteriol. 2009a doi: 10.1128/JB.00967-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romantsov T, Guan Z, Wood JM. Cardiolipin and the osmotic stress responses of bacteria. Biochim Biophys Acta. 2009b;1788:2092–2100. doi: 10.1016/j.bbamem.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romantsov T, Helbig S, Culham DE, Gill C, Stalker L, Wood JM. Cardiolipin promotes polar localization of osmosensory transporter ProP in Escherichia coli. Mol Microbiol. 2007;64:1455–1465. doi: 10.1111/j.1365-2958.2007.05727.x. [DOI] [PubMed] [Google Scholar]
- Romantsov T, Stalker L, Culham DE, Wood JM. Cardiolipin controls the osmotic stress response and the subcellular location of transporter ProP in Escherichia coli. J Biol Chem. 2008;283:12314–12323. doi: 10.1074/jbc.M709871200. [DOI] [PubMed] [Google Scholar]
- Rudner DZ, Losick R. A sporulation membrane protein tethers the prosigmaK processing enzyme to its inhibitor and dictates its subcellular localization. Genes & development. 2002;16:1007–1018. doi: 10.1101/gad.977702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffers DJ, Pinho MG. Bacterial cell wall synthesis: new insights from localization studies. Microbiol Mol Biol Rev. 2005;69:585–607. doi: 10.1128/MMBR.69.4.585-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlame M, Greenberg ML. Cardiolipin synthase from yeast. Biochim Biophys Acta. 1997;1348:201–206. doi: 10.1016/s0005-2760(97)00117-3. [DOI] [PubMed] [Google Scholar]
- Schlame M, Hostetler KY. Cardiolipin synthase from mammalian mitochondria. Biochim Biophys Acta. 1997;1348:207–213. doi: 10.1016/s0005-2760(97)00119-7. [DOI] [PubMed] [Google Scholar]
- Schlame M, Towbin JA, Heerdt PM, Jehle R, DiMauro S, Blanck TJ. Deficiency of tetralinoleoyl-cardiolipin in Barth syndrome. Ann Neurol. 2002;51:634–637. doi: 10.1002/ana.10176. [DOI] [PubMed] [Google Scholar]
- Shapiro L, McAdams HH, Losick R. Why and how bacteria localize proteins. Science. 2009;326:1225–1228. doi: 10.1126/science.1175685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorre B, Callan-Jones A, Manneville JB, Nassoy P, Joanny JF, Prost J, Goud B, Bassereau P. Curvature-driven lipid sorting needs proximity to a demixing point and is aided by proteins. Proc Natl Acad Sci U S A. 2009;106:5622–5626. doi: 10.1073/pnas.0811243106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahlberg H, Kutejova E, Muchova K, Gregorini M, Lustig A, Muller SA, Olivieri V, Engel A, Wilkinson AJ, Barak I. Oligomeric structure of the Bacillus subtilis cell division protein DivIVA determined by transmission electron microscopy. Mol Microbiol. 2004;52:1281–1290. doi: 10.1111/j.1365-2958.2004.04074.x. [DOI] [PubMed] [Google Scholar]
- Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–241. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- Szeto TH, Rowland SL, Habrukowich CL, King GF. The MinD membrane targeting sequence is a transplantable lipid-binding helix. J Biol Chem. 2003;278:40050–40056. doi: 10.1074/jbc.M306876200. [DOI] [PubMed] [Google Scholar]
- Szeto TH, Rowland SL, Rothfield LI, King GF. Membrane localization of MinD is mediated by a C-terminal motif that is conserved across eubacteria, archaea, and chloroplasts. Proc Natl Acad Sci U S A. 2002;99:15693–15698. doi: 10.1073/pnas.232590599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi S, DiLuzio WR, Weibel DB, Whitesides GM. Controlling the shape of filamentous cells of Escherichia coli. Nano Lett. 2005;5:1819–1823. doi: 10.1021/nl0507360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooij C, Eichenberger P, Losick R. Dynamic patterns of subcellular protein localization during spore coat morphogenesis in Bacillus subtilis. J Bacteriol. 2004;186:4441–4448. doi: 10.1128/JB.186.14.4441-4448.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooij C, Losick R. Subcellular localization of a small sporulation protein in Bacillus subtilis. J Bacteriol. 2003;185:1391–1398. doi: 10.1128/JB.185.4.1391-1398.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreken P, Valianpour F, Nijtmans LG, Grivell LA, Plecko B, Wanders RJ, Barth PG. Defective remodeling of cardiolipin and phosphatidylglycerol in Barth syndrome. Biochem Biophys Res Commun. 2000;279:378–382. doi: 10.1006/bbrc.2000.3952. [DOI] [PubMed] [Google Scholar]
- Wiggins P, Phillips R. Membrane-protein interactions in mechanosensitive channels. Biophys J. 2005;88:880–902. doi: 10.1529/biophysj.104.047431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KD. The selective value of bacterial shape. Microbiol Mol Biol Rev. 2006;70:660–703. doi: 10.1128/MMBR.00001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Schlame M, Rua D, Greenberg ML. Cardiolipin synthase is associated with a large complex in yeast mitochondria. J Biol Chem. 1998;273:2402–2408. doi: 10.1074/jbc.273.4.2402. [DOI] [PubMed] [Google Scholar]
- Zhou J, Zhong Q, Li G, Greenberg ML. Loss of cardiolipin leads to longevity defects that are alleviated by alterations in stress response signaling. J Biol Chem. 2009;284:18106–18114. doi: 10.1074/jbc.M109.003236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerberg J, McLaughlin S. Membrane curvature: how BAR domains bend bilayers. Curr Biol. 2004;14:R250–252. doi: 10.1016/j.cub.2004.02.060. [DOI] [PubMed] [Google Scholar]