Abstract
We explored the effect of single-nucleotide polymorphisms (SNPs) in the fibroblast growth factor 20 gene (FGF20) associated with risk for Parkinson's disease on brain structure and function in a large sample of healthy young-adult human subjects and also in elderly subjects to look at the interaction between genetic variations and age (N = 237; 116 men; 18–87 years). We analyzed high-resolution anatomical magnetic resonance images using voxel-based morphometry, a quantitative neuroanatomical technique. We also measured FGF20 mRNA expression in postmortem human brain tissue to determine the molecular correlates of these SNPs (N = 108; 72 men; 18–74 years). We found that the T allele carriers of rs12720208 in the 3′-untranslated region had relatively larger hippocampal volume (p = 0.0059) and diminished verbal episodic memory (p = 0.048) and showed steeper decreases of hippocampal volume with normal aging (p = 0.026). In postmortem brain, T allele carriers had greater expression of hippocampal FGF20 mRNA (p = 0.037), consistent with a previously characterized microRNA mechanism. The C allele matches a predicted miR-433 microRNA binding domain, whereas the T allele disrupts it, resulting in higher FGF20 protein translation. The strong FGF20 genetic effects in hippocampus are presumably mediated by activation of the FGFR1 (FGF receptor 1), which is expressed in mammalian brain most abundantly in the hippocampus. These associations, from mRNA expression to brain morphology to cognition and an interaction with aging, confirm a role of FGF20 in human brain structure and function during development and aging.
Introduction
The abundant expression of fibroblast growth factor 20 (FGF20) in the midbrain, its role in dopaminergic (DA) neuron survival, and its location on chromosome 8 p22-p21.3 close to a Parkinson's disease (PD) linkage peak (Scott et al., 2001), have made FGF20 a promising candidate gene for PD. As a neurotrophin, FGF20 plays critical roles not only in the growth and survival of neurons in early development but also in the biology of adult neurons. FGF20 appears to be specifically expressed within the brain and particularly within neurons of the substantia nigra (Ohmachi et al., 2000) and cerebellum (Jeffers et al., 2001).
FGF20 mediates its biological response by activating a cell surface tyrosine kinase FGF receptor-1 (FGFR1) (Ohmachi et al., 2003). FGF20 signaling has been studied in DA neurons of the substantia nigra, as a potential factor related to the pathogenesis and potential treatment of Parkinson's disease, where it promotes the survival of calbindin-negative DA neurons that express specifically FGFR1 (Murase and McKay, 2006). FGFR1 expression also is widely distributed in the brain from neocortex to amygdala, caudate nucleus, substantia nigra, thalamus, and hippocampus (Weickert et al., 2005), the latter structure expressing the highest level of FGFR1 in brain (Takami et al., 1998; Fu et al., 2003).
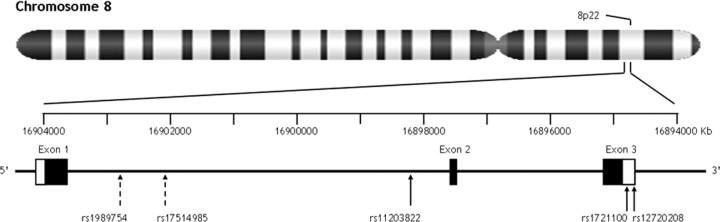
In a family-based genetic study, van der Walt et al. (2004) reported association of FGF20 single-nucleotide polymorphisms (SNPs) with increased risk for PD. Two SNPs located in the 3′-untranslated region (UTR) of the gene, rs12720208[C/T] (T allele positively associated; p = 0.0008) and rs1721100[C/G] (C allele positively associated; p = 0.02), and one intronic SNP, rs1989754[C/G] (C allele positively associated; p = 0.0006), showed association with illness (see Fig. 1). One of these SNPs, rs12720208, was subsequently shown in tissue culture to affect the translation of FGF20 via disrupting a putative microRNA binding site within the 3′-untranslated region (Wang et al., 2008). Specifically, the risk-associated allele in FGF20 removed this binding domain and resulted in relatively greater levels of FGF20 protein and presumably greater FGF20 effects.
Figure 1.
Gene structure of FGF20 (9.341 kb) on chromosome 8p22. The coding regions and the UTRs are indicated by the closed and open boxes, respectively. rs11203822 (a surrogate for rs1989754) (see Materials and Methods) is in the first intron, and rs1721100 and rs12720208 (a surrogate for rs17514985) (see Results) are in the 3′-UTR regulatory region.
The evidence from functional data of the role of a SNP in regulating expression of FGF20 makes examination of the effect of FGF20 SNPs on brain structure and function of considerable interest. We explored in this study the impact of these risk-associated genetic polymorphisms (rs12720208, rs1721100, and rs1989754) on the morphology of the brain in a large sample of healthy young adult using voxel-based morphometry (VBM), a quantitative neuroanatomical magnetic resonance imaging (MRI) technique (Good et al., 2001). A sample of elderly subjects was also included to study specifically the interaction between age and genetic variation. We also measured FGF20 mRNA expression in postmortem human brain tissue to determine the molecular correlates of these SNPs. We hypothesized that genetic variations associated with increased risk for PD and modulation of gene expression should have a direct or indirect effect on measures of brain structures and also on related cognitive functions.
Materials and Methods
Subjects.
The study sample consisted of 237 normal volunteers, including 194 healthy volunteers (18–58 years of age) recruited as part of the Clinical Brain Disorders Branch Sibling Study [National Institutes of Health (NIH) Study ID NCT00001486] described in detail previously (Egan et al., 2001) and an additional 43 healthy volunteers (60–87 years of age) recruited as part of a study of normal aging (NIH Study ID NCT0001284). The elderly subjects were included in the study to answer the question of the interaction between genetic variation and age. To reduce genetic heterogeneity, we included only Caucasian subjects of European ancestry who were free of any history of psychiatric or neurological illness, of psychiatric treatment, or of drug or alcohol abuse within the 12 months before participation in this study. All of the subjects gave written informed consent and participated in the study according to the guidelines of the National Institute of Mental Health Institutional Review Board.
DNA collection and genotyping.
DNA was extracted from whole blood using standard procedures. All genotypes were determined using the 5′-exonuclease TaqMan assay; SNP probe and primer sets were acquired as Assays on Demand from Applied Biosystems. We genotyped three FGF20 SNPs (Fig. 1) that have been originally associated with increased risk for PD (van der Walt et al., 2004): rs12720208[C/T], rs1721100[C/G], and rs11203822[A/T] [the latter a surrogate for rs1989754[C/G], with which it is in strong linkage disequilibrium (LD) (D′ = 1; r2 = 0.97)]. All genotypes were in Hardy–Weinberg equilibrium and the SNPs are not in strong LD with each other (D′ = 1; r2 < 0.38).
In an effort to identify causative mutations not previously observed in FGF20 and for which rs12720208 might represent a proxy and not the functional mutation, we resequenced the entire FGF20 gene, beginning 2 kb upstream of the transcription start site and extending 1 kb downstream of the 3′-UTR poly(A) signal in a subsample of subjects. These subjects were selected from the original sample based on their rs12720208 genotype and hippocampal phenotype to maximize the likelihood of finding causative variants in the gene related specifically to the enlarged hippocampal volume phenotype (see Results). We thus resequenced 48 individuals including all T (i.e., risk-associated) carriers (N = 20) and 28 C/C homozygotes at rs12720208 who also had hippocampal volume in the lower quartile of the entire sample. Any new SNPs found enriched in the rs12720208 T carrier subsample were then genotyped in the entire sample of subjects.
Structural image processing.
For all subjects, a three-dimensional structural MRI was acquired on a 1.5 tesla GE scanner using a T1-weighted spoiled gradient-recalled acquisition in a steady state sequence (repetition time, 24 ms; echo time, 5 ms; excitations, 1; flip angle, 45°), with 124 sagittal slices at a thickness of 1.5 mm and an in-plane resolution of 0.94 × 0.94 mm. Images were reconstructed and visually checked for major artifacts before additional processing. All of the scans were processed in SPM2 (www.fil.ion.ucl.ac.uk/spm) using an optimized voxel-based morphometry protocol as described in detail previously (Ashburner and Friston, 2000; Good et al., 2001). Briefly, each subject T1 volume was first segmented, thereby providing gray matter (GM) partition image in the subject native space. Each subject GM image was then spatially normalized (25 mm cutoff of discrete cosine transform bases, medium regularization, 16 nonlinear iterations) to a representative GM template derived from a subset of 140 subjects (70 men) statistically similar for age, intelligence quotient (IQ), and education level with the entire study group. The GM warping parameters so obtained were then used to spatially normalize the whole-brain T1 volume, the resulting volume being further interpolated as 1 mm3 isotropic voxel. Then, the normalized whole-brain T1 volume was segmented using a priori GM, white matter (WM), and CSF probability maps derived from the subset of subjects previously described. Then, a so-called “modulation” step was added, to adjust voxel intensity for the strength of the deformation performed during the spatial normalization process. This step was applied to each tissue partition to preserve the subject's original tissue quantity after its transfer to the reference space. Finally, all tissue images were smoothed with a 10 mm full-width half-maximum Gaussian kernel. For each subject, GM, WM, and CSF volumes were computed as the integral of the voxel intensities of the corresponding modulated tissue image. Total intracranial volume was defined as the sum of the GM, WM, and CSF volumes. To estimate the magnitude of the genetic effect, we estimated the hippocampal volume by calculating the integral of the voxel intensities within the hippocampal region (Lemaitre et al., 2005). The hippocampal boundary was automatically derived from an independent model of macroscopic neuroanatomical parcellation (automated anatomical labeling) (Tzourio-Mazoyer et al., 2002).
Imaging statistics.
Because of the low minor allele frequencies of rs12720208 and rs1721100, few subjects were T/T and G/G, respectively (<7% of the sample). For this reason, we combined minor allele heterozygotes and homozygotes for these two SNPs. Consequently, single SNP effects of rs12720208 and rs1721100 were examined using simple t test analysis (dominant model), whereas rs11203822 was examined using a regression analysis (additive model). Structural images were analyzed using SPM, whereas cerebral volumes were analyzed using R (http://cran.r-project.org). All of them were used within the framework of the general linear model. For the younger sample (N = 194), the following variables were included in the model to control for confounds: (1) total intracranial volume for interindividual variability of head size, (2) age, and (3) gender. At the voxel level, the exploratory full-brain statistics were performed with a statistical threshold set to p < 0.001 (uncorrected for multiple comparisons). At the cluster level, because of the nonuniform smoothness of the VBM data (Ashburner and Friston, 2000), we used a nonstationary random-field theory cluster size test (Hayasaka et al., 2004) with a cluster size threshold set to p < 0.05, familywise error (FWE) corrected for multiple comparisons based on the number of voxels contained within the whole brain. For the expanded sample including elderly subjects (total N = 237), we used the same strategy and looked more specifically at the interaction between genotypes and age on the hippocampal volume.
Cognitive evaluation.
To address the potential functional implications of genetic variation in FGF20, we measured a set of standardized neuropsychological tests that survey a broad span of cognitive functions (Egan et al., 2001). The neurocognitive battery included the Wechsler Memory Scale, Revised (WMS-R); Wechsler Adult Intelligence Scale, Revised (WAIS-R) (arithmetic, similarities, digit–symbol substitution, and picture completion); the Trail Making Test, Parts A and B; the Verbal and Category Fluency Test; the Continuous Performance Test; the N-Back Task; the California Verbal Learning Test; the Judgment of Line Orientation; and the Wisconsin Card Sorting Test. The 24 subtests from this battery were reducible via principal components and confirmatory factor analyses to a seven-factor solution (Genderson et al., 2007). These factors were more independent than the individual subtest scores and putatively closer to the individuals' underlying psychometric structure than any single subtest (Genderson et al., 2007). Factor 1 was loaded on verbal episodic memory measures from the WMS-R and California Verbal Learning Test; factor 2 on aspects of working memory from the N-Back Task; factor 3 on spatial episodic memory measures from WMS-R and Judgment of Line Orientation; factor 4 on IQ, executive cognitive control, and processing speed measures from WAIS-R, trails A and B, and letter and category fluency; factor 5 on logical reasoning measures from the Wisconsin Card Sorting Test; factor 6 on attentional measures from the Continuous Performance Test; and factor 7 on measures from the WMS-R digit span backward and forward (Genderson et al., 2007). FGF20 genetic contribution to these cognitive factors was analyzed in the group of 194 young adult subjects in whom neuropsychological data were only available (elderly subjects did not perform the same battery of tests). Individual SNP analyses were performed in R via the general linear model using dominant and additive models, respectively, as in the previous analyses to identify genetic variations associated with cognitive factor scores. Values of p were reported uncorrected for multiple comparisons.
FGF20 expression. To confirm the previously reported functional effect of SNPs in FGF20, we measured mRNA expression in postmortem human brain using quantitative PCR. Postmortem brain samples from 108 subjects (72 normal controls, 36 schizophrenic cases; 0.5 female/male ratio; 43.4 ± 15.1 years of age; 38 Caucasians, 70 African Americans) were collected at the Clinical Brain Disorders Branch (NIH Study ID NCT00001260). Detailed information about the postmortem sample has been previously described (Lipska et al., 2006). Briefly, tissue from hippocampus was pulverized and stored at −80°C. Total RNA was extracted from 300 mg of tissue with Trizol reagent (Invitrogen). The yield of total RNA was determined by spectrophotometry by measuring absorbance at 260 nm. RNA quality was assessed with high-resolution capillary electrophoresis on an Agilent Bioanalyzer 2100 (Agilent Technologies). RNA integrity number (RIN) was used as an estimate of RNA quality (only samples with RIN > 3.8 were included in the analysis). Total RNA (4 μg) was used in 50 μl of reverse transcriptase reaction to synthesize cDNA, by using a SuperScript First-Strand Synthesis System for reverse transcription (RT)-PCR (Invitrogen), according to the manufacturer's protocol. Expression levels of mRNAs were measured by quantitative real-time RT-PCR, with specific combinations of primers and probes (Hs00173929_ m1 spanning exons 3–4 of FGF20) and an ABI Prism 7900 sequence detection system with 384-well format (Applied Biosystems). Each 10–20 μl reaction contained 900 nmol/L primer, 250 nmol/L probe, and TaqMan Universal PCR Mastermix (Applied Biosystems) containing Hot Goldstar DNA Polymerase, dNTPs (deoxynucleoside triphosphates) with dUTP (deoxyuridine triphosphate), uracil-N-glycosylase, passive reference, and cDNA template generated from 100 to 200 ng of RNA. The PCR cycle parameters were 50°C for 2 min, 95°C for 10 min, 40 cycles of 95°C for 15 s, and 59 or 60°C for 1 min. The PCR data were acquired from the sequence detector software (SDS) (version 2.0; Applied Biosystems) and quantified by a standard curve method with serial dilutions of pooled cDNA derived from RNA obtained from hippocampus of 10–12 normal control subjects. The SDS software plotted real-time fluorescence intensity and selected the threshold within the exponential phase of the amplicon profiles. All measurements were performed in triplicate. FGF20 expression level was normalized to the geometric mean of three housekeeping genes (β-glucuronidase, β-actin, and β2-microglobulin). Data were analyzed in R using the general linear model and the same genetic models as before with FGF20 SNPs as predictors and pH, RNA quality, gender, age, ethnicity, and diagnosis of schizophrenia as confounding variables. Values of p were reported uncorrected for multiple comparisons. Five subjects had missing genotype data.
Results
Effect of FGF20 on brain structure
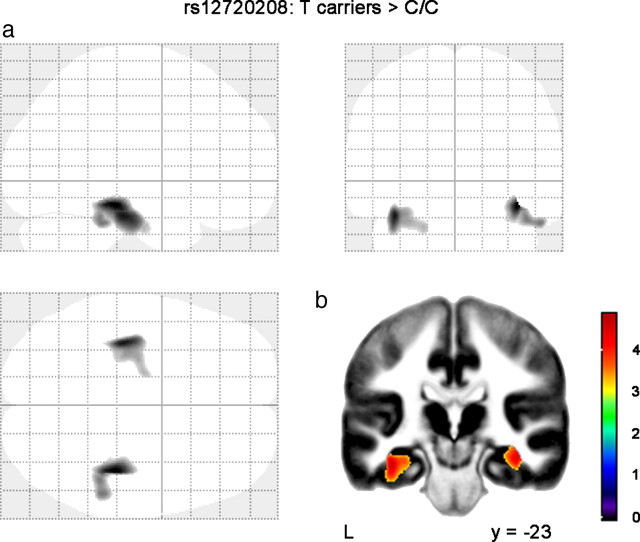
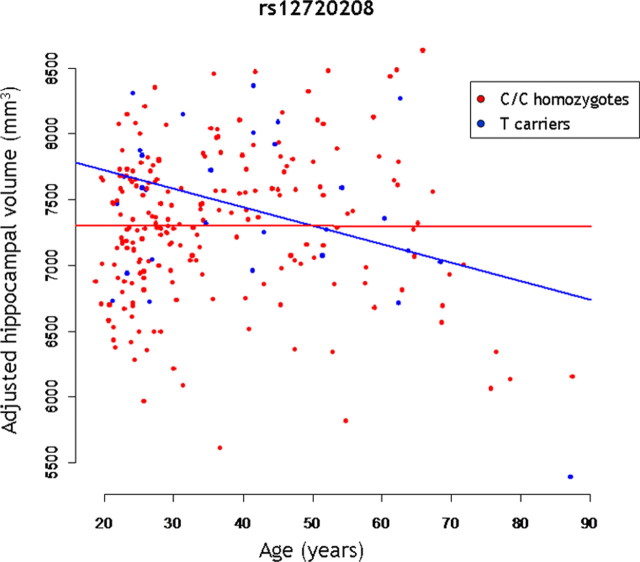
The genotype groups for each SNP did not differ significantly for age, gender, education level, IQ, and handedness either for the young adult sample or for the wide age range MRI sample (Table 1). For rs12720208, the voxel-based comparison of the GM maps in the young adult sample (N = 195) showed a selective increase in GM within the hippocampal region bilaterally in T carriers compared with C/C homozygotes (Fig. 2, Table 2). Examining the magnitude of rs12720208 effect on the adjusted volume of the hippocampal region, we found a significant volume difference (p = 0.0059), with 9909 ± 515 mm3 for T carriers against 9550 ± 580 mm3 for C/C homozygotes. The voxel-based comparison of the GM maps did not show significant regional differences across genotype groups for rs1721100 or for rs11203822. We then looked at the interaction between FGF20 genotypes and age on hippocampal volume in the wide age range sample including elderly participants (N = 237). We found a significant interaction between rs12720208 and age on hippocampal volume (p = 0.026) (Fig. 3), with a greater age-related decrease of adjusted hippocampal volume in T carriers (post hoc test, p = 0.014; −15 mm3/year) compared with C/C homozygotes (post hoc test, p = 0.97; −0.1 mm3/year). Finally, we did not find significant interactions with age for rs1721100 or rs11203822. In addition, there was no significant difference across genotype groups for rs12720208, rs1721100, and rs11203822 for any of the global cerebral volumes: gray matter, white matter, CSF, and total intracranial volumes.
Table 1.
Samples
| Young MRI sample | Full MRI sample | Postmortem sample | |
|---|---|---|---|
| No. of subjects | 194 | 237 | 108 |
| Age (years) | 32.6 ± 9.8 (18–58) | 36.7 ± 14.9 (18–87) | 43.4 ± 15.1 (18–74) |
| Sex ratio (women/men) | 1.06 | 1.04 | 0.49 |
| WAIS full-scale IQ (score) | 108.4 ± 9.1 | 109.5 ± 9.3 | — |
| Education (years) | 16.9 ± 2.9 | 17.0 ± 2.6 | — |
| Handedness (score) | 77.9 ± 52.0 | 78.9 ± 49.7 | — |
| rs12720208 | 148 C/C, 24 C/T, 1 T/T | 206 C/C, 28 C/T, 1 T/T | 93 C/C, 10 C/T |
| rs1721100 | 90 C/C, 75 C/G, 11 G/G | 123 C/C, 89 C/G, 16 G/G | 41 C/C, 45 C/G, 18 G/G |
| rs11203822a | 54 T/T, 85 T/A, 33 A/A | 73 T/T, 108 T/A, 50 A/A | 63 T/T, 32 T/A, 8 A/A |
Values represent mean ± SD, and range is shown in parentheses.
aA surrogate for rs1989754.
Figure 2.
Structural brain differences associated with FGF20 genotypes. a, Statistical t map represented on the SPM glass brain showing regional GM differences for rs12720208: T carriers > C/C homozygotes. Map is displayed with a voxel-level threshold at p < 0.001 (uncorrected for the whole brain) and a cluster-size threshold at p < 0.05 (FWE corrected for the whole brain). b, The same map is superimposed on a coronal slice of the average GM map of 194 healthy volunteers (y = −23). L, Left.
Table 2.
Regional GM difference for rs12720208: T carriers > CC homozygotes
| Non-isotropic adjusted cluster level |
Voxel level |
Location |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| pFWE-corr | k | pFWE-corr | pFDR-corr | T | Z | puncorr | x | y | z |
| 0.003 | 4439 | 0.044 | 0.069 | 4.75 | 4.60 | <0.001 | 39 | −29 | −15 |
| 0.554 | 0.075 | 3.92 | 3.83 | <0.001 | 53 | −38 | −25 | ||
| 0.608 | 0.078 | 3.87 | 3.78 | <0.001 | 45 | −39 | −23 | ||
| 0.007 | 3744 | 0.097 | 0.069 | 4.53 | 4.40 | <0.001 | −37 | −24 | −21 |
| 0.840 | 0.088 | 3.64 | 3.57 | <0.001 | −30 | −17 | −22 | ||
| 0.922 | 0.094 | 3.53 | 3.46 | <0.001 | −20 | −11 | −27 | ||
k, Cluster size in cubic millimeters; FDR, false discovery rate; corr, corrected for the whole brain; T, T score; Z, Z score; x, y, z, MNI stereotactic coordinates in millimeters.
Figure 3.
Scatterplot illustrating the age by genotype interaction on the adjusted hippocampal volume for rs12720208. The red and blue lines are regressions for C/C homozygotes (p = 0.97; −0.1 mm3/year) and T carriers (p = 0.014; −15 mm3/year), respectively.
Effect of FGF20 on cognition
From the young adult sample, SNP association tests were performed with various cognitive measures. We found association of rs12720208 T allele and rs11203822 T allele with reduced scores for factor 1, which is related to verbal episodic memory (p = 0.048 and 0.041, respectively). rs1721100 did not show association with cognition.
FGF20 expression in postmortem brain
We investigated the impact of the FGF20 genetic variations on hippocampal FGF20 mRNA expression in 108 postmortem human brains (Table 1). The genotype groups for each SNP did not differ significantly for age and gender. rs12720208 T carriers showed significantly greater expression of FGF20 mRNA than C/C homozygotes (88.3 ± 23.3 vs 67.0 ± 28.9%; p = 0.037). No difference of mRNA expression was detected for rs1721100 and rs11203822 polymorphisms.
SNP detection via resequencing
To identify causative mutations not previously observed in FGF20 and for which rs12720208 might represent a proxy and not the functional mutation, we resequenced the entire FGF20 gene (9.341 kb) in a subsample of subjects who were selected from the original sample based on their rs12720208 genotype and hippocampal volume (see Materials and Methods). Five previously undocumented SNPs with a minor allele frequency >5% were found based on this resequencing. However, only one of these SNPs was overrepresented in the subsample of 20 individuals who were rs12720208 T carriers. This SNP, rs17514985 (Fig. 1) in intron 1B, was genotyped in the entire sample, and it was in virtual allelic identity with rs12720208 (D′ = 0.99; r2 = 0.95).
Discussion
We have found that rs12720208, a FGF20 SNP shown to be associated with PD risk (van der Walt et al., 2004) and also shown to alter FGF20 expression via a microRNA mechanism (Wang et al., 2008), had an effect in healthy living subjects on the morphology of the brain, more specifically on the hippocampal region (p = 0.0059), and also on verbal episodic memory (p = 0.048) referable to hippocampal function. This SNP also affected levels of FGF20 mRNA in postmortem human brain (p = 0.037) consistent with its previously demonstrated microRNA inhibitory effects. Perhaps most strikingly, the effect of the same SNP on hippocampal volume interacted with normal aging (p = 0.026).
Of the three SNPs we tested, only rs12720208 showed a significant effect on any of the measures we examined. This 3′-UTR SNP lies within a predicted interaction site for miR-433 microRNA (Wang et al., 2008). microRNAs are small noncoding transcripts of ∼21–23 nt that negatively regulate the translation of complementary mRNA, usually by binding the 3′-UTR and triggering the degradation of the mRNA transcript. The C allele of rs12720208 matches the predicted miR-433 binding domain, whereas the T allele disrupts it, causing an inhibitory effect on microRNA-mediated repression of gene translation. In vitro experiments have shown that miR-433 does not bind to FGF20 mRNA transcripts containing the T allele, resulting in higher FGF20 protein translation (Wang et al., 2008). In a sample of 108 human postmortem brains, we observed increased levels of hippocampal FGF20 mRNA for T carriers compared with C/C homozygotes, which is consistent with the presumed molecular mechanism. Thus, the greater expression of FGF20 in the hippocampus in subjects with the T allele could account for the greater hippocampal volume we found in our voxel-based morphological approach.
The selective effect of the functional SNP rs12720208 on the hippocampus is presumably mediated by the neurotrophic activity of FGF20 on neurons through activation of FGFR1 (Ohmachi et al., 2003). FGF20 has been shown to be an important endogenous neurotrophic factor for the survival of DA neurons through MAPK (mitogen-activated protein kinase) pathway activation and tyrosine hydroxylase activation, which results in increased levels of DA release (Murase and McKay, 2006). FGF20 signaling requires the FGFR1, which is abundantly expressed on the surface of neurons in the midbrain as well as in the hippocampus (Weickert et al., 2005), where it is critical for neurogenesis in the dentate gyrus (Zhao et al., 2007). In our study, the genetic mediation of hippocampal structure could be related to FGF20 exerting its activity by binding to FGFR1 expressed on hippocampal neurons.
In our study of only normal controls, the PD risk-associated T allele of rs12720208 (van der Walt et al., 2004; Wang et al., 2008) is associated with what it appears at first sight to be a “positive” biomarker (i.e., larger hippocampal volume). Although subjects carrying the T allele had greater hippocampal volume, they also performed poorer on verbal episodic memory tests. This seemingly paradoxical effect on both structure and cognition is not easily reconciled, but it is conceivable that relatively excessive FGF20 signaling in hippocampus, although supportive of hippocampal growth and perhaps neurogenesis, may have negative functional implications even in the absence of a disease state. This is speculation that merits additional study. Many biological processes are not invariably linear; nonlinear and inverted “U”-type effects are common (Mattay et al., 2003). Likewise, increased hippocampal volume per se does not invariably mean enhanced function. For example, larger amygdala and hippocampus volumes are found in patients with autism compared with normal control (Sparks et al., 2002). In addition, the negative interaction of the rs12720208 T allele on age-related decreases of hippocampal volume reinforces the notion that increased FGF20 signaling may have deleterious consequences in certain biological contexts.
Several studies have looked at associations between FGF20 and PD; however, their results have been variable. Using a larger dataset of subjects than the original report (van der Walt et al., 2004), the same authors found the same three SNPs associated with risk of PD (Wang et al., 2008). In a Japanese sample, positive association was reported for rs1721100 and rs1989754; rs12720208 is not present in the Japanese population (Satake et al., 2007). However, in a small sample of Finnish and Greek subjects, no association was found for any of these three SNPs (Clarimon et al., 2005). In a recent study including subjects from different nationalities, no association was identified between rs12720208 and risk for PD (Wider et al., 2009). However, it is worth noting that only the studies by van der Walt et al. (2004) and Wang et al. (2008) were family-based designs (i.e., affected individuals and family members), which avoids bias from population stratification. Two genome-wide association studies to identify susceptibility genes for PD have been reported. The first study genotyped only one SNP (rs17515020) in FGF20, which was not significant (Maraganore et al., 2005), and the second study did not genotype any SNPs in FGF20 (Fung et al., 2006). Thus, additional association studies are needed to test and replicate the previously reported associations and confirm or reject the role of FGF20 in PD risk.
Our study of elderly subjects brings these results closer to the framework of PD, the incidence of PD being clearly increased with advancing age. A molecular link between FGF20 and PD has been inferred from the observations that the rs12720208 T allele leads not only to a higher level of FGF20 but also to a higher level of α-synuclein in the brain (Wang et al., 2008). This protein, although its exact role is not known, is the primary component of Lewy body fibrils, the pathological marker of diseases such as PD. As we did not study PD patients, we cannot directly address the question of the relationship between our results and the pathophysiology of PD. However, given the results in the aging sample, chronically increased levels of FGF20 could indirectly contribute to neuron death at later stages of life, which would constitute favorable conditions to the development of the disease.
Our original expectation was that we would observe an effect of FGF20 genotype in the midbrain region especially in the substantia nigra, where FGF20 is abundantly expressed compared with other brain regions (Ohmachi et al., 2000). However, a possible reason for not observing morphological differences in the substantia nigra might be the weak MRI contrast between gray and white matter in this region resulting in the poor segmentation of such a small structure. Only structural brain scans using higher spatial resolution and more specific pulse sequence could address this issue (Manova et al., 2009). Probably because of this limitation, our anatomical data highlighted effects of FGF20 genotype only in the hippocampus, a brain region expressing the highest FGFR1 density (Weickert et al., 2005) but not a primary target region for neurodegeneration in PD. It is reasonable to speculate, although we have no direct data about this, that similar molecular effects of rs12720208 genotype might also occur with FGF20 and FGFR1 signaling in substantia nigra DA neurons. Along with this work on SNPs already identified, we used a novel approach to identify other potential causative SNPs by enriching our resequencing sample based on risk-associated alleles and hippocampal volume as a brain phenotype. We found an interesting candidate in rs17514985, but this SNP is in virtual allelic identity with rs12720208, making its effect impossible to disentangle from that of rs12720208. However, it is not as likely to be functional as rs12720208, which is located in the 3′-UTR regulatory region.
Our study is based on the classic scientific method of data collection and hypothesis-driven testing and is in line with translational genetic strategies that try to find convergent evidence at different biological levels that genetic variants are functional (Meyer-Lindenberg, 2009). In this manner, we have performed a number of tests in this study, testing several SNPs on a variety of clinical and biological phenotypes. Each of these tests was based on specific hypotheses related to the basic science of FGF20 and the structure of the gene, and as such the tests were not independent from each other. Although the p values we report would not survive a biologically agnostic Bonferroni-type “correction” for all of these tests if they were treated as independent, and without previous probability of a finding, we believe that this type of p value adjustment is an overly conservative approach to deal with the problem of multiple testing (Straub et al., 2007). First, the FGF20 SNPs are in substantial LD and thus are not independent. Second, the previous probability of the 3′-UTR SNP in this gene impacting on expression of this gene is not null as it had been shown to affect FGF20 protein levels previously in cell model experiments. Finally, the likelihood that the same SNP would consistently predict variation in each of these independent intermediate biological phenotypes by chance, and always in the predicted direction of the risk-associated allele, is particularly remote.
In summary, we have found convergent evidence in healthy subjects that a functional FGF20 SNP modulated human hippocampal biology on a series of hypothesis-driven experiments including mRNA expression, verbal episodic memory, and hippocampal morphology, the latter interacting with effects of normal aging. Overall, these results support a role of FGF20 genetic variation in human brain structure and function during development and aging.
Footnotes
This work was supported by the Intramural Research Program of the National Institute of Mental Health–National Institutes of Health.
References
- Ashburner J, Friston KJ. Voxel-based morphometry—the methods. Neuroimage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Clarimon J, Xiromerisiou G, Eerola J, Gourbali V, Hellström O, Dardiotis E, Peuralinna T, Papadimitriou A, Hadjigeorgiou GM, Tienari PJ, Singleton AB. Lack of evidence for a genetic association between FGF20 and Parkinson's disease in Finnish and Greek patients. BMC Neurol. 2005;5:11. doi: 10.1186/1471-2377-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MF, Goldberg TE, Gscheidle T, Weirich M, Rawlings R, Hyde TM, Bigelow L, Weinberger DR. Relative risk for cognitive impairments in siblings of patients with schizophrenia. Biol Psychiatry. 2001;50:98–107. doi: 10.1016/s0006-3223(01)01133-7. [DOI] [PubMed] [Google Scholar]
- Fu L, Abu-Khalil A, Morrison RS, Geschwind DH, Kornblum HI. Expression patterns of epidermal growth factor receptor and fibroblast growth factor receptor 1 mRNA in fetal human brain. J Comp Neurol. 2003;462:265–273. doi: 10.1002/cne.10727. [DOI] [PubMed] [Google Scholar]
- Fung HC, Scholz S, Matarin M, Simón-Sánchez J, Hernandez D, Britton A, Gibbs JR, Langefeld C, Stiegert ML, Schymick J, Okun MS, Mandel RJ, Fernandez HH, Foote KD, Rodríguez RL, Peckham E, De Vrieze FW, Gwinn-Hardy K, Hardy JA, Singleton A. Genome-wide genotyping in Parkinson's disease and neurologically normal controls: first stage analysis and public release of data. Lancet Neurol. 2006;5:911–916. doi: 10.1016/S1474-4422(06)70578-6. [DOI] [PubMed] [Google Scholar]
- Genderson MR, Dickinson D, Diaz-Asper CM, Egan MF, Weinberger DR, Goldberg TE. Factor analysis of neurocognitive tests in a large sample of schizophrenic probands, their siblings, and healthy controls. Schizophr Res. 2007;94:231–239. doi: 10.1016/j.schres.2006.12.031. [DOI] [PubMed] [Google Scholar]
- Good CD, Johnsrude IS, Ashburner J, Henson RN, Friston KJ, Frackowiak RS. A voxel-based morphometric study of ageing in 465 normal adult human brains. Neuroimage. 2001;14:21–36. doi: 10.1006/nimg.2001.0786. [DOI] [PubMed] [Google Scholar]
- Hayasaka S, Phan KL, Liberzon I, Worsley KJ, Nichols TE. Nonstationary cluster-size inference with random field and permutation methods. Neuroimage. 2004;22:676–687. doi: 10.1016/j.neuroimage.2004.01.041. [DOI] [PubMed] [Google Scholar]
- Jeffers M, Shimkets R, Prayaga S, Boldog F, Yang M, Burgess C, Fernandes E, Rittman B, Shimkets J, LaRochelle WJ, Lichenstein HS. Identification of a novel human fibroblast growth factor and characterization of its role in oncogenesis. Cancer Res. 2001;61:3131–3138. [PubMed] [Google Scholar]
- Lemaitre H, Crivello F, Dufouil C, Grassiot B, Tzourio C, Alpérovitch A, Mazoyer B. No epsilon4 gene dose effect on hippocampal atrophy in a large MRI database of healthy elderly subjects. Neuroimage. 2005;24:1205–1213. doi: 10.1016/j.neuroimage.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Lipska BK, Deep-Soboslay A, Weickert CS, Hyde TM, Martin CE, Herman MM, Kleinman JE. Critical factors in gene expression in postmortem human brain: focus on studies in schizophrenia. Biol Psychiatry. 2006;60:650–658. doi: 10.1016/j.biopsych.2006.06.019. [DOI] [PubMed] [Google Scholar]
- Manova ES, Habib CA, Boikov AS, Ayaz M, Khan A, Kirsch WM, Kido DK, Haacke EM. Characterizing the mesencephalon using susceptibility-weighted imaging. AJNR Am J Neuroradiol. 2009;30:569–574. doi: 10.3174/ajnr.A1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraganore DM, de Andrade M, Lesnick TG, Strain KJ, Farrer MJ, Rocca WA, Pant PV, Frazer KA, Cox DR, Ballinger DG. High-resolution whole-genome association study of Parkinson disease. Am J Hum Genet. 2005;77:685–693. doi: 10.1086/496902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattay VS, Goldberg TE, Fera F, Hariri AR, Tessitore A, Egan MF, Kolachana B, Callicott JH, Weinberger DR. Catechol O-methyltransferase val158-met genotype and individual variation in the brain response to amphetamine. Proc Natl Acad Sci U S A. 2003;100:6186–6191. doi: 10.1073/pnas.0931309100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Lindenberg A. Intermediate or brainless phenotypes for psychiatric research? Psychol Med First View. 2009:1–6. doi: 10.1017/s0033291709991929. [DOI] [PubMed] [Google Scholar]
- Murase S, McKay RD. A specific survival response in dopamine neurons at most risk in Parkinson's disease. J Neurosci. 2006;26:9750–9760. doi: 10.1523/JNEUROSCI.2745-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmachi S, Watanabe Y, Mikami T, Kusu N, Ibi T, Akaike A, Itoh N. FGF-20, a novel neurotrophic factor, preferentially expressed in the substantia nigra pars compacta of rat brain. Biochem Biophys Res Commun. 2000;277:355–360. doi: 10.1006/bbrc.2000.3675. [DOI] [PubMed] [Google Scholar]
- Ohmachi S, Mikami T, Konishi M, Miyake A, Itoh N. Preferential neurotrophic activity of fibroblast growth factor-20 for dopaminergic neurons through fibroblast growth factor receptor-1c. J Neurosci Res. 2003;72:436–443. doi: 10.1002/jnr.10592. [DOI] [PubMed] [Google Scholar]
- Satake W, Mizuta I, Suzuki S, Nakabayashi Y, Ito C, Watanabe M, Takeda A, Hasegawa K, Sakoda S, Yamamoto M, Hattori N, Murata M, Toda T. Fibroblast growth factor 20 gene and Parkinson's disease in the Japanese population. Neuroreport. 2007;18:937–940. doi: 10.1097/WNR.0b013e328133265b. [DOI] [PubMed] [Google Scholar]
- Scott WK, Nance MA, Watts RL, Hubble JP, Koller WC, Lyons K, Pahwa R, Stern MB, Colcher A, Hiner BC, Jankovic J, Ondo WG, Allen FH, Jr, Goetz CG, Small GW, Masterman D, Mastaglia F, Laing NG, Stajich JM, Slotterbeck B, et al. Complete genomic screen in Parkinson disease: evidence for multiple genes. JAMA. 2001;286:2239–2244. doi: 10.1001/jama.286.18.2239. [DOI] [PubMed] [Google Scholar]
- Sparks BF, Friedman SD, Shaw DW, Aylward EH, Echelard D, Artru AA, Maravilla KR, Giedd JN, Munson J, Dawson G, Dager SR. Brain structural abnormalities in young children with autism spectrum disorder. Neurology. 2002;59:184–192. doi: 10.1212/wnl.59.2.184. [DOI] [PubMed] [Google Scholar]
- Straub RE, Lipska BK, Egan MF, Goldberg TE, Callicott JH, Mayhew MB, Vakkalanka RK, Kolachana BS, Kleinman JE, Weinberger DR. Allelic variation in GAD1 (GAD67) is associated with schizophrenia and influences cortical function and gene expression. Mol Psychiatry. 2007;12:854–869. doi: 10.1038/sj.mp.4001988. [DOI] [PubMed] [Google Scholar]
- Takami K, Matsuo A, Terai K, Walker DG, McGeer EG, McGeer PL. Fibroblast growth factor receptor-1 expression in the cortex and hippocampus in Alzheimer's disease. Brain Res. 1998;802:89–97. doi: 10.1016/s0006-8993(98)00552-6. [DOI] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- van der Walt JM, Noureddine MA, Kittappa R, Hauser MA, Scott WK, McKay R, Zhang F, Stajich JM, Fujiwara K, Scott BL, Pericak-Vance MA, Vance JM, Martin ER. Fibroblast growth factor 20 polymorphisms and haplotypes strongly influence risk of Parkinson disease. Am J Hum Genet. 2004;74:1121–1127. doi: 10.1086/421052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, van der Walt JM, Mayhew G, Li YJ, Züchner S, Scott WK, Martin ER, Vance JM. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. Am J Hum Genet. 2008;82:283–289. doi: 10.1016/j.ajhg.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weickert CS, Kittell DA, Saunders RC, Herman MM, Horlick RA, Kleinman JE, Hyde TM. Basic fibroblast growth factor and fibroblast growth factor receptor-1 in the human hippocampal formation. Neuroscience. 2005;131:219–233. doi: 10.1016/j.neuroscience.2004.09.070. [DOI] [PubMed] [Google Scholar]
- Wider C, Dachsel JC, Soto AI, Heckman MG, Diehl NN, Yue M, Lincoln S, Aasly JO, Haugarvoll K, Trojanowski JQ, Papapetropoulos S, Mash D, Rajput A, Rajput AH, Gibson JM, Lynch T, Dickson DW, Uitti RJ, Wszolek ZK, Farrer MJ, et al. FGF20 and Parkinson's disease: No evidence of association or pathogenicity via alpha-synuclein expression. Mov Disord. 2009;24:455–459. doi: 10.1002/mds.22442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Li D, Shimazu K, Zhou YX, Lu B, Deng CX. Fibroblast growth factor receptor-1 is required for long-term potentiation, memory consolidation, and neurogenesis. Biol Psychiatry. 2007;62:381–390. doi: 10.1016/j.biopsych.2006.10.019. [DOI] [PubMed] [Google Scholar]