Abstract
Crystal structures of A-form and B-form DNA duplexes containing 2′-S-methyl-uridines reveal that the modified residues adopt a RNA-like C3′-endo pucker, illustrating that the replacement of electronegative oxygen at the 2′-carbon of RNA by sulfur does not appear to fundamentally alter the conformational preference of the sugar in the oligonucleotide context and sterics trump stereoelectronics.
The conformation of nucleosides is subject to various stereoelectronic and steric effects.1 In the case of the 2′-deoxyribose moiety the Southern and Northern regions in the pseudorotational cycle represent preferred ring puckers.2,3 Thus, conformations corresponding to the C2′-endo and C3′-endo phase angle (P) ranges adjacent to the South and North poles, respectively, give rise to the canonical B- and A-DNA duplex forms at the oligodeoxynucleotide level.4 By comparison, the presence of a hydroxyl group at the ribose 2′-position shifts the conformational equilibrium to the Northern half at the mono-nucleoside level and locks the RNA duplex in the A-form. Using molecular dynamics simulations, Ferguson and coworkers proposed that the sugar moieties in an oligo-2′-deoxy-2′-S-methyl-ribonucleotide (2′-SMe-RNA) paired to RNA preferentially adopt the C2′-endo conformation.[5] This is in contrast to the 2′-deoxy-2′-O-methyl RNA analog (2′-OMe-RNA) paired to RNA, in that its sugars are limited to the C3′-endo range by the electronegative substituent.5 The latter observation is supported by structural data.6,7 Moreover, the conformational preorganization of the 2′-OMe-RNA strand for the RNA target is consistent with the increased thermodynamic stability of 2′-OMe-RNA:RNA duplexes compared to the corresponding 2′-SMe-RNA:RNA duplexes (ref. 5 and cited refs.). Although sulfur is less electronegative than oxygen, the switch from the North to the South conformation of the ribose in the oligonucleotide context as a result of replacing oxygen with sulfur at the 2′-position is surprising. In order to analyze the conformational properties of 2′-SMe-RNA and to establish the conformational boundaries of 2′-thiomethyl-ribonucleosides, we determined crystal structures of two DNA duplexes with incorporated 2′-SMe-rU residues.
The conformational preference of the sugar furnished by electronegative and/or bulky 2′-substituents has been widely used to preorganize oligonucleotides for RNA targets in antisense and RNAi applications.8–10 For example, the 2′-deoxy-2′-fluororibo- (2′-F-RNA11) and 2′-deoxy-2′-[(2-methoxy)ethyl]-ribonucleic acid modifications (2′-MOE-RNA12) exhibit a strong preference for the C3′-endo sugar conformation and their duplexes with RNA are restricted to the A-form. Conversely, the 2′-deoxy-2′-arabinonucleic acid modification (2′-FANA) shows a preference for the C2′-endo pucker at the level of the mono-nucleoside.13 But in duplexes, the 2′-FANA sugar has been observed to preferably adopt the O4′-endo (East; in B-form DNA or 2′-FANA/RNA hybrids) 14–16 or even the C3′-endo or C4′-exo conformations (Northern; in A-form DNA).17
Individual nucleosides can adopt a range of conformations within duplexes without significantly affecting the overall geometry of the latter (i.e. O4′-endo in B-DNA).14 But neighboring residues whose P values differ more drastically (i.e. >120°) can cause deformations of the duplex that include kinking and groove widening/narrowing.17,18 Therefore, it is evident that the conformational preferences of nucleosides affect the geometry and stability of oligonucleotide duplexes and, in turn, that the duplex geometry can have an effect on the conformation of individual nucleosides. However, in reality it is often not straightforward to predict the conformational properties of nucleosides, or to model the geometric features of duplexes containing a mix of residues that have deviating sugar puckers. In addition to the intrinsic sequence-dependent, conformational preferences of oligonucleotides, interactions with cations,19,20 water21,22 and proteins4,23,24 impinge on their geometry.
We chose two DNA oligomers to study the conformational properties of 2′-SMe-RNA, the decamer d(GCGTATACGC) and the dodecamer d(CGCGAATTCGCG) (the Dickerson-Drew dodecamer or DDD). The 10mer normally adopts an A-form conformation when at least one nucleoside is replaced by a 2′-modified residue25 and the DDD constitutes a classic example of a B-form DNA.22,26 Here, we report crystal structures for duplexes of sequence GCGTAU*ACGC and CGCGAAU*U*CGCG containing 2′-SMe-rU (U*) residues at 1.04 and 1.25 Å resolution, respectively.‡ Examples of the quality of the final electron density are shown in Fig. 1 and final refinement parameters are listed in Table S1 (see ESI†).
Fig. 1.
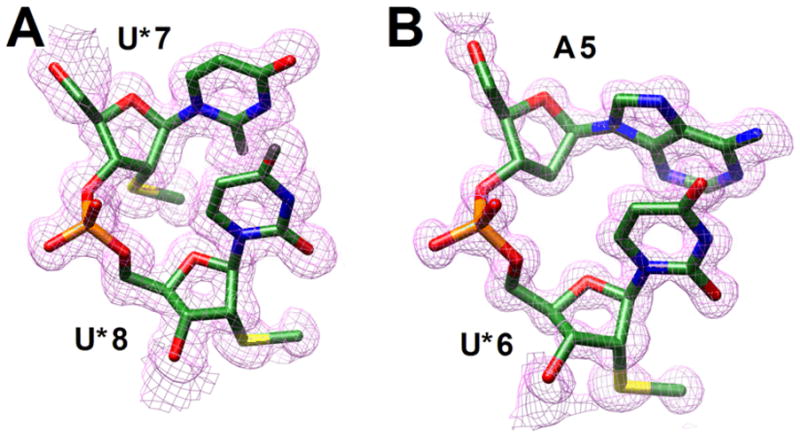
Quality of the final structures. Fourier (2Fo-Fc) sum electron densities around the (A) U*7pU*8 step in the modified dodecamer and the (B) A5pU*6 step in the modified decamer.
The 10mer duplex adopts a regular A-form conformation with all residues including U*6 and U*16 (nucleosides are numbered 1–10 in strand 1 and 11–20 in strand 2) exhibiting a C3′-endo pucker (Fig. S1). Although it is possible that the 2′-SMe-ribofuranose is driven towards North as a result of its surroundings, this observation provides an indication that sulfur in place of oxygen (either in the form of 2′-OH or 2′-OMe) does not appear to fundamentally affect the conformational preferences of the sugar. The situation of U* embedded in a more or less canonical A-DNA duplex likely does not emulate the constraints put on a chimeric 2′-SMe-RNA/DNA or all-2′-SMe-RNA strand opposite RNA. However, it is noteworthy that 2′-arabino-T nucleosides displayed a C1′-exo (South-Eastern) pucker inside the same A-form duplex, 17 indicating that the overall geometry of the 10mer does not necessarily limit the sugar conformation to North.
Unlike the A-form duplex that shows a regular overall geometry, the [CGCGAAU*U*CGCG]2 duplex with U* residues replacing dT at positions 7 and 8 of the first and positions 18 and 19 of the second strand (nucleosides are numbered 1–12 in strand 1 and 13–24 in strand 2) undergoes a significant expansion in the central minor groove compared to the native 12mer (Fig. 2). At the ApU* step (underlined in the above sequence), the roll amounts to −20° (−1°; values at ApT in the native DDD are in parentheses), the rise is 4.0 Å (3.34 Å), the twist is 41.5° (33°) and the slide is 0.44Å (−0.30 Å). These distortions are a direct consequence of the C3′-endo sugar pucker of all four U* residues. In addition to the Northern sugar conformation of these residues, the deoxyriboses of 3′-adjacent C9 in strand 1 and C21 in strand 2 also adopt the C3′-endo pucker. Both cytidines display a C2′-endo pucker in the structure of the native DDD,22 indicating that the change is brought about by their vicinity to the U* residues. All adenosines display a C2′-endo pucker and the transition from A (South) to U* (North) is accompanied by a subtle change in the backbone conformations of U*7 and U*19. The β and γ orsion angles of both residues flip from the standard ap/+sc to the −ac/ap ranges.
Fig. 2.
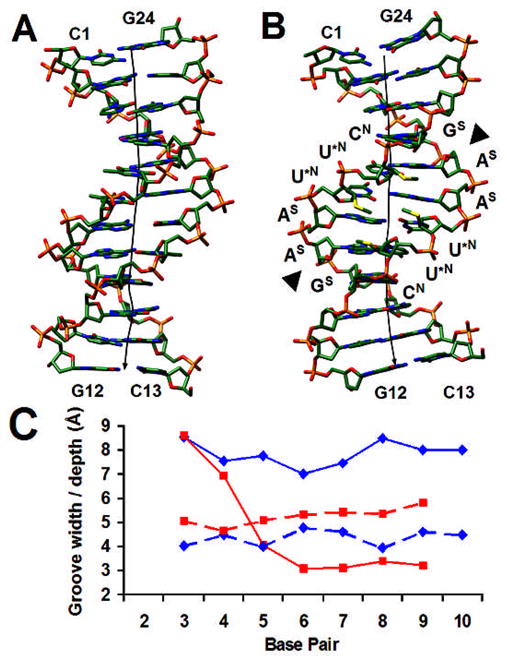
(A) The native DDD and (B) the 2′-SMe-modified DDD viewed into the minor groove. Sulfur atoms in the modified duplex are highlighted in yellow, the helical axes are depicted as solid black lines, terminal and the six central residues are labeled (with the symbol in superscript next to the latter indicating the sugar pucker), and EcoRI cutting sites are marked by a triangle. (C) Minor groove width (solid line) and depth (dashed line) in the native22 (red) and modified (blue) DDDs.
Compared to the structural changes observed in the central tetramer, the outer G-tract tetramers show helical geometries that are similar to those found in the native DDD (Fig. S2). Thus, the strong roll observed at the ApU* step does not lead to a kink of the duplex as indicated by the relatively straight global helix axis (Fig. 2B). Moreover, there are no significant geometrical deviations between the two independent duplexes per crystallographic asymmetric unit. As a result of the wider minor groove there are no short contacts between 2′-SMe-substituents from opposite strands in the modified DDD duplex. A close-up view into the central minor groove reveals intra- and inter-strand van der Waals interactions between 2′-SMe moieties and an interrupted minor groove hydration spine (Fig. 3).
Fig. 3.
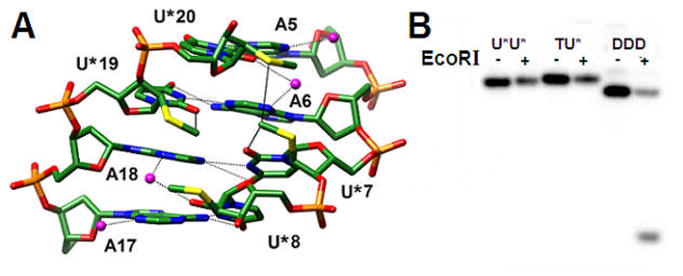
(A) The U*-modified DDD viewed into the central minor groove. Van der Waals interactions involving 2′-SMe-substituents and hydrogen bonds are indicated by solid and dashed lines, respectively, and water molecules are magenta spheres. Note the strong roll of uracils at the central base-pair step and the altered tip of the two inner compared with the two outer A:U* pairs. (B) PAGE assay of EcoRI cleavage experiments with the native DDD and modified dodecamers with either one (TU*) or both Ts (U*U*) replaced by 2′-SMe-U (from right to left). The − and + signs indicate absence and presence of the restriction enzyme, respectively.
The finding that 2′-SMe-RNA residues adopt a C3′-endo (North) pucker inside both A- and B-form duplexes argues strongly against the earlier conclusion based on modeling[5] that the sugars of an oligo-2′-SMe-RNA strand paired to RNA, unlike oligo-2′-OMe-RNA, prefer a C2′-endo (South) conformation. The 2′-substituents of U* residues in our crystal structures are pointing into the minor groove (Fig. 3A). Based on the conformational preferences predicted earlier, one would expect the substituents to lodge inside the major groove. The latter orientation would not interfere with RNase H, an endonuclease that cleaves the RNA portion of RNA:DNA hybrids,27 binding the latter from the minor groove side. However, it was recently reported that 2′-SMe-RNA:RNA hybrids do not elicit RNase H,27 consistent with our above structural observations. That the DDD duplex undergoes a drastic conformational change as a result of the substitution of T residues by U* is also demonstrated by assays with a restriction enzyme (Fig. 3B). EcoRI cleaves between G and A (underlined) in the recognition sequence 5′-GAATTC-3′/3′-CTTAAG-5′ (contained in the DDD, Fig. 2B). Accordingly, neither the DDD with one T or both Ts replaced by U* (labeled as TU* and U*U* respectively) is subject to cleavage by EcoRI.
Supplementary Material
Acknowledgments
Support by the National Institutes of Health is gratefully acknowledged (Grant R01 GM55237). We thank Dr. Z. Wawrzak for help with X-ray diffraction data collection. Vanderbilt University is a member institution of the LS-CAT at the Advanced Photon Source, Argonne, IL. Use of the APS was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
Footnotes
Electronic Supplementary Information (ESI) for this article is available: Crystal data and refinement parameters (Table S1), illustrations of the 10- and 12-mer duplexes.
Oligonucleotide synthesis: Both modified DNA oligonucleotides GCGTAU*ACGC and CGCGAAU*U*CGCG (U*=2′-SMe-rU) were synthesized via the solid-phase phosphoramidite approach and following published procedures.27 The 10mer and 12mer were purified by reverse phase HPLC and characterized by ES-MS and their purities were >95% as evaluated by capillary gel electrophoresis.
Crystallization and data collection: Crystals were grown by the hanging-drop vapor diffusion technique using the Nucleic Acid Miniscreen28 (Hampton Research, Aliso Viejo, CA). Crystals were mounted in nylon loops without further cryo-protection and frozen in liquid nitrogen. Diffraction data were collected on insertion beamlines at sector 21 of the Life Sciences Collaborative Access Team (LS-CAT) at the Advanced Photon Source (Agonne National Laboratory, Argonne, Illinois). Diffraction data were processed with the programs HKL200029 (RNase-H complex; see below) or XDS30 (10 and 12-mer duplexes). Both the 10mer and 12mer crystals are of space group P212121, whereby the asymmetric unit contained a single duplex and two independent duplexes, respectively. A summary of crystal data and data collection parameters is provided in Table S1.
Structure solution and refinement: The structure of the 10mer was determined by the Molecular Replacement (MR) technique using an A-form DNA (PDB ID 411D) as the search model.31,32 All attempts to phase the 12mer data by the same approach failed, as did crystallization experiments for subsequent heavy atom phasing with 12mers containing either Br5C in place of C or, either one of the two, or both U* residues replaced by 2′-SeMe-U.33–35 The structure was ultimately determined by co-crystallizing the U*-modified DDD with B. halodurans RNase H36 and then phasing the structure of the complex by MR, using the coordinates of the enzyme alone as the search model. The coordinates of the 12mer obtained from the complex structure then served as the model to determine the crystal structure of the duplex alone by MR. Initial crystallographic and B-factor refinements of the 10- and 12mer structures was carried out with the program CNS,37 setting aside 10% randomly chosen reflections for calculating the R-free. After the addition of water molecules and metal ions as well as incorporation of the U* residues and adaptation of the dictionary files, refinements were continued with the program SHELX,38 using anisotropic temperature factors for all nucleic acid and solvent atoms. Final refinement parameters for the two duplex structures as well as the RNase H complex are listed in Table S1 (supp. information). All helical parameters were calculated with the program CURVES.39
EcoRI cleavage assay: 500 nM DNA labeled with 32P were mixed with 2 μL of 10X NEB EcoRI buffer (New England Biolabs), 2 μL of EcoRI (20K units/mL; New England Biolabs) and diluted to a reaction volume of 20 μL to obtain a final buffer concentration of 50 mM NaCl, 100 mM Tris-HCl pH 7.5, 10 mM MgCl2, and 0.025% Triton X-100. The samples were incubated at 37°C for 90 minutes, followed by PAGE.
Data Deposition: Final coordinates and structure factor files have been deposited in the Brookhaven Protein Data Bank (http://www.rcsb.org). The entry codes are 3EY1 (RNase H:dodecamer complex), 3EY2 (decamer) and 3EY3 (dodecamer).
Notes and references
- 1.Thibaudeau C, Acharya P, Chattopadhyaya J. Stereoelectronic Effects in Nucleosides and Nucleotides and Their Structural Implications. Uppsala University Press; Uppsala, SE: 1999. [Google Scholar]
- 2.Altona C, Sundaralingam M. J Am Chem Soc. 1972;94:8205–8212. doi: 10.1021/ja00778a043. [DOI] [PubMed] [Google Scholar]
- 3.Sun G, Voigt JH, Fillippov IV, Marquez VE, Nicklaus MC. J Chem Inf Comput Sci. 2004;44:1752–1762. doi: 10.1021/ci049881+. [DOI] [PubMed] [Google Scholar]
- 4.Neidle S. Principles of Nucleic Acid Structure. Academic Press; London, UK: 2008. [Google Scholar]
- 5.Venkateswarlu D, Lind KE, Mohan V, Manoharan M, Ferguson DM. Nucleic Acids Res. 1999;27:2189–2195. doi: 10.1093/nar/27.10.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lubini P, Zürcher W, Egli M. Chem Biol. 1994;1:39–45. doi: 10.1016/1074-5521(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 7.Adamiak DA, Milecki J, Popenda M, Adamiak RW, Dauter Z, Rypniewski WR. Nucleic Acids Res. 1997;25:4599–4607. doi: 10.1093/nar/25.22.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egli M. Antisense Nucl Acid Drug Dev. 1998;8:123–128. doi: 10.1089/oli.1.1998.8.123. [DOI] [PubMed] [Google Scholar]
- 9.Manoharan M. Biochim Biophys Acta. 1999;1489:117–130. doi: 10.1016/s0167-4781(99)00138-4. [DOI] [PubMed] [Google Scholar]
- 10.Manoharan M. Curr Opin Chem Biol. 2004;8:570–579. doi: 10.1016/j.cbpa.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 11.Kawasaki AM, Casper MD, Freier SM, Lesnik EA, Zounes MC, Cummins LL, Gonzalez C, Cook PD. J Med Chem. 1993;36:831–841. doi: 10.1021/jm00059a007. [DOI] [PubMed] [Google Scholar]
- 12.Teplova M, Minasov G, Tereshko V, Inamati G, Cook PD, Manoharan M, Egli M. Nat Struct Biol. 1999;6:535–539. doi: 10.1038/9304. [DOI] [PubMed] [Google Scholar]
- 13.Barchi JJ, Jr, Jeong LS, Siddiqui MA, Marquez VE. J Biochem Biophys Meth. 1997;34:11–29. doi: 10.1016/s0165-022x(96)00032-2. [DOI] [PubMed] [Google Scholar]
- 14.Berger I, Tereshko V, Ikeda H, Marquez VE, Egli M. Nucleic Acids Res. 1998;26:2473–2480. doi: 10.1093/nar/26.10.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minasov G, Teplova M, Nielsen P, Wengel J, Egli M. Biochem. 2000;39:3525–3532. doi: 10.1021/bi992792j. [DOI] [PubMed] [Google Scholar]
- 16.Trempe JF, Wilds CJ, Denisov AY, Pon RT, Damha MJ, Gehring K. J Am Chem Soc. 2001;123:4896–4903. doi: 10.1021/ja003859p. [DOI] [PubMed] [Google Scholar]
- 17.Li F, Sarkhel S, Wilds CJ, Wawrzak Z, Prakash TP, Manoharan M, Egli M. Biochem. 2006;45:4141–4152. doi: 10.1021/bi052322r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Z, Maderia M, Barchi JJ, Jr, Marquez VE, Bax A. Proc Natl Acad Sci USA. 2005;102:24–28. doi: 10.1073/pnas.0408498102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Egli M. Chem Biol. 2002;9:277–286. doi: 10.1016/s1074-5521(02)00116-3. [DOI] [PubMed] [Google Scholar]
- 20.Egli M, Tereshko V. Curvature and Deformation of Nucleic Acids: Recent Advances, New Paradigms. In: Stellwagen N, Mohanty U, editors. ACS Symp Ser. Vol. 884. 2004. pp. 87–109. [Google Scholar]
- 21.Egli M, Portmann S, Usman N. Biochem. 1996;35:8489–8494. doi: 10.1021/bi9607214. [DOI] [PubMed] [Google Scholar]
- 22.Tereshko V, Minasov G, Egli M. J Am Chem Soc. 1999;121:470–471. [Google Scholar]
- 23.Olson WK, Gorin AA, Lu XJ, Hock LM, Zhurkin VB. Proc Natl Acad Sci USA. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarai A, Kono H. Annu Rev Biophys Biomol Struct. 2005;34:379–398. doi: 10.1146/annurev.biophys.34.040204.144537. [DOI] [PubMed] [Google Scholar]
- 25.Egli M, Usman N, Rich A. Biochem. 1993;32:3221–3237. [PubMed] [Google Scholar]
- 26.Wing R, Drew H, Takano T, Broka C, Tanaka S, Itakura K, Dickerson RE. Nature. 1980;287:755–758. doi: 10.1038/287755a0. [DOI] [PubMed] [Google Scholar]
- 27.Lima WF, Nichols JG, Wu H, Prakash TP, Migawa MT, Wyrzykiewicz TK, Bhat B, Crooke ST. J Biol Chem. 2004;279:36317–36326. doi: 10.1074/jbc.M405035200. [DOI] [PubMed] [Google Scholar]
- 28.Berger I, Kang CH, Sinha N, Wolters M, Rich A. Acta Cryst D. 1996;52:465–468. doi: 10.1107/s0907444995013564. [DOI] [PubMed] [Google Scholar]
- 29.Otwinowski Z, Minor W. Meth Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 30.Kabsch W. J Appl Cryst. 1993;26:795–800. [Google Scholar]
- 31.Vagin A, Teplyakov A. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 32.Collaborative Computational Project, Number. Acta Cryst D. 1994;50:760–763. [Google Scholar]
- 33.Du Q, Carasco N, Teplova M, Wilds CJ, Kong X, Egli M, Huang Z. J Am Chem Soc. 2002;124:24–25. doi: 10.1021/ja0171097. [DOI] [PubMed] [Google Scholar]
- 34.Teplova M, Wilds CJ, Wawrzak Z, Tereshko V, Du Q, Carrasco N, Huang Z, Egli M. Biochim. 2002;84:849–858. doi: 10.1016/s0300-9084(02)01440-2. [DOI] [PubMed] [Google Scholar]
- 35.Pallan PS, Egli M. Nat Protocols. 2007;2:647–651. doi: 10.1038/nprot.2007.75. [DOI] [PubMed] [Google Scholar]
- 36.Pallan PS, Egli M. Cell Cycle. 2008;7:2562–2569. doi: 10.4161/cc.7.16.6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Cryst D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 38.Sheldrick GM, Schneider TR. Meth Enzymol. 1997;277:319–343. [PubMed] [Google Scholar]
- 39.Lavery R, Sklenar H. J Biomol Struct Dyn. 1989;6:655–667. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


