Abstract
Taspase1 (TASP-1), the MLL and TFIIAα-β cleaving protease, enables cell proliferation and permits oncogenic initiation. Here, we demonstrate its critical role in cancer maintenance and thus offer a new anticancer target. Taspase1 is over-expressed in primary human cancers and deficiency of Taspase1 in cancer cells not only disrupts proliferation but also enhances apoptosis. Mechanistically, loss of Taspase1 induces the levels of CDKIs (p16, p21, and p27) and reduces the level of anti-apoptotic MCL-1. Therapeutically, deficiency of Taspase1 synergizes with chemotherapeutic agents and ABT-737, an inhibitor of BCL-2/BCL-XL, to kill cancer cells. Taspase1 alone or in conjunction with MYC, RAS, or E1A fail to transform NIH/3T3 cells or primary MEFs, respectively, and yet plays critical roles in cancer initiation and maintenance. Therefore, Taspase1 is better classified as a “non-oncogene addiction” protease, of which inhibition may offer a novel anticancer therapeutic strategy. The reliance of oncogenes on subordinate non-oncogenes during tumorigenesis underscores the “non-oncogene addiction” hypothesis in which a large class of non-oncogenes functions to maintain cancer phenotypes and presents attractive anti-cancer therapeutic targets. The emergence of successful cancer therapeutics targeting non-oncogenes to which cancers are addicted supports the future development and potential application of small molecule Taspase1 inhibitors for cancer therapy.
Introduction
Cancer is a genetic disease that evolves over years with accumulating genetic and epigenetic abnormalities that confer an enhanced capacity to proliferate and evade apoptosis, eventually compromising the host (1-2). Although cancer exhibits complex genetic aberrations and functional deregulations, it has been shown that abrogation of certain apical oncogenes can effectively inhibit cancer cell growth and prolong patient survival–a phenomenon termed “oncogene addiction” (3). In accordance with this concept, molecularly targeted drugs have been developed and successfully utilized in treating cancer patients, exemplified by the use of imatinib for chronic myelogenous leukemia, erlotinib for lung cancer, and sunitinib for kidney cancer (4). Despite these recent strides against cancer, most end-stage cancer patients still succumb to their diseases, highlighting the urgent need for novel anti-cancer therapeutic strategies. Recently, the “non-oncogene addiction” hypothesis was proposed based on the heavy reliance of cancer cells on certain non-oncogenes for their continuous growth and survival, which offers a new class of therapeutic targets that are not conventional oncogenes (5). Thus far, well-characterized non-oncogene addiction factors include the 26S proteasome, HSF1 (6), and IRF4 (7), which handles the rapid protein turnover, mediates the stress response, and maintains expression of the MYC oncogene, respectively. The fact that the proteasome inhibitor bortezomib is effective against multiple myeloma in human patients substantiates this new anti-cancer therapeutic concept (8).
Uncontrolled proliferation and increased resistance to apoptosis are two cardinal features of cancer, and consequently, inhibiting cancer cell division and enhancing cancer cell death constitute two effective anti-cancer therapeutic strategies (2, 9). Cancer cell cycle is typified by constitutively activated Cyclin/CDK complexes, resulting from either over-expression of Cyclins or down-regulation of CDK inhibitors (CDKIs) that are oncogenes and tumor-suppressor genes, respectively (10-11). By analogy, impaired apoptosis in cancer cells can result from either up-regulation of anti-apoptotic (such as BCL-2, BCL-XL, and MCL-1) or down-regulation of pro-apoptotic BCL-2 family proteins (such as BAX, BAK, BIM, and PUMA) (12). Targeted anti-cancer drugs have been developed and are on clinical trials based on these frameworks, and hence continued elucidation of the molecular control of cancer cell proliferation and cell death should unveil new anti-cancer targets and offer novel treatment options.
Taspase1 (threonine aspartase 1) encodes a highly conserved 50kD α-β proenzyme which undergoes intramolecular autoproteolysis, generating a mature α28/β22 heterodimeric protease that displays an overall α/β/β/α structure (13-14). Taspase1 is the only protease within the family of enzymes that possesses an asparaginase_2 (PF01112) homology domain, while other members, including L-asparaginase and glycosylasparaginase, participate in the metabolism of asparagine and the ordered breakdown of N-linked glycoproteins, respectively (13). Taspase1-mediated cleavage follows distinct aspartate residues of conserved QXD/GXDD motifs (15), suggesting that Taspase1 evolved from hydrolyzing asparagine and glycosylasparagine to cleaving polypeptides. The cloning of Taspase1 founded a novel class of endopeptidases which utilizes the N-terminal threonine of mature β subunit to cleave protein substrates after P1 aspartate. Taspase1 was initially purified as the protease which cleaves MLL to regulate HOX gene expression (13). Subsequent studies identified additional Taspase1 substrates, including MLL2, TFIIAα-β, ALF (TFIIAτ), and Drosophila HCF (dHCF) (16-18). Collectively, all of the known Taspase1 substrates are nuclear factors that control gene expression, suggesting that Taspase1 cleaves nuclear factors to orchestrate genetic programs.
Our prior genetic study of Taspase1−/− mice uncovered a critical role of Taspase1 in cell cycle control (16). In the absence of Taspase1, cell cycle is disrupted with decreased expression of Cyclins, including E, A, and B, and increased expression of CDK inhibitors (CDKIs), including p16, p21, and p27 (16). Hence, Taspase1 may play a permissive role in tumorigenesis, which is supported by the demonstrated resistance of Taspase1−/− mouse embryonic fibroblasts (MEFs) to oncogenic transformation induced by MYC, RAS, DNp53, and E1A. Although the absence of Taspase1 poses a profound roadblock in cancer initiation (16), whether Taspase1 is required for cancer maintenance remains unknown. Here, we demonstrate that Taspase1 is required to maintain a full cancer phenotype and is over-expressed in primary human cancer tissues. Deficiency of Taspase1 in human cancer cells results in impaired proliferation and enhanced susceptibility to death stimuli. Taspase1 is not a classical oncogene, yet enables full cancer characteristics, and thus better classified as a “non-oncogene addiction” protease. As proteases are ideal drug targets, small molecule inhibitors of Taspase1 may be developed for cancer therapy.
Materials and Methods
Cell Culture, Virus Production, and Retroviral Transduction
All cancer cell lines were provided by the NCI Developmental Therapeutics Program (Bethesda, MD) and cultured according to the provided instructions. Amphotrophic retroviruses were generated by transfection of 293T cells with a helper-free packaging system as described (19).
Tumor Xenograft Assay
U251 glioblastoma lines transduced with control- or Taspase1-shRNA retroviruses were selected for 3 days in 1.5μg/mL puromycin. Cells were then harvested and suspended in RPMI 1640 media in a 2:1 ratio with growth factor reduced Matrigel (BD Biosciences, San Jose, CA). Ten million cells were engrafted into each flank of male NOD-scid IL2Rγ−/− mice (Jackson Lab) between 6-8 weeks of age. Tumor size was measured with calipers and volume determined as described previously (20). Tumor volumes were compared using a two-tailed Student’s t test. For in vivo ABT-737 treatment, mice were injected with vehicle (30% 1,2-propanediol and 5% Tween-80 in D5W, pH 4.65) or ABT-737 (50 mg/kg) when tumor reached a volume of 300-500 mm3.
Results
Taspase1 is required to maintain MYC-RAS transformed MEFs
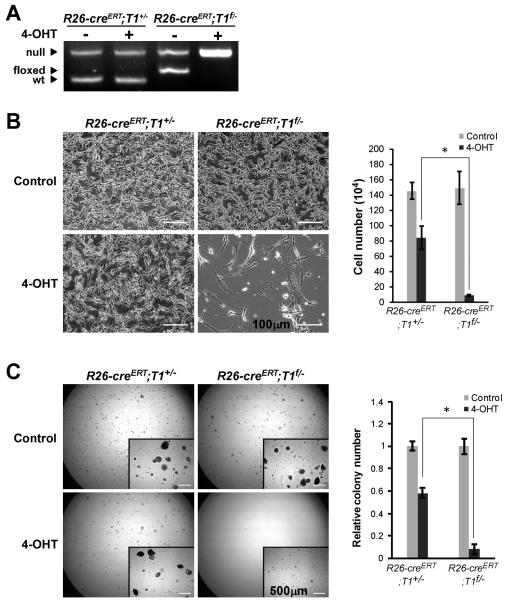
Our prior study demonstrated that Taspase1−/− primary MEFs are defective in proliferation and thus resistant to in vitro transformation induced by well-characterized oncogene pairs including MYC-RASG12V, DNp53-RASG12V, and E1A-RASG12V (16). These data highlight a critical participation of Taspase1 in cancer initiation, whereas the role of Taspase1 in cancer maintenance remains unanswered. To address this question, we employed a tamoxifen-inducible cre-lox system that allows for a precisely controlled genomic deletion of the conditional “floxed” (f) Taspase1 allele. Specifically, MYC-RAS-transformed Rosa26-creERT;Taspase1f/− (R26-creERT;T1f/−) and R26-creERT;T1+/− MEFs were treated with a 6-hour pulse of 500 nM 4-hydroxytamoxifen (4-OHT), which efficiently activated creERT to excise the conditional allele of Taspase1 (Fig. 1A). Deletion of Taspase1 in MYC-RAS transformed R26-creERT;T1f/− MEFs resulted in a profound proliferation block and disruption of colony formation on soft agar (Fig. 1B and C), indicating the requirement of Taspase1 for the proliferation of transformed cells. Of note, in agreement with the known toxicity of cre recombinase in MEFs (21), the pulse activation of creERT in control (MYC-RAS transformed R26-creERT;T1+/−) MEFs generated minor phenotypes in both assays (Fig. 1B and C). Taken together, our data support a permissive role of Taspase1 in cancer maintenance apart from cancer initiation.
Figure 1.
Taspase1 is Required for the Maintenance of MYC-RAS Transformed Mouse Embryonic Fibroblasts (MEFs). A, MYC-RAS transduced MEFs of the indicated genotypes were treated with 500 nM 4-hydroxytamoxifen (4-OHT) for 6 hours to activate R26-creERT. Deletion of the conditional (floxed; f) Taspase1 allele was confirmed by PCR at day 3. B, 105 MYC-RAS transduced MEFs of the indicated genotypes were mock (DMSO) or 4-OHT treated for 6 hours before plating on 6 cm dishes. Cells were photographed and counted at day 5. Scale bar equals 100 μm. Data presented are mean ± SD of duplicates of three independent experiments. C, 5×104 MYC-RAS transduced MEFs of the indicated genotypes were treated as B and plated on soft agar. Positive clones (>200 μm) were scored 1.5-2 weeks after the initial plating. Insets are higher-magnification images. Scale bar equals 500 μm. Data presented are mean ± SD of duplicates of two independent experiments. The average of colony numbers in mock treated plates was assigned as 1. Asterisk indicates P < 0.0005, determined by Fisher’s exact test.
Taspase1 is not a classical oncogene
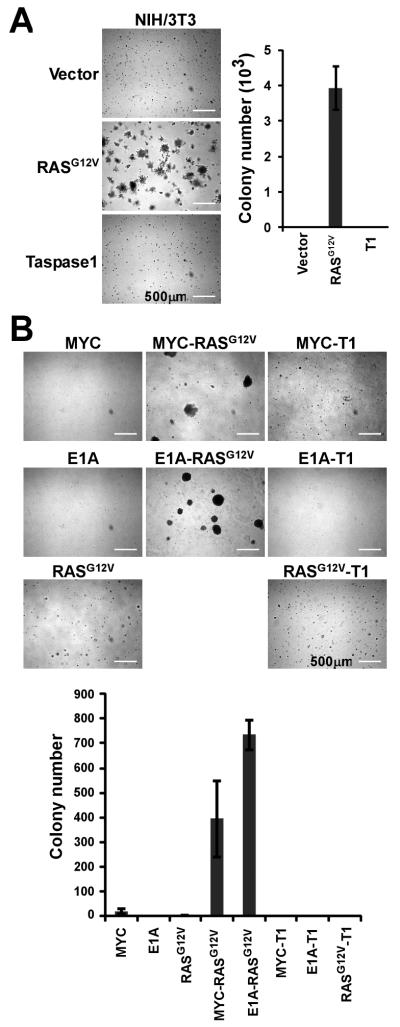
To further define the role of Taspase1 in tumorigenesis, we asked whether Taspase1 functions as an oncogene. To directly assess the oncogenic potential of Taspase1, we employed two different sets of in vitro transformation assays that are commonly utilized to characterize newly discovered oncogenes. First, we stably introduced Taspase1 into NIH/3T3 cells by retrovirus-mediated gene transduction, and determined the ability of transduced NIH/3T3 cells to grow on soft agar. Taspase1-transduced NIH/3T3 cells were unable to produce discernible colonies on soft agar, whereas RASG12V, as a positive control, efficiently rendered anchorage-independent growth (Fig. 2A and Supplementary Fig. S1A). Second, we tested whether Taspase1 can work in concert with RASG12V, MYC, or E1A to transform primary MEFs, which indicated that Taspase1 was unable to complement respective oncogenes to induce colonies on soft agar (Fig. 2B and Supplementary Fig. S1B). Thus, Taspase1 appears not to fit the criteria for a classical oncogene despite its requirement for cancer initiation and maintenance. The fact–Taspase1 is not an oncogene and yet enables oncogenesis–suggests that Taspase1 is better classified as a “non-oncogene addiction” protease.
Figure 2.
Taspase1 is not a Classical Oncogene. A, NIH/3T3 cells were transduced with the indicated genes and plated on soft agar. 5×104 cells were plated in each 6 cm dish. Positive clones (>200 μm) were scored 10 days after the initial plating. Data presented are mean ± SD of duplicates of two independent experiments. B, Wild-type primary MEFs were transduced with the indicated pairs of genes and plated on soft agar. 5×104 cells were plated in each 6 cm dish. Positive clones (>200 μm) were scored 2-3 weeks after the initial plating. Data presented are mean ± SD of triplicates of two independent experiments. Inserts are higher-magnification images. Scale bar equals 500 μm. T1 denotes Taspase1.
Deficiency in Taspase1 inhibits proliferation of human cancer cell lines
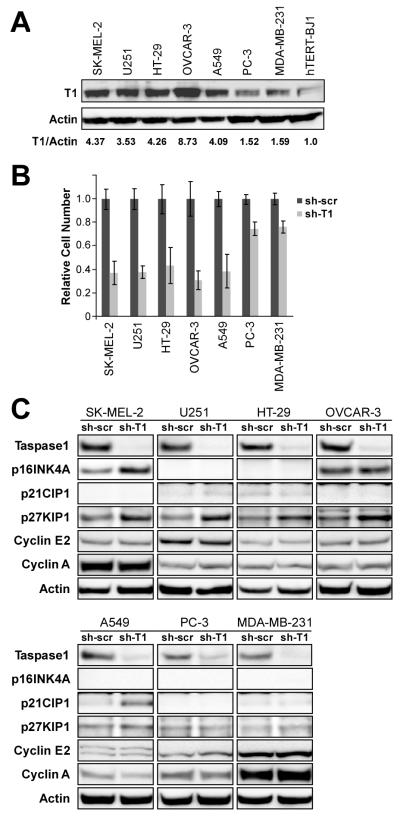
Given the requirement of Taspase1 in murine oncogenesis in vitro, we chose a wide variety of human cancer cell lines from the well-characterized NCI-60 panel to study the role of Taspase1 in human carcinogenesis. Human melanoma (SK-MEL-2), glioblastoma (U251), colon (HT-29), ovary (OVCAR-3), lung (A549), prostate (PC3), and breast (MDA-MB-231) cancer cell lines, representing major tissue types of human cancers, were examined. The expression of Taspase1 in these cell lines was determined (Fig. 3A). Knockdown of Taspase1 in these human cancer cell lines compromised their proliferative capacity but to a varying degree (Fig. 3B). Interestingly, cancer cell lines with higher Taspase1 levels appear to exhibit a more profound proliferation block upon knockdown of Taspase1 (Fig. 3B and Supplementary Figs. S2 & S3), supporting the notion that Taspase1 is co-opted by oncogenes to enforce cancer characteristics. Therefore, increased Taspase1 expression in cancer cells likely reflects an inherently elevated dependence on Taspase1 for maintaining a full cancer phenotype. The molecular basis underlying the proliferation defect induced by the deficiency in Taspase1 was further interrogated with a primary focus on the regulation of Cyclin E, A, and CDKIs–known downstream targets of Taspase1 in the cell cycle control (16). Analyses of p16, p21, p27, Cyclin E2, and Cyclin A in these cancer cell lines revealed that the induction of p27 is most correlative with the growth inhibition induced by the Taspase1 loss (Fig. 3B), for example, PC-3 and MDA-MB-231 cells exhibited minimal growth inhibition upon Taspase1 knockdown and no induction of p27 was observed in these Taspase1-knockdown cancer cells. Additionally, p16 and p21 were induced in SK-MEL-2 and A549 cell lines, respectively. The role of p27 in inhibiting cancer cell cycle dominates over p16 and p21, likely due to frequent deletions of the p16INK4A/p14ARF locus, common mutations in p53, the upstream activator of p21, and the extremely rare incidence of p27 mutations in human cancers (10). The mechanisms by which Taspase1 regulates p27 protein level were further investigated in U251 cells. Upon Taspase1 knockdown, p27 transcription was increased ~1.5 fold (Supplementary Fig. S4A), consistent with the known regulation of p27 transcription by Taspase1 (16). Interestingly, the protein half-life of p27 was also increased (Supplementary Fig. S4B), indicating that Taspase1 can modulate p27 protein expression through transcription and protein degradation. Taspase1 activates the transcription of Cyclins through MLL proteolysis (16, 22). However, we did not observe a reduction of Cyclin E2 or A expression in these cancer cell lines (Fig. 3C), suggesting that the Taspase1-MLL axis in regulating Cyclins may have been disrupted during the development of these cancers. Of note, analysis of known genetic mutations in oncogenes and tumor suppressors in these cancer cell lines did not identify a causal mutation explaining the Taspase1 dependence (Supplementary Table S1), highlighting the potential, general application of Taspase1 inactivation to inhibit cancer cell growth.
Figure 3.
Taspase1 is Required for the Proliferation of Human Cancer Cells. A, The protein levels of Taspase1 and β-actin of the indicated human cancer cell lines were assessed by anti-Taspase1 α28 and anti-β-actin antibodies. The relative expression of Taspase1 versus β-actin in hTERT-BJ-1 cells was assigned as 1. B, Human cancer cell lines with control- or Taspase1-knockdown were plated on 6-well plates and cell numbers were counted four days after the initial plating. The average cell number of control-knockdown cells was assigned as 1. Data presented are mean ± SD of duplicates of three independent experiments. C, The stable knockdown of Taspase1 in the indicated human cancer cell lines was determined by anti-Taspase1 α28 immunoblots. The expression of p16, p21, p27, Cyclin E2, and Cyclin A was determined by respective antibodies. The anti-β-actin immunoblots indicate equal protein loading.
Taspase1 deficiency in SK-MEL-2 melanoma and U251 glioblastoma cells enhances anoikis
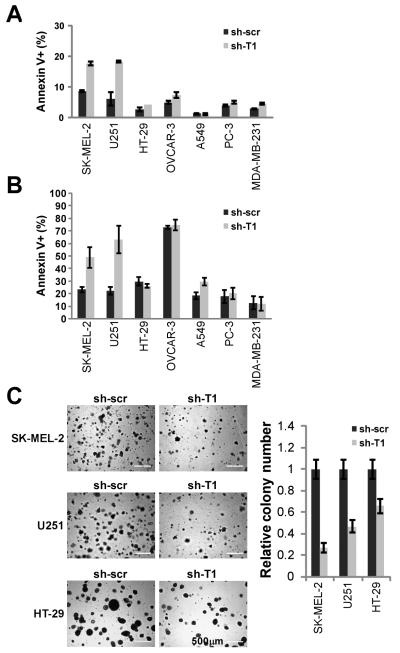
As Taspase1 plays a critical role in sustaining cancer cell proliferation, we investigated whether Taspase1 is also involved in modulating cancer cell death–another principal mechanism often deregulated in cancers. Interestingly, Taspase1 deficiency resulted in a minor increase in spontaneous apoptosis in both SK-MEL-2 and U251 cells which express high levels of Taspase1 (Fig. 4A). The increase in spontaneous apoptosis due to Taspase1 deficiency suggests that Taspase1 governs a fundamental process in cellular survival and that deregulation of this process, in the absence of Taspase1, would sensitize these cell lines to further death stimuli. The initiation of cell death by detachment from its substrata is an important, physiological initiator of apoptosis, termed anoikis, and the cellular resistance to anoikis constitutes a primary determinant of metastatic potential. Accordingly, we determined whether the loss of Taspase1 in U251 and SK-MEL-2 would affect anoikis sensitivity. Taspase1 deficiency sensitized SK-MEL-2 and U251 cells to anoikis, which was not evident in other tested human cancer cell lines except to a lesser degree in A549 lung cancer cells (Fig. 4B). The degree to which Taspase1-sustained proliferation and enhanced anoikis resistance contribute to tumorigenesis was further interrogated with soft agar assays. The ability of cells to form colonies on soft agar is a stringent in vitro predictor of in vivo tumorigenicity as soft agar imitates tumor microenvironment and thereby simultaneously assesses cellular proliferation and anoikis resistance. Taspase1 deficiency in SK-MEL-2 and U251 cancer cells resulted in a more pronounced reduction in colony formation on soft agar than HT-29 cells (Fig. 4C). Altogether, our data reveal a novel capacity of Taspase1 in regulating cell death in human cancers.
Figure 4.
Deficiency of Taspase1 Results in Increased Anoikis and Decreased Anchorage-independent Growth of SK-MEL-2 Melanoma and U251 Glioblastoma Cells. A, Human cancer cell lines with control- or Taspase1-knockdown were stained with annexin V and analyzed by FACS. B, Human cancer cell lines with control- or Taspase1-knockdown were cultured in PolyHEMA pre-treated 6-well plates for 3 days, stained with annexin V, and analyzed by FACS. C, SK-MEL-2, U251, or HT-29 cells with control- or Taspase1-knockdown were plated on soft agar. Positive clones (>100 μm) were scored 2-3 weeks after the initial plating of 105 cells. Scale bar equals 500 μm. Annexin V stains positive for apoptotic cells. Data presented in A, B, and C are mean ± SD duplicates of three independent experiments.
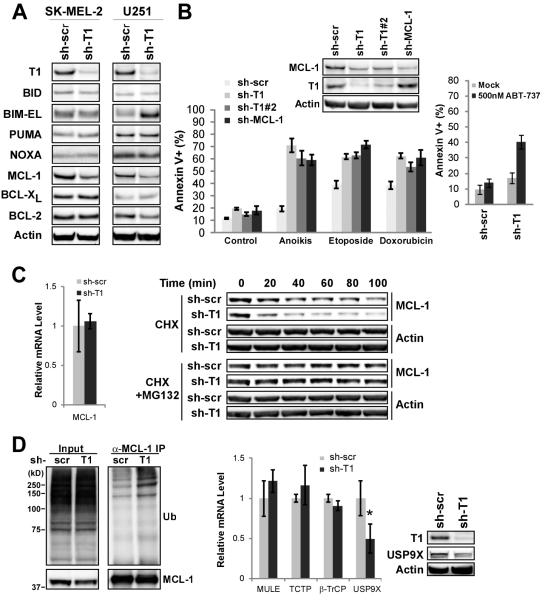
Taspase1 deficiency results in decreased MCL-1 level and increased sensitivity to chemotherapeutic agents and ABT-737
Melanoma and glioblastoma multiforme (GBM) are two highly aggressive human cancers that portend dismal outcomes when presenting at advanced stages. Their increased sensitivity to death upon Taspase1 knockdown prompted us to investigate the mechanism by which Taspase1 regulates apoptosis in these cancers. The BCL-2 family constitutes the core apoptotic machinery which operates at the mitochondria, controlling cytochorome c release and thereby caspase activation (12). The activator BH3-only molecules BID, BIM, and PUMA directly activate the apoptotic effectors BAX and BAK, whereas BCL-2, BCL-XL, and MCL-1 inhibit apoptosis by sequestering BH3s (Supplementary Fig. S5A) (23-27). Based on this framework, we examined the expression of core regulators of mitochondrial apoptosis in control- or Taspase1-knockdown SK-MEL-2 and U251 cells, which identified decreased MCL-1 as the common alteration (Fig. 5A). Therefore, Taspase1 may function to maintain the MCL-1 level and thus modulate the apoptotic threshold in GBM and melanoma (Supplementary Fig. S5B). To examine this hypothesis, we first knocked down MCL-1 to a comparable level as observed in Taspase1-knockdown cells. When comparing the propensity of Taspase1- or MCL-1-knockdown cells to undergo anoikis or DNA damage-induced apoptosis, we observed a comparably enhanced apoptotic sensitivity in both U251 and SK-MEL-2 cells (Fig. 5B and Supplementary Fig. S6A). Notably, the Taspase1 deficiency-induced down-regulation of MCL-1 is of potential clinical significance. As ABT-737, a specific inhibitor of BCL-2/BCL-XL, is ineffective at killing MCL-1 over-expressing cancer cells (28-31), inactivation of Taspase1 may offer an effective strategy to sensitize these resistant cancer cells to ABT-737-induced apoptosis. Accordingly, we treated control- or Taspase1-knockdown U251 and SK-MEL-2 cells with ABT-737 and demonstrated increased apoptosis in Taspase1 deficient cancer cells (Fig. 5B and Supplementary Fig. S6B). In summary, deficiency of Taspase1 confers sensitivity to apoptosis in GBM and melanoma cancer cells through down-regulation of MCL-1, suggesting a potential therapeutic benefit from combining Taspase1 inhibitors with currently-used cytotoxic agents as well as ABT-737 for cancer treatment.
Figure 5.
Taspase1 Deficiency Leads to Decreased MCL-1 Protein and Increased Cancer Cell Death upon Treatment with Chemotherapeutic Agents and ABT-737. A, Cellular extracts of SK-MEL-2 or U251 cells with control- or Taspase1-knockdown were subjected to Western blot analyses using the indicated antibodies. B, U251 cells with the indicated knockdown were treated with various apoptotic stimuli, stained with annexin-V, and analyzed by FACS. For anoikis assays, cells were plated in PolyHEMA coated plates for 3 days. For chemotherapeutic agent-induced cell death, cells were treated with 100 μg/mL of etoposide or 10 μM of doxorubicin for 30 hours. Data presented are mean ± SD of duplicates of two independent experiments. U251 cells with indicated knockdown were mock (DMSO) or ABT-737 treated for 24 hours, stained with annexin-V, and analyzed by FACS. Data presented are mean ± SD of triplicates of two independent experiments. C, The transcript levels of MCL-1 in control- or Taspase1-knockdown U251 cells were determined by quantitative RT-PCR analysis (left). U251 cells with indicated knockdown were subjected to cyclohexamide or cyclohexamide plus MG132 treatment for the indicated periods of time, and the protein levels of MCL-1 and β-actin were determined by immunoblot (right). D, The ubiquitination status of MCL-1 in U251 cells with control- or Taspase1-knockdown were treated with MG132, and subjected to anti-MCL-1 immunoprecipitation followed by anti-ubiquitin immunoblots. Transcript levels of known MCL-1 stability regulators were analyzed and the protein expression of USP9X was determined. Asterisk indicates P < 0.01. Data presented in C and D are mean ± SD of duplicates of three independent experiments and the transcript level in control-knockdown cells was assigned as 1.
Taspase1 regulates the protein half-life of MCL-1
The MCL-1 protein level can be titrated through transcriptional, translational, and post-translational mechanisms, which explains its unusual capacity among BCL-2 family members to rapidly respond to environmental cues for cell death control (32). As Taspase1 functions mainly through processing nuclear factors and thus orchestrating transcription, we determined if Taspase1 directly regulates the transcription of MCL-1. Interestingly, there was no decrease of the MCL-1 transcript level in Taspase1 deficient U251 and SK-MEL-2 cells (Fig. 5C and Supplementary Fig. S6C). On the contrary, Taspase1 deficiency shortened the protein half-life of MCL-1 from ~1 hour to ~20 minutes in U251 cells (Fig. 5C). Furthermore, treatment with proteasome inhibitor MG132 prevented the degradation of MCL-1 in both control- and Taspase1-knockdown U251 cells (Fig. 5C). The resultant half-life of MCL-1 is comparable between control- and Taspase1-knockdown cells upon MG132 treatment, suggesting that Taspase1 sustains the MCL-1 protein by enhancing its protein stability. Indeed, Taspase1 deficiency resulted in increased ubiquitination of MCL-1 (Fig. 5D). The control of MCL-1 degradation is complex and context dependent, which at least involves the inactivator BH3 NOXA, the GSK3β signaling, the molecular chaperone TCTP, the E3 ubiquitin ligases MULE and β-TrCP, and the deubiquitinase USP9X (33-38). We determined the expression of NOXA and did not detect an induction of NOXA protein in Taspase1-deficient cells to account for the increased degradation of MCL-1 (Fig. 5A). We investigated whether Taspase1 disrupts the GSK3β signaling and thereby extends the MCL-1 half-life. Inhibition of GSK3β by TZDZ-8 in Taspase1 deficient cells did not result in increased MCL-1 protein (Supplementary Fig. S7). To provide further mechanistic details regarding how Taspase1 controls the MCL-1 degradation, we determined the transcript levels of TCTP, MULE, β-TrCP, and USP9X in Taspase1-knockdown cells (Fig. 5D). In Taspase1 deficient cells, we detected a statistically significant decrease of USP9X transcript and a resulting low expression of USP9X protein (Fig. 5D), suggesting that Taspase1 modulates the MCL-1 half-life in part through the transcriptional control of USP9X.
Taspase1 is over-expressed in primary human GBM and melanoma tissues and its deficiency disrupts tumor growth in a xenograft model
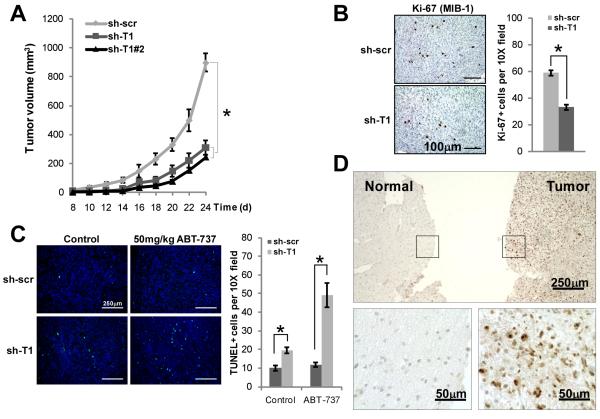
Our in vitro assays indicate that Taspase1 enables a full cancer phenotype by facilitating cancer cell cycle progression and suppressing cancer cell death. To probe the in vivo requirement of Taspase1 in human cancer growth, we performed a tumor xenograft assay using immunocompromised NOD;scid;IL2Rγ−/− mice. Importantly, loss of Taspase1 significantly inhibited the growth of U251 tumors in NOD;scid;IL2Rγ−/− mice (Fig. 6A). Histological examination of these Taspase1 knockdown tumors demonstrated decreased Ki67+ (Fig. 6B) and increased TUNEL+ cells (Fig. 6C), indicating a lower proliferation index and a higher rate of cell death, respectively. Importantly, the percentage of cells undergoing apoptosis was greatly increased in Taspase1 knockdown tumors upon the treatment with one dose of ABT-737 (Fig. 6C). Several lines of evidence suggest that Taspase1 is recruited by oncogenes to maintain cancer characteristics during tumorigenesis. First, our in vitro and in vivo studies indicate that Taspase1 functions as a critical node in the oncogenic network that sustains proliferation and suppresses cell death. Second, Taspase1 is over-expressed in most of the NCI-60 human cancer cell lines (16). Third, there is a positive correlation between the level of Taspase1 expression and the degree of disruption in cancer phenotypes upon its inactivation in examined human cancer cell lines. Theoretically, Taspase1 may be highly-expressed in certain human cancers, and based on our data, its over-expression would indicate a favorable therapeutic outcome upon its inhibition. To enable assessment of Taspase1 expression in primary tissues, we established an immunohistochemistry (IHC) assay using a newly raised anti-Taspase1 monoclonal antibody. We focused on GBM and melanoma as our cell line data suggested a central role played by Taspase1 in these cancers. Remarkably, Taspase1 is abundantly expressed in primary human GBM tissues (n = 19), whereas no or weak staining of Taspase1 is observed in adjacent or control brain sections (n = 13) that mainly consist of astroglial cells–the cellular origin of GBM (Fig. 6D and Supplementary Fig. S9). The specificity of Taspase1 staining was confirmed as no signal was detected when the anti-Taspase1 antibody was pre-incubated with the Taspase1 immunogen (Supplementary Fig. S10). Of note, a recent gene expression profiling analysis demonstrated an over-expression of Taspase1 in primary human GBM tissues (39). Furthermore, Taspase1 is preferentially expressed in melanoma compared to melanocytic nevus (Supplementary Fig. S11A). Similar expression of Taspase1 between melanocytic nevus and adjacent keratinocytes serves as an internal reference for Taspase1 expression, since normal melanocytes cannot be differentiated from malignant melanocytes in the same section. To quantify the preferential expression of Taspase1 in melanoma, we compared the relative immunoflourescence signal between melanoma and keratinocytes versus that of nevus and keratinocytes, which confirmed the over-expression of Taspase1 in melanomas (Supplementary Fig. S11B). In summary, IHC and IF assays were established to assess the expression of Taspase1 in primary human tissues, which demonstrated over-expression of Taspase1 in primary human GBM and melanoma. This suggests that Taspase1 may have an important role in the pathogenesis of these human cancers and that it may serve as a new therapeutic target for these currently intractable diseases.
Figure 6.
Taspase1 Loss Disrupts Growth of Tumor Xenografts and Taspase1 is Over-expressed in Primary Human Cancers. A, U251 cells with control- or Taspase1-knockdown were injected into the flanks of NOD;scid;IL2Rγ−/− mice and tumor growth was measured. B, Proliferative index of U251 xenografts was determined by anti-Ki-67 immunohistochemistry. Sections shown at 20X magnification. Bar graph represents mean ± SEM of Ki-67+ cells per 10X fields (10 fields per tumor, n=2 tumors each). C, Baseline and ABT-737-induced tumor cell death were determined by TUNEL staining 12 hours after a single vehicle control or ABT-737 intraperitoneal injection. Tumors Sections shown are 10X fields of FITC-TdT (green) or DAPI (blue). Bar graph represents mean ± SEM of TUNEL+ cells per 10X fields (10 fields per tumor, n=2 tumors each). D, Immunohistochemistry of primary human GBM and adjacent normal brain was examined with an anti-Taspase1 monoclonal antibody (10H2F6). High magnification pictures of boxed areas are provided. Asterisks indicate P < 0.005.
Discussion
Taspase1 plays an essential role in regulating embryonic cell cycle, as evidenced by the smaller body size of Taspase1−/− mice (16). Molecularly, Taspase1 functions to activate transcription of Cyclin E, A and B, and to suppress that of CDKIs p16, p21 and p27 in primary MEFs for cellular proliferation. In contrast, the role of Taspase1 in adult tissue homeostasis is less clear. As Taspase1 coordinates the expression of Cyclins and CDKIs that are known oncogenes and tumor suppressors, respectively, inactivation of Taspase1 may offer a novel mechanism to inhibit cancer cell proliferation. In agreement with this hypothesis, deficiency of Taspase1 in human cancer cells impedes proliferation. In contrast to our prior observation that the loss of Taspase1 has no impact on the baseline cell death of primary MEFs which express lower levels of Taspase1 (16), GBM and melanoma cancer cells that express high levels of Taspase1 exhibited increased sensitivity to death stimuli upon Taspase1 inactivation. Mechanistically, Taspase1 deficiency in U251 and SK-MEL-2 cells resulted in an increased degradation of MCL-1, accounting for the enhanced death phenotype. MCL-1 is unique among anti-apoptotic BCL-2 family members as it rapidly responds to external signals and can be regulated by transcriptional, translational, and post-translational mechanisms (32). Regulation of MCL-1 protein turnover is itself complex and implicates multiple pathways. MULE/ARFBP-1 was the first described E3-ligase required for MCL1 degradation, though in MULE deficient cells, MCL-1 is still degraded upon initiation of apoptosis, suggesting the existence of additional mechanisms for MCL-1 degradation (33). For example, the PI3K/AKT pathway inhibits GSK3β-mediated phosphorylation of MCL-1 and thereby prevents β-TrCP-mediated ubiquitylation and proteasomal degradation of MCL-1 (34-35). Additionally, MCL-1 protein stability can be regulated by direct protein-protein interactions, as its interaction with NOXA and TCTP is known to modulate MCL-1 turnover (36, 38). Recently, USP9X was found to be an MCL-1 deubiquitinase which promotes tumor cell survival through stabilizing MCL-1. Among these known MCL-1 regulators, we identified USP9X as a downstream effector of Taspase1. When Taspase1 was knocked down, the level of USP9X reduced, correlating with the increased ubiquitination of MCL-1 and the resulting shortened protein half-life. Although additional contributing mechanisms concerning how Taspase1 regulates the protein stability of MCL-1 may be present. Nevertheless, down-regulation of MCL-1 by Taspase1 knockdown in cancer cell lines likely has important implications in cancer therapy. The small molecule inhibitor of BCL2/BCL-XL, ABT-737, is currently on clinical trials. Although ABT-737 is quite potent in killing certain types of cancer cells, it is ineffective against MCL-1 over-expressing tumors due to its inability to inhibit MCL-1 (28-30). Accordingly, Taspase1 inhibitors may synergize with ABT-737 to induce apoptosis in human cancers.
Taspase1 is positioned to regulate a broad array of biological functions, including development, cellular proliferation, and cell death, as all of the Taspase1 substrates described to date are broad-acting transcription regulators, including TFIIA and MLL. Although Taspase1 plays a critical role in embryonic development, acute deletion of Taspase1 in mice did not incur obvious organismal distress and changes in blood counts (data not shown), supporting a potential application of Taspase1 inhibitors in treating cancers. Though the mechanisms whereby Taspase1 is co-opted to promote tumorigenesis remain to be examined, the increased requirement for Taspase1 in certain cancers represents a possibility for a therapeutic window in which Taspase1 inhibition can work in conjunction with other forms of targeted and non-targeted cancer treatments. Taspase1, as a protease, allows for rational design of substrate mimetic inhibitors as well as expedient high-throughput screening for potential small molecule therapeutic leads (40). Although targeting the non-oncogene addiction network for cancer therapeutics is in its rudimentary stage, the successful utilization of this novel strategy is likely to add effective chemotherapeutic agents to our current armamentarium for combating cancers.
Supplementary Material
Acknowledgements
We thank Abbott Laboratories for kindly providing ABT-737 and thank Drs. Jeffrey Arbeit and Raleigh Kladney for technical advices on immunohistochemistry.
Grant Support
USA National Institute of Health CA R01-119008, American Cancer Society, and American Society of Hematology.
Footnotes
Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
References
- 1.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nature medicine. 2004;10:789–99. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 3.Weinstein IB, Joe A. Oncogene addiction. Cancer Res. 2008;68:3077–80. doi: 10.1158/0008-5472.CAN-07-3293. discussion 80. [DOI] [PubMed] [Google Scholar]
- 4.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PubMed] [Google Scholar]
- 5.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–37. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130:1005–18. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaffer AL, Emre NC, Lamy L, Ngo VN, Wright G, Xiao W, et al. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–31. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams J. The proteasome: a suitable antineoplastic target. Nat Rev Cancer. 2004;4:349–60. doi: 10.1038/nrc1361. [DOI] [PubMed] [Google Scholar]
- 9.Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–15. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 10.Besson A, Dowdy SF, Roberts JM. CDK inhibitors: cell cycle regulators and beyond. Dev Cell. 2008;14:159–69. doi: 10.1016/j.devcel.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 11.Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–7. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 12.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–19. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 13.Hsieh JJ, Cheng EH, Korsmeyer SJ. Taspase1: a threonine aspartase required for cleavage of MLL and proper HOX gene expression. Cell. 2003;115:293–303. doi: 10.1016/s0092-8674(03)00816-x. [DOI] [PubMed] [Google Scholar]
- 14.Khan JA, Dunn BM, Tong L. Crystal structure of human Taspase1, a crucial protease regulating the function of MLL. Structure. 2005;13:1443–52. doi: 10.1016/j.str.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 15.Hsieh JJ, Ernst P, Erdjument-Bromage H, Tempst P, Korsmeyer SJ. Proteolytic cleavage of MLL generates a complex of N- and C-terminal fragments that confers protein stability and subnuclear localization. Mol Cell Biol. 2003;23:186–94. doi: 10.1128/MCB.23.1.186-194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takeda S, Chen DY, Westergard TD, Fisher JK, Rubens JA, Sasagawa S, et al. Proteolysis of MLL family proteins is essential for taspase1-orchestrated cell cycle progression. Genes Dev. 2006;20:2397–409. doi: 10.1101/gad.1449406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou H, Spicuglia S, Hsieh JJ, Mitsiou DJ, Hoiby T, Veenstra GJ, et al. Uncleaved TFIIA is a substrate for taspase 1 and active in transcription. Mol Cell Biol. 2006;26:2728–35. doi: 10.1128/MCB.26.7.2728-2735.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Capotosti F, Hsieh JJ, Herr W. Species selectivity of Mixed Lineage Leukemia/Trithorax and HCF proteolytic maturation pathways. Mol Cell Biol. 2007;27:7063–72. doi: 10.1128/MCB.00769-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu H, Cheng EH, Hsieh JJ. Bimodal degradation of MLL by SCFSkp2 and APCCdc20 assures cell cycle execution: a critical regulatory circuit lost in leukemogenic MLL fusions. Genes Dev. 2007;21:2385–98. doi: 10.1101/gad.1574507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tu HC, Ren D, Wang GX, Chen DY, Westergard TD, Kim H, et al. The p53-cathepsin axis cooperates with ROS to activate programmed necrotic death upon DNA damage. Proc Natl Acad Sci U S A. 2009;106:1093–8. doi: 10.1073/pnas.0808173106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loonstra A, Vooijs M, Beverloo HB, Allak BA, van Drunen E, Kanaar R, et al. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc Natl Acad Sci U S A. 2001;98:9209–14. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tyagi S, Chabes AL, Wysocka J, Herr W. E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol Cell. 2007;27:107–19. doi: 10.1016/j.molcel.2007.05.030. [DOI] [PubMed] [Google Scholar]
- 23.Cheng EH, Wei MC, Weiler S, Flavell RA, Mak TW, Lindsten T, et al. BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol Cell. 2001;8:705–11. doi: 10.1016/s1097-2765(01)00320-3. [DOI] [PubMed] [Google Scholar]
- 24.Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ, et al. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–30. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Letai A, Bassik MC, Walensky LD, Sorcinelli MD, Weiler S, Korsmeyer SJ. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2002;2:183–92. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 26.Kim H, Rafiuddin-Shah M, Tu HC, Jeffers JR, Zambetti GP, Hsieh JJ, et al. Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat Cell Biol. 2006;8:1348–58. doi: 10.1038/ncb1499. [DOI] [PubMed] [Google Scholar]
- 27.Kim H, Tu HC, Ren D, Takeuchi O, Jeffers JR, Zambetti GP, et al. Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol Cell. 2009;36:487–99. doi: 10.1016/j.molcel.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–81. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 29.van Delft MF, Wei AH, Mason KD, Vandenberg CJ, Chen L, Czabotar PE, et al. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell. 2006;10:389–99. doi: 10.1016/j.ccr.2006.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konopleva M, Contractor R, Tsao T, Samudio I, Ruvolo PP, Kitada S, et al. Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell. 2006;10:375–88. doi: 10.1016/j.ccr.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 31.Deng J, Carlson N, Takeyama K, Dal Cin P, Shipp M, Letai A. BH3 profiling identifies three distinct classes of apoptotic blocks to predict response to ABT-737 and conventional chemotherapeutic agents. Cancer Cell. 2007;12:171–85. doi: 10.1016/j.ccr.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 32.Opferman JT. Unraveling MCL-1 degradation. Cell Death Differ. 2006;13:1260–2. doi: 10.1038/sj.cdd.4401978. [DOI] [PubMed] [Google Scholar]
- 33.Zhong Q, Gao W, Du F, Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2005;121:1085–95. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 34.Ding Q, He X, Hsu JM, Xia W, Chen CT, Li LY, et al. Degradation of Mcl-1 by beta-TrCP mediates glycogen synthase kinase 3-induced tumor suppression and chemosensitization. Mol Cell Biol. 2007;27:4006–17. doi: 10.1128/MCB.00620-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maurer U, Charvet C, Wagman AS, Dejardin E, Green DR. Glycogen synthase kinase-3 regulates mitochondrial outer membrane permeabilization and apoptosis by destabilization of MCL-1. Mol Cell. 2006;21:749–60. doi: 10.1016/j.molcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 36.Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 37.Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010;463:103–7. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- 38.Liu H, Peng HW, Cheng YS, Yuan HS, Yang-Yen HF. Stabilization and enhancement of the antiapoptotic activity of mcl-1 by TCTP. Mol Cell Biol. 2005;25:3117–26. doi: 10.1128/MCB.25.8.3117-3126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scrideli CA, Carlotti CG, Jr., Okamoto OK, Andrade VS, Cortez MA, Motta FJ, et al. Gene expression profile analysis of primary glioblastomas and non-neoplastic brain tissue: identification of potential target genes by oligonucleotide microarray and real-time quantitative PCR. J Neurooncol. 2008;88:281–91. doi: 10.1007/s11060-008-9579-4. [DOI] [PubMed] [Google Scholar]
- 40.Lee JT, Chen DY, Yang Z, Ramos AD, Hsieh JJ, Bogyo M. Design, syntheses, and evaluation of Taspase1 inhibitors. Bioorganic & medicinal chemistry letters. 2009;19:5086–90. doi: 10.1016/j.bmcl.2009.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.