Abstract
Myotonic dystrophy (DM1), the most common muscular dystrophy in adults, is caused by an expanded (CTG)n tract in the 3′ UTR of the gene encoding myotonic dystrophy protein kinase (DMPK)1, which results in nuclear entrapment of the ‘toxic’ mutant RNA and interacting RNA-binding proteins (such as MBNL1) in ribonuclear inclusions2. It is unclear if therapy aimed at eliminating the toxin would be beneficial. To address this, we generated transgenic mice expressing the DMPK 3′ UTR as part of an inducible RNA transcript encoding green fluorescent protein (GFP). We were surprised to find that mice overexpressing a normal DMPK 3′ UTR mRNA reproduced cardinal features of myotonic dystrophy, including myotonia, cardiac conduction abnormalities, histopathology and RNA splicing defects in the absence of detectable nuclear inclusions. However, we observed increased levels of CUG-binding protein (CUG-BP1) in skeletal muscle, as seen in individuals with DM1. Notably, these effects were reversible in both mature skeletal and cardiac muscles by silencing transgene expression. These results represent the first in vivo proof of principle for a therapeutic strategy for treatment of myotonic dystrophy by ablating or silencing expression of the toxic RNA molecules.
Common features of adult-onset DM1 include myotonia, progressive skeletal muscle loss, cardiac conduction defects, smooth muscle dysfunction, cataracts and insulin resistance2. The normal number of CTG repeats (n = 5 to ~30) is higher (n = 50 to >3,000) in individuals with DM1 (ref. 1). Unlike the wild-type transcript, mutant DMPK mRNA forms nuclear aggregates3,4 and is thought to trigger dominant effects by aberrant interactions with or altered activity of RNA splicing factors, principally members of the muscleblind-like (MBNL) family (such as MBNL1) and the CUG-BP and ETR3-like factor (CELF) family (such as CUG-BP1), leading to abnormal splicing of specific RNAs such as chloride channel (Clcn1), insulin receptor and troponin-T isoforms2. Similar findings have been reported in DM2, a rarer form, caused by an expanded (CCTG)n tract (n = 75 to ~11,000) in the first intron of ZNF9 (ref. 2). Myoblast cell culture models5,6 and subsequently a transgenic mouse model7 have provided strong evidence for the involvement of RNA containing expanded CUG repeat tracts in aspects of DM1 skeletal muscle pathology. However, there is no clear model of RNA toxicity in the heart, and instead it has been suggested that DM1 cardiac pathology may be due to misexpression of DMPK8,9.
One potential therapeutic approach in DM1 is to get rid of the toxic RNA from cells. However, it is unclear if this will alleviate the effects of the disease. We used the tetracycline (Tet) inducible system with the reverse tetracycline transactivator (rtTA) to generate double transgenic mice harboring (i) a Tet-responsive, DMPK promoter10,11–driven transgene (named GFP-DMPK 3′ UTR) expressing the DMPK 3′ UTR mRNA as part of a GFP transcript, and (ii) a constitutively expressed rtTA transgene (Fig. 1a). The transgene does not encode DMPK protein, allowing a clear delineation of the contribution of the DMPK 3′ UTR mRNA to myotonic dystrophy pathophysiology when transgene expression is induced.
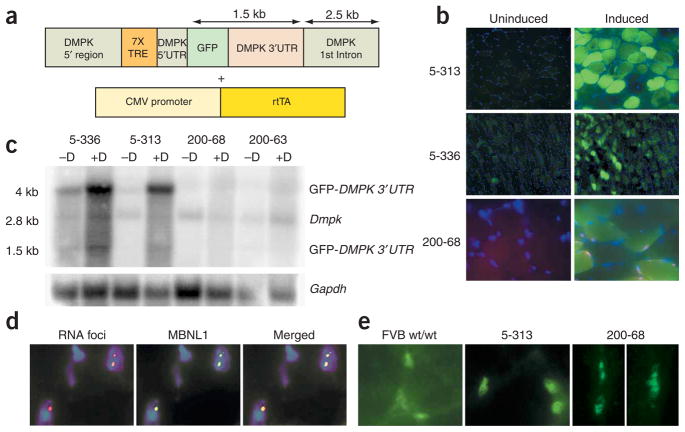
Figure 1.
Transgene expression. (a) Transgenes used to create double transgenic tetracycline-inducible mice. (b) RNA-FISH and fluorescence microscopy of skeletal muscle in mice in which transgene expression was induced demonstrates GFP expression in all mice and RNA foci only in mice expressing the GFP-DMPK 3′ UTR (CTG)200. (c) RNA blot of skeletal muscle RNA showing significant transgene induction in (CTG)5 mice relative to endogenous Dmpk expression. Gapdh was used as loading control. (d) RNA-FISH and immunofluorescence for MBNL1 demonstrating colocalization in (CTG)200 mice. (e) Antibodies to MBNL1 show that wild-type mice (FVB) and (CTG)5 mice have diffuse nuclear staining, whereas (CTG)200 mice have distinct MBNL foci.
We created two sets of transgenic founder mice: six founders with the wild-type (CTG)5 DMPK 3′ UTR and nine founders with the mutant (CTG)200 DMPK 3′ UTR. Three founders for the (CTG)200 and two for the (CTG)5 transgenes (5-313 and 5-336) showed evidence of induced transgene expression as assessed by GFP fluorescence and/or RNA-FISH for DMPK 3′ UTR (Fig. 1b), RNA blotting (Fig. 1c and Supplementary Fig. 1 online) and real time RT-PCR (Supplementary Fig. 1). Notably, RNA blots of skeletal muscle RNA showed two major species due to alternative use of polyadenylation signals located at either the end of the DMPK 3′ UTR or after the DMPK first intron (Supplementary Fig. 1). The transgene expression levels were relatively low in the heterozygous (CTG)200 mice (Fig. 1c), resulting in a lack of phenotypic effects. We are in the process of generating homozygotes for further analysis. Nevertheless, we saw formation of RNA foci in all muscle lineages in the transgenic lines expressing the mutant DMPK 3′ UTR RNA (Supplementary Fig. 2 online), and we observed MBNL colocalization with the RNA foci (Fig. 1d) analogous to results seen in individuals with DM1 (ref. 12). However, neither RNA foci nor MBNL foci were evident in the (CTG)5 mice (Fig. 1b,e).
We were surprised to find, on repeated attempts, that the 5-336 transgenic mice (expressing the wild-type DMPK 3′ UTR with (CTG)5) died within 3 to 4 weeks (range, 5–22 days) after induction of transgene expression. Most mice were overtly normal in appearance and behavior, with lethargy and pallor 1 or 2 d before death. Given the sudden death, we suspected a cardiac problem. Therefore, we induced transgene expression with doxycycline in 20 mice and performed daily electromyography (EMG) tests for myotonia and electrocardiograms (ECGs) for cardiac conduction studies. First, all the mice developed profound myotonia, a hallmark of myotonic dystrophy, within 9 d, some as early as 2 d post-induction (Fig. 2a). Subsequently, the mice had abnormal ECGs that progressed variably (2 d to 2 months) from a mild conduction abnormality (prolonged PR interval) to complete heart block and sudden death (Fig. 2b). All but one mouse died within 2–3 weeks after induction. Other 5-336 mice (>20 at the time this paper was written) in which the induction was stopped after 1 week (when they had both myotonia and first- or second-degree heart block) have lived for more than 2 months and some for more than 6 months. This lifespan is similar to 5-336 mice, in which transgene expression was not induced, which, owing to inherently leaky expression of the transgene (see Fig. 1c), spontaneously develop myotonia and die by about 9 months because of progressive heart block and sudden death. The cardiac conduction abnormalities observed in these mice are exactly like those seen in up to 70% of individuals with DM1: namely, heart block and atrioventricular node dysfunction13. This is the first transgenic mouse model with cardiac conduction abnormalities clearly associated with the expression of the DMPK 3′ UTR mRNA. The combination of myotonia and ECG abnormalities is seen only in myotonic dystrophy.
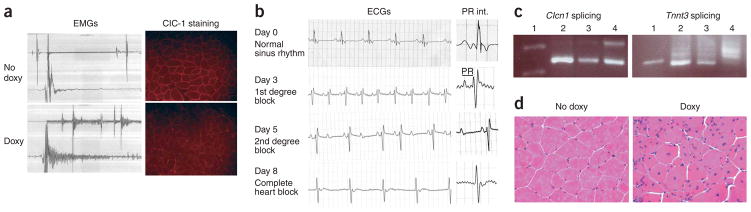
Figure 2.
Myotonic dystrophy phenotypes in transgenic mice. (a) Electromyography in skeletal muscles of mice in which transgene expression was not induced (‘No doxy’) shows a quiet baseline and classic myotonia in mice in which transgene expression was induced (‘Doxy’). ClC-1 immunohistochemistry showing normal sarcolemmal pattern of ClC-1 and loss of ClC-1 in myotonic mice. (b) Progressive cardiac conduction disturbances detected by serial ECGs on transgenic mice in which transgene expression was induced PR interval (PR int.) is prolonged by day 3. Note irregular and dropped beats by day 5 and lack of P waves by day 8. (c) Clcn1 and Tnnt3 RNA splicing abnormalities in skeletal muscle of mice in which transgene expression was induced. Lane 1: DNA marker; lane 2: wild-type; lane 3: uninduced 5-313; lane 4: induced 5-313. Note increased amounts of larger splice products. (d) H&E staining of skeletal muscle demonstrating increased numbers of central nuclei, fiber size variation and nuclear clumping in mice in which transgene expression was induced.
Myotonia in DM1 and DM2 is associated with reduced chloride channel (Clcn1, also known as Clc-1) expression and splicing abnormalities of Clcn1 mRNA14,15. We found ClC-1 protein significantly reduced or absent from the muscle membranes in mice in which transgene expression was induced (Fig. 2a). Furthermore, we performed RT-PCR for Clcn1 and Tnnt3 in our mice and uncovered splicing abnormalities (Fig. 2c) similar to those in transgenic mice overexpressing CUG repeats15, in the Mbnl1ΔE3 knockout mouse16 and in individuals with myotonic dystrophy14–16. Skeletal muscle histology also showed induction of central nuclei, nuclear clumping and fiber size variation as seen in individuals with DM1 (Fig. 2d). Thus, mechanistically, the underlying basis for phenotypes in our mice seems to be analogous to that found in individuals with myotonic dystrophy.
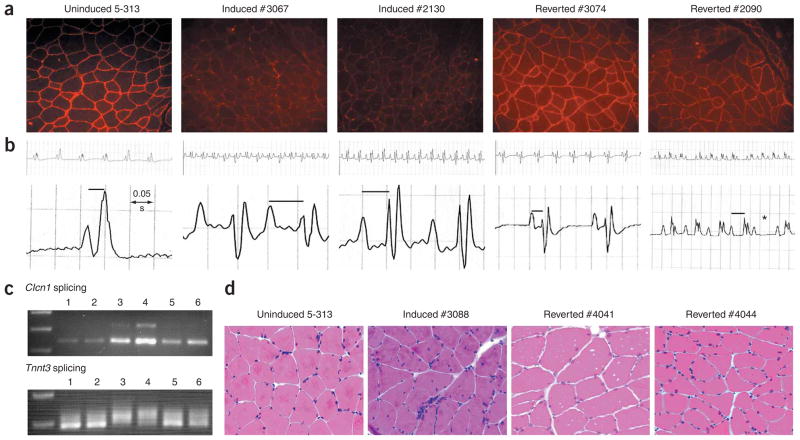
We also identified another line (5-313) with similar abnormalities. 5-313 mice in which transgene expression was not induced showed minimal expression of the GFP-DMPK 3′ UTR transgene, were completely asymptomatic regardless of age (1 to >18 months) and developed myotonia and cardiac conduction abnormalities when expression was induced (n = 20). All the mice developed robust myotonia within 3–4 weeks and subsequently varying degrees of heart block. Six mice died from higher-degree heart block. We stopped doxycycline administration in the remaining mice (n = 14). Notably, in most mice (n = 11), EMG myotonia was absent by 20 d, and in the others, we observed one or two runs of myotonia occasionally but not consistently. The absence of myotonia has persisted for at least five months after reversion, as of the writing of this paper. Molecular analyses of tissues from mice that reverted showed restoration of Clc1 staining in skeletal muscle (Fig. 3a). We were even more surprised to find that the mice showed marked stabilization or even reversal of their cardiac conduction abnormalities (Fig. 3b). This was particularly notable in the ten mice with first-degree heart block (1:1 ratio of atrial and ventricular contractions) in which the PR interval (a measure of cardiac conduction from the atria to the ventricle) was prolonged from a normal range of 0.025–0.035 s to >0.045 s. Of these mice, seven reverted to the normal range. The remainder with more severe heart block (second or third degree, in which the ratio of atrial to ventricular contractions is dissociated) did not revert to normal. Also, RNA splicing returned to more normal patterns for Clcn1 and Tnnt3 transcripts in skeletal muscle (Fig. 3c), as did muscle histology (Fig. 3d).
Figure 3.
Reversal of RNA toxicity. (a) ClC-1 immunohistochemistry shows loss of ClC-1 in mice in which transgene expression was induced and reversion to normal patterns with transgene silencing. (b) ECGs demonstrating examples of first-degree heart block (prolonged PR interval) in mice in which transgene expression was induced. Mouse #3074 reverted to normal from first-degree heart block. Mouse #2090 had second-degree heart block (Wenkebach-type) refractory to reversal (* indicates dropped beat). (c) Clcn1 and Tnnt3 RNA splicing abnormalities in skeletal muscle were reversed. Lanes 1,2: uninduced; lanes 3,4: induced; lanes 5,6: mice that reverted. (d) H&E staining of skeletal muscle showing reversal of histopathology. Mice reverted for over two months. Note obvious reductions in small fibers, central nuclei and nuclear clumping in mice that have reverted.
MBNL1 sequestration12 and increased CUG-BP1 (refs. 17,18) (Supplementary Fig. 3 online) have been observed in muscles from individuals with DM1. As noted earlier (Fig. 1b,e), the mice do not form ribonuclear inclusions, nor do they show obvious sequestration of MBNL proteins. Furthermore, there were no changes in MBNL1 expression in skeletal muscle or heart tissues (Fig. 4). However, we found increased CUG-BP1 in skeletal muscle, but not heart, in mice in which transgene expression was induced (Fig. 4 and Supplementary Fig. 4 online). Notably, we also did not find any obvious splicing abnormalities in the heart for previously reported targets19 (data not shown). Furthermore, CUG-BP1 in mice that reverted tended to return to levels found in mice in which transgene expression was not induced, correlating with the correction of molecular and phenotypic defects in skeletal muscle (Fig. 4).
Figure 4.
CUG-BP1 levels elevated by toxic RNA. Protein blots showing expression of CUG-BP1, MBNL1 and GFP in mouse skeletal muscle and heart. GAPDH was used as a loading control. Note induction of CUB-BP1 in skeletal muscle and reversion with transgene silencing. Also, there was no change in MBNL1 expression in mouse tissues in which transgene expression was induced. The 5-313 mice used in this figure had been induced continuously for over 10 months, and tissues from mice that reverted were collected within 20 d after stopping doxycycline.
A conundrum remains: it is still not clear why mice expressing a normal DMPK 3′ UTR develop myotonic dystrophy. Two previous reports have recently suggested that RNAs with the normal DMPK 3′ UTR may cause myotonic dystrophy–like effects9,20. However, the phenotype in the first instance is negligible and, in the second instance, is reported in aged mice (>10 months of age), and it is not clear whether the effects are due to DMPK or the DMPK mRNA. We compared transgene expression and found robust transgene RNA expression in our mice in which transgene expression was induced relative to other reported mouse models of DM9,20,21 (Supplementary Fig. 5 online). The effects of overexpression of many DMPK 3′ UTR transcripts with only five CUGs in our mice may be pathogenically equivalent to expressing mutant transcripts with hundreds of CUGs. Clearly, the sequence elements required for toxicity and that induce higher CUG-BP1 levels exist within the normal transcript. Of note, although individuals with DM1 or DM2 and transgenic mice over-expressing CUG repeats have RNA-MBNL1 foci12, only individuals with DM1 have increased CUG-BP1 (ref. 22). The RNA foci in DM2 contain only the (CCUG)n RNA23, whereas in DM1 the entire processed mutant DMPK mRNA is present3,4, suggesting that the context in which the (CUG) repeats reside (that is, the DMPK 3′ UTR) is important with respect to induction of CUG-BP1. Although speculative, this may prove to be important in understanding differences between DM1 and DM2, such as congenital myotonic dystrophy, which is seen only in DM1.
Given that expression of DMPK and concomitant RNA foci formation occurs from early fetal stages, it is notable that adult DM1 is such a late-onset disease. This could be related to developmentally regulated reductions in CUG-BP1 or MBNL1 (ref. 22). CUG-BP1 is highly expressed in myoblasts and developing muscle and markedly reduced in postnatal skeletal muscle. Thus, DMPK 3′ UTR–induced elevations in CUG-BP1 may be more consequential in adult muscle owing to the relative change in CUG-BP1 levels. Alternatively, it is possible that RNA foci formation is protective. Perhaps saturation of the RNA retention mechanisms in differentiated, post-mitotic tissues (due to reduced MBNL1 levels in adult tissues) eventually leads to RNA toxicity; that is, ‘free’ mutant DMPK transcripts are the toxins. The (CTG)5 transcripts are not retained, so overexpression may represent an accelerated version of this process. Nevertheless, the present findings, our previous findings in a myoblast cell culture model24 and, more recently, Drosophila melanogaster models25 and a study showing that RNA foci formation and splicing defects are separable26 highlight the need to reexamine the role of RNA-protein sequestration in myotonic dystrophy pathogenesis.
Alterations in the dynamic balance between MBNL1 and CUG-BP1 can affect the mutual antagonism of these two splicing factors on targets such as Clcn1 and Tnnt3 and may have a key role in the disease process27. In individuals with DM1, MBNL1 sequestration in foci and elevated CUG-BP1 presumably affect both arms of the balance (Supplementary Fig. 6 online). In (CTG)5 mice in which expression of GFP-DMPK 3′ UTR is induced, the elevated CUG-BP1 levels in the absence of MBNL1 changes still results in an imbalance, albeit one that affects only one side of the equation. This is analogous to results from the CUG-BP1 transgenic mouse28 and the Mbnl1ΔE3 knockout mouse16, in which perturbations of only one (CUG-BP1) or the other (MBNL-1) protein results in DM1-like splicing defects and DM1 pathology. Therapeutic strategies aimed at addressing only one side of the balance (that is, correction of MBNL1 sequestration or decreasing CUG-BP1) may not be sufficient, as both these proteins may have other functions in addition to their roles as splicing factors17.
The fact that the course of the disease can be reversed both overtly and at the molecular level suggests that the toxic RNA functions as a reversible metabolic toxin. Furthermore, the experiments described here and the reversibility of the myotonic dystrophy phenotype provide proof of principle for therapeutic strategies aimed at silencing the expression of or destroying the mutant DMPK transcript.
METHODS
Transgenic mice
The GFP-DMPK 3′ UTR transgene was driven by regulatory elements consisting of contiguous DNA starting from a BamHI site approximately 2.5 kb 5′ of the start codon and extending to the end of the first intron of DMPK. We included the intron because it has been shown to act as a muscle-specific enhancer that significantly increased the basal promoter activity10. These regulatory elements had also been used previously to generate a DMPK transgenic mouse and had been shown to emulate expression patterns of the endogenous mouse Dmpk gene11. Proximal to a putative transcriptional start site29, at an MfeI site, we inserted seven copies of the tetracycline responsive element (TRE). We then cloned an enhanced GFP (eGFP) gene modified to have a Kozak consensus start codon at the exact location of the start codon of the DMPK gene. After the eGFP stop codon, we cloned the human DMPK 3′ UTR with either (CTG)5 or (CTG)>200. We created double transgenic FVB mice using a second gene expressing the rtTA (reverse tetracycline transactivator) protein, driven by a CMV promoter (Clontech). Transgene expression was induced with 0.2% doxycycline in their drinking water.
EMGs and ECGs
We anesthetized mice with intraperitoneal valium (5 mg/kg body weight) and ketamine (100–150 mg/kg body weight) and kept them under a warming lamp during the entire procedure. Standard mouse anesthetics were cardiotoxic, especially in mice with preexisting conduction defects, and after extensive experimentation we settled on the valium and ketamine combination. We performed and scored EMGs as previously reported7. We performed three lead ECGs using a BioAmp/Powerlab from ADInstruments and collected data on a computer for later analysis. Mice revived in about 30–40 min without complications. All protocols were approved by institutional committees for animal care and use of the University of Virginia.
Histology, immunohistochemistry and RNA-FISH
We performed all microscopy using an Olympus IX 50 inverted microscope with fluorescent attachments and captured images with a CCD camera and presented them using Photoshop 5.5. Hematoxylin and eosin (H&E) staining was done using standard techniques. ClC-1 and MBNL1 immunohistochemistry and RNA-FISH were done as previously described12. Also, RNA-FISH using CY3-labeled oligonucleotides binding to DMPK 3′ UTR sequences flanking the (CUG)n repeats did not show any ribonuclear inclusions in the (CTG)5 mice. We performed ClC-1 immunofluorescence using an antibody from ADI (ClC11-A) at a 1:100 dilution (in PBS and 1% bovine serum albumin (BSA)). Secondary antibodies were from Molecular Probes and were used at 1:400 dilutions.
RNA analyses
We extracted total RNA from tissues collected in isopentane and flash-frozen in liquid nitrogen30. We performed RT-PCR for Clcn1 and Tnnt3 as described16. All RT-PCR products were cloned and their identity confirmed by DNA sequencing. All RNAs were treated with DNAse I (Ambion) and checked by PCR for beta-actin to ensure no DNA contamination before analysis. We probed RNA blots of 10 μg of total RNAs separated on 1% glyoxal gels with an equal mix of DMPK and Dmpk-3′ UTR fragments or Gapdh using standard procedures. We scanned blots and quantified DMPK-3′ UTR RNA expression relative to endogenous Dmpk expression using Image-Quant 5.1. See Supplementary Figure 1 for RNA blot quantification and Supplementary Methods online for quantitative RT-PCR of transgene expression and confirmation of transgene identity.
Protein blotting
We made total protein extracts from frozen tissues using standard protocols in RIPA buffer (50 mM Tris-HCl, pH. 7.4; 150 mM NaCl; 1% NP-40; 0.5% sodium deoxycholate; 0.1% SDS) and protease inhibitor (Roche). We used protein blots of 50 μg of total protein to detect CUG-BP1 using monoclonal antibody 3B1 (Upstate, cat.# 05-621) at a 1:4,000 dilution. We detected MBNL1 as previously described16. Glyceraldehyde phosphate dehydrogenase (GAPDH) levels were detected using a GAPDH antiserum (Ambion) and were used as a loading control. We used GFP antiserum (Invitrogen, cat.# R970-01) to detect transgene expression. Blots were scanned and relative protein expression was quantified using ImageQuant 5.1.
Supplementary Material
Acknowledgments
We wish to thank P. Mahadevan and A. Tucker for their insights and continued support. MBNL antibodies were provided by M. Swanson and C. Thornton. Human tissues were provided by J. Puymirat and C. Thornton and purchased from the University of Miami Brain and Tissue Bank. Mouse tissues from other myotonic dystrophy models were provided by J. Puymirat, B. Wieringa and G. Gourdon. Transgenic mice were generated by the University of Wisconsin-Madison Transgenic Core Facility. All studies were done under the auspices of the University of Virginia Animal Care and Use Committee and Institutional Review Board. This work was supported by the Muscular Dystrophy Association and the US National Institute of Arthritis and Musculoskeletal and Skin Diseases.
Footnotes
AUTHOR CONTRIBUTIONS
M.S.M., R.S.Y., Q.Y., C.D.F.-M., T.D.B. and L.H.P. performed experimental work and data analysis. S.B. generated the transgene constructs. M.S.M. was responsible for conceptual design and execution.
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Mahadevan M, et al. Myotonic dystrophy mutation: an unstable CTG repeat in the 3′ untranslated region of the gene. Science. 1992;255:1253–1255. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- 2.Day JW, Ranum LP. RNA pathogenesis of the myotonic dystrophies. Neuromuscul Disord. 2005;15:5–16. doi: 10.1016/j.nmd.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 3.Taneja KL, McCurrach M, Schalling M, Housman D, Singer RH. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J Cell Biol. 1995;128:995–1002. doi: 10.1083/jcb.128.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis BM, McCurrach ME, Taneja KL, Singer RH, Housman DE. Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc Natl Acad Sci USA. 1997;94:7388–7393. doi: 10.1073/pnas.94.14.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amack JD, Paguio AP, Mahadevan MS. Cis and trans effects of the myotonic dystrophy (DM) mutation in a cell culture model. Hum Mol Genet. 1999;8:1975–1984. doi: 10.1093/hmg/8.11.1975. [DOI] [PubMed] [Google Scholar]
- 6.Furling D, Lemieux D, Taneja K, Puymirat J. Decreased levels of myotonic dystrophy protein kinase (DMPK) and delayed differentiation in human myotonic dystrophy myoblasts. Neuromuscul Disord. 2001;11:728–735. doi: 10.1016/s0960-8966(01)00226-7. [DOI] [PubMed] [Google Scholar]
- 7.Mankodi A, et al. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 8.Berul CI, Maguire CT, Gehrmann J, Reddy S. Progressive atrioventricular conduction block in a mouse myotonic dystrophy model. J Interv Card Electrophysiol. 2000;4:351–358. doi: 10.1023/a:1009842114968. [DOI] [PubMed] [Google Scholar]
- 9.O’Cochlain DF, et al. Transgenic overexpression of human DMPK accumulates into hypertrophic cardiomyopathy, myotonic myopathy and hypotension traits of myotonic dystrophy. Hum Mol Genet. 2004;13:2505–2518. doi: 10.1093/hmg/ddh266. [DOI] [PubMed] [Google Scholar]
- 10.Storbeck CJ, Sabourin LA, Waring JD, Korneluk RG. Definition of regulatory sequence elements in the promoter region and the first intron of the myotonic dystrophy protein kinase gene. J Biol Chem. 1998;273:9139–9147. doi: 10.1074/jbc.273.15.9139. [DOI] [PubMed] [Google Scholar]
- 11.Jansen G, et al. Abnormal myotonic dystrophy protein kinase levels produce only mild myopathy in mice. Nat Genet. 1996;13:316–324. doi: 10.1038/ng0796-316. [DOI] [PubMed] [Google Scholar]
- 12.Mankodi A, et al. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum Mol Genet. 2001;10:2165–2170. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- 13.Melacini P, et al. Correlation between cardiac involvement and CTG trinucleotide repeat length in myotonic dystrophy. J Am Coll Cardiol. 1995;25:239–245. doi: 10.1016/0735-1097(94)00351-p. [DOI] [PubMed] [Google Scholar]
- 14.Charlet BN, et al. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol Cell. 2002;10:45–53. doi: 10.1016/s1097-2765(02)00572-5. [DOI] [PubMed] [Google Scholar]
- 15.Mankodi A, et al. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol Cell. 2002;10:35–44. doi: 10.1016/s1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- 16.Kanadia RN, et al. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 17.Timchenko NA, et al. Overexpression of CUG triplet repeat-binding protein, CUGBP1, in mice inhibits myogenesis. J Biol Chem. 2004;279:13129–13139. doi: 10.1074/jbc.M312923200. [DOI] [PubMed] [Google Scholar]
- 18.Savkur RS, Philips AV, Cooper TA. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 19.Mankodi A, Lin X, Blaxall BC, Swanson MS, Thornton CA. Nuclear RNA foci in the heart in myotonic dystrophy. Circ Res. 2005;97:1152–1155. doi: 10.1161/01.RES.0000193598.89753.e3. [DOI] [PubMed] [Google Scholar]
- 20.Storbeck CJ, et al. Inhibition of myogenesis in transgenic mice expressing the human DMPK 3′-UTR. Hum Mol Genet. 2004;13:589–600. doi: 10.1093/hmg/ddh064. [DOI] [PubMed] [Google Scholar]
- 21.Seznec H, et al. Mice transgenic for the human myotonic dystrophy region with expanded CTG repeats display muscular and brain abnormalities. Hum Mol Genet. 2001;10:2717–2726. doi: 10.1093/hmg/10.23.2717. [DOI] [PubMed] [Google Scholar]
- 22.Lin X, et al. Failure of MBNL1-dependent postnatal splicing transitions in myotonic dystrophy. Hum Mol Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- 23.Margolis JM, Schoser BG, Moseley ML, Day JW, Ranum LP. DM2 intronic expansions: evidence for CCUG accumulation without flanking sequence or effects on ZNF9 mRNA processing or protein expression. Hum Mol Genet. 2006;15:1808–1815. doi: 10.1093/hmg/ddl103. [DOI] [PubMed] [Google Scholar]
- 24.Amack JD, Mahadevan MS. The myotonic dystrophy expanded CUG repeat tract is necessary but not sufficient to disrupt C2C12 myoblast differentiation. Hum Mol Genet. 2001;10:1879–1887. doi: 10.1093/hmg/10.18.1879. [DOI] [PubMed] [Google Scholar]
- 25.Houseley JM, et al. Myotonic dystrophy associated expanded CUG repeat muscleblind positive ribonuclear foci are not toxic to Drosophila. Hum Mol Genet. 2005;14:873–883. doi: 10.1093/hmg/ddi080. [DOI] [PubMed] [Google Scholar]
- 26.Ho TH, et al. Colocalization of muscleblind with RNA foci is separable from mis-regulation of alternative splicing in myotonic dystrophy. J Cell Sci. 2005;118:2923–2933. doi: 10.1242/jcs.02404. [DOI] [PubMed] [Google Scholar]
- 27.Ho TH, et al. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho TH, Bundman D, Armstrong DL, Cooper TA. Transgenic mice expressing CUG-BP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Hum Mol Genet. 2005;14:1539–1547. doi: 10.1093/hmg/ddi162. [DOI] [PubMed] [Google Scholar]
- 29.Mahadevan MS, et al. Structure and genomic sequence of the myotonic dystrophy (DM kinase) gene. Hum Mol Genet. 1993;2:299–304. doi: 10.1093/hmg/2.3.299. [DOI] [PubMed] [Google Scholar]
- 30.Langlois MA, Lee NS, Rossi JJ, Puymirat J. Hammerhead ribozyme-mediated destruction of nuclear foci in myotonic dystrophy myoblasts. Mol Ther. 2003;7:670–680. doi: 10.1016/s1525-0016(03)00068-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.