Abstract
Purpose
The lens assembles two systems of intermediated filaments—vimentin intermediate filament (IF) and highly divergent, lens-specific beaded filament (BF)—sequentially as epithelial cells differentiate into fiber cells. The goal of this study was to identify linker proteins that integrate the different lens IF into the biology of the lens fiber cells.
Methods
Antibodies to periplakin were used in coimmunoprecipitation studies to identify proteins that complex with BF and IF in detergent extracts of mouse lens. GST-periplakin fusion proteins were used to confirm coimmunoprecipitation results. Yeast two-hybrid analysis was used to establish direct linkage between periplakin and BF/IF proteins and to narrow down binding domains. Immunocytochemistry was used to establish spatial and temporal coexpression of periplakin and BF/IF.
Results
Periplakin is found complexed to BF and IF in the lens. The COOH terminus of periplakin was shown to have a strong affinity for the CP49 rod 2 domain but not its head or rod 1 domains. Low-level affinity was seen between the filensin rod domain and periplakin. Periplakin localization in lens overlapped with BF and IF.
Conclusions
Despite divergence in primary sequence, predicted secondary structure, and filament structure, CP49 has conserved the capacity to bind a common IF linker protein, periplakin, and shares that binding capacity with the other major lens IF protein, vimentin. This suggests that mutations in periplakin have the potential to emulate the cataract seen in lenses with defective BF proteins.
The vertebrate lens consists predominantly of elongated fiber cells assembled with a high degree of precision and long-range order. Covering the anterior surface of the lens, and representing a tiny fraction of the total lens mass, is a single layer of flattened cuboidal lens epithelial cells. Fiber cells arise by differentiation of lens epithelial cells, a process that represents a remarkable transformation of the epithelial cell, approximately 20 μm in diameter, into a fiber cell that measures 2 × 6 × 10,000 μm, depending on the species. As each generation of epithelial cells undergoes this differentiation process, a new layer of fiber cells is added to the lens, giving rise to a series of concentric shells arranged chronologically by “birth date.” This differentiation and addition of new layers goes on throughout life, albeit more slowly in the adult organism. Because older lens cells do not die, the lens contains a full spectrum of cells ranging from those at the center, created in the embryonic period, to those at the surface that have just recently differentiated. This process is all the more remarkable in that a fiber cell will dismantle all its membranous organelles shortly after completing the elongation process, a process that occurs approximately 50 fiber cells from the lens equatorial surface.1–6 The other thousand or more fiber cells lack organelles and, therefore, the repair and renewal processes associated with them. Despite this, these nonrenewing fiber cells last the life of the organism, which can span several decades.
As with some other tissue types, such as epidermis, lens cells undergo a differentiation-dependent switch in the expression of intermediate filament (IF) proteins.7–9 The lens epithelium predominantly expresses vimentin, a well-conserved type 3 IF protein common to many mesenchymal tissues.10–14 However, in contrast to epidermis, which switches from one keratin pair to another, the lens fiber cells express an entirely novel, fiber cell–specific pair of IF proteins, CP49 and filensin.15–20 These are considered the most divergent IF proteins yet described and are often assigned to their own class because of the low levels of identity with existing classes.18–22 Moreover, CP49 and filensin are not found in canonical 11-nm IFs but rather in the structurally unique beaded filament (BF).15,16,23
Targeted genomic deletion of either BF protein in the mouse eliminates the assembly of the BF in the lens fiber cell.24–27 The resultant change in phenotype is subtle, showing a slight light scatter in the lens that is detectable at approximately 3 weeks of age but that grows progressively worse over the lifespan of the animal. Electron microscopy of the knockout lens suggests that the elaborate differentiation of fiber cells proceeds in a manner not identifiably different from that of the wild type. This phenotype gradually is lost, as is the exquisite long-range stacking of fiber cells, suggesting that the BF is not required to achieve the differentiated phenotype but is required to maintain cellular and tissue-level phenotypes with aging.27,28 Thus, despite the very high degree of divergence in the primary sequence, in the predicted secondary structure and in the filament that is ultimately assembled, the BF remains true to its roots as an IF protein in that it serves to provide mechanical support and to resist changes in the differentiated phenotype.29–34
IF function is clearly dependent on integration with other cellular structures and other cytoskeletal elements, in part through IF-associated proteins (IFAPs). As a result, mutations in IFAPs would be predicted to have the potential to result in cataracts that emulate the absence of an IF by breaking the link that connects IFs to other cell structures. The plakins are a major class of IFAP that have been identified as crucial to IF function.35–38 To date, seven plakins have been identified, desmoplakin, plectin, BPAG-1, envoplakin, periplakin, epiplakin, and MACF-1.37 With the exception of epiplakin, they share a common structure: a central coiled-coil domain, flanked by globular domains at the N- and C termini.35,37,39,40 The IF-binding domain has been localized to the C terminus, whereas the N-terminus mediates binding to cell adhesion sites at the plasma membrane or other cytoskeletal networks.41–45
Previous reports have identified periplakin as a constituent of lens fiber cells.46 Periplakin has been shown to bind to IF proteins, specifically vimentin and K8. The binding is mediated by periplakin's C-terminal Linker domain, which targets rod domain 2 on the IF protein. In this report, we examine the relationships formed between periplakin and lens IF and BF proteins. We were particularly interested in establishing whether the high degree of sequence and structural divergence of the BF proteins mandated the emergence of a novel IFAP adapted to BF structure or whether the BF could make use of established IFAPs.
Materials and Methods
All procedures involving animals complied with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
Antibodies
Polyclonal antibody to recombinant periplakin was raised using a flag-tagged C-terminal periplakin fragment that included the periplakin Linker (ppl-L; Fig. 1, schematic of constructs) region. pT7-FLAG vector was generated by inserting the flag tag sequence into the pT7–7 vector between the enzyme sites NdeI and EcoRI. The sequence ppl-L was inserted using the EcoRI enzyme site; thus, the FLAG tag was fused to the N-terminal end of the ppl-L. The pl-L consisted of the C-terminal 209 amino acids. The resultant plasmid was transformed into BL21 AI and was induced with arabinose. Induction was performed overnight at 20°C. Protein was enriched by size-exclusion and ion-exchange chromatography. Purified proteins were injected into rabbits in four separate immunizations at 10-day intervals. Antibodies were affinity-purified using immobilized ppl-L (Affigel 15; Bio-Rad Laboratories, Hercules, CA) and eluted with 50 mM glycine, pH 2.3. Polyclonal and affinity-purified rabbit antibodies to CP49, filensin, and vimentin were raised and characterized previously.16,23,47,48
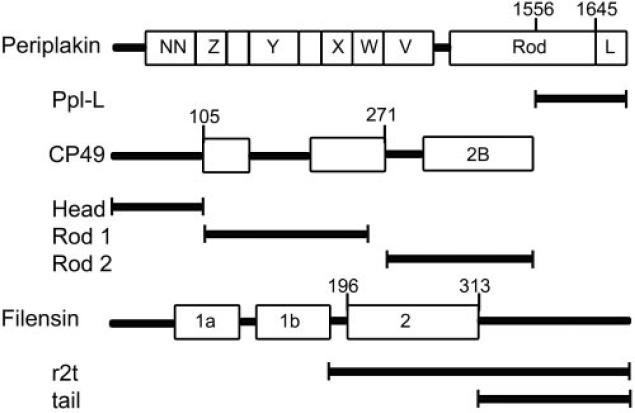
Figure 1.
Schematic representation of cDNA constructs used in the yeast two-hybrid assay. ppl-L, periplakin C-terminal linker domain; r2t, rod 2 plus tail domain of mouse filensin.
Immunocytochemistry
Mouse eyes were removed and immediately frozen by immersion in dry ice-cooled propane. Frozen lenses were transferred to −80°C methanol/acetic acid (97:3) for at least 48 hours to exchange methanol for water. Eyes were then removed to −20°C for 24 hours and finally to room temperature. For paraffin embedding, eyes were transferred to 100% ethanol and processed into paraffin using routine methods. Sections (6-μm thick) were harvested, deparaffinized, and rehydrated in phosphate-buffered saline (PBS). Background suppression was achieved by incubating sections in 5% normal goat serum (NGS) for 10 minutes. Affinity-purified primary antibodies were diluted in PBS-NGS, and sections were incubated for 2 hours at room temperature, followed by three 10-minute washes in PBS. Bound primary antibody was localized by incubation with Alexa Fluor-488–conjugated goat anti–rabbit antibody (Invitrogen, Carlsbad, CA) for 2 hours. After washing with PBS as described, sections were rinsed with water and cover-slipped using glycerol as a mounting medium. Alternatively, primary antibody was visualized with peroxidase-conjugated goat anti–rabbit antibodies and imaged with diaminobenzidine. Eyes were removed from methanol/acetic acid to PBS for 1 hour to remove the fixative and then transferred to 2.3 M sucrose in PBS for 2 hours before they were frozen in OCT medium for cryosectioning.
Generation of cDNA Constructs
To generate mouse lens cDNA, mRNA was isolated from rat or mouse lens with the RNeasy kit (Qiagen, Valencia, CA). With oligo-dT as a primer, mRNA was reverse-transcribed into cDNA (Superscript II; Invitrogen). All constructs were PCR-amplified from the mouse lens cDNA and were cloned into clone vector (TOPO; Invitrogen). The ppl-L fragment was made by cutting the EcoRI enzyme site in periplakin, located toward the C-terminal end of the periplakin rod domain. This yielded an approximately 650-bp fragment that included the whole linker region and the C-terminal end of the rod domain, which could then be ligated into subsequent vectors.
Lens Extracts
Whole lens protein extract was prepared by lysis in SDS-PAGE sample buffer. For coimmunoprecipitation (Co-IP) studies and GST pull-downs, a lens detergent extract was prepared using OG buffer according to Walker et al.49 (60 mM octylglucoside in 10 mM imidazole, 100 mM NaCl, 1 mM MgCl2, 5 mM EDTA, pH 7.4, and protease inhibitors; Complete Mini [Roche, Indianapolis, IN]). Extracts were centrifuged at 12,000g for 10 minutes at 4°C.
GST Pull-downs
The ppl-L fragment was cloned into pGEX-5X-1 vector, transformed into BL21 cells, and induced with 0.1 M isopropylthiogalactoside at 20°C. Cells were sonicated in PBS containing protease inhibitors and were centrifuged at 12,000g for 10 minutes at 4°C. Triton-X was added to the supernatant at 1% and was incubated with glutathione-agarose beads for 1 hour at room temperature. Control beads were incubated in PBS + 1% Triton-X for the same amount of time. Beads were washed four times with the same buffer and incubated in lens OG extract for 4 hours at room temperature. After four washes in OG buffer and PBS, the bound proteins were solubilized in SDS-PAGE sample buffer and resolved by SDS PAGE. Western blots were conducted as described previously.50,51
Coimmunoprecipitation
Affinity-purified periplakin antibody was immobilized on beads as directed in a Co-IP kit (Pierce, Rockford, IL). Immobilized antibody beads were incubated in lens OG extract for 4 hours at room temperature. Washing and elution steps were carried out as directed, and an additional wash with high-salt phosphate buffer (PBS + 1 M NaCl) was added to reduce nonspecific binding. Eluted sample was resolved by SDS-PAGE and analyzed by Western blotting.
Yeast Two-Hybrid Analysis
Full-length mouse vimentin and CP49 and the filensin rod two-tail fragment were excised from TOPO vectors and ligated into pAS2-1 vectors as a GAL4 DNA binding-domain fusion protein. Protein expression in yeast was confirmed by Western blot, and ppl-L was cloned into pGAD vector as a GAL4 activation domain fusion protein. Bait and prey plasmids were cotransfected into PJ4 - 09A yeast strain containing three reporter genes (lacZ, HIS3, and ADE2). Cotransformants were grown in liquid SD-Leu/-Trp media, and 3-μL droplets of 0.1 OD/mL and 0.05 OD/mL cells were grown on SD-Leu/-Trp and SD-Leu/-Trp/-His plates to compare growth rates and to assess interaction.
To test interactions with different domains in CP49, three fragments of head, rod1, and rod2 were PCR amplified, then cloned into pAS2-1. All plasmids were tested for self-activation by growing on SD-Trp/-His or SD-Leu/-His plates.
Results
Coimmunoprecipitation
To determine whether periplakin was found in the same complexes as vimentin, CP49, and filensin, we conducted Co-IP studies on detergent extracts of whole mouse lens.
We first developed an antiserum to the recombinant periplakin C-terminal domain, which includes the Linker region, as described in Materials and Methods. A schematic that identified the region and boundaries of this fragment (ppl-L) is included in Figure 1. Using immobilized recombinant ppl-L, antibodies were affinity purified and tested for reactivity and specificity by Western blotting against whole lens extract (Fig. 2A). A single band at approximately 195 kDa, the relative molecular weight of periplakin, was identified, showing specificity of the antibody and a lack of detectable cross-reactivity with other lens proteins.
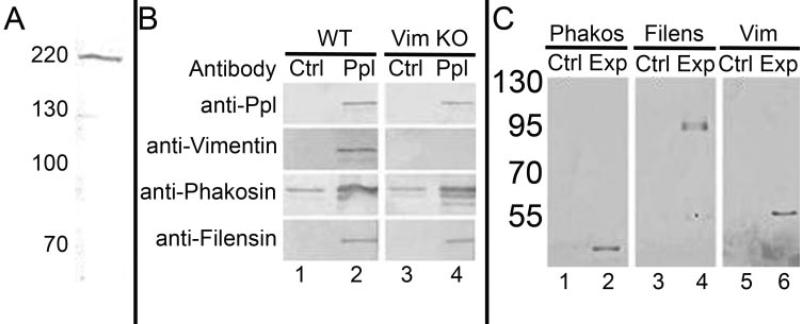
Figure 2.
Affinity-based assays. (A) Western blot of total lens extract, probed with rabbit antibody to the recombinant, purified ppl-L, characterizing the affinity-purified antibody. (B) Total lens detergent extracts of wild-type lenses (lanes 1, 2) and vimentin knockout lenses (lanes 3, 4). Lanes 1 and 3 are controls probed with irrelevant rabbit IgG. Lanes 2 and 4 are probed with the antibodies indicated at the left.(C) GST pull-down assays. Immobilized GST-ppl-L fusion protein was used to probe detergent-extracted lens. The washed pellet was resolved by SDS-PAGE and transferred to nitrocellulose for probing with antibodies to (lane 1) CP49, (lane 2) filensin, (lane 3) vimentin.
Immobilized anti–ppl-L antibody was then used in Co-IP studies of detergent extracts of whole lens. After washing, the pelleted material was resolved by SDS-PAGE and was transferred to nitrocellulose for Western blot analysis using antibodies to periplakin, vimentin, CP49, and filensin. Figure 2B, lane 2, is a composite of four different blots showing a positive reaction for each of the four proteins. Reactive lower molecular weight bands are breakdown products of the parent proteins, as shown by the use of monoclonal antibodies and by their absence in the knockout animals.24,26,50 As a control, immunoprecipitation was conducted in parallel with irrelevant rabbit IgG. With the exception of CP49, the controls were negative. The CP49 control showed a persistent reaction, though always at a lower level than the experimental lane. These results established that all three proteins coimmunoprecipitate with periplakin, suggesting that periplakin, either directly or indirectly, is complexed with the IF and BF proteins.
BFs and vimentin IFs overlap in expression during fiber cell elongation and are physically intertwined. This raised concerns that pelleting one filament system might artifactually pellet the other because of mechanical trapping. To control for this, we repeated the Co-IP on lenses from the vimentin knockout mouse, which lacks the vimentin IFs (Fig. 2B, lanes 3, 4). The absence of vimentin IFs did not change the results of the Co-IP/periplakin antibody-pelleted CP49 and filensin.
GST Pull-down Assays
To confirm the results of Co-IP and to ask whether the periplakin C-terminal domain was responsible for the interactions established by Co-IP, we conducted GST pull-down assays. A fusion protein consisting of GST, linked to the ppl-L, was expressed in bacteria and purified. The purified fusion protein was bound to glutathione beads and was incubated with mouse lens detergent extract. The GST pull-down complex was washed, as described, then subjected to Western blot analysis to probe for the presence of vimentin, CP49, and filensin. Figure 2C, lanes 2, 4, and 6, respectively, establish the presence of CP49, filensin, and vimentin in the complexes affinity purified by the GST-ppl-L fusion protein. These data confirmed the Co-IP findings and further established that the periplakin C-terminal linker domain is responsible for the interaction.
Yeast Two-Hybrid Assays
Co-IP and the GST pull-down assays do not rule out the possibility that periplakin interaction with BF/IF proteins is indirect, mediated by an intermediary protein. To test for direct interaction between periplakin and the lens IF proteins and to further refine the binding domains, we conducted yeast two-hybrid screening of several different constructs.
Previous studies have shown direct interactions between the periplakin linker domain and vimentin and K8 but not K18. Therefore, we used vimentin and K8 as positive controls and K18 as a negative control, based on studies by Kazerounian et al.40 All IF/BF proteins were expressed in full-length and were fused to the GAL4-binding domain, with the exception of filensin, for which only the C-terminal half of the protein (r2t fragment; Fig. 1) was expressed.
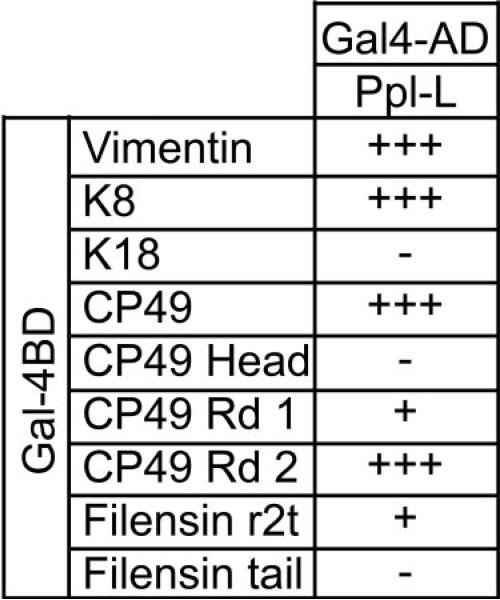
Figure 3 summarizes the results. In accordance with published data, vimentin and K8 gave robust positive results, whereas K18 did not reveal signs of interaction. Full-length CP49 showed a robust interaction with the periplakin linker domain and had a growth rate comparable to that of the positive controls vimentin and K8.
Figure 3.
Yeast two-hybrid assay results. Two-hybrid assay testing the interaction of BF proteins with the ppl-L domain. CP49 shows strong interaction with periplakin, and the region of interaction could be narrowed down to the rod 2 region. Filensin showed little interaction in the corresponding region included in the r2t fragment. Vimentin and K8 served as positive controls, with known rod 2 binding activity with periplakin. K18 served as a negative control.
Earlier work had shown that periplakin interacted with the rod 2 domain of IF proteins.41–43 To establish whether this pattern of binding was conserved in the highly divergent CP49, we subdivided CP49 into three regions (head, rod 1, and rod 2, described in Fig. 1) and tested each against the periplakin linker domain. The head domain of CP49 did not show any interaction with the periplakin linker domain. The rod1 region showed a very weak interaction. However, the rod 2 domain showed high levels of interaction, indistinguishable from those of positive controls. Interestingly, this is consistent with some of the other IFAPs that bind to the 2B region of IF proteins. Thus, despite the high degree of divergence in the CP49 amino acid sequence, predicted secondary structure, and filament structure, periplakin is able to bind to CP49, and interaction occurs at the same region reported for other IFAP interactions.
The filensin r2t and the tail fragments (Fig. 1) were also tested for interaction. However, the tail domain showed almost no interaction, and the r2t fragment showed very weak interaction. This suggests that periplakin is able to bind only one of the two principal proteins in the BF, analogous the binding of periplakin to only one of the two keratins in the K8/K18 filament.
Immunocytochemistry
Co-IP, GST pull-down, and yeast two-hybrid data are all consistent with a direct interaction between periplakin and vimentin IFs and BFs in the lens. To establish whether periplakin was coexpressed temporally and spatially with the IF and BF proteins, we conducted immunocytochemical localizations of all four proteins in the lens.
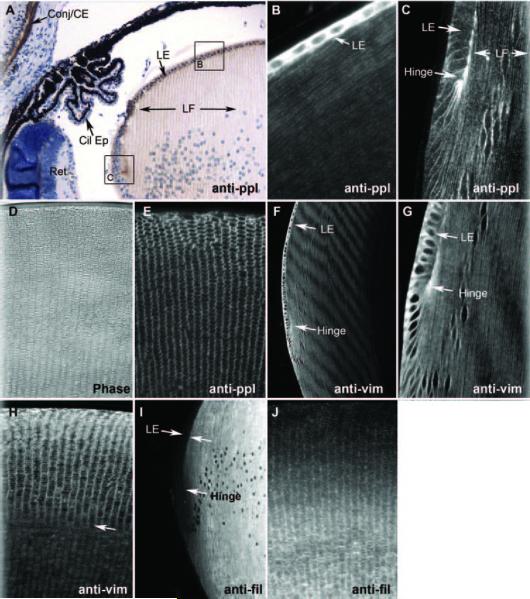
Figures 4A–C, and 4E show immunocytochemical localizations of periplakin in the 3-week-old wild-type mouse eye. Figure 4A is an immunoperoxidase-labeled paraffin section, counterstained to reveal the histology of this region of the eye. The brown reaction product marks the location of periplakin. Periplakin reactivity is strong, as expected, in the corneal epithelium and conjunctiva (Conj/CE). Neither the ciliary epithelium (Cil Ep) nor the retina (Ret) shows an obvious reaction product. In the lens, both the lens epithelium (LE) and the lens fiber cell (LF) are labeled, though the pattern of labeling is different. Figure 4B, a higher magnification view of an area comparable to the boxed “B” in Figure 4A but labeled with fluorescent secondary antibodies, shows very heavy labeling of the cytoplasm of the more anterior lens epithelial cells but lighter, predominantly membrane, labeling in the underlying fiber cells. The narrow, highly elongated shape of the fiber cells is evident. The gain was lowered in Figure 4B to reveal the presence of the nuclei in the epithelial cells. Figure 4C is an area comparable to the boxed “C” in 4A, again labeled by immunofluorescence. This area includes the lens coronal equator, in which epithelial cells are starting to elongate into fiber cells. At this point, the intensity of the epithelial cytoplasmic labeling drops dramatically, leaving a predominantly membranous labeling in epithelial and fiber cells. Note the particularly intense labeling of the “hinge” region, where the newly elongating fiber cells are attached to the apical surfaces of the overlying epithelial cells. Figure 4E shows an immunofluorescence-labeled coronal section of the mid-lens, similar to the phase-contrast section in Figure 4D, which displays the flattened-hexagon shape of the fiber cells in cross-section. The labeling pattern is predominantly membranous and appears to label all surfaces of the cell.
Figure 4.
Immunocytochemistry. Six-micron-thick paraffin sections probed with antibodies to (A) periplakin, then localized with HRP-goat anti-rabbit antibodies. Brown: reaction product can be seen in the corneal epithelium and conjunctiva (conj/CE) but not in ciliary epithelium (Cil Ep) or retina (Ret). The lens epithelium (LE) is very heavily reacted, as is the “hinge” region. Labeling of fiber cells (LF) is principally at the membrane. Areas comparable to boxed areas “B” and “C” are shown in (B) and (C). (B) Periplakin immunofluorescence labeling of more anterior epithelium and underlying fiber cells. Epithelial reactivity is very high, but fiber cell membrane labeling is also evident. (C) Periplakin labeling of lens “bow” or equatorial region, where fiber cell elongation begins. Epithelial cytoplasmic labeling is sharply reduced, and most labeling is at the plasma membrane. (D) Phase-contrast image to demonstrate histology of a coronal section of mid-lens. (E) Coronal section. Periplakin immunofluorescence showing cross-sectioned fiber cells. (F-H) Vimentin immunofluorescence of sagittal (F, G) and coronal (G) sections. (I, J) Filensin immunofluorescence in sagittal (I) and coronal (J) sections. (I) Note the complete absence of the labeling of lens epithelium.
Figures 4F–H are sections similar to Figures 4B and 4C but are labeled with antibodies to vimentin. Figure 4F shows an intensely labeled anterior epithelium (LE) similar to that seen with periplakin antibodies. However, as seen in Figure 4G, at higher magnification, the cytoplasmic labeling does not diminish in the elongating cells at the equator, though it shows a preferential distribution to the basal end of the elongating cell. As differentiation proceeds, the labeling becomes predominantly, though not exclusively, membranous, as seen for periplakin. This is evident in the longitudinal view of fiber cells in Figures 4F and 4G and in the cross-sectioned fiber cells in Figure 4H. Note in Figure 4H that the pattern of labeling changes near the arrow from what seems to be mostly membrane to what looks more cytoplasmic. These figures establish similar, but not identical, patterns of labeling for periplakin and vimentin
Figures 4I and 4J show sections labeled with antibodies to the BF protein filensin. Figure 4I shows no labeling of the lens epithelium, as previously reported. Filensin signal begins to emerge as differentiation progresses and becomes intense as the cells elongate. Cross-sectioned fiber cells (Fig. 4J) show filensin labeling is initially more intense at the membrane but increases in the cytoplasm as cells mature and the BF network increases in abundance.
These data collectively show that periplakin overlaps in temporal and spatial dimensions with vimentin IF and BF, consistent with the hypothesis that it serves as an IFAP for both.
Discussion
CP49 and filensin are lens fiber cell-specific IF proteins. Although they are members of the IF family, they comprise the beaded filament, a cytoskeletal element that is structurally distinct from the canonical 10- to 11-nm IF. The closest IF relative to CP49 is the type 1 cytokeratin, K19, with which it shares 34% identity. This is less than half the level of intraclass sequence identity typical among IF proteins. Despite this, CP49 shares an essentially identical gene structure with the type 1 cytokeratins,19,20 suggesting that CP49 is indeed a type 1 cytokeratin but a uniquely divergent one.
Gene knockout studies indicate that despite sequence and structural differences, BF serves a typically IF-like function: it stabilizes cellular and tissue-level phenotypes. In the absence of BFs, lens fiber cells cannot maintain their highly differentiated structure or their exquisite tissue-level organization.28 This loss of long-range stacking and alignment of fiber cells results in an initially subtle cataract that grows progressively worse with age.24,26 Why the BF proteins must diverge so greatly and why the BF structure must be different from the IF to accomplish this task are unknown.
In this report we asked whether the dramatic divergence in BF primary sequence from the rest of the IF family precluded the ability of BF proteins to use conventional IFAPs. The presence of vimentin in the lens epithelium and early fiber cells has been well established. Also established in the literature is the presence of periplakin in the lens, an IFAP with known vimentin-binding properties. We tested whether BF proteins could interact with periplakin using several approaches. We established that periplakin is capable of binding directly to CP49 and that this interaction occurs between the periplakin linker region and the CP49 rod domain 2, similar to what has been described for other IF proteins such as K8. Interestingly, lengsin, a nonfunctional glutamate synthetase, also binds to CP49 through the rod 2B region and may serve as another IFAP.52 Thus, despite the high degree of sequence divergence in CP49, this plakin-binding property is conserved.40,53
GST pull-down and Co-IP approaches demonstrated that filensin was also part of this complex. However, direct testing of periplakin binding through yeast two-hybrid studies showed, at best, a weak interaction between periplakin and filensin. This disparity could be attributed simply to the fact that periplakin directly binds only CP49 and filensin simply copellets because it is bound to CP49. Alternatively, the incorporation of full-length filensin into the yeast two-hybrid vector was not achieved; hence, a possible filensin binding site might have been excluded from the assay. We note that in the case of lengsin, similar results were reported: CP49 showed interaction with lengsin but not filensin.52
Although the C-terminus of periplakin interacts with IF proteins, the N-terminus has typically been implicated as responsible for localizing periplakin to the plasma membrane or to many other candidate cellular structures. Two proteins have been identified to date to directly interact with the periplakin N-terminus. Kazrin is a desmosomal component that was identified through a yeast two-hybrid screen; thus far, expression has been shown in the epidermal cells and in embryos.54 Plectin was also found to interact with the N-terminus of periplakin and was found to colocalize in the MCF-7 and the HaCaT cell line.55 Although the nature of the interaction is unknown, periplakin localizes to cortical actin in keratinocytes through its N-terminus.56 In MCF-7 cells, however, periplakin does not localize with cortical actin,57 suggesting that the interaction in keratinocytes might be mediated or modulated through another protein. However, the plakin family as a whole is proving to be remarkably versatile in the constellation of interactions it can form, many of which are not at the plasma membrane. Linkages to organelles and other cytoskeletal elements have been shown, possibly accounting for our demonstration of strong cytoplasmic labeling in the lens epithelium.37–39,54,55
Recently, it has been shown that the lens has a junctional complex of N-cad/γ-cat/desmoplakin/vimentin complex.58 It is possible that periplakin is part of the same complex because it localizes to desmosomes in the epidermis, where desmoplakin is also localized. However, periplakin has also been shown to form a complex with ezrin, desmoyokin, and periaxin in the lens, a complex that has a separate localization from the N-cadherin complex.46 A study by Rose et al.59 showed that aquaporin 0/MIP interacts with BF. We note that aquaporin did not coimmunoprecipitate with periplakin (not shown), and thus it seems that periplakin's role of connecting the BF to the plasma membrane is independent of the interaction between BFs and aquaporin 0/MIP at the plasma membrane.
Periplakin has a wide tissue expression similar to that of plectin, distinguishing it from other plakin proteins such as envoplakin or BPAG-1, which have a more restricted tissue expression.40 Corresponding to its wide tissue expression, the short C-terminus was found to bind to many different proteins other than the IF, suggesting that periplakin may function in more roles than simply IF binding. The list of interactions includes structural proteins such as hemidesmosomal trans-membrane protein type XVII collagen/BPAG2/BP180,60 periphilin,61 and proteins that are involved in signaling such as G-protein receptor MOP-1,62 protein kinase B/c-Akt,63 and melanin-concentrating hormone receptor-1,64 modulating their signaling capacity. Thus, though we feel this evidence clearly implicates periplakin as an IFAP that contributes to the biology of vimentin IFs and BFs, it is not unreasonable to expect that the complexity of its role in lens biology will expand significantly before the story is complete. Specifically, others and we have established that the elimination of BF function leads to a degradation of optical quality and to light scatter.24,26,27 Thus, if periplakin plays a critical role in mediating BF function, one would expect that loss of periplakin would emulate the phenotype of the BF knockout. However, periplakin clearly serves more than one role in the lens. We establish here that it contributes to vimentin biology and show that it plays a role in the cytoplasm and at the membrane. Thus, it is likely that a loss of periplakin function will present a compound phenotype with a loss or perturbation of multiple lens functions.
Acknowledgments
Supported by National Eye Institute Grants EY08747 (PGF) and P30 EY12576. This work was conducted in a facility constructed with support from Research Facilities Improvement Program Grant Number C06 RR-12088-01 from the National Center for Research Resources, National Institutes of Health.
Footnotes
Disclosure: K. Yoon, None; P.G. FitzGerald, None
References
- 1.Bassnett S. The fate of the Golgi apparatus and the endoplasmic reticulum during lens fiber cell differentiation. Invest Ophthalmol Vis Sci. 1995;36:1793–1803. [PubMed] [Google Scholar]
- 2.Bassnett S. Fiber cell denucleation in the primate lens. Invest Ophthalmol Vis Sci. 1997;38:1678–1687. [PubMed] [Google Scholar]
- 3.Bassnett S. Lens organelle degradation. Exp Eye Res. 2002;74:1–6. doi: 10.1006/exer.2001.1111. [DOI] [PubMed] [Google Scholar]
- 4.Bassnett S, Beebe DC. Coincident loss of mitochondria and nuclei during lens fiber cell differentiation. Dev Dyn. 1992;194:85–93. doi: 10.1002/aja.1001940202. [DOI] [PubMed] [Google Scholar]
- 5.Kuwabara T. The maturation of the lens cell: a morphologic study. Exp Eye Res. 1975;20:427–443. doi: 10.1016/0014-4835(75)90085-8. [DOI] [PubMed] [Google Scholar]
- 6.Kuwabara T, Imaizumi M. Denucleation process of the lens. Invest Ophthalmol. 1974;13:973–981. [PubMed] [Google Scholar]
- 7.Fuchs E, Green H. The expression of keratin genes in epidermis and cultured epidermal cells. Cell. 1978;15:887–897. doi: 10.1016/0092-8674(78)90273-8. [DOI] [PubMed] [Google Scholar]
- 8.Fuchs E, Green H. Changes in keratin gene expression during terminal differentiation of the keratinocyte. Cell. 1980;19:1033–1042. doi: 10.1016/0092-8674(80)90094-x. [DOI] [PubMed] [Google Scholar]
- 9.Fuchs EV, Coppock SM, Green H, Cleveland DW. Two distinct classes of keratin genes and their evolutionary significance. Cell. 1981;27:75–84. doi: 10.1016/0092-8674(81)90362-7. [DOI] [PubMed] [Google Scholar]
- 10.Ellis M, Alousi S, Lawniczak J, Maisel H, Welsh M. Studies on lens vimentin. Exp Eye Res. 1984;38:195–202. doi: 10.1016/0014-4835(84)90103-9. [DOI] [PubMed] [Google Scholar]
- 11.Bagchi M, Caporale MJ, Wechter RS, Maisel H. Vimentin synthesis by ocular lens cells. Exp Eye Res. 1985;40:385–392. doi: 10.1016/0014-4835(85)90151-4. [DOI] [PubMed] [Google Scholar]
- 12.Bloemendal H, Benedetti EL, Ramaekers F, Dunia I. The lens cytoskeleton: intermediate-sized filaments, their biosynthesis and association with plasma membranes. Mol Biol Rep. 1981;7:167–168. doi: 10.1007/BF00778749. [DOI] [PubMed] [Google Scholar]
- 13.Quax WJ, Dodemont HJ, Lenstra JA, et al. Genes coding for vimentin and actin in mammals and birds. Adv Exp Med Biol. 1982;158:349–357. doi: 10.1007/978-1-4899-5292-9_36. [DOI] [PubMed] [Google Scholar]
- 14.Bloemendal H, Willemsen M, Groenewoud G, Oomen P. Isolation of the intermediate filament protein vimentin by chromatofocusing. FEBS Lett. 1985;180:181–184. doi: 10.1016/0014-5793(85)81067-x. [DOI] [PubMed] [Google Scholar]
- 15.Ireland M, Maisel H. A cytoskeletal protein unique to lens fiber cell differentiation. Exp Eye Res. 1984;38:637–645. doi: 10.1016/0014-4835(84)90182-9. [DOI] [PubMed] [Google Scholar]
- 16.FitzGerald PG. Methods for the circumvention of problems associated with the study of the ocular lens plasma membrane-cytoskeleton complex. Curr Eye Res. 1990;9:1083–1097. doi: 10.3109/02713689008997582. [DOI] [PubMed] [Google Scholar]
- 17.Gounari F, Merdes A, Quinlan R, et al. Bovine filensin possesses primary and secondary structure similarity to intermediate filament proteins. J Cell Biol. 1993;121:847–853. doi: 10.1083/jcb.121.4.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hess JF, Casselman JT, FitzGerald PG. cDNA analysis of the 49 kDa lens fiber cell cytoskeletal protein: a new, lens-specific member of the intermediate filament family? Curr Eye Res. 1993;12:77–88. doi: 10.3109/02713689308999499. [DOI] [PubMed] [Google Scholar]
- 19.Hess JF, Casselman JT, FitzGerald PG. Gene structure and cDNA sequence identify the beaded filament protein CP49 as a highly divergent type I intermediate filament protein. J Biol Chem. 1996;271:6729–6735. doi: 10.1074/jbc.271.12.6729. [DOI] [PubMed] [Google Scholar]
- 20.Hess JF, Casselman JT, Kong AP, FitzGerald PG. Primary sequence, secondary structure, gene structure, and assembly properties suggests that the lens-specific cytoskeletal protein filensin represents a novel class of intermediate filament protein. Exp Eye Res. 1998;66:625–644. doi: 10.1006/exer.1998.0478. [DOI] [PubMed] [Google Scholar]
- 21.Remington SG. Chicken filensin: a lens fiber cell protein that exhibits sequence similarity to intermediate filament proteins. J Cell Sci. 1993;105:1057–1068. doi: 10.1242/jcs.105.4.1057. [DOI] [PubMed] [Google Scholar]
- 22.Oshima RG. Intermediate filaments: a historical perspective. Exp Cell Res. 2007;313:1981–1994. doi: 10.1016/j.yexcr.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.FitzGerald PG, Gottlieb W. The Mr 115 kd fiber cell-specific protein is a component of the lens cytoskeleton. Curr Eye Res. 1989;8:801–811. doi: 10.3109/02713688909000870. [DOI] [PubMed] [Google Scholar]
- 24.Alizadeh A, Clark J, Seeberger T, Hess J, Blankenship T, FitzGerald PG. Targeted deletion of the lens fiber cell-specific intermediate filament protein filensin. Invest Ophthalmol Vis Sci. 2003;44:5252–5258. doi: 10.1167/iovs.03-0224. [DOI] [PubMed] [Google Scholar]
- 25.Alizadeh A, Clark J, Seeberger T, Hess J, Blankenship T, FitzGerald PG. Characterization of a mutation in the lens-specific CP49 in the 129 strain of mouse. Invest Ophthalmol Vis Sci. 2004;45:884–891. doi: 10.1167/iovs.03-0677. [DOI] [PubMed] [Google Scholar]
- 26.Alizadeh A, Clark JI, Seeberger T, et al. Targeted genomic deletion of the lens-specific intermediate filament protein CP49. Invest Ophthalmol Vis Sci. 2002;43:3722–3727. [PubMed] [Google Scholar]
- 27.Sandilands A, Prescott AR, Wegener A, et al. Knockout of the intermediate filament protein CP49 destabilises the lens fibre cell cytoskeleton and decreases lens optical quality, but does not induce cataract. Exp Eye Res. 2003;76:385–391. doi: 10.1016/s0014-4835(02)00330-5. [DOI] [PubMed] [Google Scholar]
- 28.Yoon KH, Blankenship T, Shibata B, Fitzgerald PG. Resisting the effects of aging: a function for the fiber cell beaded filament. Invest Ophthalmol Vis Sci. 2008;49:1030–1036. doi: 10.1167/iovs.07-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pekny M, Lane EB. Intermediate filaments and stress. Exp Cell Res. 2007;313:2244–2254. doi: 10.1016/j.yexcr.2007.04.023. [DOI] [PubMed] [Google Scholar]
- 30.Lai Cheong JE, Wessagowit V, McGrath JA. Molecular abnormalities of the desmosomal protein desmoplakin in human disease. Clin Exp Dermatol. 2005;30:261–266. doi: 10.1111/j.1365-2230.2005.01736.x. [DOI] [PubMed] [Google Scholar]
- 31.Li Z, Mericskay M, Agbulut O, et al. Desmin is essential for the tensile strength and integrity of myofibrils but not for myogenic commitment, differentiation, and fusion of skeletal muscle. J Cell Biol. 1997;139:129–144. doi: 10.1083/jcb.139.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLean WH, Pulkkinen L, Smith FJ, et al. Loss of plectin causes epidermolysis bullosa with muscular dystrophy: cDNA cloning and genomic organization. Genes Dev. 1996;10:1724–1735. doi: 10.1101/gad.10.14.1724. [DOI] [PubMed] [Google Scholar]
- 33.Schroder R, Vrabie A, Goebel HH. Primary desminopathies. J Cell Mol Med. 2007;11:416–426. doi: 10.1111/j.1582-4934.2007.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whittock NV, Smith FJ, Wan H, et al. Frameshift mutation in the V2 domain of human keratin 1 results in striate palmoplantar keratoderma. J Invest Dermatol. 2002;118:838–844. doi: 10.1046/j.1523-1747.2002.01750.x. [DOI] [PubMed] [Google Scholar]
- 35.Green KJ, Parry DA, Steinert PM, et al. Structure of the human desmoplakins: implications for function in the desmosomal plaque. J Biol Chem. 1990;265:11406–11407. [PubMed] [Google Scholar]
- 36.Green KJ, Stappenbeck TS, Parry DA, Virata ML. Structure of desmoplakin and its association with intermediate filaments. J Dermatol. 1992;19:765–769. doi: 10.1111/j.1346-8138.1992.tb03777.x. [DOI] [PubMed] [Google Scholar]
- 37.Leung CL, Green KJ, Liem RK. Plakins: a family of versatile cytolinker proteins. Trends Cell Biol. 2002;12:37–45. doi: 10.1016/s0962-8924(01)02180-8. [DOI] [PubMed] [Google Scholar]
- 38.Sonnenberg A, Liem RK. Plakins in development and disease. Exp Cell Res. 2007;313:2189–2203. doi: 10.1016/j.yexcr.2007.03.039. [DOI] [PubMed] [Google Scholar]
- 39.Jefferson JJ, Leung CL, Liem RK. Plakins: goliaths that link cell junctions and the cytoskeleton. Nat Rev Mol Cell Biol. 2004;5:542–553. doi: 10.1038/nrm1425. [DOI] [PubMed] [Google Scholar]
- 40.Kazerounian S, Uitto J, Aho S. Unique role for the periplakin tail in intermediate filament association: specific binding to keratin 8 and vimentin. Exp Dermatol. 2002;11:428–438. doi: 10.1034/j.1600-0625.2002.110506.x. [DOI] [PubMed] [Google Scholar]
- 41.Kouklis PD, Hutton E, Fuchs E. Making a connection: direct binding between keratin intermediate filaments and desmosomal proteins. J Cell Biol. 1994;127:1049–1060. doi: 10.1083/jcb.127.4.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith EA, Fuchs E. Defining the interactions between intermediate filaments and desmosomes. J Cell Biol. 1998;141:1229–1241. doi: 10.1083/jcb.141.5.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stappenbeck TS, Green KJ. The desmoplakin carboxyl terminus coaligns with and specifically disrupts intermediate filament networks when expressed in cultured cells. J Cell Biol. 1992;116:1197–1209. doi: 10.1083/jcb.116.5.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Svitkina TM, Verkhovsky AB, Borisy GG. Plectin sidearms mediate interaction of intermediate filaments with microtubules and other components of the cytoskeleton. J Cell Biol. 1996;135:991–1007. doi: 10.1083/jcb.135.4.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Y, Dowling J, Yu QC, Kouklis P, Cleveland DW, Fuchs E. An essential cytoskeletal linker protein connecting actin microfilaments to intermediate filaments. Cell. 1996;86:655–665. doi: 10.1016/s0092-8674(00)80138-5. [DOI] [PubMed] [Google Scholar]
- 46.Straub BK, Boda J, Kuhn C, et al. A novel cell-cell junction system: the cortex adherens mosaic of lens fiber cells. J Cell Sci. 2003;116:4985–4995. doi: 10.1242/jcs.00815. [DOI] [PubMed] [Google Scholar]
- 47.Blankenship TN, Hess JF, FitzGerald PG. Development- and differentiation-dependent reorganization of intermediate filaments in fiber cells. Invest Ophthalmol Vis Sci. 2001;42:735–742. [PubMed] [Google Scholar]
- 48.FitzGerald PG. Immunochemical characterization of a Mr 115 lens fiber cell-specific extrinsic membrane protein. Curr Eye Res. 1988;7:1243–1253. doi: 10.3109/02713688809033228. [DOI] [PubMed] [Google Scholar]
- 49.Walker JL, Menko AS. α6 Integrin is regulated with lens cell differentiation by linkage to the cytoskeleton and isoform switching. Dev Biol. 1999;210:497–511. doi: 10.1006/dbio.1999.9277. [DOI] [PubMed] [Google Scholar]
- 50.FitzGerald PG. Age-related changes in a fiber cell-specific extrinsic membrane protein. Curr Eye Res. 1988;7:1255–1262. doi: 10.3109/02713688809033229. [DOI] [PubMed] [Google Scholar]
- 51.FitzGerald PG, Casselman J. Immunologic conservation of the fiber cell beaded filament. Curr Eye Res. 1991;10:471–478. doi: 10.3109/02713689109001754. [DOI] [PubMed] [Google Scholar]
- 52.Wyatt K, Gao C, Tsai JY, Fariss RN, Ray S, Wistow G. A role for lengsin, a recruited enzyme, in terminal differentiation in the vertebrate lens. J Biol Chem. 2008;283:6607–6615. doi: 10.1074/jbc.M709144200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karashima T, Watt FM. Interaction of periplakin and envoplakin with intermediate filaments. J Cell Sci. 2002;115:5027–5037. doi: 10.1242/jcs.00191. [DOI] [PubMed] [Google Scholar]
- 54.Groot KR, Sevilla LM, Nishi K, DiColandrea T, Watt FM. Kazrin, a novel periplakin-interacting protein associated with desmosomes and the keratinocyte plasma membrane. J Cell Biol. 2004;166:653–659. doi: 10.1083/jcb.200312123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boczonadi V, McInroy L, Maatta A. Cytolinker cross-talk: periplakin N-terminus interacts with plectin to regulate keratin organisation and epithelial migration. Exp Cell Res. 2007;313:3579–3591. doi: 10.1016/j.yexcr.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 56.DiColandrea T, Karashima T, Maatta A, Watt FM. Subcellular distribution of envoplakin and periplakin: insights into their role as precursors of the epidermal cornified envelope. J Cell Biol. 2000;151:573–586. doi: 10.1083/jcb.151.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Long HA, Boczonadi V, McInroy L, Goldberg M, Maatta A. Periplakin-dependent re-organisation of keratin cytoskeleton and loss of collective migration in keratin-8-downregulated epithelial sheets. J Cell Sci. 2006;119:5147–5159. doi: 10.1242/jcs.03304. [DOI] [PubMed] [Google Scholar]
- 58.Leonard M, Chan Y, Menko AS. Identification of a novel intermediate filament-linked N-cadherin/gamma-catenin complex involved in the establishment of the cytoarchitecture of differentiated lens fiber cells. Dev Biol. 2008;319:298–308. doi: 10.1016/j.ydbio.2008.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lindsey Rose KM, Gourdie RG, Prescott AR, Quinlan RA, Crouch RK, Schey KL. The C-terminus of lens aquaporin 0 interacts with the cytoskeletal proteins filensin and CP49. Invest Ophthalmol Vis Sci. 2006;47:1562–1570. doi: 10.1167/iovs.05-1313. [DOI] [PubMed] [Google Scholar]
- 60.Aho S, McLean WH, Li K, Uitto J. cDNA cloning, mRNA expression, and chromosomal mapping of human and mouse periplakin genes. Genomics. 1998;48:242–247. doi: 10.1006/geno.1997.5188. [DOI] [PubMed] [Google Scholar]
- 61.Kazerounian S, Aho S. Characterization of periphilin, a widespread, highly insoluble nuclear protein and potential constituent of the keratinocyte cornified envelope. J Biol Chem. 2003;278:36707–36717. doi: 10.1074/jbc.M303896200. [DOI] [PubMed] [Google Scholar]
- 62.Feng GJ, Kellett E, Scorer CA, Wilde J, White JH, Milligan G. Selective interactions between helix VIII of the human mu-opioid receptors and the C-terminus of periplakin disrupt G protein activation. J Biol Chem. 2003;278:33400–33407. doi: 10.1074/jbc.M305866200. [DOI] [PubMed] [Google Scholar]
- 63.van den Heuvel AP, de Vries-Smits AM, van Weeren PC, et al. Binding of protein kinase B to the plakin family member periplakin. J Cell Sci. 2002;115:3957–3966. doi: 10.1242/jcs.00069. [DOI] [PubMed] [Google Scholar]
- 64.Murdoch H, Feng GJ, Bachner D, et al. Periplakin interferes with G protein activation by the melanin-concentrating hormone receptor-1 by binding to the proximal segment of the receptor C-terminal tail. J Biol Chem. 2005;280:8208–8220. doi: 10.1074/jbc.M405215200. [DOI] [PubMed] [Google Scholar]