SUMMARY
We recently identified a novel class of pimelic diphenylamide histone deacetylase (HDAC) inhibitors that show promise as therapeutics in the neurodegenerative diseases Friedreich’s ataxia (FRDA) and Huntington’s disease. Here we describe chemical approaches to identify the HDAC enzyme target of these inhibitors. Incubation of a trifunctional activity-based probe with a panel of class I and class II recombinant HDAC enzymes, followed by click chemistry addition of a fluorescent dye and gel electrophoresis, identifies HDAC3 as a unique high-affinity target of the probe. Photoaffinity labeling in a nuclear extract prepared from human lymphoblasts with the trifunctional probe, followed by biotin addition through click chemistry, streptavidin enrichment and western blotting also identifies HDAC3 as the preferred cellular target of the inhibitor. Additional inhibitors with different HDAC specificity profiles were synthesized and results from transcription experiments in FRDA cells point to a unique role for HDAC3 in gene silencing in Friedreich’s ataxia.
INTRODUCTION
Histone deacetylase (HDAC) inhibitors have received considerable attention as potential therapeutics for cancer (Marks and Breslow, 2007) and for a variety of neurological and neurodegenerative diseases (Kazantsev and Thompson, 2008). In this latter context, we recently described a series of pimelic diphenylamide HDAC inhibitors that reverse heterochromatin-mediated silencing of the frataxin (FXN) gene in the neurodegenerative disease Friedreich’s ataxia (FRDA) (Herman et al., 2006) and also show efficacy in a mouse model for Huntington’s disease (HD) (Thomas et al., 2008). FRDA is caused by the expansion of the simple triplet repeat DNA sequence GAA·TTC within intron 1 of the FXN gene, encoding the essential mitochondrial protein frataxin. Repeats over a threshold level of ~70 induce heterochromatin formation (Herman et al., 2006), and concomitant gene silencing, resulting in decreased amounts of frataxin protein. Importantly for therapeutic development, the pimelic diphenylamides cross the blood brain barrier, cause global increases in histone acetylation in cells and in the mouse brain, and show good tolerance in murine models of disease (Rai et al., 2008; Thomas et al., 2008). These molecules also directly affect the histone acetylation status of FXN gene chromatin in FRDA patient cells and in the mouse brain, increasing acetylation at particular lysine residues on histones H3 and H4, and increase FXN gene expression in the brain and heart in a mouse model for FRDA (Rai et al., 2008). Strikingly, gene expression microarray analysis indicates that most of the differentially expressed genes in FRDA mice revert towards wild-type levels on treatment with the pimelic diphenylamide HDAC inhibitor (Rai et al., 2008). Similar results have been obtained in a mouse model for HD, where one of these compounds ameliorated the disease phenotype and reversed many of the transcriptional abnormalities found in the brain of R6/2 HD mice (Thomas et al., 2008).
While the pimelic diphenylamides show considerable promise for clinical development, we unexpectedly found that only compounds related to the commercial product BML-210 are effective activators of the FXN gene in FRDA cells, and none of the common HDAC inhibitors, such as valproic acid, trichostatin A (TSA) and suberoylanilide hydroxamic acid (SAHA), have a positive effect on FXN mRNA levels (at their reported IC50 concentrations). This was surprising since many of these common HDAC inhibitors are more potent inhibitors than BML-210 and our derivatives when assayed in vitro or in cell culture. These findings suggest either a unique cellular target for activation of FXN gene expression that is not inhibited in the context of cellular chromatin by the potent HDAC inhibitors, or some unusual mode of action of the pimelic diphenylamides compared to the hydroxamic acids SAHA and TSA. In a recent study, we reported that one of our pimelic diphenylamides (compound 106, Figure 1) is specific for class I histone deacetylases (comprising HDACs 1, 2, 3 and 8), with no apparent inhibitory activity against class II enzymes (Chou et al., 2008). We found that 106 exhibits a Ki of 14 nM for HDAC3, compared to ~10- to 15-fold higher Kis for HDAC1 and HDAC2, and is only weakly active against HDAC8. Inhibition of HDAC3 with 106 is through a slow, tight-binding mechanism, and cells treated with this compound show prolonged histone acetylation and frataxin protein expression, even after removal of the inhibitor (Chou et al., 2008). These properties contrast with a rapid-on/rapid-off inhibition mechanism observed for the hydroxamates SAHA and TSA, both in vitro and in cell culture. We now report the identification of HDAC3 as the preferred cellular target of the pimelic diphenylamide inhibitor 106. A derivative of 106 that specifically targets HDAC1, and inhibitors of class II and III HDACs, fail to increase FXN gene expression in FRDA lymphocytes, pointing to a unique role of HDAC3 in FXN gene silencing in FRDA, and suggest that this enzyme is a valid therapeutic target in this disease.
Figure 1.
Structures of HDAC inhibitors and activity-profiling probes: 106; the trifunctional probe 1-BP and its control derivative 2-BP, lacking a 2-amino group; the HDAC1/2-specific inhibitor 3 and the activity-profiling probe 3-BP; and, the class II HDAC inhibitor 4.
RESULTS
Activity Profiling Approach for Target Identification
To provide evidence as to the cellular target(s) of our inhibitors, and the possible role of these targets in gene silencing in FRDA, we synthesized an activity-profiling probe for proteomic studies (Evans and Cravatt, 2006; Hagenstein et al., 2003; Hagenstein and Sewald, 2006). This approach has recently been employed for the identification of the HDAC targets of SAHA in cancer cells (Salisbury and Cravatt, 2007; Salisbury and Cravatt, 2008), and HDACs 1 and 2 were identified, as would be expected from previous studies on this compound (Marks et al., 2001). Our trifunctional probe (1-BP, Figure 1) consists of a benzophenone photolabeling group, which is attached through a flexible ethylene glycol linker to HDAC inhibitor 106 (Rai et al., 2008), and an alkyne for subsequent attachment of an azide-linked reporter dye or biotin for affinity capture. Prior to using 1-BP for target identification, it was important to demonstrate that this compound retained HDAC inhibitory activity, and hence was capable of binding HDAC enzymes. 1-BP had a lower IC50 value for HDAC inhibition using a HeLa cell nuclear extract as a source of HDACs than did 106 (IC50 for 106 = 1.3 μM; IC50 for 1-BP = 0.3 μM; Supplemental Figure 1a and Supplemental Table 1), while comparable IC50 values were obtained with recombinant HDAC3/NcoR2 (IC50 for 106 = 0.79 μM; IC50 for 1-BP = 0.86 μM; Supplemental Figure 1c and Supplemental Table 1). In the case of recombinant HDAC1, a small loss in activity was noted for 1-BP compared to 106 (IC50 for 106 = 0.24 μM; IC50 for 1-BP = 0.57 μM; Supplemental Figure 1b and Supplemental Table 1); however, 1-BP still inhibited this enzyme at concentrations required for target identification.
Having established that the probe retains HDAC inhibitory activity, we next used 1-BP for target identification. We incubated 1-BP with identical amounts of each of the recombinant class I HDAC enzymes (HDACs 1, 2, 3 and 8) and with two representative class II enzymes (HDACs 4 and 5). Note that each of these enzymes is catalytically active ((Chou et al., 2008) and data not shown). After irradiation to effect photocrosslinking, a fluorescent dye (rhodamine)-azide was attached to the probe by solution-phase Cu(I)-catalyzed click chemistry (Salisbury and Cravatt, 2007). After SDS-PAGE, the gels were exposed to the excitation wavelength of rhodamine, and fluorescent images are presented in Figure 2a. These experiments clearly show that, at least among these six enzymes, 1-BP exhibits a clear preference for HDAC3. The probe is crosslinked to both HDAC3 (49 kDa) and its co-factor NcoR2 (~40 kDa fragment (Yang et al., 2002)). Low levels of crosslinking to recombinant HDAC1 are observed at higher input concentrations of enzyme (not shown). To establish the specificity of the 1-BP/HDAC3 crosslinking reaction, we performed a competition experiment with the parent compound 106 (Figure 2b), where the reactivity of 1-BP with HDAC3 is strongly competed by pre-incubation with an equal concentration 106 prior to the addition of 1-BP. The intensity of the band corresponding to NcoR2 was also diminished by pre-incubation with 106, but not as significantly as HDAC3, suggesting that some reactivity with NcoR2 may be non-specific.
Figure 2.
Photoaffinity labeling of recombinant HDAC enzymes. (a) 5 μg of each of the indicated recombinant HDAC enzymes were incubated with 1-BP (at 4 μM) for 5 min, followed by UV irradiation for 1 h, and rhodamine azide was added by click chemistry. A fluorescent image of an SDS-PAGE is shown and fluorescent dye markers are shown at the left of the gel. Note that HDAC3 consists of the enzyme plus its required cofactor NcoR2, which is a recombinant fragment that is also crosslinked by 1-BP. Minor bands at ~80 kDa and above represent multimers of HDAC3/NcoR2. Reactions for the class I HDACs 1, 2, 3 and 8 were analyzed on a separate gel from the reactions with the class II HDACs 4 and 5. (b) Competition with 106. HDAC3/NcoR2 was incubated with or without 106 at 10 μM, for 2 h at RT, prior to the addition of 1-BP (10 μM), followed by photocrosslinking and click chemistry as in a. (c) Determination of the half-life of the 1-BP/HDAC3 complex. 1-BP and recombinant HDAC3/NcoR2 were pre-incubated for 2 h prior to the addition of a 20-fold molar excess of 106, and samples were withdrawn at the indicated times, UV crosslinked and a rhodamine-azide was added by click chemistry. The inset shows a fluorescence image of an SDS-PAGE analysis of these samples, and the graph is a plot of the natural log of the fraction 1-BP/HDAC3 remaining at each time point, relative to the zero time point, versus time. ImageQuant software was used to quantify the data, which were normalized for HDAC3 protein concentration in each sample (determined by western blotting, not shown). A least-squares fit of the data (solid line, R2 = 0.934) yields a t1/2 of ~4 h.
We previously reported that 106 exhibits a slow-on/slow-off inhibition mechanism with HDAC3, likely involving a conformational change in the enzyme on binding the inhibitor, and kinetic studies suggested that the half-life of the 106-HDAC3 complex was on the order of ~6 h (Chou et al., 2008). To provide a physical estimate of this half-life for the photoaffinity version of 106, we performed a competition experiment where we first incubated recombinant HDAC3/NcoR2 with 1-BP for 2 h (in the absence of UV irradiation) prior to the addition of a 20-fold molar excess of 106. Samples were then taken at various times, subjected to UV crosslinking and click chemistry addition of the rhodamine-azide, as described above. We expect that upon dissociation of 1-BP from HDAC3, the excess of 106 will largely prevent re-association of 1-BP with the enzyme, and the rate of disappearance of the fluorescent signal can be used to determine the half-life of the 1-BP/HDAC3 complex. Figure 2c shows that the intensity of the fluorescent band corresponding to the 1-BP-HDAC3 complex disappears slowly, and quantification of these data suggest a half-life of ~4 h for this complex, in reasonable agreement with our previous kinetic estimate for the half-life of the 106-HDAC3 complex (Chou et al., 2008).
Cellular Target of the Activity Probe
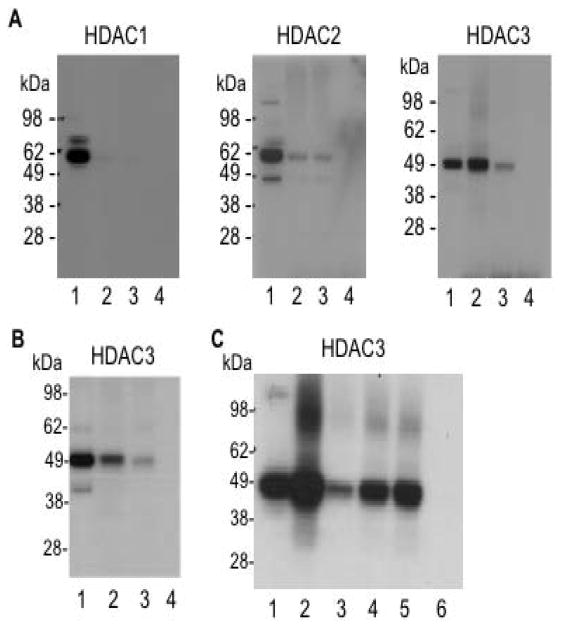
We next sought to identify targets of 1-BP in nuclear extracts. We prepared a nuclear extract from an Epstein Barr virus transformed lymphoid cell line derived from an FRDA patient (line GM15850, alleles with 650 and 1030 GAA·TTC repeats in FXN, from the NIGMS Human Genetic Cell Repository, Coriell Institute, Camden, NJ). After incubation of 1-BP in the nuclear extract and photocrosslinking, a biotin tag was appended to the probe through Cu(I)-mediated click chemistry (Salisbury and Cravatt, 2007). Streptavidin beads were then used for affinity capture of probe-labeled protein targets, and western blotting with antibodies to the class I HDAC enzymes HDAC1, 2 and 3, was used for target identification. Since the parent compound 106 is only weakly active against recombinant HDAC8 (see above), we did not pursue this enzyme further. Differences in band intensity for the input lanes (lane 1 for each blot) could reflect differences in either HDAC protein abundance or antibody avidity as equivalent amounts of nuclear extract protein were used for each of the blots shown in Figure 3a. In agreement with results for the recombinant HDACs (Figure 2), HDAC3 is indeed cross-linked by 1-BP and retained by the streptavidin beads using this approach. In contrast, the other class I HDACs 1 and 2 failed to react with the probe and were not retained by the streptavidin beads, again in accord with results for the recombinant HDACs. Quantification of these data reveal that as much as ~2 – 4% of the input HDAC3 protein is retained by the streptavidin beads in this experiment. Importantly, pre-incubation of the nuclear extract with the parent compound 106 significantly reduces the HDAC3 signal in this experiment (Figure 3a, lane 3), demonstrating the specificity of the reaction. Omission of the biotin-azide also abolished retention of HDAC3 on streptavidin beads (lane 4), showing that retention of HDAC3 on the streptavidin beads requires the addition of biotin, and is not due to non-specific background binding of HDAC3 to the beads. A low, but detectable level of crosslinking to HDAC2 was observed (Figure 3a), but this crosslinking was not competed with 106, suggesting that this might be non-specific binding of HDAC2 by the probe.
Figure 3.
(a) Photoaffinity crosslinking of proteins in a nuclear extract from FRDA lymphoblasts with 1-BP followed by addition of a biotin-azide by click chemistry, streptavidin binding, and western blotting with antibodies to the indicated HDACs. Lane 1, input (2% of the amount of total protein corresponding to lanes 2 – 4 used for affinity capture); lane 2, proteins retained on streptavidin beads; lane 3, same as lane 2 but with pre-incubation of a 20-fold excess of 106 prior to the addition of 1-BP to the extract; lane 4, no click chemistry control. (b) Photoaffinity crosslinking and capture with 1-BP or 2-BP (each at 4 μM) as above, and western blotting with antibody to HDAC3. Lane 1, input (2% of lanes 2 - 3); lane 2, proteins retained on streptavin beads after incubation with 1-BP; lane 3, proteins retained on streptavin beads after incubation with 2-BP, lacking a 2-amino group; lane 4, no click chemistry control. (c) Competition with 106, TSA and SAHA. Affinity capture with 1-BP (at 4 μM) as in panel a, and western blotting with antibody to HDAC3. Lane 1, input (2% of lanes 2 – 6); lane 2, proteins retained on streptavidin beads; lane 3, same as lane 2 but with pre-incubation with 106 (80 μM) for 1.5 h prior to the addition of 1-BP to the extract; lane 4, same as lane 3 but with pre-incubation with TSA (308 nM); lane 5, same as lane 3 but with pre-incubation with SAHA (3 μM); lane 6, no click chemistry control. In lanes 3 – 5, the amounts of competitor compounds correspond to 60-times the reported IC50 value for each inhibitor.
As a control, we synthesized a derivative of 1-BP lacking a 2-amino group in the HDAC inhibitor portion of the molecule, which should not chelate zinc in the HDAC catalytic pocket and hence should be far less active as an HDAC inhibitor (Butler and Kozikowski, 2008). Indeed, this compound, 2-BP (Figure 1), is at least 200-fold less active as an HDAC inhibitor compared to 1-BP (IC50 = 0.8 μM for 1-BP; IC50 > 180 μM for 2-BP, using the HeLa nuclear extract as a source of HDACs). The parent compound N1-phenyl-N7-phenylheptanediamide (termed 5b in (Herman et al., 2006)) is a poor HDAC inhibitor (IC50 ~200 μM) and also fails to activate FXN gene expression in the FRDA lymphoblast cell line (Herman et al., 2006). When these two photoaffinity reagents were compared for their ability to retain HDAC3 on the streptavidin beads, 1-BP was found to be significantly more active in this regard than 2-BP (Figure 3b).
As additional controls for the specificity of HDAC3 capture by 1-BP, we performed pre-incubation experiments with either TSA or SAHA (at 60 times their reported IC50 values) prior to the addition of 1-BP to the nuclear extract, and compared the ability of these compounds to compete with 1-BP, to a similar excess of 106 (Figure 3c, lanes 3 – 5). Recall that neither TSA nor SAHA is an activator of FXN transcription in cell culture (Herman et al., 2006) and both of these compounds exhibit fast-on/fast-off kinetics with class I HDACs while 106 has slow-on/slow-off kinetics with these enzymes (Chou et al., 2008). Clearly, 106 is far more effective in competing with 1-BP for HDAC3 than either TSA or SAHA. Taken together, our results suggest that HDAC3 is the primary cellular target of the pimelic diphenylamide HDAC inhibitors, exemplified by 106 and related compounds (Herman et al., 2006), and show that 106 forms a far more stable complex with this enzyme than the hydroxamates TSA and SAHA.
Role of Other Class I HDACs in FXN Gene Regulation
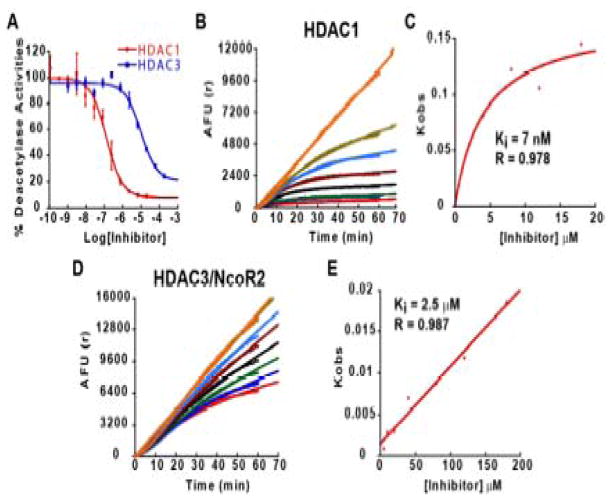
While the results described above and previous studies (Chou et al., 2008; Herman et al., 2006; Rai et al., 2008) strongly suggest that HDAC3 is involved in FXN gene silencing, this conclusion does not exclude a possible role for other HDACs in FXN gene silencing in FRDA. We therefore chose to design and synthesize an HDAC inhibitor that would have a different enzyme specificity profile than 106, and to test such a compound for effects on FXN transcription in FRDA cells. Recent studies identified compounds with potent inhibition of HDAC1 and 2, but with substantially reduced activity towards HDAC3 (Methot et al., 2008). While these compounds contained an aryl linker, we hypothesized that appending a phenyl group at the 5-position of the “right” hand ring of 106 would generate a compound with specificity for HDAC1/2 over HDAC3 (compound 3, N1-(4-aminobiphenyl-3-yl)-N7-phenylheptanediamide; Figure 1). IC50 and Ki measurements for 3 with recombinant class I HDACs were performed to determine the selectivity of such a pimelic acid derivative (Figure 4 and Supplemental Table 1). IC50 measurements for HDACs 1 and 3 with compound 3 are shown in Figure 4a, and enzyme progression curves in the presence of increasing concentrations of 3 are shown in Figures 4b and 4d, along with plots of Kobs versus inhibitor concentration (Figures 4c and 4e). HDACi 3 has a ~75-fold preference for HDAC1 over HDAC3 comparing IC50 values (Figure 4a) and a ~350-fold preference for HDAC1 over HDAC3 from Ki measurements (Figures 4c and 4e). HDACi 3 is also similarly active against recombinant HDAC2 (data not shown). Thus, HDACi 3 and 106 are both active inhibitors of HDAC1/2 (Chou et al., 2008), but addition of the 5-phenyl group has a pronounced deleterious effect on the activity of 3 against HDAC3/NcoR2. Similar to 106, the kinetic data for 3 and HDAC1 are best fit to a slow-on/slow-off inhibition mechanism (Figure 4c and (Chou et al., 2008)).
Figure 4.
IC50 and Ki determinations for 3 determined with recombinant HDAC1 or HDAC3/NcoR2. (a) IC50 determinations were performed as described (Chou et al., 2008) with a 1 h pre-incubation of HDAC1 or HDAC3/NcoR2 and inhibitor prior to adding substrate. Enzyme progression curves for HDAC1 (b) or HDAC3/NcoR2 (d) in the presence of increasing concentrations of 3. In panel b, the curves, starting at the top, represent the following final concentrations of inhibitor: no inhibitor, 0.5 μM, 1 μM, 2 μM, 4 μM, 8 μM, 12 μM, and 18 μM. In panel d, the curves represent the following final concentrations of inhibitor: no inhibitor, 5 μM, 10 μM, 20 μM, 40 μM, 80 μM, 120 μM, and 180 μM. Plots of Kobs versus inhibitor concentration for HDAC1 (c) and HDAC3/NcoR2 (e), as in (Chou et al., 2008). For HDAC1, the data are best fit to a slow-on/slow-off inhibition mechanism involving a stable intermediate, while for HDAC3/NcoR2 a simple slow-on/slow-off mechanism provides the best fit to the data. Ki and R values (from the least-squares fit to the data) are shown in the figure, while IC50 values are given in Table 1 (Supplemental material).
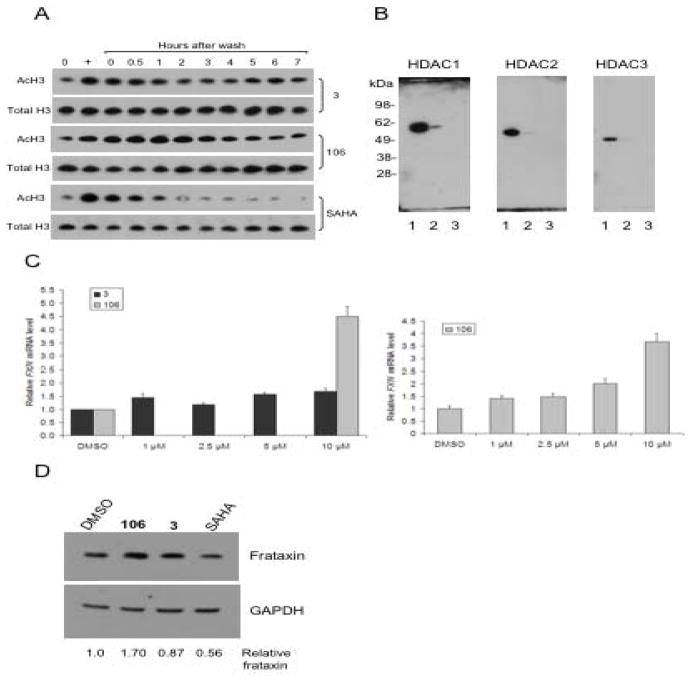
Since 106 and 3 are both slow-on/slow-off inhibitors, we asked whether HDACi 3 would cause prolonged acetylation of endogenous histones in the FRDA lymphoblast cell line, as we have previously documented for 106 (Chou et al., 2008). Cells were treated with 3 or 106, each at 10 μM, or with SAHA at 2 μM, for 24 h in separate cultures, and the cells were then washed and resuspended in fresh media lacking inhibitors. Aliquots of cells were taken prior to washing and at time points ranging from 0 to 7 h after removal of the inhibitor. Levels of total histone H3 and acetylated histone H3 were monitored by western blotting (Figure 5a). Hyperacetylation of histone H3 was clearly seen with each inhibitor in the cell cultures where inhibitors were present, and at the zero time points, compared to the no inhibitor control cultures (lanes marked “0”). Hyperacetylation of histone H3 due to the HDAC inhibitors 3 and 106 decreased only slightly after inhibitor removal, and did not even fully return to basal levels 6 – 7 h after the removal of the inhibitor. In contrast, histone H3 hyperacetylation due to SAHA disappeared rapidly after washing the cells free of inhibitor. The level of acetylation returned to no-inhibitor levels within 2 h of removing SAHA from the medium, in agreement with our previous study (Chou et al., 2008). Thus, both 3 and 106 cause prolonged histone acetylation in FRDA cells. We also performed a titration experiment varying the exposure of FRDA lymphoblasts to increasing concentrations of 3 or 106 and determined the levels of histone acetylation by western blotting. These two compounds were found to be comparably active HDAC inhibitors in FRDA cells (Supplemental Figure 2). To insure that compound 3 is not an inhibitor of class II HDACs, we monitored tubulin acetylation after incubation of the FRDA lymphoblasts. Tubulin acetylation is highly indicative of inhibition of class II HDAC6 in cells (Wong et al., 2003). Neither 106 nor 3 caused tubulin acetylation, while a potent class II HDAC inhibitor (compound 4, see below) was highly active in inducing tubulin acetylation (Supplementary Figure 3).
Figure 5.
Effects of HDAC inhibitor 3 in FRDA cells. (a) Histone acetylation in FRDA cells. FRDA lymphoblasts were either untreated (DMSO vehicle control, marked 0 at top) or treated with HDACi 3 (top panels) or 106 (middle panels) at 10 μM, or with SAHA at 2 μM for 24 h (lane marked with “+” at top), washed to remove the inhibitors, and the cells were suspended in fresh medium lacking inhibitors. Aliquots of cells were harvested at the indicated times (lanes marked 0 – 7 h), protein extracts prepared and subjected to western blotting with antibody to unacetylated histone H3 as a loading control (indicated Total H3) or antibody to acetylated histone H3 (K9 + K14; indicated Ac-H3) for each of the inhibitors. (b) Photoaffinity crosslinking of proteins in a nuclear extract from FRDA lymphoblasts with 3-BP followed by addition of a biotin-azide by click chemistry, streptavidin binding, and western blotting with antibodies to the indicated HDACs. Lane 1, input (4% of the amount of total protein corresponding to lanes 2 – 3 used for affinity capture); lane 2, proteins retained on streptavidin beads; lane 3, same as lane 2 but no click chemistry control (omission of the Cu(I) regent). (c) Effects of HDACi 3 and 106 on FXN gene expression in primary lymphocytes from FRDA patients. Lymphocytes were isolated from donor blood from a FRDA patient and were incubated in culture media containing either 0.4% DMSO, as a control, or 106 or the 5-phenyl compound 3, each at the indicated concentrations in 0.4% DMSO, for 48 h prior to determination of mRNA levels by qRT-PCR, using GAPDH mRNA as an internal control. The y-axis denotes FXN mRNA levels, normalized to GAPDH mRNA, relative to the DMSO controls, set to 1.0. Each determination was done in triplicate, and the SEM is shown. A separate dose response experiment for 106 is shown at the right. (d) Effects of HDACi 3, 106 and SAHA on frataxin protein expression in FRDA lymphoblasts. Cells were incubated with each inhibitor (at 10 μM for 106 or 3 and at 2 μM for SAHA, in culture media plus 0.1% DMSO or media plus DMSO as a control) for 48 h prior to analysis by western blotting for frataxin or GAPDH, as indicated. The x-ray films were scanned and quantified, and the relative levels of frataxin protein, normalized to GAPDH protein, are shown at the bottom of the figure.
We also prepared a photoaffinity version of HDACi 3 (3-BP) and used this compound to identify cellular HDAC targets. 3-BP was incubated in the FRDA lymphoblast nuclear extract, subjected to photocrosslinking, and a biotin tag was appended to the probe through Cu(I)-mediated click chemistry. Streptavidin beads were again used for affinity capture, and western blotting with antibodies to the class I HDAC enzymes HDAC1, 2 and 3, was used for target identification, with the result that HDAC1 was identified as the likely cellular target of this probe (Figure 5b). Much lower extents of recovery of HDACs 2 and 3 were found with this probe, compared to HDAC1. While the extent of recovery of HDAC1 with 3-BP is far less than that observed for HDAC3 and 1-BP in the same nuclear extract (Figure 3), the observed signal for HDAC1 is significant over background (compared to a no click chemistry control, lane 3) and reproducible in several experiments (not shown). Quantification of these experiments reveals that 9-times more HDAC1 than HDAC2, and 5-times more HDAC1 than HDAC3 are retained by 3-BP. We thus conclude that HDAC1 is the primary class I HDAC target of 3-BP.
Having established that HDACi 3 is a potent HDAC inhibitor in FRDA cells, we next asked whether this HDAC1-selective compound would increase FXN mRNA in primary FRDA lymphocytes. Lymphocytes from FRDA patients were incubated for 48 h with either 3 or 106, and FXN mRNA levels were quantified by qRT-PCR, using GAPDH mRNA as an internal control for each determination. Patient lymphocytes have FXN mRNA levels of ~10 – 30% of those in lymphocytes from unaffected individuals (Herman et al., 2006), and previous studies have shown that 106 increases FXN gene expression in patient lymphocytes (Rai et al., 2008). While 106 is highly active in up-regulation of FXN mRNA levels, 3 fails to reproducibly up-regulate FXN mRNA levels in these cells (Figure 5c). We have observed small levels of up-regulation of FXN mRNA in some experiments (up to ~1.7-fold); however, this degree of up-regulation is never as large as that observed for 106 (generally ~3 – 4-fold) and is not consistently observed (data not shown). To verify that HDACi 3 is indeed active in cells, we monitored p21WAF1 mRNA levels in HEK293 cells after treatment with SAHA or 3, and found that both compounds increased the levels of this mRNA (data not shown), as expected for a class I HDAC inhibitor (Gui et al., 2004). We also examined frataxin protein levels by western blotting after incubation of FRDA lymphoblasts with HDACi 3, 106 or SAHA, using GAPDH protein as a recovery standard (Figure 5d). Quantification of these data reveals that only 106 had a positive effect on frataxin protein levels, 3 was without significant effect and SAHA actually decreased frataxin levels in these cells. We thus conclude that among the class I HDACs 1, 2 and 3, increases in FXN gene expression and frataxin protein are only observed with an inhibitor of HDAC3.
Class II HDACs and Sirtuin 1 in FXN Gene Silencing
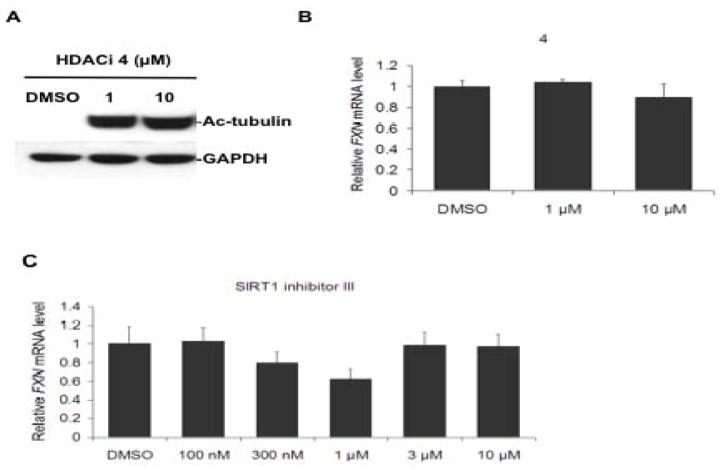
To assess any potential contribution of class II HDACs to FXN gene regulation in FRDA cells, we tested whether inhibitors of these enzymes would have a positive effect on FXN mRNA levels. Based on previous studies (Jones et al., 2008), we synthesized 4 (tert-butyl 4-(6,6,6-trifluoro-5-oxohexanamido)phenylcarbamate, Figure 1), which we expected would have a preference for class II HDACs over class I HDACs. IC50 determinations with recombinant enzymes representing class I (HDAC1) and class II (HDAC7) confirm this expectation: the IC50 of 4 with HDAC7 is 4 μM, while the IC50 of this compound with HDAC1 is 30 μM, showing a 7.5-fold preference for a class II HDAC over a class I HDAC (Supplemental Figure 4). Additionally, HDACi 4 is a potent inducer of tubulin acetylation in FRDA lymphoblast cells (at 1 μM concentration in the culture medium for 24 h; Figure 6a). FRDA lymphoblasts were incubated with this compound, but no positive effect was found on the levels of FXN mRNA after treatment with concentrations up to 10 μM for 24 h, suggesting that inhibition of class II HDACs has no effect on FXN gene expression (Figure 6b). Similar experiments with primary FRDA patient lymphocytes also failed to show up-regulation of FXN transcription with HDACi 4 at concentrations up to 40 μM (Supplemental Figure 5).
Figure 6.
A class II HDAC inhibitor and a Sirt1 inhibitor fail to activate FXN gene expression. (a) Class II HDAC inhibitor 4 causes tubulin acetylation in FRDA cells. FRDA lymphoblasts were incubated with the indicated concentrations of 4 or DMSO alone (at 0.1%) for 24 h in culture medium, prior to SDS-PAGE and western blotting with anti-ac-tubulin antibody, or antibody to GAPDH, as a loading control. (b) HDAC inhibitor 4 fails to activate FXN gene expression. FRDA lymphoblasts were incubated in culture media containing either 0.1% DMSO, as a control, or 4, at the indicated concentrations in 0.1% DMSO, for 24 h prior to determination of FXN mRNA levels by qRT-PCR, using GAPDH mRNA as an internal control. The y-axis denotes FXN mRNA levels, normalized to GAPDH mRNA, relative to the DMSO control, set to 100%. Each determination was done in triplicate, and the SEM is shown. (c) The Sirt1 inhibitor 6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1-carboxamide fails to up-regulate FXN gene expression in primary lymphocytes from a FRDA patient. Lymphocytes were incubated in culture media containing either 0.4% DMSO, as a control, or with the indicated concentrations of inhibitor in 0.4% DMSO, for 48 h prior to determination of mRNA levels by qRT-PCR, using GAPDH as an internal control, as in panel b.
We also tested the Sirt1-specific inhibitor 6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1-carboxamide (Napper et al., 2005) for effects on FXN transcription. This compound is a potent Sirt1-specific inhibitor with a reported IC50 of 100 nM, compared to IC50s of 20 and 50 μM for Sirt2 and Sirt3, respectively. At concentrations ranging up to 10 μM, the Sirt1 inhibitor had no effect on FXN mRNA levels in FRDA patient lymphocytes (Figure 6c) or in either the FRDA lymphoblast cell line or patient fibroblasts (not shown), although this compound has been shown to be cell permeable, and to function as a Sirt1 inhibitor in several cell lines (Solomon et al., 2006). Our results suggest that inhibition of Sirt1 does not positively affect FXN gene expression, at least in FRDA lymphocytes and fibroblasts.
DISCUSSION
Previous studies have shown that the pimelic diphenylamide 106 is a class I HDAC inhibitor with little or no activity against class II HDACs (IC50s >180 μM for class II enzymes) (Chou et al., 2008). Among the class I HDACs, 106 shows a ~10-fold preference for HDAC3 over HDACs 1 and 2 (comparing Ki values, Supplementary Table 1), with only weak inhibitory activity against HDAC8. Our present results pointing to HDAC3 as the target of 106 are in agreement with these inhibition experiments with recombinant enzymes. However, since 106 is a reasonable inhibitor of HDACs 1 and 2, it was somewhat surprising that our activity-profiling probe 1-BP failed to crosslink these enzymes to any appreciable extent, either as recombinant enzymes (Figure 2a) or in a cell-free extract (Figure 3a). Moreover, 1-BP is an effective inhibitor of HDAC1 in vitro (IC50 = 0.57 μM), demonstrating that this compound can indeed interact with HDAC1. We speculate that the remarkable stability of the 106-HDAC3/NcoR2 complex (Figure 2c) may account for this difference in crosslinking activity of 1-BP for these enzymes. This stability has also been demonstrated in dilution experiments and in cell-based histone acetylation assays (Figure 5a and (Chou et al., 2008)). Histone H3 acetylation in the FRDA lymphoblast cell line persists for many hours after removal of 106 from the culture medium. While SAHA is also capable of inducing histone H3 acetylation in cells, removal of SAHA from the culture medium caused a rapid loss in acetylated histones (Figure 5a) and SAHA fails to activate the FXN gene in these cells (Chou et al., 2008; Herman et al., 2006). Our previous kinetic study showed that SAHA is a rapid-on/rapid-off inhibitor of class I HDACs (Chou et al., 2008), and so we speculate that the stability of the 106-HDAC3 complex may account for the ability of this compound to act as a positive regulator of FXN gene expression. Similarly, our kinetic measurements indicated a difference in the inhibition mechanism of 106 for HDACs 1 and 3, respectively. While 106 inhibited both enzymes through slow-on/off mechanisms, our data suggest that the 106/HDAC3/NcoR2 complex forms a stable intermediate that is not observed for HDAC1. This difference may well explain why we fail to observe crosslinking of 1-BP to either recombinant HDAC1 or to HDAC1 in nuclear extracts. Structural studies of these inhibitor/HDAC enzyme complexes will be needed to elucidate the molecular basis for this difference in stability, and why the pimelic diphenylamides differ from hydroxamates in their inhibitory mechanisms for class I HDACs.
Although the experiments shown in Figure 3 were performed with a nuclear extract from human lymphoblasts, HDAC3 is known to be present in the brain (see Allen Brain Atlas: www.brain-map.org; and (Broide et al., 2007)), attesting to the potential relevance of HDAC3 in neurological disease. Other studies have clearly shown that the pimelic diphenylamides function as HDAC inhibitors in the mouse brain, causing increases in global histone acetylation in various brain regions, and reversing the gene expression changes associated with FRDA and HD (Rai et al., 2008; Thomas et al., 2008). We are currently exploring the ability of 1-BP to target HDAC3 in the mouse brain. Our results with the 5-phenyl derivative, HDACi 3, suggest that only targeting HDAC3 can lead to up-regulation of FXN mRNA levels in FRDA lymphoid cells. Similar to 106 and its mechanism of inhibition of HDAC3/NcoR2 (Chou et al., 2008), inhibition of HDAC1 by 3 is also due to a slow-on/slow-off mechanism, with a stable intermediate formed by the enzyme-inhibitor complex (Figure 4). While both 3 and 106 show prolonged histone H3 acetylation in cells (Figure 5a), only 106 and other HDAC3-specific derivatives (such as 4b and other derivatives (Herman et al., 2006)) are effective inducers of FXN gene expression (Figure 5c), resulting in increased levels of frataxin protein (Figure 5d). Microarray studies comparing the transcriptional profile of mouse embryonic stem cells and HDAC1 knockout stem cells also fail to show an increase in Fxn gene expression ((Zupkovitz et al., 2006) and see http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE5583), suggesting that HDAC1 is not involved in regulation of the wild-type Fxn gene. Similar knockout studies in FRDA cells or FRDA mouse models may provide additional evidence for the role of particular class I HDACs in FXN gene expression.
Although our current chemical approaches suggest that HDAC3 plays an important role in gene silencing in FRDA, our results do not distinguish between a direct or indirect role for this enzyme. We showed that the pimelic diphenylamides cause increases in histone acetylation at particular lysine residues at the FXN locus in both FRDA lymphoid cells (Herman et al., 2006) and in the brain of an FRDA mouse (Rai et al., 2008), clearly pointing to the action of a histone acetyltransferase at the FXN locus upon inhibition of an HDAC. Neither SAHA nor TSA had such a positive effect on FXN histone acetylation (Herman et al., 2006). Remarkably, the pattern of FXN histone H4 acetylation observed on treatment of FRDA cells with the related pimelic diphenylamide 4b (which differs from 106 by a single methyl group in the “left” phenyl ring) is similar to that observed after genetic knock down of HDAC3 (Hartman et al., 2005), where H4K5 acetylation predominates (with the order of acetylated residues: H4K5 > K8 > K12 > K16). This pattern of acetylation contrasts with the reported cellular effects of TSA, where H4K16 is the most affected lysine residue (H4K16 > K12 > K8 > K5) (Ren et al., 2005), and the general effects of SAHA at H3 and H4 lysine residues, at least on the p21WAF1 promoter (Gui et al., 2004). Taken together, these results are supportive of a role of HDAC3 in causing increased histone acetylation on the FXN gene. However, such acetylation events at the FXN locus could be the consequence of inhibition of another HDAC enzyme, through the indirect inhibition of HDAC3. To assess this possibility, we have used chromatin immunoprecipitation methods to examine occupancy of class I HDACs on FRDA FXN alleles, but have failed to obtain convincing evidence for such occupancy. This may be related to either a transient association of HDACs with the FXN gene, or to the indirect scenario suggested above. In another approach, we attempted to knock down class I HDACs in cells with various siRNA and shRNA approaches to see if such knock down would cause FXN gene activation. While we are able to reduce class I HDAC mRNA levels using these methods, no significant reduction in HDAC3 protein was observed in lymphoid cells, and no effect on FXN mRNA was observed. We are currently deriving a neuronal cell model for FRDA based on human induced puripotent stem (iPS) cells (Ebert et al., 2008), in the hope that these molecular biology approaches will be more amenable in neuronal cells. Notwithstanding these caveats, our current chemical data clearly indicate that inhibition of HDAC3 leads to FXN gene activation in FRDA lymphoid cells, and suggests that this enzyme is a valid therapeutic target for FRDA. It will be important to fully examine the mechanisms responsible for FXN gene silencing in neuronal models, such as iPS-derived cells.
SIGNIFICANCE
Numerous studies have pointed to HDAC inhibitors as potential therapeutics for various neurological and neurodegenerative diseases, and clinical trials with several HDAC inhibitors have been performed or are underway. However, the HDAC inhibitors that have been tested to date are either highly cytotoxic or have very low specificities for different HDAC enzymes. In the course of studies on Friedreich’s ataxia (FRDA), our laboratory identified a novel class of HDAC inhibitors (pimelic diphenylamides) that reverse heterochromatin-mediated silencing of the frataxin (FXN) gene in this disease. Recent studies show that these HDAC inhibitors cross the blood brain barrier in mice, exhibit no acute or chronic toxicity at potential therapeutic doses, and act as HDAC inhibitors in the mouse brain. Importantly, our compounds increase FXN mRNA levels in the brain and heart in a mouse model for FRDA. We have now identified HDAC3 as the likely cellular target of the pimelic diphenylamides. We find that a 5-phenyl derivative of our lead compound, which preferentially inhibits HDACs 1 and 2, fails to fully activate FXN gene expression in FRDA cells. While both the HDAC3- and HDAC1/2-specific compounds share a similar mechanism of inhibition of their target enzymes, namely, a slow-on/slow-off binding of the inhibitor, generating a stable inhibitor/HDAC enzyme complex, only HDAC3-specific compounds increase FXN gene expression and frataxin protein in cells. Additionally, a potent inhibitor of class II HDACs and a Sirt1 inhibitor fail to show activation of FXN gene expression in similar assays, again pointing to HDAC3 as a target for therapeutic intervention in FRDA.
EXPERIMENTAL PROCEDURES
Cell Culture
The human Friedreich’s ataxia lymphoblast cell line GM15850 (Coriell Institute, New Jersey) was grown in RPMI medium 1640 containing 10% FBS, 1% HEPES and 1% PMSF. Nuclear extracts were prepared by first adding cold 10 mM HEPES pH7.9, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT and 0.2 mM PMSF to washed cell pellets, and after incubation on ice for 10 min; the lysed cells were centrifuged at 3000xg for 15 min and the soluble fractions removed. The pellet was resuspended in a 1:1 mixture of low salt buffer (20 mM HEPES pH7.9, 25% glycerol, 20 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT and 0.2 mM PMSF) and high salt buffer (20 mM HEPES pH7.9, 25% glycerol, 1.2 M KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT and 0.2 mM PMSF), homogenization, followed by stirring at 4°C for 30 min. The lysed nuclear pellet solution was centrifuged at 14000xg for 30 min at 4°C to provide the nuclear fractions (supernatant) and a membrane pellet. All fractions were stored at −80°C until use.
Chemical Synthesis
HDAC inhibitors were synthesized as described (Herman et al., 2006) with modifications described in detail in Supplemental material. The strategy for synthesis of trifunctional probes 1-BP, 2-BP and 3-BP are presented in the schemes shown in Supplemental material, along with detailed procedures and analytical data for all compounds. The synthesis of the class II inhibitor 4 is also described in Supplementary material. Sirt1 inhibitor 6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1-carboxamide was purchased from Calbiochem (Sirt1 inhibitor III).
HDAC Inhibition Assays
Recombinant human HDAC1, HDAC 2, HDAC3/NcoR2, and HDAC8, expressed in baculovirus, were purchased from BPS Bioscience (San Diego, CA). The class II HDACs 4 and 5 were tagged with the Flag epitope from their respective cDNAs (obtained from Addgene, Cambridge, MA), expressed in HEK293t cells, and purified on Flag-M2 affinity resin (Sigma-Aldrich, MO), as described (Chou et al., 2008). The trifunctional probes, 106 and other HDAC inhibitors were assayed with the BioMol AK500 kit to determine IC50 values with recombinant HDACs or with a HeLa nuclear extract. Since our previous studies indicated a slow on-rate for 106, these IC50 measurements include a prolonged incubation time (1 h) to insure that the inhibitor-enzyme complex came to equilibrium before initiation of the enzyme assay (Chou et al., 2008). Samples were processed as described by BioMol and read with a 96-well fluorescence plate reader. For class I HDACs, the synthetic substrate acetyl-Lys(Ac)-AMC (from BioMol) was used and deacetylated lysine-AMC was released by trypsin treatment and free flourogenic 4-methylcoumarin-7-amide (MCA) was generated. The flourogenic MCA could then be read with an excitation wavelenghth of 370 nm and emission wavelength of 460 nm. Assays for class II HDACs were done using acetyl-Lys(trifluoroacetyl)-AMC (Lahm et al., 2007) under the same conditions. A semilogarithmic plot of the data was analyzed with Kaleidagraph software (Synergy software) to obtain the IC50 value. Ki values were determined from enzyme progression curves, performed at various inhibitor concentrations, as described (Chou et al., 2008).
Photoaffinity Labeling of Recombinant HDAC Enzymes
Equal amounts (5 μg) of HDAC1, HDAC2, HDAC3, HDAC8, HDAC4 and HDAC5 were added to a 96-well plate, followed by addition of the trifunctional probe to give a final concentration of 4 μM in 50 μL. After incubation on ice for 5 min, the solutions were photo-crosslinked with UV at 366 nm for 1.5 h on ice. Then click reagent (rhodamine-azide (RdN3), Tris[(1-benzyl-1-H-1,2,3-triazol-4-yl)methyl]amine (ligand), Tris(2-carboxy-ethyl)phosphine hydrochloride (TCEP), and CuSO4) was added to the solution to give final concentrations of 86 μM RdN3, 53 μg/mL ligand, 0.29 mg/mL TCEP and 1 mM CuSO4, respectively, and rotated at RT for 1.5 h. 2 – 4 μL was taken from each reaction for SDS-PAGE analysis. Fluorescent protein markers were a generous gift of B. Cravatt and colleagues (Scripps). The gel was scanned with a Hitachi FMBI011 instrument.
Streptavidin Bead Enrichment and Western Blotting
300 μL nuclear extract (3.8 μg/μL protein) in 2100 μL PBS was added to different wells in a 6-well plate, and in some experiments along with competitor HDAC inhibitors, at the concentrations indicated in the figure legends. In competition experiments, the competing HDAC inhibitor was incubated with the extract for 2 h at RT prior to addition of 1-BP. 240 μL trifunctional probe was added to give a final concentration of 4 μM, and incubation continued on ice for 5 min. Samples were then cross-linked with UV at 365 nm for one h on ice. 360 μL click reagent were added to the wells, as described above except using biotin-azide in place of rhodamine-azide, or 360 μL click reagent with no biotin-azide as a control, and the resulting solutions were rotated at RT for one h. 1000 μL PBS was added to each well and the solution was kept at −20°C overnight. The next day, the solutions from each well were transferred to separate Eppendorf tubes and centrifuged to precipitate proteins, which were then washed with cold methanol (1 mL, twice), dried and resuspended in 1 mL 0.2% SDS in PBS, and then incubated with 0.8 ml magnetic streptavidin beads (Invitrogen) for 2 h. (The supernatant was removed from the original bead solution, and the beads were washed with PBS (1 mL, twice, prior to use.) The supernatant was removed and the beads were washed with 0.2% SDS in PBS (1 mL, twice), 6M urea (1 mL, twice) and PBS (1 mL, three times), and the resulting beads were eluted with 60 μL 2x SDS loading buffer at 90° and loaded onto three separate SDS polyacrylamide gels, and subjected to western blotting. Each membrane was immunostained with antibodies to HDAC1, HDAC2 and HDAC3 (all from Abcam), respectively, followed by anti-rabbit IgG-horseradish peroxidase conjugated secondary antibody (Cell Signaling, MA). Other antibodies used in western blotting experiments were anti-acetylated histone H3 (Upstate Biotechnology), histone H3 (Abcam), acetylated tubulin (Abcam), frataxin (Mitosciences) and GAPDH (Abcam). For quantification of western blots, the x-ray films, with exposures within the linear range of the film, were scanned and converted into digital images, which were then analyzed with ImageQuant software (Molecular Dynamics). After background correction, the levels of the protein of interest were normalized to a reference protein as a recovery or input standard.
Human Subjects, Primary Lymphocyte Isolation, and FXN mRNA Determinations
All experiments were conducted with appropriate informed consent, under a protocol approved by the Scripps Health Human Subjects Committee. Primary lymphocytes were isolated from donor blood by ficoll density gradient centrifugation (GE Healthcare), and mRNA determinations after incubation with various HDAC inhibitors were carried out as previously described (Herman et al., 2006).
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institute of Neurological Disorders and Stroke (NIH), the Friedreich’s Ataxia Research Alliance, GoFAR, Ataxia UK and by Repligen Corporation. We also thank Cleo Salisbury and Ben Cravatt (Scripps) for helpful discussions and for the generous gifts of reagents.
Footnotes
Author Contributions
C.X., C.J.C., J.R.R., and J.M.G. designed research; C.X., C.J.C., D.H., L.S., and H.P. performed research; all authors analyzed data; and J.M.G. wrote the paper with the assistance of all authors.
Competing Interests
J.M.G. is a consultant to Repligen Corporation, and has a competing financial interest in this work. H.P. and J.R.R. are employees of Repligen Corporation.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Broide RS, Redwine JM, Aftahi N, Young W, Bloom FE, Winrow CJ. Distribution of histone deacetylases 1-11 in the rat brain. J Mol Neurosci. 2007;31:47–58. doi: 10.1007/BF02686117. [DOI] [PubMed] [Google Scholar]
- Butler KV, Kozikowski AP. Chemical origins of isoform selectivity in histone deacetylase inhibitors. Curr Pharm Des. 2008;14:505–528. doi: 10.2174/138161208783885353. [DOI] [PubMed] [Google Scholar]
- Chou CJ, Herman D, Gottesfeld JM. Pimelic diphenylamide 106 is a slow, tight-binding inhibitor of class I histone deacetylases. J Biol Chem. 2008;283:35402–35409. doi: 10.1074/jbc.M807045200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert AD, Yu J, Rose FF, Mattis VB, Lorson CL, Thomson JA, Svendsen CN. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2008;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Cravatt BF. Mechanism-based profiling of enzyme families. Chem Rev. 2006;106:3279–3301. doi: 10.1021/cr050288g. [DOI] [PubMed] [Google Scholar]
- Gui CY, Ngo L, Xu WS, Richon VM, Marks PA. Histone deacetylase (HDAC) inhibitor activation of p21WAF1 involves changes in promoter-associated proteins, including HDAC1. Proc Natl Acad Sci USA. 2004;101:1241–1246. doi: 10.1073/pnas.0307708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenstein MC, Mussgnug JH, Lotte K, Plessow R, Brockhinke A, Kruse O, Sewald N. Affinity-based tagging of protein families with reversible inhibitors: a concept for functional proteomics. Angew Chem Int Ed Engl. 2003;42:5635–5638. doi: 10.1002/anie.200352084. [DOI] [PubMed] [Google Scholar]
- Hagenstein MC, Sewald N. Chemical tools for activity-based proteomics. J Biotechnol. 2006;124:56–73. doi: 10.1016/j.jbiotec.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Hartman HB, Yu J, Alenghat T, Ishizuka T, Lazar MA. The histone-binding code of nuclear receptor co-repressors matches the substrate specificity of histone deacetylase 3. EMBO Rep. 2005;6:445–451. doi: 10.1038/sj.embor.7400391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman D, Jenssen K, Burnett R, Soragni E, Perlman SL, Gottesfeld JM. Histone deacetylase inhibitors reverse gene silencing in Friedreich’s ataxia. Nature Chem Biol. 2006;2:551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- Jones P, Altamura S, De Francesco R, Gallinari P, Lahm A, Neddermann P, Rowley M, Serafini S, Steinkuhler C. Probing the elusive catalytic activity of vertebrate class IIa histone deacetylases. Bioorg Med Chem Lett. 2008;18:1814–1819. doi: 10.1016/j.bmcl.2008.02.025. [DOI] [PubMed] [Google Scholar]
- Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov. 2008;7:854–868. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- Lahm A, Paolini C, Pallaoro M, Nardi MC, Jones P, Neddermann P, Sambucini S, Bottomley MJ, Lo Surdo P, Carfí A, et al. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc Natl Acad Sci USA. 2007;104:17335–17340. doi: 10.1073/pnas.0706487104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks P, Breslow R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat Biotechnol. 2007;25:84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- Marks P, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK. Histone deacetylases and cancer: causes and therapies. Nat Rev Cancer. 2001;1:194–202. doi: 10.1038/35106079. [DOI] [PubMed] [Google Scholar]
- Methot JL, Chakravarty PK, Chenard M, Close J, Cruz JC, Dahlberg WK, Fleming J, Hamblett CL, Hamill JE, Harrington P, et al. Exploration of the internal cavity of histone deacetylase (HDAC) with selective HDAC1/HDAC2 inhibitors (SHI-1:2) Bioorg Med Chem Lett. 2008;18:973–978. doi: 10.1016/j.bmcl.2007.12.031. [DOI] [PubMed] [Google Scholar]
- Napper AD, Hixon J, McDonagh T, Keavey K, Pons JF, Barker J, Yau WT, Amouzegh P, Flegg A, Hamelin E, et al. Discovery of indoles as potent and selective inhibitors of the deacetylase SIRT1. J Med Chem. 2005;48:8045–8054. doi: 10.1021/jm050522v. [DOI] [PubMed] [Google Scholar]
- Rai M, Soragni E, Jenssen K, Burnett R, Herman D, Gottesfeld JM, Pandolfo M. HDAC inhibitors correct frataxin deficiency in a Friedreich ataxia mouse model. PLoS ONE. 2008;3:e1958. doi: 10.1371/journal.pone.0001958. doi: 1910.1371/journal.pone.0001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C, Zhang L, Freitas MA, Ghoshal K, Parthun MR, Jacob ST. Peptide mass mapping of acetylated isoforms of histone H4 from mouse lymphosarcoma cells treated with histone deacetylase (HDACs) inhibitors. J Am Soc Mass Spectrom. 2005;16:1641–1653. doi: 10.1016/j.jasms.2005.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury CM, Cravatt BF. Activity-based probes for proteomic profiling of histone deacetylase complexes. Proc Natl Acad Sci USA. 2007;104:1171–1176. doi: 10.1073/pnas.0608659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury CM, Cravatt BF. Optimization of activity-based probes for proteomic profiling of histone deacetylase complexes. J Am Chem Soc. 2008;130:2184–2194. doi: 10.1021/ja074138u. [DOI] [PubMed] [Google Scholar]
- Solomon JM, Pasupuleti R, Xu L, McDonagh T, Curtis R, DiStefano PS, Huber LJ. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol Cell Biol. 2006;26:28–38. doi: 10.1128/MCB.26.1.28-38.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas EA, Coppola G, Desplats PA, Tang B, Soragni E, Burnett R, Gao F, Fitzgerald KM, Borok JF, Herman D, et al. The HDAC inhibitor 4b ameliorates the disease phenotype and transcriptional abnormalities in Huntington’s disease transgenic mice. Proc Natl Acad Sci USA. 2008;105:15564–15569. doi: 10.1073/pnas.0804249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JC, Hong R, Schreiber SL. Structural biasing elements for in-cell histone deacetylase paralog selectivity. J Am Chem Soc. 2003;125:5586–5587. doi: 10.1021/ja0341440. [DOI] [PubMed] [Google Scholar]
- Yang WM, Tsai SC, Wen YD, Fejer G, Seto E. Functional domains of histone deacetylase-3. J Biol Chem. 2002;277:9447–9454. doi: 10.1074/jbc.M105993200. [DOI] [PubMed] [Google Scholar]
- Zupkovitz G, Tischler J, Posch M, Sadzak I, Ramsauer K, Egger G, Grausenburger R, Schweifer N, Chiocca S, Decker T, Seiser C. Negative and positive regulation of gene expression by mouse histone deacetylase 1. Mol Cell Biol. 2006;26:7913–7928. doi: 10.1128/MCB.01220-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.