Abstract
Multipotent stem cells have the potential to establish a new field of promising regenerative medicine to treat tissue damage, genetic disorders, and degenerative diseases. However, limited resource of stem cells has turned to be an evitable obstacle in clinical applications. We utilized a simple in vitro epigenetic reprogramming approach to convert skin fibroblasts into multipotent cells. After transient reprogramming, stem cell markers, including Oct4 and Nanog, became activated in the treated cells. The reprogrammed cells were multipotent as demonstrated by their ability to differentiate into a variety of cells and to form teratomas. Genomic imprinting of insulin-like growth factor II (Igf2) and H19 was not affected by this short period of cell reprogramming. This study may provide an alternative strategy to efficiently generate patient-specific stem cells for basic and clinical research, solving major hurdles of virally-induced pluripotent stem (iPS) cells that entail the potential risks of mutation, gene instability, and malignancy.
Keywords: Induced multipotent stem cells, iMS cells, epigenetic reprogramming, multipotency, fish oocyte extracts, differentiation
INTRODUCTION
Patient-specific pluripotent stem cells may someday be useful for treating various chronic diseases and injuries. However, the use of currently available human ES cell lines in patients risks rejection by patients' immune system. Creating “autologous” stem cells specific to individual patients by therapeutic cloning[1-7] has offered an alternative methodology, but ethical concerns and technical challenges remain. The procedure for producing these stem cells requires human oocytes, and the successful rate of cell cloning is extremely low, baring its practical use.
Several other approaches attempt to generate stem cells by cell reprogramming, including cell fusion[8-11] and trans-differentiation[12], have not been successful in producing stem cells for clinical studies. The generation of personalized stem cells from somatic cells, so called “induced pluripotent stem” (iPS) cells from viral transduction of defined factors, marks an important breakthrough in stem cell research[13-20]. Remarkably, these iPS cells are indistinguishable from embryonic stem (ES) cells, as they are able to differentiate into multiple cell types[13-16] and to produce germline-competent chimeras. The induced cell can express stem cell marker antigens and have the ability of differentiating into mature cells which are the source of the three germ layers under suitable conditions. Thus, reprogramming of somatic cells to generate iPS cells using one's own skin may represent a simple approach to circumvent the technical difficulties and ethical concerns of therapeutic cloning.
However, numerous hurdles need to be resolved before iPS cells can be translated to the clinic. First, the risk of tumors from iPS cells is of great concern because up to a third of the mice produced from iPS cells have tumors associated with c-Myc reactivation[21]. Secondly, the potential risk of mutation and genomic instability derived from as many as 20 copies of random viral insertions[18] casts another safety concern. Finally, the efficiency of forming iPS cells is still very low[16, 18, 22], probably reflecting the poor transduction of four viral vectors simultaneously into a single cell to achieve cell reprogramming.
Recently, several approaches have been explored to create virus-free iPS cells[23-26]. However, the whole induction process of iPS clones is still time-consuming and very inefficient. In some cases, the residual of retroviral sequences in the genome is still a concern when applied to clinics.
We have proposed to test a simple approach to create induced multipotent stem (iMS) cells from somatic cells. Unlike iPS cells, these iMS cells are transiently reprogrammed cells and still keep original cell characteristics. However, they have the potential to be differentiated into therapeutic cells, but in theory, unlike pluripotent stem cells, they do not impose a risk of tumor formation from undifferentiated cells. In this study, we explored a very simple approach to create induced multipotent cells by utilizing in vitro epigenetic reprogramming.
MATERIALS AND METHODS
Experiment reagents
DMEM/F12 medium was purchased from Hyclone (Logan, UT); fetal bovine serum (FBS) was from Invitrogen (Carlsbad, CA); mouse anti-human Oct3/4 monoclonal antibody (sc-5279) was from SANTA CRUZ Biotechnology; and DAB chromogenic liquid was from MaiXin Bio-company (Fuzhou, China).
Culture of fibroblasts
Mouse fibroblasts were cultured from the abdominal skins of 3-day old Sprague-Dawley mice (SD mouse) or M. spretus × M. musculus F1 mice using the method as previously described[27, 28]. Briefly, skin tissues (1×1 cm2) were collected under sterile conditions. The skin was cut with scissors into small pieces (1 mm2) in phosphate buffered solution (PBS) containing 100U/ml ampicillin and 100mg/ml streptomycin, and was digested with DMEM containing 200U/ml collagenase and 300U/ml hyaluronidase for 24 hours at 4 C. After centrifugation, tissues were treated with PBS containing 0.25% trypsogen and 0.02% EDTA for 20 minutes in a 37 C shaking bath. The reaction was terminated by adding 10 ml DMEM/F12 medium containing 10% fetal bovine serum (FBS). The cells were collected by centrifugation for 10 minutes at 1000 rpm. After rinsing twice with PBS, 2×105/ml cells were seeded in 6 well plates in DMEM at 37 C, with 5% CO2 and 90% humidity. The medium was changed every 2 days. After reaching confluency, cells were trypsinized and used for studies. The experiment was conducted according to the protocol (#0000083) approved by the Animal Committee of Animal Center of Kunming General Hospital.
Preparation of fish egg extracts
Fish oocytes were acquired from local fish production farms. Fish oocytes, collected under sterile condition, were ground by mortar and extracted with equal volumes of 0.9% sodium chloride. Egg extracts were centrifuged at a speed of 1000 rpm for 10 minutes and followed by 3000 rpm for another 10 minutes. Supernatants were collected and filtrated by 0.2 μm filters. Protein levels of the extracts were determined as described[29]. The extracts were diluted using DMEM/F12 medium into 10 mg protein/ml and preserved at −80 C for studies as stock extracts.
In vitro cell reprogramming
The third passage of cultured primary fibroblasts was used for cell treatment studies. Exponentially growing fibroblasts were digested with trypsin-EDTA (Invitrogen, CA) and 4×105 cells were inoculated in new 6-well plates for cell reprogramming.
Two approaches were utilized to reprogram fibroblasts. The first was to directly reprogram fibroblasts with fish oocyte extracts without cell membrane permeabilization. In this approach, cells were directly treated for varied time with fish oocyte extracts at concentrations of 10μ-10mg protein/ml. The second approach was to permeabilize cell membrane before cell reprogramming using Streptolysin O (STO) as previously described[12, 30, 31]. Briefly, fibroblasts were first reversibly membrane-premeabilized with 500-800 ng/mL streptolysin O (SLO, Sigma) at 37° C for 1 hr, and were reprogrammed for 1 hr at 37° C in rejuvenating buffer containing 1 mg/mL BSA, 1 mM ATP, 5 mM phosphocreatine, 25 μg/ml creatine kinase (Sigma), 0.4 U/mL RNase inhibitor (Invitrogen), and 1 mM each of the four dNTPs (nucleotides triphosphate), and fish oocyte extracts at varying concentrations. The cell membrane was then resealed with DMEM containing 2 mM CaCl2.
After treatment, cells were seeded in 6-well plates coated with laminin 1 (5μg/ml,) and cultured in DMEM/F12 supplemented with 20% Knockout Serum Replacer (KSR), 2 mM non-essential amino acids (NAA), 0.1 μM β-mercaptoethanol (all from Invitrogen), and 4 ng/mL bFGF (Sigma), and 10 ng/ml Leukemia inhibitory factor LIF (Sigma). Low oxygen (3%) was used to culture the induced cells using Forma Series II 3140 Water Jacketed CO2/O2 incubator. Cells were collected at different time points for the analysis of pluripotent cell markers.
As a first step to identify reprogramming factors, 180 μl fish oocyte extracts (1 mg/ml) were pretreated with 60U DNase I and/or 6 mg RNase A to remove the RNA and DNA components. The pretreated extracts were then used to induce fibroblast reprogramming using the same approach as described above.
Quantitation of stem cell marker gene expression
Total RNA was extracted from cells by Tri-Reagent (Sigma, St. Louis, MO) and treated with DNase I to eliminate contamination of genomic DNA[27, 29, 32]. cDNA was synthesized with RNA reverse transcriptase and gene expression was examined by PCR. The cDNAs and primers were heated to 95° C for 5 min, then amplified by 26 cycles at 95° C for 30 sec, 57° C for 45 sec and 72° C for 45 sec. PCR primers used for mRNA quantitation included Oct4-sense: AAA AAG CAG GCT CCA CCT TCC CCA TGG CTG GAC ACC and antisense: AGA AAG CTG GGT TGA TCA ACA GCA TCA CTG AGC TTC; Nanog-sense: AAA AAG CAG GCT CTG ACA TGA GTG TGG GTC TT and antisense: AGA AAG CTG GGT AAG TCT CAT ATT TCA CCT GG; and internal control 28S-sense: GCG GCT TTG GTG ACT CTA and antisense: CTG CCT CCT TGG ATG TG.
Immunohistochemical staining
Cells were fixed by 4% paraformaldehyde and were washed with PBS three times for five minutes each. The cells were incubated with 0.3% Triton-100 for 5 minutes, washed for 3 times with PBS, and blocked with endogenous peroxidase with 3% H2O2 for 5 minutes. After PBS wash, anti-mouse antibody Oct3/4 was added to the fixed cells of both induced and control groups and incubated at 4 C for 12 hours followed with PBS washing for three times. Biotinylation di-antibody was added and was incubated at 37 C for 20 minutes, followed by the same PBS washing process. After staining with DAB working solution, cell morphology was observed with an optical microscope.
Differentiation of induced cells into other type of cells
The cells were induced with repeated treatment of fish oocyte extracts. The induced cells were cultured in KO-DMEM medium with 15% FBS at 37° C in the presence of 1000 U/mL of LIF, and the embryoid bodies (EBs) were formed in hanging droplets as previously described[33]. To promote EB formation, induced cells were cultured in low oxygen (3% O2 in a Forma Series II 3140 Water Jacketed CO2/O2 incubator).
For adipocyte differentiation, the attached EBs were treated with 10−6 M of all-trans-retinoic acid (ATRA) for 3 d followed by 10−7 M of insulin and 2 × 10−9 M of triiodothyronine (T3)[33]. Differentiated adipocytes were stained with Oil-Red-O and counterstained with hematoxylin.
For muscle differentiation, induced cells were differentiated in Iscove's MEM supplemented with 20% FBS, 2 mM L-glutamine, 1x nonessential amino acids, 450μM monothioglycerol (Sigma) and antibiotics[34]. Skeletal muscle was induced in DMEM supplemented with 2% inactivated horse serum and 30% C1C12 cell conditional medium. Myotube formation during the differentiation was examined by microscopic observation, using immunostaining with mouse anti-MHC primary antibody (Sigma, 1:500).
For cardiomyocyte differentiation, EBs were collected and induced in 10% FBS/KO-DMEM containing 100 ng/mL activin A, 10−6M ATRA, 10 ng/mL bFGF for 3 d and were switched to N2 medium containing DMEM/F12 (1:1) supplemented with B27 (Invitrogen, CA), 1 μg/mL of laminin, 10 mM nicotinamide and 10 ng/mL bFGF. Beating cardiomyocytes were examined under an inverted microscope and cells were immunostained using a 1:100 Troponin T mouse monoclonal antibody (Lab Vision, Fremont, CA).
A three-step method[35] was used with minor modifications to differentiate insulin-secreting cells. The insulin-secreted cell clusters were stained by primary antibodies of insulin AB-6 mouse monoclonal antibody (1:200, Lab Vision, Fremont, CA), and anti-C-Peptide antibody (1:100, LINCO Research, Inc., St. Louis, MO). DAB (3,3′-diaminobenzidine) was used as the reaction substrate.
Generation of neuroectodermal cells from reprogrammed cells was performed by the method described previously by Zhang et al[36]. Briefly, aggregated EBs were differentiated to form large numbers of neural tube-like structures in the presence of FGF-2. Neural precursors were isolated and purified on the basis of their differential adhesion. Following the replacement of FGF-2 with brain-derived neurotrophic factor (BDNF), the cells were differentiated into neurons, astrocytes, and oligodendrocytes. Immunohistochemical staining of neural cells was performed using polyclonal antibodies against nestin (Chemicon, Temecula, CA, 1:750) and βIII-tubulin (Covance Research Products).
Teratoma formation in animals
We further examined the ability of the induced cells to generate teratomas in animals. Induced cells were suspended at 2.5×107 cells/ml in 0.9% normal saline. Nude mice were anesthetized with diethylether. We injected 200μl of the cell suspension (5.0×106cells) subcutaneously into the dorsal flank. Two weeks after the injection, tumors were surgically dissected from the mice. Samples were fixed in PBS containing 4% formaldehyde, and embedded in paraffin. Sections were stained with hematoxylin and eosin. Neural tissues were immunostained by S-100 antibody.
Imprinting status of Igf2 and H19 in reprogrammed and differentiated cells
To examine genomic imprinting, we created iMS cells from MBW2 cells that were cultured from an F1 newborn mouse derived from breeding a M. spretus male with a C57B/6 female[27, 29, 32]. MBW2 cells contained several polymorphisms, with which it is easy to study the imprinting status of several known imprinted genes[27, 37, 38]. As previously described, total RNA was extracted from cells by TRI-REAGENT (Sigma, St. Louis, MO), according to the manufacturer's guide, and cDNA was synthesized with RNA reverse transcriptase. Genomic imprinting was examined by PCR in cDNA samples using primers specific for polymorphic restriction enzymes in Igf2 and H19. PCR primers used to measure allelic expression of H19 were #4025 (sense): TAAGTCGATTGCACTGGTTTGGAGT, and #4026 (antisense): TGATGGAACTGCTTCCAGACTAGG; and Igf2 were #MII84 (sense): CTTGTGCTGGATCGCTGCTTACGG, and #MII219 (antisense): CTGCGACGGTTGGCACGGCTTGA. Allelic expression of H19 was assessed by polymorphic restriction enzyme Fok1 and Igf2 by Mbo1[32, 37, 38]. PCR products were digested with polymorphic restriction enzymes and were electrophoresed on 5% polyacrylamide-urea gel and scanned by PhosphoImage Scanner (Molecular Dynamics, Sunnyvale, CA).
RESULTS
In vitro cell reprogramming
In this study, we used two approaches to reprogram fibroblasts. The first approach was to directly reprogram fibroblasts without cell membrane permeabilization. Skin fibroblasts were directly treated with varying concentrations of fish oocyte extract and cells were collected at different time points for the analysis of pluripotent cell markers.
Depending upon the concentration of fish oocyte extracts used, treated fibroblasts underwent a change in cell morphology. Following a short exposure to low concetrations of extracts, cells grew more rapidly than did control cells (Fig.1A, middle panel). After repeated exposure of cells for more than three passages, we found that cells formed colonies, especially when seeded on MEF feeder cells (Fig.1A, right panel). When exposed to high concentrations of oocyte extracts, however, the fibroblasts became enlarged with an irregular cell morphology.
Figure 1. Enhanced cell proliferation after in vitro reprogramming with fish oocyte extracts.
A. Morphological change of fibroblasts treated with fish occyte extracts. Fibroblasts were treated with PBS control (left panel), fish oocyte extracts (10μg protein/ml) for 12 hours (middle panel), and repeat treatment of fish oocyte extracts (10μg protein/ml) for three passages (right panel).
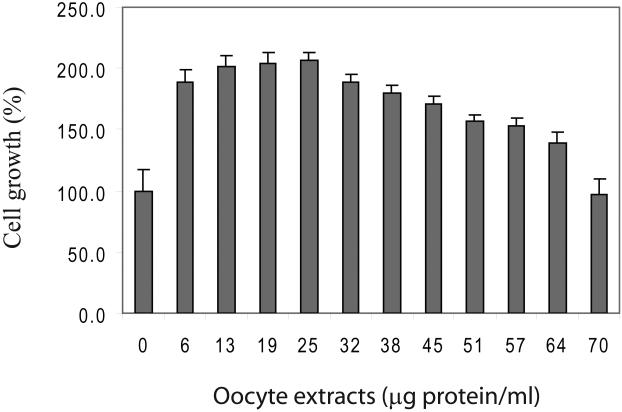
B. Cell growth of fibroblasts following the exposure of varying concentrations of fish occyte extracts. Forth-eight hours following the treatment, cells were measured with the MTT assay and standardized using the PBS-treated control group as 100%.
Treatment with fish oocyte extracts also significantly promoted cell proliferation. Skin fibroblasts were treated with varying concentrations of extracts. Forty-eight hours after the treatment, cells number was measured using the MTT assay. At low concentrations, fish oocyte extracts increased cell growth (Figure 1B). Cell proliferation gradually decreased from the peak when the extract concentration was over 25μg protein/ml. Therefore, in subsequent studies, we exposed cells to low concentration of the fish oocyte extracts for a relatively short period of time.
We also tested cell reprogramming following Streptolysin O cell membrane permeabilization. However, in most cases with high doses of extracts we observed more cell death than the direct incubation approach, probably due to cell toxicity (data not shown). Thus, permeabilization with Streptolysin O greatly sensitized cells to the extract treatment. As a result, low doses of extracts were used in this approach.
Expression of stem cell marker genes
After exposure to the fish oocyte extract, we collected the reprogrammed cells and used RT-PCR to examine expression of several marker genes related to cell pluripotency. As expected, Oct4 and Nanog were not expressed in primary fibroblasts. However, both pluripotent markers became activated in treated cells as early as 48 hr following reprogramming (Fig.2A).
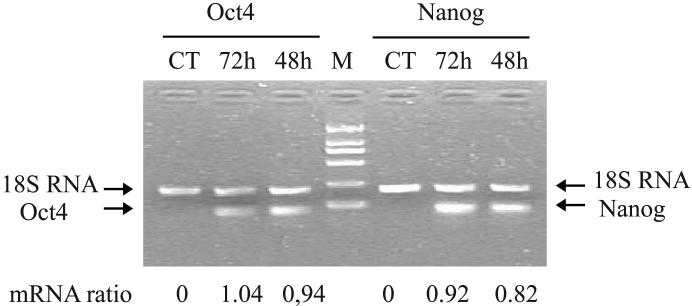
Figure 2. Activation of pluripotent cell marker genes following cell reprogramming.
A. Measurement of Oct4 and Nanog expression by RT-PCR. Treated cells were collected at 48 and 72 hours following extract treatment and were used for PCR quantitation of Oct4 and Nnaog mRNAs. CT: PBS control. 18S RNA was used as the internal control for PCR. mRNA ratio: PCR band density of Oct4 and Nanog over that of 18S RNA. Note the activation of both pluripotent genes in treated cells.
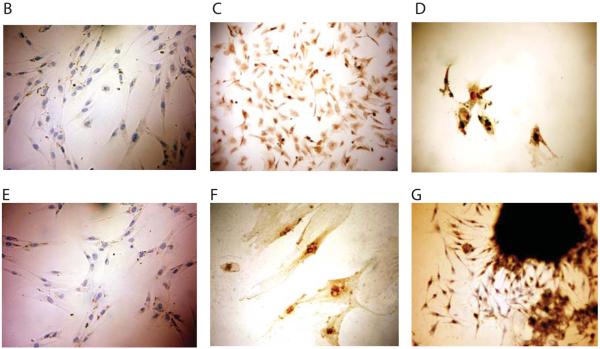
B-G. Immunohistochemical staining of Oct4. Skin fibroblasts were treated with PBS control (B, E, ×200) or varying concentrations of fish occyte extracts: 10 μg/ml for 12 hours (C, ×200); 2 mg/ml for 12 hours and then 1 mg/ml for another 12 hours (D, ×200); 10mg/ml for 12 hours (F, ×400); and clone proliferation from repeated treatment with 10mg/ml (G, ×200).
We then used immunohistochemical staining to identify Oct4 protein in fibroblasts treated with varied concentrations of fish oocyte extracts. Untreated fibroblasts did not stain for Oct4 (Figs 2B, 2E), but Oct4 protein was expressed in the reprogrammed cells treated with 10μg protein/ml extracts (Fig.2C). Cells induced with high concentrations of fish oocyte extracts expressed more Oct4 protein, but cell growth was also affected (Figs.2D, 2F). However, with extended induction, colony proliferation was more obvious in the group treated with very high concentrations of fish oocyte extracts (10mg protein/ml). These colonies expressed abundant Oct4 proteins (Fig.2G).
Multipotency of the reprogrammed cells
To confirm the multipotency of established iMS cells, we assessed their ability to differentiate into other types of cells. We followed the conventional two-step procedures used for differentiating embryonic stem cells. We found that these shortly reprogrammed cells were able to be differentiated into other types of cells.
In the early stage of differentiation, iMS cells secreted insulin and formed small insulin-positive clusters (Fig.3A). After a longer period of differentiation, the clusters expanded and joined together to form larger islands of insulin-secreted cells (Fig.3B), and secreted C-peptide (Fig.3C).
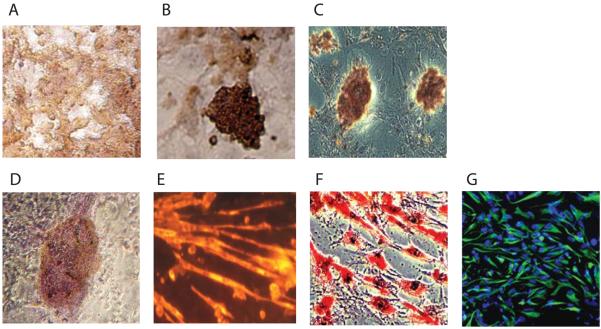
Figure 3.
Differentiation of induced cells into other cell types. After repeated exposure to fish occyte extracts under 3% O2 culturing, cell clones were expanded and induced into other cell types. A: immunostaining for insulin in early differentiation; B: immunostaining of insulin in cell clusters; C: C-peptide-positive clusters; D: immunostaining of beating cardiocytes with anti-troponin; E: immunostaining of Troponin T; F: adipocytes stained with oil red O; and G: neural precursor cells immunostained with anti-nestin.
In the presence of media used to differentiate skeletal muscle, the iMS cells aggregated and fused, and ultimately formed typical skeletal myocyte tubes with multiple nuclei and synthesized skeletal muscle-specific proteins (Fig.3E). In a separate study, we were able to differentiate iMS cells into cardiomyocytes that immunostained with anti-troponin serum (Fig.D). Interestingly, the cardiomyocytes were functional, beating in the medium at a rate of 26-30 beats/min.
Similarly, iMS cells were able to form precursor adipocytes and then mature adipocytes in the presence of retinoic acid. In this study, we found that almost all iMS cells were positive for lipid staining with a much higher frequency than E14 ES cells. The synthesized lipids accumulated in the cytoplasm (Fig.3F). We also differentiated IMS cells into neural precursors (Fig.3G). After selection and induction, these neural precursors were further differentiated into mature neurons.
These data demonstrated that these in vitro reprogramming-derived iMS cells possess similar potency as other stem cells in differentiating into a variety of cell types that are useful for cell therapy.
Teratoma formation from reprogrammed cells
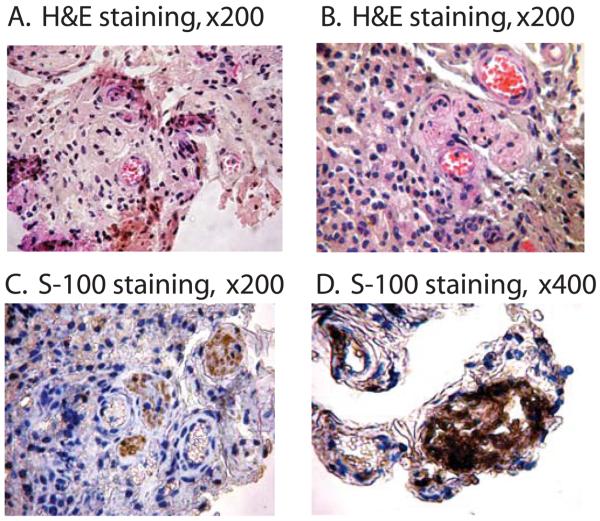
The induced cells were transplanted subcutaneously into dorsal flanks of immunodeficient mice. Two weeks after injection, we observed tumor formation. Histological examination showed that the tumor included various tissues containing blood vessels, neuronal, muscle and adipose tissues (Figs.4A, 4B). Immunohistochemical staining showed the presence of neural tissues that were positive for the glial marker S100 (Figs.4C, 4D).
Figure 4.
Teratoma formation from the induced multipotent cells. Skin fibroblasts were exposed to fish occyte extracts and were transplanted subcutaneously into the dorsal flank of nude mice. Two weeks after the injection, tumors were surgically dissected and stained with hematoxylin and eosin (A, B). The presence of neural structures was identified by immunostainign with S-100 antibody (C, D).
Imprinting status of Igf2/H19
Nuclear cloning is often accompanied by the alteration of epigenetics in offspring, resulting in the low efficiency of animal cloning[39]. We thus investigated whether the in vitro nuclear reprogramming by nuclear extracts also caused the similar abnormalities in formed iMS cells and differentiated cells. We created iMS cells from FSKN4 fibroblasts that were derived from the skin of a F1 mouse hybridized between a M. spretus male and a C57BL/c female. It was thus easy to study the epigenetic modifications in several known imprinted genes using the known polymorphisms[29].
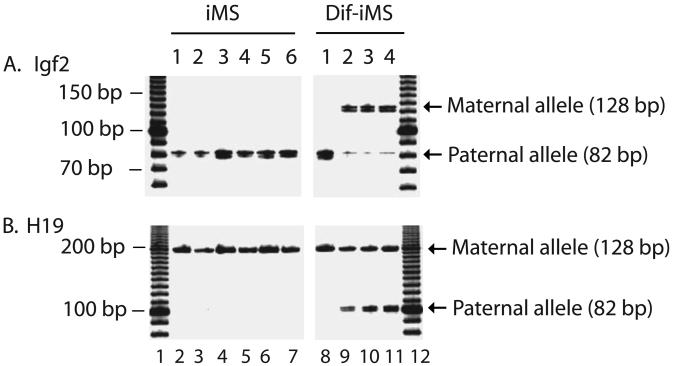
Imprinting status of two common imprinted genes was tested in iMS cells and differentiated cells using primers specific for insulin-like growth factor II (Igf2) and H19. Igf2 is normally expressed from the paternal allele and H19 is using their maternal allele for transcription. As seem in Figure 5, untreated control FSKN4 fibroblasts maintained imprinting for Igf2 and H19, with only the one of the parental alleles expressed. The same imprinting pattern was observed in the reprogrammed iMS clones. These data thus indicate that epigenetics and imprinting status of these two genes are maintained well during this in vitro nuclear reprogramming, thus providing a potential advantage over those procedures that often causes aberrant imprinting in the offspring or tissues.
Figure 5.
Status of genomic imprinting of insulin-like growth factor II (Igf2) and H19 in induced cells and differentiated adipocytes. iMS: induced multipotent cells; Di-iMS: differentiated iMS cells. Allelic expression of Igf2 was distinguished by restriction digestion of PCR products with Dpn2 and H19 by Fok1. Note the maintenance of mono-allelic expression of both Igf2 and H19 in both fibroblast controls and induced cells. Biallelic expression of both Igf2 and H19 was observed in some differentiated cells.
However, in some differentiated cells, especially in insulin-secreting and neural precursor cells, H19 and Igf2 showed the biallelic expression in differentiated cells (Fig.5).
Reprogramming with DNA/RNA-removed extracts
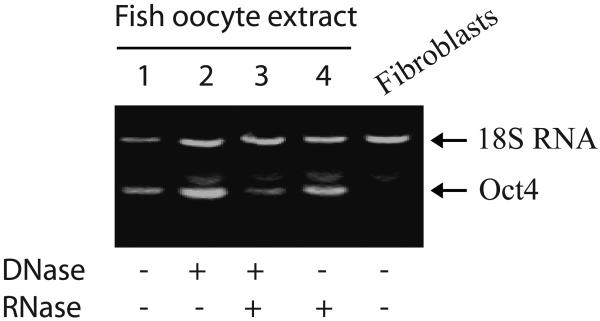
As an initial step towards exploring the mechanism underlying cell reprogramming, we pretreated fish oocyte extracts with DNase I and RNase A, aiming to exclude the role of DNA and RNA in the extracts. The pretreated extracts were then used to reprogram fibroblasts. Expression of pluripotent cell marker Oct4 was measured using RT-PCR. As seen in Figure 6, pretreatment with either DNase or RNase does not significantly affect the expression of Oct4 in treated cells (lanes 2 and 4), although there was a slight decrease in Oct4 expression in cells treated with both DNA and RNA removed extracts (lane 3). These data suggest that the DNA and RNA components in the extract contribute very little to the induction of cell multipotency.
Figure 6.
Oc4 expression in cells treated with fish oocyte extracts in which DNA and RNA were removed. Fibroblasts were induced for 48 hours with extracts that were pretreated with DNase I and/or RNase. Expression of Oct4 was measured with RT-PCR as described as in Figure 2.
DISCUSSION
This study describes an economic and efficient approach to generate multipotent cells useful for cell replacement therapy. After a brief exposure to fish oocyte extracts, skin fibroblasts become at least partially reprogrammed as demonstrated by 1) enhanced cell proliferation and 2) the activation of a set of genes characteristic of undifferentiated embryonic stem (ES) cells. The alterations in cell growth and gene expression profiles implicate global remodeling of chromatin by this short period of cell reprogramming. Notably, Oct4 and Nanog, which are expressed in ES cells to maintain pluripotency[40-44], were activated in these reprogrammed cells. Oct4 and Nanog regulate the ES potency network by acting on promoters of thousands of ES genes[45-47], which collectively define ES cell identity through their essential roles in early development.
We have demonstrated that the induced cells produced under low oxygen culturing are able to be differentiated into multiple functional cell types, including insulin-secreting islets, beating myoblasts, skeletal muscle, adipocytes, and neural cells (Fig.3), suggesting a potential for multiple lineage differentiation and acquisition. The capacity of forming teratoma in nude mice also demonstrated that the induced cells, at least partially, exhibit differentiation potency (Fig.4). Thus, the transient reprogramming by fish oocyte extracts was able to form a functional transcriptional network between these factors in a similar manner towards cell multipotency as in other stem cells, like mesenchymal stem cells from bone marrow and adipose tissue[48-52]. We found that the reprogrammed cells showed similarity in cell morphology to bone marrow stromal cells.
Most published reports that use xenopus oocyte extracts or embryonic stem cell extracts to reprogram cells required cell membrane pore opening with either detergents or streptolysin O[9, 12, 30, 31]. With cell permeabilization, the presumed cellular factors in the extracts enter the cell to reprogram the nucleus. After reprogramming, the cell membrane needs to be resealed by incubation in a low concentration of calcium in the medium. In addition, cell reprogramming usually required the sufficient supply of energy from ATP, creatine phosphate, and creatine kinase to be provided in the reaction mixture. However, in this study we demonstrated that skin fibroblasts were able to be reprogrammed simply by adding fish oocyte extracts in the cell medium without adding any permeabilizing agents.
In our experience, a short exposure of the fish oocyte extracts at low concentrations was a gentle reprogramming process that did not induce any noticeable cell damage. Cell growth became retarded only after the exposure to very high concentrations of extract. In a separate study, using this strategy we also demonstrated the generation of multipotent cells useful for cell therapy from human fibroblasts cultured from forearm skin (Zhu et al, unpublished data). The activation of ES marker genes was in parallel with CpG DNA demethylation of gene promoters in treated human fibroblasts. The induced cells formed teratomas in nude mice and were able to be differentiated into heamatopoitic precursor cells. When implanted in the kidney and liver of the monkey fetus, the BrdU-labeled human iMS cells were able to differentiate into functional hepacytes and renal tubules (Zhu et al, unpublished data). Collectively, these data demonstrate that the approach used in this study may be useful to efficiently reprogram somatic cells and greatly simplify the process of cell reprogramming.
The success in generating induced multipotent cells using this approach led us to explore the possible mechanism underlying genomic reprogramming. We showed that fibroblasts were induced to express pluripotent markers no matter whether cell membrane was permeabilized or not. Without cell membrane permeabilization, it is hard for the passive entry of macromolecules in the extracts into the cell, like ES transcription factor proteins Oct4, Sox2, and Nanog. Theoretically, only small molecules can freely penetrate into the cell for reprogramming. It is thus assumed that small peptides or non-peptide molecules in the extract initiate cell reprogramming or that large molecules are endocytosed or otherwise actively transported into the cell. There is evidence for the regulation of multiple target genes by a single small interfering RNA[53, 54], which functions at a posttranscription level or through a control of DNA methylation[54-59]. It is possible that small non-coding RNAs in the oocyte extract could participate in nuclear reprogramming in this model. However, our study using occyte extracts pretreated with RNase and DNase did not significantly alter the efficiency of cell reprogramming (Fig.6).
Alternatively, it is possible that fish oocyte extracts may be different from other extracts, like xenopus oocyte extracts, in that they may contain other soluble factors that interact with cellular surface receptors and activate one or more signal pathways involved in cell reprogramming. Further studies are needed to address the specific mechanisms underlying cell reprogramming by this short exposure of fish occyte extracts.
It should also be noted that there are many cases in which cells are reprogrammed without altering cell membrane permeabilization. Cell microenvironment (or cell niche) is able to significantly modify epigenotypes of a group of functional genes, leading to the altered cell phenotype. As a typical example, intact somatic cells can be reprogrammed when transferred into the enucleated oocytes[60, 61]. Tumor cells significantly alter their phenotypes when co-cultured with bone marrow stromal cells[62] or fetal fibroblasts[63]. Another notable case is that pretreatment of skin keratinocytes with ES conditional medium epigenetically changes gene expression, including pluripotent markers Oct4, Sox2, and Nanog[64]. Thus, it could be possible that fish oocyte extracts may create a reprogramming environment that turns on the expression of multipotent genes in skin fibroblasts.
Nevertheless, the in vitro cell reprogramming approach as described in this study offers many advantages over existing strategies in generating patient-specific stem cells for clinical applications. First, the reprogramming process is very simple, consisting of a short exposure of cells to fish oocyte extracts in the medium. No viral delivery or construction of vectors is required in this approach. The method can be easily adopted in any lab that is equipped with cell biology facility. Potentially, this methodology can be for large scale production of cells for clinical studies. Second, the method appears to be free of many safety concerns. The fish eggs are themselves edible. Third, the methodology is very simple. The multipotent cells can be generated with an overnight incubation or with a repeat treatment of second passage cells. The generation of iPS cells, on the other hand, usually requires months of cell culture. Finally, the approach used in this study is very efficient in yielding multipotent cells. We found that after reprogramming, nearly all of the treated cells stained positive for Oct4. In contrast, the existing strategies using defined factors are generally inefficient and usually generate iPS cells from millions of starting cells[16, 18, 22].
It is also worth noting that the induced cells generated from in vitro programming behave very much like bone marrow stromal cells (BMSCs) and umbilical cord blood mesenchymal stem cells (UBMSCs). Both BMSCs and UBMSCs have been broadly tested in both animal models and human clinical trials. They are recognized as a safe recipe for treating patients with a variety of diseases. In clinical applications, it is not necessary to fully differentiate these stem cells. In contrast, complete differentiation is sometimes required for clinical testing of embryonic stem (ES) cells. Undifferentiated ES cells may pose a risk of tumorigenesis. Thus, from this point of view, we predict that the induced potent cells from this study will also be proved safe in treating patients without concerning of the tumor risk.
CONCLUSIONS
This study demonstrates that a simple method of in vitro reprogramming with fish oocyte extracts may be sufficient to epigenetically remodel somatic cells to a state that resembles multipotent stem cells such as bone marrow stromal cells and umbilical cord blood mesenchymal stem cells. The reprogrammed cells exhibit the potential to be differentiated into other cell types useful for cell replacement therapy. Further studies are needed to examine whether these induced cells can be safely utilized to replace embryonic stem cells or iPS cells in treating diseases in animal models.
ACKNOWLEDGMENTS
The authors thank Jie Wu and Zhihong Sun for their help and support in conducting part of studies. This work was supported by The Key Program of Science and Technology Commission of Yunnan (2003013Z) to X.H.P; and NIH grant (1R43 CA103553-01) and The Department of Defense Grant (W81XWH-04-1-0597) to J.F.H, and the Research Sources of the Department of Veterans affairs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Yang X, Smith SL, Tian XC, Lewin HA, Renard JP, Wakayama T. Nuclear reprogramming of cloned embryos and its implications for therapeutic cloning. Nat Genet. 2007;39:295–302. doi: 10.1038/ng1973. [DOI] [PubMed] [Google Scholar]
- 2.Tweedell KS. New paths to pluripotent stem cells. Curr Stem Cell Res Ther. 2008;3:151–162. doi: 10.2174/157488808785740361. [DOI] [PubMed] [Google Scholar]
- 3.Han Z, Vandevoort CA, Latham KE. Therapeutic cloning: status and prospects. Curr Opin Mol Ther. 2007;9:392–327. [PubMed] [Google Scholar]
- 4.Hochedlinger K, Jaenisch R. Nuclear reprogramming and pluripotency. Nature. 2006;441:1061–1067. doi: 10.1038/nature04955. [DOI] [PubMed] [Google Scholar]
- 5.Tamada H, Kikyo N. Nuclear reprogramming in mammalian somatic cell nuclear cloning. Cytogenet Genome Res. 2004;105:285–291. doi: 10.1159/000078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colman A. Somatic cell transfer in mammals: Progress and applications. Cloning. 2000;1:185–200. doi: 10.1089/15204559950019825. [DOI] [PubMed] [Google Scholar]
- 7.Shi W, Zakhartchenko V, Wolf E. Epigenetic reprogramming in mammalian nuclear transfer. Differentiation. 2003;71:91–113. doi: 10.1046/j.1432-0436.2003.710201.x. [DOI] [PubMed] [Google Scholar]
- 8.Tada M, Tada T, Lefebvre L, Barton SC, Surani MA. Embryonic germ cells induce epigenetic reprogramming of somatic nucleus in hybrid cells. Embo J. 1997;16:6510–6520. doi: 10.1093/emboj/16.21.6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tada M, Takahama Y, Abe K, Nakatsuji N, Tada T. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr Biol. 2001;11:1553–1558. doi: 10.1016/s0960-9822(01)00459-6. [DOI] [PubMed] [Google Scholar]
- 10.Do JT, Scholer HR. Nuclei of embryonic stem cells reprogram somatic cells. Stem Cells. 2004;22:941–949. doi: 10.1634/stemcells.22-6-941. [DOI] [PubMed] [Google Scholar]
- 11.Cowan CA, Atienza J, Melton DA, Eggan K. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science. 2005;309:1369–1373. doi: 10.1126/science.1116447. [DOI] [PubMed] [Google Scholar]
- 12.Taranger CK, Noer A, Sorensen AL, Hakelien AM, Boquest AC, Collas P. Induction of dedifferentiation, genomewide transcriptional programming, and epigenetic reprogramming by extracts of carcinoma and embryonic stem cells. Mol Biol Cell. 2005;16:5719–5735. doi: 10.1091/mbc.E05-06-0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 14.Meissner A, Wernig M, Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat Biotechnol. 2007;25:1177–1781. doi: 10.1038/nbt1335. [DOI] [PubMed] [Google Scholar]
- 15.Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 16.Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 17.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 18.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 19.Kim JB, Zaehres H, Wu G, Gentile L, Ko K, Sebastiano V, et al. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;29:29. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- 20.Aoi T, Yae K, Nakagawa M, Ichisaka T, Okita K, Takahashi K, et al. Generation of Pluripotent Stem Cells from Adult Mouse Liver and Stomach Cells. Science. 2008;14:14. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- 21.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 22.Mali P, Ye Z, Hommond HH, Yu X, Lin J, Chen G, et al. Improved Efficiency and Pace of Generating Induced Pluripotent Stem Cells from Human Adult and Fetal Fibroblasts. Stem Cells. 2008;29:29. doi: 10.1634/stemcells.2008-0346. [DOI] [PubMed] [Google Scholar]
- 23.Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T, et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, et al. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu JF, Vu TH, Hoffman AR. Genomic deletion of an imprint maintenance element abolishes imprinting of both insulin-like growth factor II and H19. J Biol Chem. 1997;272:20715–20720. doi: 10.1074/jbc.272.33.20715. [DOI] [PubMed] [Google Scholar]
- 28.Hu JF, Oruganti H, Vu TH, Hoffman AR. Tissue-specific imprinting of the mouse insulin-like growth factor II receptor gene correlates with differential allele-specific DNA methylation. Mol Endocrinol. 1998;12:220–232. doi: 10.1210/mend.12.2.0062. [DOI] [PubMed] [Google Scholar]
- 29.Chen HL, Li T, Qiu XW, Wu J, Ling JQ, Sun ZH, et al. Correction of aberrant imprinting of IGF2 in human tumors by nuclear transfer-induced epigenetic reprogramming. Embo J. 2006;25:5329–5338. doi: 10.1038/sj.emboj.7601399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martys JL, Shevell T, McGraw TE. Studies of transferrin recycling reconstituted in streptolysin O permeabilized Chinese hamster ovary cells. J Biol Chem. 1995;270:25976–25984. doi: 10.1074/jbc.270.43.25976. [DOI] [PubMed] [Google Scholar]
- 31.Hansis C, Barreto G, Maltry N, Niehrs C. Nuclear reprogramming of human somatic cells by xenopus egg extract requires BRG1. Curr Biol. 2004;14:1475–1480. doi: 10.1016/j.cub.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 32.Hu JF, Vu TH, Hoffman AR. Promoter-specific modulation of insulin-like growth factor II genomic imprinting by inhibitors of DNA methylation. J Biol Chem. 1996;271:18253–18262. doi: 10.1074/jbc.271.30.18253. [DOI] [PubMed] [Google Scholar]
- 33.Chen TL, Shen WJ, Qiu XW, Li T, Hoffman AR, Kraemer FB. Generation of novel adipocyte monolayer cultures from embryonic stem cells. Stem Cells Dev. 2007;16:371–380. doi: 10.1089/scd.2006.0037. [DOI] [PubMed] [Google Scholar]
- 34.Darabi R, Gehlbach K, Bachoo RM, Kamath S, Osawa M, Kamm KE, et al. Functional skeletal muscle regeneration from differentiating embryonic stem cells. Nat Med. 2008;14:134–143. doi: 10.1038/nm1705. [DOI] [PubMed] [Google Scholar]
- 35.Shi Y, Hou L, Tang F, Jiang W, Wang P, Ding M, et al. Inducing embryonic stem cells to differentiate into pancreatic beta cells by a novel three-step approach with activin A and all-trans retinoic acid. Stem Cells. 2005;23:656–662. doi: 10.1634/stemcells.2004-0241. [DOI] [PubMed] [Google Scholar]
- 36.Zhang SC, Wernig M, Duncan ID, Brustle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol. 2001;19:1129–1133. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- 37.Hu J, Vu T, Hoffman A. Differential biallelic activation of three insulin-like growth factor II promoters in the mouse central nervous system. Mol Endocrinol. 1995;9:628–636. doi: 10.1210/mend.9.5.7565809. [DOI] [PubMed] [Google Scholar]
- 38.Yang Y, Hu JF, Ulner G, Li T, Yao X, Vu TH, et al. Epigenetic regulation of Igf2/H19 imprinting at CTCF insulator binding sites. J Cell Biochem. 2003;90:1038–1055. doi: 10.1002/jcb.10684. [DOI] [PubMed] [Google Scholar]
- 39.Humpherys D, Eggan K, Akutsu H, Hochedlinger K, Rideout WM, 3rd, Biniszkiewicz D, et al. Epigenetic instability in ES cells and cloned mice. Science. 2001;293:95–97. doi: 10.1126/science.1061402. [DOI] [PubMed] [Google Scholar]
- 40.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 41.Cavaleri F, Scholer HR. Nanog: a new recruit to the embryonic stem cell orchestra. Cell. 2003;113:551–552. doi: 10.1016/s0092-8674(03)00394-5. [DOI] [PubMed] [Google Scholar]
- 42.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 43.Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 44.Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, et al. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 45.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 48.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 49.Woodbury D, Reynolds K, Black IB. Adult bone marrow stromal stem cells express germline, ectodermal, endodermal, and mesodermal genes prior to neurogenesis. J Neurosci Res. 2002;69:908–917. doi: 10.1002/jnr.10365. [DOI] [PubMed] [Google Scholar]
- 50.Zuk PA, Zhu M, Mizuno H, Huang J, Futrell JW, Katz AJ, et al. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng. 2001;7:211–228. doi: 10.1089/107632701300062859. [DOI] [PubMed] [Google Scholar]
- 51.Barry FP, Murphy JM. Mesenchymal stem cells: clinical applications and biological characterization. Int J Biochem Cell Biol. 2004;36:568–584. doi: 10.1016/j.biocel.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 52.Boquest AC, Shahdadfar A, Brinchmann JE, Collas P. Isolation of stromal stem cells from human adipose tissue. Methods Mol Biol. 2006;325:35–46. doi: 10.1385/1-59745-005-7:35. [DOI] [PubMed] [Google Scholar]
- 53.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mathieu O, Bender J. RNA-directed DNA methylation. J Cell Sci. 2004;117:4881–4888. doi: 10.1242/jcs.01479. [DOI] [PubMed] [Google Scholar]
- 55.Petersen CP, Bordeleau ME, Pelletier J, Sharp PA. Short RNAs repress translation after initiation in mammalian cells. Mol Cell. 2006;21:533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 56.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 57.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 58.Cannell IG, Kong YW, Bushell M. How do microRNAs regulate gene expression? Biochem Soc Trans. 2008;36:1224–1231. doi: 10.1042/BST0361224. [DOI] [PubMed] [Google Scholar]
- 59.Barroso-del Jesus A, Lucena-Aguilar G, Menendez P. The miR-302-367 cluster as a potential stemness regulator in ESCs. Cell Cycle. 2009;8:394–398. doi: 10.4161/cc.8.3.7554. [DOI] [PubMed] [Google Scholar]
- 60.Lin TA, Tsay C, Chen CH, Tang PC, Ju JC. Nuclear and cytoskeletal dynamics during oocyte maturation and development of somatic cell cloned pig embryos injected with membrane disintegrated donor cells. Anim Reprod Sci. 2008;103:107–119. doi: 10.1016/j.anireprosci.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 61.Chang KH, Lim JM, Kang SK, Lee BC, Moon SY, Hwang WS. Blastocyst formation, karyotype, and mitochondrial DNA of interspecies embryos derived from nuclear transfer of human cord fibroblasts into enucleated bovine oocytes. Fertil Steril. 2003;80:1380–1387. doi: 10.1016/j.fertnstert.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 62.Zhang S, Wang J, Bilen MA, Lin SH, Stupp SI, Satcher RL. Modulation of prostate cancer cell gene expression by cell-to-cell contact with bone marrow stromal cells or osteoblasts. Clin Exp Metastasis. 2009;29:29. doi: 10.1007/s10585-009-9289-0. [DOI] [PubMed] [Google Scholar]
- 63.Bouziges F, Simo P, Simon-Assmann P, Haffen K, Kedinger M. Altered deposition of basement-membrane molecules in co-cultures of colonic cancer cells and fibroblasts. Int J Cancer. 1991;48:101–108. doi: 10.1002/ijc.2910480119. [DOI] [PubMed] [Google Scholar]
- 64.Grinnell KL, Bickenbach JR. Skin keratinocytes pre-treated with embryonic stem cell-conditioned medium or BMP4 can be directed to an alternative cell lineage. Cell Prolif. 2007;40:685–705. doi: 10.1111/j.1365-2184.2007.00464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]