Abstract
Arachidonic acid (AA), an essential polyunsaturated fatty acid, is one of the major components of neural membranes, which show an altered phospholipid composition in schizophrenia. Arachidonate 12-lipoxygenase (ALOX12), an important enzyme, metabolizes AA to 12-HPETE, which affects catecholamine synthesis. However, research has yet to show the genetic association between ALOX12 and schizophrenia. Therefore, we investigated single nucleotide polymorphisms (SNP) of the ALOX12 gene in schizophrenia, recruiting patients with schizophrenia (n = 289) and normal controls (n = 306) from a Korean population. We selected three SNPs (rs1126667, rs434473, and rs1042357) of the ALOX12 gene and genotyped them by direct sequencing. We reviewed the schizophrenic patients' medical records and assessed them clinically using the Brief Psychiatric Rating Scale (BPRS), the Scale for the Assessment of Negative Symptoms (SANS), and the Operational Criteria Checklist (OPCRIT). Then we statistically analyzed the genetic associations between the SNPs and schizophrenia, finding a genetic association between both rs1126667 and rs1042357 and schizophrenia, in the recessive model (p = 0.015 and 0.015, respectively). We also found an association between rs434473 and negative symptoms, defined through a factor analysis of the OPCRIT data (p = 0.040). Consequently, we suggest that SNPs of the ALOX12 gene might be associated with schizophrenia and negative symptoms in this Korean population. These weak positives require additional study.
Findings
Essential polyunsaturated fatty acids (EPUFAs) are major components of neural membranes. These membranes show an altered composition in schizophrenia patients [1]. Arachidonic acid (AA) and docosahexaenoic acid (DHA) constitute 80-90% of EPUFAs in neuronal tissue, but there is reduced AA in schizophrenia [2]. The enzyme ALOX12 oxygenates the C-12 of arachidonic acid, producing 12-hydroperoxy-5, 8, 10, 14- eicosatetraenoic acid (12-HPETE) [3], which, research has shown, inhibits protein kinase II [4]. Protein kinase II phosphorylates tyrosine hydroxylase, which is the rate-limiting enzyme in catecholamine synthesis [5]. Accordingly, 12-HPETE might affect dopamine synthesis. In addition, 12- HPETE inhibits neurotransmitter release per se [6]. However, no research has yet shown the association between ALOX12 and schizophrenia. Therefore, we hypothesized genetic variances of the ALOX12 gene might be associated with schizophrenia. To test this hypothesis, we investigated the single nucleotide polymorphisms (SNPs) of the ALOX12 gene in schizophrenia patients.
We recruited patients meeting the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV) criteria for schizophrenia [7], resulting in a study population consisting of 289 Korean patients with schizophrenia [189 men and 100 women; age 42.85 ± 10.84 (mean ± S.D., in years)]. We also recruited 306 Korean control subjects (148 men and 158 women; age 36.06 ± 6.79). We reviewed the medical records of each patient and assessed the schizophrenia patients using the Brief Psychiatric Rating Scale (BPRS), the Scale for the Assessment of Negative Symptoms (SANS), and the operational criteria (OPCRIT) checklist [8]. All studies were carried out according to the guidelines of the Declaration of Helsinki [9]. Written informed consent was obtained from each subject, and the study was approved by the Institutional Review Board of Kyung Hee University Hospital, Seoul, Republic of Korea. We selected three coding SNPs with validated heterozygosity (>0.2; http://www.ensembl.org, http://www.ncbi.nlm.nih.gov/SNP, http://www.hapmap.org): coding region SNPs rs1126667, rs434473, and rs1042357 (SNP database, BUILD 129, Table 1). We used direct sequencing to conduct SNP genotyping and amplified Genomic DNA was using the following primers for each SNP: rs1126667 (sense, 5'- TCAACTCAGAGAGGCCTTGAGAA-3'; antisense, 5'- AAGTGAGGA AGTGCCATCAGGTG-3'; 622 bp), rs434473 and rs1042357 (sense, 5'- CTCC TTCACATTCCACCACCATC-3'; antisense, 5'- GTGAGTGAAGAGGAGACTGT CTC-3'; 625 bp). Then, we sequenced the samples using an ABI Prism 377 automatic sequencer (PE Applied Biosystems, Foster City, CA, USA) and analyzed sequence data using SeqMan II software (DNASTAR Inc., Madison, WI, USA). The genotyping completion rates of our samples were 96.4% (rs1126667), 95.0% (rs434473), and 98.1% (rs1042357). We excluded samples with 1 or more misread or unreadable SNPs. Therefore, 95.0% of the original samples were analyzed. We analyzed Hardy-Weinberg equilibrium (HWE) and genotype frequencies using SNPStats [10] and used multiple logistic regression models to calculate odds ratios (OR), 95% confidence intervals (CI), and corresponding p values; to control age and gender as covariables; and to analyze the association between SNPs and schizophrenia. To calculate the power of the sample size, we used a genetic power calculator http://pngu.mgh.harvard.edu/~purcell/gpc. In our case-control study, the powers were 0.8142 (rs1126667; effective sample size; number of cases for 80% power = 292), 0.8105 (rs434473, n = 292), and 0.8141 (rs1042357, n = 296). However, the sample power in subgroup-divided schizophrenia was not sufficient (data not shown). We also tested clinical variables, including OPCRIT checklist data. The significance level for all statistical tests was 0.05, and we applied Bonferroni corrections for multiple tests in the association study (number of tests = 18).
Table 1.
Genotype distribution of SNPs of the ALOX12 gene among schizophrenia patients in a Korean population.
| SNP | Amino acid | Genotype | Control N = 306 |
Schizophrenia N = 289 |
Model | OR | 95% CI | p | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Freq. | % | Freq. | % | LCL | UCL | ||||||
| rs1126667 | R261Q | G/G | 96 | 31.4 | 77 | 26.6 | Codominant | 1.09 | 0.75 | 1.59 | 0.048 |
| (Exon 6) | A/G | 151 | 49.4 | 132 | 45.7 | Dominant | 1.26 | 0.88 | 1.80 | 0.200 | |
| A/A | 59 | 19.3 | 80 | 27.7 | Recessive | 1.60 | 1.09 | 2.35 | 0.015* | ||
| rs434473 | S322N | A/A | 96 | 31.4 | 78 | 27.0 | Codominant | 1.09 | 0.74 | 1.58 | 0.073 |
| (Exon 8) | A/G | 153 | 50.0 | 135 | 46.7 | Dominant | 1.24 | 0.87 | 1.76 | 0.240 | |
| G/G | 57 | 18.6 | 76 | 26.3 | Recessive | 1.56 | 1.06 | 2.30 | 0.025* | ||
| rs1042357 | T364T | C/C | 95 | 31.1 | 77 | 26.6 | Codominant | 1.07 | 0.73 | 1.57 | 0.050* |
| (Exon 8) | A/C | 152 | 49.7 | 132 | 45.7 | Dominant | 1.24 | 0.87 | 1.77 | 0.240 | |
| A/A | 59 | 19.3 | 80 | 27.7 | Recessive | 1.60 | 1.09 | 2.35 | 0.015* | ||
ALOX12, arachidonate 12-lipoxygenase; R, arginine; Q, glutamine; S, serine; N, asparagine; T, threonine; Freq, frequency; OR, odds ratio; CI, confidence interval; LCL, lower confidence limit; UCL, upper confidence limit; p, probability value.
*These values are not significant after the Bonferroni correction (number of tests = 18).
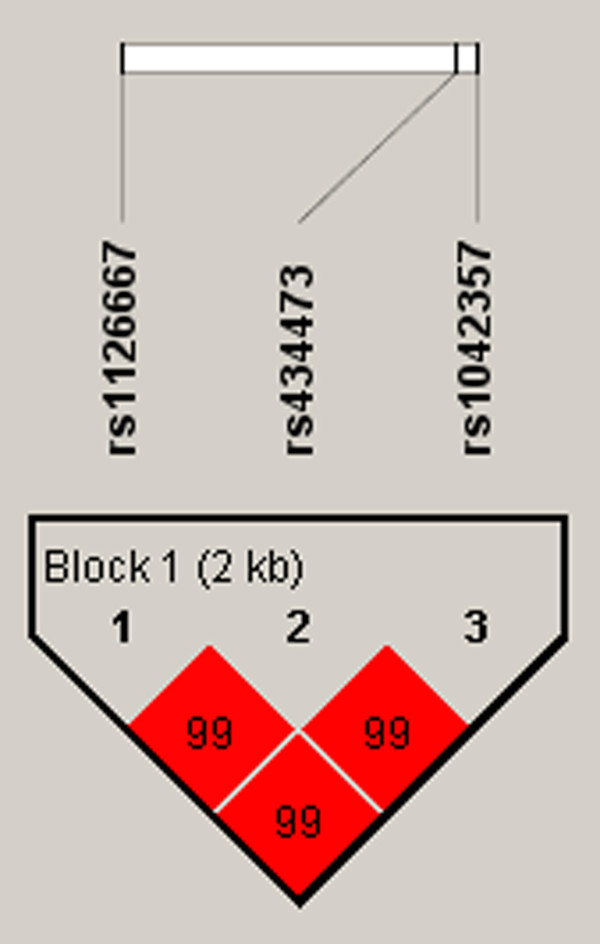
We found no deviation from the Hardy-Weinberg equilibrium for the selected SNPs. The SNPs rs1126667 and rs1042357 showed significant associations in the recessive model after Bonferroni correction (p = 0.015, OR = 1.60, 95% CI = 1.09-2.35; p = 0.015, OR = 1.60, 95% CI = 1.09-2.35, Table 1). We found no genetic association with gender, schizophrenia subtypes, BPRS, SANS, or any of the OPCRIT items. However, when we categorized the OPCRIT items into four symptom subgroups (excitement, negative symptoms, hallucinations, and delusions) by factor analysis, we found an association between rs434473 and the negative symptom subgroup (Table 2). While our genetic data is statistically significant, the p values of our data are only weakly positive. We characterized the LD block between the three ALOX12 SNPs in the total subjects, using the pairwise D' values (Figure 1). The D' values from rs4730775 to rs32024233 and from rs37795488 to rs17132543 ranged over 0.99, indicating a strong LD between each pair of markers (Figure 1). This revealed an LD block, comprised of rs1126667, rs434473, and rs1042357.
Table 2.
Genotype distribution of ALOX12 gene SNPs among schizophrenia patients in a Korean population.
| SNP | Amino acid | Genotype |
Negative† (+) N = 168 |
Negative† (-) N = 121 |
χ2 | df | P | ||
|---|---|---|---|---|---|---|---|---|---|
| Freq. | % | Freq. | % | ||||||
| rs1126667 | R261Q | G/G | 41 | 24.4 | 36 | 29.8 | |||
| (Exon 6) | A/G | 86 | 51.2 | 46 | 38.0 | 4.984 | 2 | 0.083 | |
| A/A | 41 | 24.4 | 39 | 32.2 | |||||
| rs434473 | S322N | A/A | 41 | 24.4 | 37 | 30.6 | |||
| (Exon 8) | A/G | 89 | 53.0 | 46 | 38.0 | 6.428 | 2 | 0.040* | |
| G/G | 38 | 22.6 | 38 | 31.4 | |||||
| rs1042357 | T364T | C/C | 41 | 24.4 | 36 | 29.8 | |||
| (Exon 8) | A/C | 86 | 51.2 | 46 | 38.0 | 4.984 | 2 | 0.083 | |
| A/A | 41 | 24.4 | 39 | 32.2 | |||||
ALOX12, arachidonate 12-lipoxygenase; R, arginine; Q, glutamine; S, serine; N, asparagine; T, threonine; Freq, frequency; df, degree of freedom; p, probability value.
*The value is not significant after the Bonferroni correction (number of test = 18).
†Negative symptom subgroup identified by factor analysis of OPCRIT data.
Figure 1.
Linkage disequilibrium (LD) block of ALOX12 gene. LD coefficient (|D'|) and LD blocks among ALOX12 SNPs. Block consists of rs1126667, rs434473, and rs1042357.
Lipoxygenases are non-heme iron dioxygenase enzymes that insert molecular oxygen into free and esterified polyunsaturated fatty acids [11]. Research has suggested that ALOX12 might be involved with neurodegeneration [12]. Pratico et al. [13] showed that ALOX12 is higher in Alzheimer's disease, suggesting that it correlates with brain oxidative stress. In our data, rs1126667 and rs1042357 showed associations with schizophrenia in the recessive model. A/A carriers of rs1126667 and A/A carriers of rs1042357 had significantly higher odds ratios, of 1.60 (95% CI = 1.09-2.35), for schizophrenia. The SNP rs1126667 is a missense SNP that substitutes glutamine for arginine at amino acid position 261. This SNP is located in the lipoxygenase domain (amino acid position: from 124 to 655) in the ALOX12 protein. However, no research has yet shown a functional change in catalytic ability due to rs1126667. Although rs1042357 is synonymous, it can make an exonic splicing enhancer (5'- AGAAGC-3' and 5'- CAGAAG-3') with its A allele, whereas its C allele does not make one (genes.mit.edu/burgelab/rescue-ese). Thus, the A allele of rs1042357 in the lipoxygenase domain of the ALOX12 protein may lead to inaccurate splicing of pre-mRNA of the ALOX12 gene, which can produce an abnormal ALOX12 enzyme. In addition, rs434473 was associated with the negative symptom subgroup in OPCRIT items, which comprise slower activity, loss of energy/tiredness, loss of pleasure, and diminished libido (Table 2). There is evidence that 12-HPETE, the metabolite of arachidonate made by ALOX12 correlates with synthesis of catecholamine. High 12-HPETE inhibits protein kinase II [4]. In addition, research has shown protein kinase II is responsible for the phosphorylation of tyrosine hydroxylase, which is the rate-limiting step in the synthesis of (something is missing) [5]. The relatively less-phosphorylated tyrosine hydroxylase leads to reduced dopamine levels. In addition, 12-HPETE inhibits neurotransmitter release [6]. Thus, hypodopaminergia caused by decreased synthesis and release of dopamine might be present in deficit schizophrenia [14]. ALOX12 has been studied in connection with various malignancies, including colorectal cancer [11,12,15], breast cancer [16,17], prostate cancer [18-20], bladder cancer [21], and testicular cancer [22]. Research has shown that schizophrenia patients have an increased malignancy risk regarding colon cancer, marginally increased risk of breast cancer, and decreased risk of respiratory cancer [23]. ALOX12 might be one of the factors explaining the high risk of colorectal cancer in patients with schizophrenia. Thus, the relationship between ALOX12 and the higher incidence of colorectal cancer in schizophrenia patients needs future study. We conclude that rs1126667 and rs1042357 of the ALOX12 gene correlate with schizophrenia in a Korean population, and rs434473 correlates with negative symptoms in schizophrenia. These weak positives require additional replication study. These results suggest that future studies should regard ALOX12 as an important gene in the pathology of schizophrenia.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
Authors JWK and JHC designed and directed the entire project. HJK managed the literature searches and analyses. JKP carried out schizophrenia assessments and gave advice on patient selection. Author TK performed most of the statistical analyses and the genotyping and contributed substantially to the first draft of the manuscript. All authors contributed to, and have approved, the final manuscript
Contributor Information
Tae Kim, Email: tae_kim@hms.harvard.edu.
Hak-Jae Kim, Email: hj4165@gmail.com.
Jin Kyung Park, Email: parkdawit@naver.com.
Jong Woo Kim, Email: psyjongwoo@freechal.com.
Joo-Ho Chung, Email: jhchung@khu.ac.kr.
Acknowledgements
This work was supported by a 2006 grant from the Kyung Hee University (KHU-20060427).
References
- Horrobin DF. The membrane phospholipid hypothesis as a biochemical basis for the neurodevelopmental concept of schizophrenia. Schizophr Res. 1998;30:193–208. doi: 10.1016/S0920-9964(97)00151-5. [DOI] [PubMed] [Google Scholar]
- Ward PE, Sutherland J, Glen EM, Glen AI. Niacin skin flush in schizophrenia: a preliminary report. Schizophr Res. 1998;29:269–74. doi: 10.1016/S0920-9964(97)00100-X. [DOI] [PubMed] [Google Scholar]
- Yoshimoto T, Yamamoto Y, Arakawa T, Suzuki H, Yamamoto S, Yokoyama C, Tanabe T, Toh H. Molecular cloning and expression of human arachidonate 12-lipoxygenase. Biochem Biophys Res Commun. 1990;172:1230–5. doi: 10.1016/0006-291X(90)91580-L. [DOI] [PubMed] [Google Scholar]
- Piomelli D, Wang JK, Sihra TS, Nairn AC, Czernik AJ, Greengard P. Inhibition of Ca2+/calmodulin-dependent protein kinase II by arachidonic acid and its metabolites. Proc Natl Acad Sci USA. 1989;86:8550–4. doi: 10.1073/pnas.86.21.8550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa H, Okuno S. Regulation of the activity of tyrosine hydroxylase in the central nervous system. Adv Enzyme Regul. 1989;28:93–110. doi: 10.1016/0065-2571(89)90066-6. [DOI] [PubMed] [Google Scholar]
- Piomelli D, Greengard P. Lipoxygenase metabolites of arachidonic acid in neuronal transmembrane signalling. Trends Pharmacol Sci. 1990;11:367–73. doi: 10.1016/0165-6147(90)90182-8. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders. 4. Washington, DC: American Psychiatric Press; 1994. [Google Scholar]
- McGuffin P, Farmer A, Harvey I. A polydiagnostic application of perational criteria in studies of psychotic illness. Development and reliability of the OPCRIT system. Arch Gen Psychiatry. 1991;48:764–70. doi: 10.1001/archpsyc.1991.01810320088015. [DOI] [PubMed] [Google Scholar]
- Rickham PP. Human Experimentation. Code of Ethics of the World Medical Association. Declaration of Helsinki. Br Med J. 1964;2:177. doi: 10.1136/bmj.2.5402.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sole X, Guino E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22:1928–9. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- Brash AR. Lipoxygenases: occurrence, functions, catalysis, and acquisition of substrate. J Biol Chem. 1999;274:23679–82. doi: 10.1074/jbc.274.34.23679. [DOI] [PubMed] [Google Scholar]
- Li Y, Maher P, Schubert D. A role for 12-lipoxygenase in nerve cell death caused by glutathione depletion. Neuron. 1997;19:453–63. doi: 10.1016/S0896-6273(00)80953-8. [DOI] [PubMed] [Google Scholar]
- Pratico D, Zhukareva V, Yao Y, Uryu K, Funk CD, Lawson JA, Trojanowski JQ, Lee VM. 12/15-lipoxygenase is increased in Alzheimer's disease: possible involvement in brain oxidative stress. Am J Pathol. 2004;164:1655–62. doi: 10.1016/S0002-9440(10)63724-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis KL, Kahn RS, Ko G, Davidson M. Dopamine in schizophrenia: a review and reconceptualization. Am J Psychiatry. 1991;148:1474–86. doi: 10.1176/ajp.148.11.1474. [DOI] [PubMed] [Google Scholar]
- Goodman JE, Bowman ED, Chanock SJ, Alberg AJ, Harris CC. Arachidonate lipoxygenase (ALOX) and cyclooxygenase (COX) polymorphisms and colon cancer risk. Carcinogenesis. 2004;25:2467–72. doi: 10.1093/carcin/bgh260. [DOI] [PubMed] [Google Scholar]
- Gong Z, Hebert JR, Bostick RM, Deng Z, Hurley TG, Dixon DA, Nitcheva D, Xie D. Common polymorphisms in 5-lipoxygenase and 12-lipoxygenase genes and the risk of incident, sporadic colorectal adenoma. Cancer. 2007;109:849–57. doi: 10.1002/cncr.22469. [DOI] [PubMed] [Google Scholar]
- Tan W, Wu J, Zhang X, Guo Y, Liu J, Sun T, Zhang B, Zhao D, Yang M, Yu D, Lin D. Associations of functional polymorphisms in cyclooxygenase-2 and platelet 12-lipoxygenase with risk of occurrence and advanced disease status of colorectal cancer. Carcinogenesis. 2007;28:1197–1201. doi: 10.1093/carcin/bgl242. [DOI] [PubMed] [Google Scholar]
- Jiang WG, Douglas-Jones A, Mansel RE. Levels of expression of lipoxygenases and cyclooxygenase-2 in human breast cancer. Prostaglandins Leukot Essent Fatty acids. 2003;69:275–81. doi: 10.1016/S0952-3278(03)00110-8. [DOI] [PubMed] [Google Scholar]
- Mohammad AM, Abdel HA, Abdel W, Ahmed AM, Wael T, Eiman G. Expression of cyclooxygenase-2 and 12-lipoxygenase in human breast cancer and their relationship with HER-2/neu and hormonal receptors: impact on prognosis and therapy. Indian J Cancer. 2006;43:163–8. doi: 10.4103/0019-509X.29421. [DOI] [PubMed] [Google Scholar]
- Kandouz M, Nie D, Pidgeon GP, Krishnamoorthy S, Maddipati KR, Honn KV. Platelet-type 12-lipoxygenase activates NF-kappaB in prostate cancer cells. Prostaglandins Other Lipid Mediat. 2003;71:189–204. doi: 10.1016/S1098-8823(03)00042-X. [DOI] [PubMed] [Google Scholar]
- Nie D, Nemeth J, Qiao Y, Zacharek A, Li L, Hanna K, Tang K, Hillman GG, Cher ML, Grignon DJ, Honn KV. Increased metastatic potential in human prostate carcinoma cells by overexpression of arachidonate 12-lipoxygenase. Clin Exp Metastasis. 2003;20:657–63. doi: 10.1023/A:1027302408187. [DOI] [PubMed] [Google Scholar]
- Matsuyama M, Yoshimura R, Mitsuhashi M, Hase T, Tsuchida K, Takemoto Y, Kawahito Y, Sano H, Nakatani T. Expression of lipoxygenase in human prostate cancer and growth reduction by its inhibitors. Int J Oncol. 2004;24:821–7. [PubMed] [Google Scholar]
- Yoshimura R, Matsuyama M, Tsuchida K, Kawahito Y, Sano H, Nakatani T. Expression of lipoxygenase in human bladder carcinoma and growth inhibition by its inhibitors. J Urol. 2003;170:1994–9. doi: 10.1097/01.ju.0000080296.54262.c8. [DOI] [PubMed] [Google Scholar]