Abstract
N1-meA and N3-meC are cytotoxic DNA base methylation lesions that can accumulate in the genomes of various organisms in the presence of SN2 type methylating agents. We report here the structural characterization of these base lesions in duplex DNA using a cross-linked protein–DNA crystallization system. The crystal structure of N1-meA:T pair shows an unambiguous Hoogsteen base pair with a syn conformation adopted by N1-meA, which exhibits significant changes in the opening, roll and twist angles as compared to the normal A:T base pair. Unlike N1-meA, N3-meC does not establish any interaction with the opposite G, but remains partially intrahelical. Also, structurally characterized is the N6-meA base modification that forms a normal base pair with the opposite T in duplex DNA. Structural characterization of these base methylation modifications provides molecular level information on how they affect the overall structure of duplex DNA. In addition, the base pairs containing N1-meA or N3-meC do not share any specific characteristic properties except that both lesions create thermodynamically unstable regions in a duplex DNA, a property that may be explored by the repair proteins to locate these lesions.
INTRODUCTION
Genomic DNA is constantly subjected to modifications caused by exogenous environmental chemicals and cellular metabolites. Among various DNA damages, nucleobase damage is a common type that can be induced by oxidation, hydrolysis and alkylation (1,2). If left unrepaired, these lesions can cause deleterious effects on nearly all aspects of cellular functions (3,4). Therefore, guarding genetic integrity by repairing DNA damage is one of the most fundamental processes of life (5,6). Most DNA base damage is repaired through four biochemically and mechanistically distinct pathways: direct reversal repair (DRR), base excision repair (BER), nucleotide excision repair (NER) and mismatch repair (MMR) (7).
Structural characterization of base lesions in double-stranded DNA (dsDNA) is important to understand the origin of their mutagenic or cytotoxic effects. The structural information may also help to reveal how the lesions are detected and repaired by DNA repair proteins. Despite considerable advances in DNA structure characterization (8), high-quality crystals of naked dsDNA containing base lesions are hard to obtain. Even with the Dickerson–Drew dodecamer sequence (9–12), base lesions can introduce unstable base pairs, which prevent the formation of well-ordered 3D crystal lattice. In order to aid DNA crystallization, host–guest systems employing various DNA-binding proteins have been used to obtain high-quality crystals (13–17). Although this method has helped to solve the structures of some important lesions (16,17), many more remain to be elucidated. The host–guest approach has also been shown to be effective for RNA crystallization (18–22).
Alkylated nucleobases are produced mostly by endogenous and environmental alkylation agents (23). N1-methyladenine (N1-meA) and N3-methylcytosine (N3-meC) are two forms of base methylations generated usually in single-stranded DNA (ssDNA). These lesions are unable to form normal Watson–Crick base pairs, and consequently block DNA replication, resulting in cytotoxicity to cells. Escherichia coli alkB is an inducible gene of the adaptive response to alkylating agents. The alkB gene encodes a DNA-repair protein that counteracts toxic alkylation damages, such as N1-meA, N3-meC and 1,N6-ethenoadenine (24–28). Human homologues of AlkB can process similar base modifications and exhibits profound functional roles in human cells (29–31). Solving the structures of these base modifications will be important to visualize how these lesions affect the local dsDNA structure, thereby shedding light on the origin of the cytotoxic effects of the damage. The structure of 1, N6-ethenoadenine:T pair was recently characterized using the DNA glycosylase AlkA bound to the ends of dsDNA (17). The crystal structures of the other two lesions, N1-meA and N3-meC in dsDNA, remain unknown. We have solved the structure of hABH2, the human protein that repairs N1-meA and N3-meC in ssDNA and dsDNA (32,33), cross-linked to dsDNA (34). As most of the DNA bases have no direct contact with the protein residues in the ABH2–dsDNA structure (PDB 3BTX, Supplementary Figure 1), this complex seems to be a suitable host–guest-like system for characterizing lesion-containing dsDNA structures. In our design, the protein–dsDNA complex is stabilized through covalent cross-linking in the protein active site, whereas the lesioned base is installed to the locations without any direct contact to the protein (Supplementary Figure S2). A noticeable advantage of this approach versus other host–guest-like systems is its robustness since the complex is stitched together with a covalent bond. The complex cannot be crystallized in the absence of this cross-linking.
This host–guest-like system is validated through the structural characterization of the methylation base modifications N1-meA, N3-meC and N6-meA in dsDNA cross-linked to ABH2. In these structures, N1-meA forms an unambiguous anti-Hoogsteen base pair to the opposite T; N3-meC does not form any hydrogen-bonding interaction with the opposite G, but stays partially intrahelical inside the duplex DNA.
MATERIALS AND METHODS
Oligonucleotide synthesis and purification
Oligonucleotides containing disulfide-tethered cytosine and specific base lesions were synthesized using the phosphoramidite derivative of O4-triazoyl-dU (Glen Research), and functionalized with cystamine. The DNA oligomers were synthesized on a Biosystems Expedite Nucleic Acid Synthesis System using standard reagents. Following synthesis, the resin was dried under vacuum and then incubated with 200 µl of solution containing 50% cystamine in water at room temperature for 16 h. The aqueous solution was collected and the resin was washed twice with 200 µl water. The washes were combined and neutralized to pH 6.0–6.5 with glacial acetic acid on ice and desalted by a NAP-10 column (GE healthcare). The aqueous solution was then lyophilized and finally purified by reverse-phase HPLC. The dsDNA used in the crystallization of N1-meA and N6-meA is DNA1: 5′-CTGTATC*AT(meA)GCG-3′ paired with 5′-TCGCTATAATACA-3′. The dsDNA used for N3-meC is DNA 2: 5′-CTGT(meC)TC*ATTGCG-3′ paired with 5′-TCGCAATAAGACA-3′. Masses of the synthesized oligonucleotides were verified by MALDI-TOF mass spectrometry.
Cloning, expression and purification of ABH2-ΔN55 E175C
The abh2-ΔN55 gene was cloned between the NdeI and HindIII sites of a pET28a vector (Novagen). ABH2-ΔN55 E175C mutants were generated by using QuikChange Site-Directed Mutagenesis Kit (Stratagene) and this plasmid was transformed into E. coli BL21(DE3) cells (Stratagene) for protein overexpression. The protein was purified using HisTrap HP column (GE Healthcare) with an elution buffer of 50 mM sodium phosphate (pH 8.0), 300 mM NaCl and 400 mM imidazole. The N-terminal His-tag was removed by an overnight thrombin (MP Bio) digestion at 4°C and the tag-free protein was purified the next day with another round of HisTtrap chromatography. About 4–6 mg of protein could be obtained from 1 l of bacterial culture.
Preparing cross-linking, crystallization and data collection of the ABH2–DNA complexes
DNA duplexes were annealed by mixing 1 mM thiol-tether containing strand with the corresponding complimentary strand in buffer containing 10 mM Tris (pH 7.4) and 100 mM NaCl, incubating at 75°C for 10 min and cooling to 4°C by a step gradient of −1°C/min. The cross-linked complexes of protein ABH2-ΔN55 E175C with synthetic oligonucleotides were achieved by incubating dsDNA (1 mM, 50 µl) with protein (1.2 eq.) in 5 ml of buffer [100 mM NaCl, 10 mM Tris–HCl (pH 7.4)] at 14°C for 16 h. The covalently linked ABH2–DNA complexes were purified using MonoQ anion exchange chromatography (GE healthcare), then buffer exchanged to a solution containing 100 mM NaCl, 10 mM Tris–HCl (pH 8.0) (Supplementary Figure S3). Finally, the complexes were concentrated to 5 mg/ml for use in crystallization experiments.
Protein–dsDNA complex crystals were grown by hanging drop vapor diffusion crystallization at 4°C in drops containing 1 µl of complex solution and 1 µL of reservoir solution of 100 mM NaCl, 50 mM MgCl2, 100 mM cacodylate (pH 6.5) and 12% 5 K PEG. Hexagonal rod-shaped crystals grew in 1–2 weeks at 4°C. Subsequently, crystals were transferred to a cryoprotectant solution composed of 80% reservoir solution and 20% glycerol and frozen in liquid nitrogen before data collection. An X-ray data set (diffracted to 2.0 Å) for the N1-meA-containing complex (ABH2–N1-meA) crystals and an X-ray data set (diffracted to 1.8 Å) for the N6-meA-containing complex (ABH2–N6-meA) were collected at beamline 23ID-B (General Medicine and Cancer Institutes Collaborative Access Team [GM/CA-CAT]) of the Advanced Photon Source at Argonne National Laboratory. The N3-meC-containing complex (ABH2–N3-meC) gave the best crystals that diffracted to 2.0 Å (Table 1). Data were integrated and processed with the HKL2000 package.
Table 1.
Data collection and refinement statistics
| Crystal | ABH2–N1-meA:T | ABH2–N6-meA:T | ABH2–N3-meC:G |
|---|---|---|---|
| Data quality | |||
| Resolution (Å) | 20–2.0 (2.05–2.0) | 20–1.8 (1.77–1.8) | 20–2.0 (1.95–2.0) |
| Unique reflections | 29687 | 38371 | 31492 |
| Completenessa (%) | 99.9 (99.9) | 99.9 (100) | 98.3 (78.6) |
| Redundancy | 20.0 | 20.3 | 21.1 |
| <I/σ>a | 24.3 (2.6) | 32.6 (1.6) | 36.2 (2.2) |
| Crystal parameters | |||
| Space group | P6522 | P6522 | P6522 |
| Cell constants (Å) | a = 79.11 | a = 78.08 | a = 79.11 |
| b = 79.11 | b = 78.08 | b = 79.11 | |
| c = 242.27 | c = 228.71 | c = 242.30 | |
| α, β, γ (°) | α = 90 | α = 90 | α = 90 |
| β = 90 | β = 90 | β = 90 | |
| γ = 120 | γ = 120 | γ = 120 | |
| Refinement | |||
| Resolution (Å) | 20–2.0 | 20–1.77 | 20–1.95 |
| Rworkb (%) | 20.2 | 21.0 | 20.5 |
| Rfreec (%) | 23.7 | 23.6 | 23.6 |
| Model quality | |||
| R.m.s. deviation bond (Å) | 0.015 | 0.010 | 0.0107 |
| R.m.s. deviation angle (°) | 1.58 | 1.53 | 1.57 |
| Average B factor | 27.7 | 29.2 | 36.5 |
| Average B factor of Protein | 27.1 | 27.9 | 34.6 |
| Average B factor of DNA | 23.1 | 29.0 | 39.6 |
| Average B factor of water | 39.4 | 42.5 | 45.3 |
| Model content | |||
| Protein residues | 56–258 | 56–258 | 56–258 |
| Nucleotides | 26 | 26 | 26 |
| Water atoms | 197 | 222 | 179 |
| PDB accession code | 3H8O | 3H8R | 3H8X |
aValues in parentheses refer to the highest resolution bins.
bRwork = Σ |Fo – Fc|/Σ Fo, where Fo and Fc are observed and calculated structure factor amplitudes, respectively.
cRfree was calculated based on a percentage of data (5%) randomly selected and omitted through the structure refinement procedure.
Structure determination and refinement
The ABH2–dsDNA complex structures were phased by molecular replacement [using Phaser (35)], using the previously published ABH2–dsDNA structure as a search model [PDB 3BTX (34)]. The model was built by using COOT and refinement was carried out with the program REFMAC5 from the CCP4 suite. Atomic coordinates for the three structures reported have been deposited in Protein Data Bank under accession number 3H8O (ABH2–N1-meA), 3H8R (ABH2–N6-meA) and 3H8X (ABH2–N3-meC).
RESULTS
Overall structures
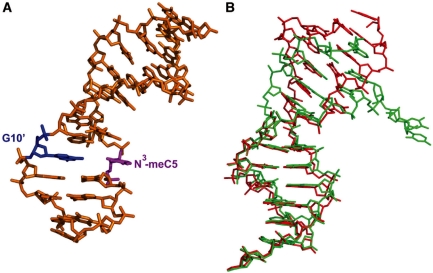
In an attempt to crystallize duplex DNAs containing the methylated base N1-meA and N3-meC, we employed the Dickerson–Drew dodecamer sequence (9–12), which failed to yield crystals. Then, we decided to use the cross-linked ABH2-dsDNA complex (34) as a host–guest-like system for crystallization since the covalent cross-linking stabilizes the protein–dsDNA complexes. The lesion-containing dsDNA is cross-linked to the ABH2 protein with an engineered Cys at position 175 (ABH2-ΔN55 E175C) (Supplementary Figure S2). Out of multiple DNA sequences screened, a 13-mer dsDNA1 (Figure 1B), with a central C*:A base pair (C* represents a disulfide-tethered cytosine), yielded the best quality crystals of the ABH2–dsDNA complexes containing N1-meA (Figure 1A) or N6-meA (Figure 1E). Another sequence, dsDNA2 (Figure 1D), with N3-meC at a different location, gave the best crystals for ABH2–dsDNA complex with N3-meC (Figure 1C). All of these complexes bearing modified nucleotides are crystallized in the hexagonal space group P6522, and the crystals diffract to 1.8 Å for the N6-meA-containing complex (ABH2–N6-meA), and 2.0 Å for both N1-meA- and N3-meC-containing complexes (ABH2–N1-meA and ABH2–N3-meC, Table 1).
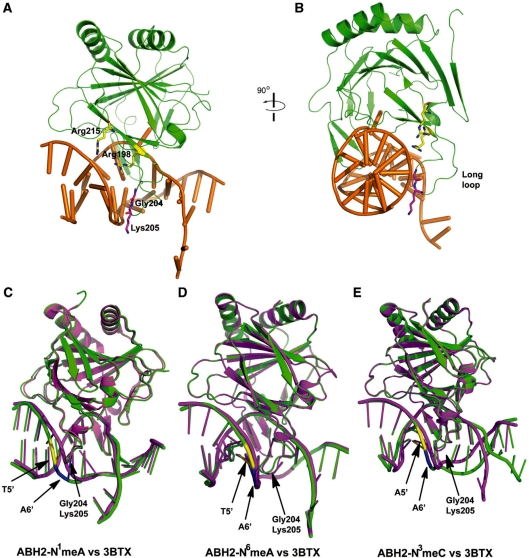
Figure 1.
Crystal structure of the ABH2–dsDNA complex. (A) Cartoon of the ABH2–dsDNA1 complex with N1-meA. The protein is shown in green, DNA in orange, N1-meA in magenta and complementary T in blue. (B) Schematic diagram showing the interaction between ABH2 and dsDNA 1 containing N1-meA. Solid arrows stand for direct interactions and dashed arrows for water mediated contacts. (C) Cartoon trace of the ABH2–dsDNA2 complex with N3-meC. The same color coding in A is used with N3-meC in magenta and the opposite G in blue. (D) Schematic diagram showing the interaction between ABH2 and dsDNA 2 containing N3-meC. (E) Cartoon of the ABH2–dsDNA1 complex with N6-meA. The same color coding in (A) is used with N6-meA in magenta and complementary T in blue. (F) Schematic diagram showing the interaction between ABH2 and dsDNA 1 containing N6-meA.
The structures of ABH2–N1-meA and ABH2–N6-meA overlap well with the original structure 3BTX (Supplementary Figure S5), with a root mean square deviation (RMSD) of 0.49 Å and 0.21 Å, respectively. Except for the site of the flipped-out C*7, the DNA adopts a right-handed conformation and its interactions with the protein are mainly through the phosphodeoxyribose backbones, which are mostly located in the middle of the lesion-containing strand and 5′-end of the complementary strand. Interestingly, the structure of ABH2–N1-meA complex shows reduced contacts between protein residues and the backbone of dsDNA at the 5′-end of the complementary strand. For instance, the backbone region around T5′ (which pairs with N1-meA) and its flanking nucleotides, A6′ and C4′ (Figure 2), do not form hydrogen bonds with the protein residues Arg198, Gly204, Lys205 and Arg215; such hydrogen-bonding contacts exist in the original structure of 3BTX. Besides Phe102, the finger residue that helps to flip C*7 out of the duplex DNA, no other protein residue affects base pairing in both structures of ABH2–N1-meA and ABH2–N6-meA as well as in 3BTX (34).
Figure 2.
Interaction of ABH2 protein to the dsDNA backbone. (A) Cartoon diagram of the ABH2–DNA1 complex with N1-meA. The protein is shown in green, DNA in gold, Arg198 and Arg215 in yellow and two residues on the loop (Gly204, Lys205) in magenta. (B) Same structure as in (A), rotated 90° to the right to show the side view of the residues and dsDNA. (C) Overall superposition of ABH2–N1-meA complex with 3BTX to show the backbone distortion caused by N1-meA. ABH2–N1-meA complex in green with backbone of T10' in yellow and A9′ in blue. 3BTX in magenta. (D) Overall superposition of ABH2–N6-meA complex with 3BTX to show the backbone distortion caused by N6-meA. The color coding is same as in (C). (E) Overall superposition of ABH2–N3-meC complex with 3BTX to show the backbone distortion caused by N3-meC. Same color coding in (C) is used.
A superposition of the structure of ABH2–N3-meC to 3BTX shows an apparent DNA backbone shift toward the 3′-end of the complementary strand (Figure 2E), which is most likely caused by different packing of the DNA ends. The original structure (3BTX) has the terminal bases twisted out of the duplex DNA to interact with the neighboring oligonucleotide; the terminal bases in ABH2–N3-meC remain intrahelical and form coaxial stacking with the neighboring duplex. As a result, the DNA bending induced by DNA end packing is slightly released when compared to the original structure (3BTX). Similar to ABH2–N1-meA, a loss of contact between ABH2 and the backbone region around the nucleotides C4′-A6′ was observed in the structure of ABH2–N3-meC.
Structures of ABH2–N1-meA and ABH2–N6-meA
Besides minor shift and rotation, the packing of the dsDNA helix in ABH2–N1-meA is similar to the unlesioned DNA in 3BTX (Figure 3). The N1-meA-containing dsDNA shows two slightly different backbone conformations, which do not seem to cause noticeable changes in the base-pairing mode of the DNA structure (Supplementary Figure S6). As N1-meA can be converted to N6-meA via Dimroth rearrangement (36), we also crystallized ABH2–N6-meA for comparison. The DNA structure with N6-meA and its alignment to the unlesioned DNA in 3BTX are shown in Supplementary Figure S7. Besides an ∼15° bending of the DNA caused by the end packing of DNA, the overall DNA conformation in these two structure remains basically B-form with an average rise per base pair along axis of around 3.2 Å, a slide of −0.5 Å and a roll of 3.6°. Because N6-meA forms Watson–Crick base pair with the complementary T (Figure 4E), this N6-meA-containing dsDNA basically resembles an unmodified B-form-like duplex structure.
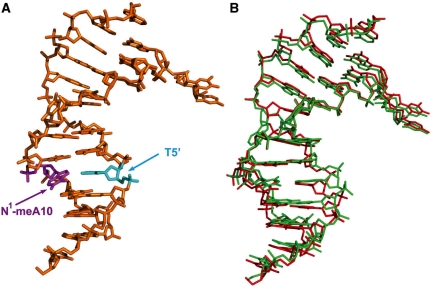
Figure 3.
Diagram of the dsDNA1 structure containing N1-meA:T. (A) Structure of dsDNA1 with N1-meA:T. N1-meA10 in magenta, opposite T5′ in cyan. dsDNA 1: 5′-CTGTATC*AT(1meA)GCG-3′ paired with 5′-TCGCTATAATACA-3′. (B) Structure of dsDNA1 with N1-meA:T aligned with the unlesioned dsDNA in structure 3BTX. DNA containing N1-meA in red, unlesioned DNA in green. dsDNA in 3BTX: 5′-CTGTATC*ATT GCG-3′ paired with 5′-TCGCAATAATACA-3′.
Figure 4.
Structure of DNA1 with N1-meA or N6-meA lesion opposite with T. (A) Side view of N1-meA:T lesion and neighboring base pairs. Atoms are colored as follows: carbon, green; nitrogen, blue; oxygen, red; phosphate, orange. (B) Fo – Fc simulated annealing omit electron density map of N1-meA:T pair. The Fo – Fc simulated annealing omit electron density map is represented as an orange mesh contoured at 3σ and the 2Fo – Fc map in gray mesh. The color scheme is same as in (A). Black dotted lines represent hydrogen bonds. (C) The N1-meA:T base pair as viewed from the major groove, illustrating an overall dihedral around 30°. Same color coding in (A) is applied. (D) Side view of N6-meA:T and neighboring base pairs. (E and F) Fo – Fc simulated annealing omit electron density map of N6-meA:T pair from top and major groove, respectively. Color scheme and dotted lines are same as in (B).
To perform a detailed analysis of the distortion of the duplex structure caused by the N1-meA lesion, the structure of the N1-meA-containing oligonucleotide was superimposed onto that of the N6-meA-containing DNA (Supplementary Figure S8). The base pair parameters of these two DNA structures were calculated using the program 3DNA v1.5 (37) and the changes for propeller twist, opening, buckle, roll and twist are shown in Figure 5. Major changes occur around the lesion site, where the positions of N1-meA10, complementary T5′, and the flanking base pair G11:C4′ shift relative to their positions in the structure of the N6-meA-containing duplex. These differences are results from the N1-methylation of A10, which blocks the normal Watson–Crick base pairing (Figures 3 and 4).
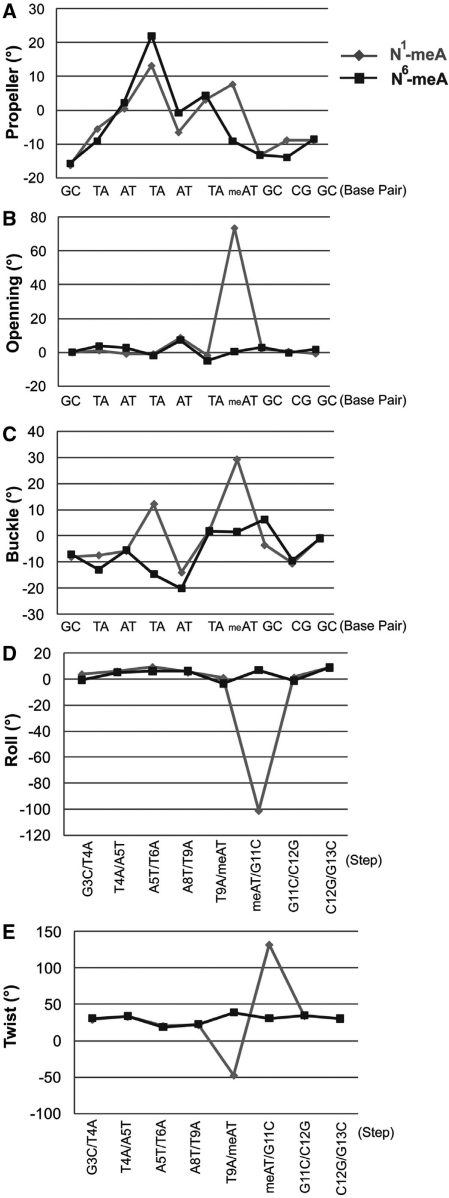
Figure 5.
Helix parameters of the dsDNA with N1-meA:T pair (light gray) and the one with N6-meA:T (dark grey), as calculated by program 3DNA v1.5. The base pairs are numbered from G3·C12′ to G13·C2′ in the absence of the C*7·A8′ mismatch and the steps are numbered from G3·C12′/T4·A11′ to C12·G3′/G13·C2′ without two steps neighbored to the C*7·A8′ mismatch. Most of the remarkable changes are at the local position of the N1-meA:T pair.
A Hoogsteen base pair between N1-meA10 and the opposite base T5′ is clearly observed with two hydrogen bonds formed between these two bases (N2_ N1-meA10 … O4_T5′, 3.1 Å; N7_ N1-meA10 … N3_ T5′, 3.2 Å; Figure 4B). Although the Hoogsteen base-pair mode has been reported by the NMR study of the N1-meA:T pair (38), the distances we observed here are much shorter than what have been assigned previously (around 4.3 Å). To accommodate both the methylated adenine and the pyrimidine ring of T within the duplex, the purine ring of N1-meA adopts a syn conformation with respect to the N9 atom around the N-glycosidic bond. In addition, a distinct dihedral angle of about 30° is found between the plane of N1-meA10 and the opposite T5′ (Figures 4C and 6), which is significantly larger than that in the ABH2–N6-meA structure (Figure 4D). Interestingly, despite dramatic changes occurring on the N1-meA10:T5′ base pair, the neighboring base pair T9:A6′ (5′ to the lesion) is almost unaffected, but the base pair G11:C4′ (3′ to N1-meA), shifts slightly towards the 3′ direction of the lesion-containing strand (Figure 6A). Compared to the N6-meA structure, the rise of the N1-meA:T pair to its flanking base pairs was decreased, which was caused by the inclination of the N1-meA base. In spite of this, the overall distance from T9:A6′ base pair to G11:C4′ is similar to that in a typical B-form DNA, which may indicate that the influence of the lesion is mainly limited to the local structure, but not the overall duplex.
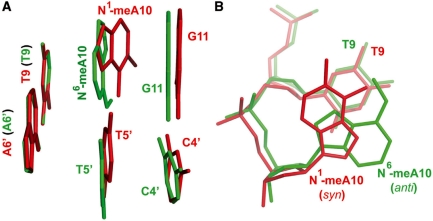
Figure 6.
Structure comparison of N1-meA:T lesion with N6-meA:T base pair. (A) Side view structure of N1-meA10:T5′ with neighboring base pairs aligned with N6-meA10:T5’. N1-meA10:T5′ with neighboring base pairs appears in red, and N6-meA10:T5′ with neighboring base pairs in green. The figure illustrates a significant dihedral between N1-meA10 and N6-meA10 when two bases are overlaid together. (B) Overlay of N1-meA10 and adjacent T9 (in red) with N6-meA10 and neighbor T9 (in green) as viewed from top. The purine ring of N1-meA10 is in syn conformation, and the N6-meA10 base is in anti-conformation.
Since the protein has no direct contact to the base pairing region around N1-meA10 or N6-meA10, an alignment of the two duplex DNAs in this region (Figure 6) allows a clear comparison of the properties of N1-meA with those of N6-meA, and reveals impacts on the local DNA structure by these modifications. It can be seen that N1-meA adopts a syn conformation to form the Hoogsteen base pair instead of the anti conformation adopted by N6-meA in a Watson–Crick base pair. A comparison of N1-meA:T pair with the N6-meA:T (Table 2) reveals that the shear of N1-meA:T pair is bigger (0.7 Å compared to −0.05 Å), and so is the stretch value (−3.8 Å in N1-meA:T versus −0.2 Å in N6-meA:T). The stagger of N1-meA:T amounts to −1.1 Å (0.01 Å in N6-meA:T), which is mainly caused by a shift of the N1-methylated adenine into the major groove by ∼3.0 Å and a shift of the opposite T by ∼1.7 Å. The propeller twist of the methylated base pair is varied from −9.2° in N6-meA:T pair to 7.7° in N1-meA:T, while the two flanking base pairs are almost unaffected. The dramatically increased base opening of N1-meA:T pair is observed (73.6° compared to 0.3° in N6-meA:T, Figure 5B), as well as the greater buckle angle (29.1° versus 1.5°). However, the buckle value of G11:C4′ in the N1-meA structure, 3′ to the lesion pair, is decreased to −3.6°, which is 6.3° for the corresponding base pair in the N6-meA structure. Apparent changes are also observed in the N1-meA structure compared to the N6-meA and the unlesioned ones when calculating the base pair step parameters as shown in Table 2. The tilt and twist values of the N1-meA:T pair to both flanking base pairs show significant changes. With respect to the roll angles, only the value of the N1-meA:T pair to 3′ adjacent G11:C4′ pair is altered dramatically to −101.2° (7.0° in N6-meA structure), but that of the 5′ neighbored T9:A9′ pair is only slightly affected. Furthermore, the C1′–C1′ distance between N1-meA and the opposite T decrease to 9.0 Å (10.5 Å in N6-meA structure), and the distances of RN9–YN1 and RC8–YC6 are also shortened by 1.7 Å and 3.1 Å, respectively, compared to those in the N6-meA:T pair. These changes might be induced by the rotation of N1-meA base around the glycosidic bond, which ‘drags’ the complementary thymine further to the center of the helix (Supplementary Figure 9). A slight DNA distortion of the backbone is indicated by an elongation of the P_T9 … P_A′10 from 7.2 Å in the N6-meA-containing structure to 7.4 Å in the case of N1-meA, which probably explains the lack of interaction between the local backbone and the protein.
Table 2.
Comparison of base pair parameters of N1-meA:T, N6-meA:T and T:Aa
| Local base-pair parametersb | |||||||
| Shear | Stretch | Stagger | Buckle | Propeller | Opening | ||
| N1-meA10:T10 | 0.7 | −3.8 | −1.1 | 29.1 | 7.7 | 73.6 | |
| N6-meA10:T10 | −0.05 | −0.2 | 0.01 | 1.5 | −9.2 | 0.3 | |
| T10:A10 | −0.05 | −0.2 | −0.2 | 1.4 | −21.0 | −0.5 | |
| Local base-pair step parametersc | |||||||
| Shift | Slide | Rise | Tilt | Roll | Twist | ||
| N1-meA | T9A/ N1-meA10T | −1.7 | −3.8 | 2.4 | −175.7 | 1.1 | −47.6 |
| N1-meA10T/G11C | −0.8 | −3.5 | −1.7 | 135.6 | −101.2 | 132.3 | |
| N6-meA | T9A/ N6-meA10T | −0.2 | −0.2 | 3.4 | −0.9 | −3.3 | 38.9 |
| N6-meA10T/G11C | 0.4 | −0.02 | 3.2 | −2.8 | 7.0 | 31.3 | |
| 3BTX | T9A/T10A | −0.3 | −0.04 | 3.2 | 2.9 | −3.7 | 40.2 |
| T10A/G11C | 0.8 | 0.9 | 3.6 | 0.4 | 6.0 | 37.3 | |
aAll data are calculated by Program 3DNA (v1.5) (ref. 37).
bParameters for Shear, Stretch and Stagger are distances (Å). Parameters for Buckle, Propeller and Opening are angles (°).
cParameters for Shift, Slide and Rise are distances (Å). Parameters for Tilt, Roll and Twist are angles (°).
A comparison of the structure of N1-meA:T to that of the Hoogstein base-paired A:T reveals that the same syn conformation of N1-meA or adenine is observed in both structures. A similar shortening of the C1′-C1′ distance is also observed (9.0 Å for N1-meA:T pair, 8.5 Å for A:T Hoogsteen base pair, as opposed to 10.5 Å for A:T Watson–Crick base pair) (39,40). However, the distinct dihedral angle of ∼30° between the plane of N1-meA10 and the opposite T5′ as well as the major changes in twist angles in this region, are only found in the N1-meA:T structure, which could be explained by the steric effect of the N1-methylation.
Structure of dsDNA-containing N3-meC
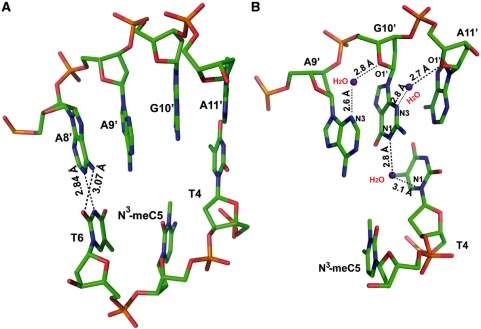
Due to the lack of hydrogen bonding between N3-meC and the opposite G, the crystal structure of ABH2–N3-meC shows a distorted backbone compared to the complex without the base lesion. The DNA ends are packed differently in two structures (Figure 7) that helps to release the bending of the dsDNA caused by DNA end packing. The absence of any interaction between N3-meC to the opposite G leads to a shift of N3-meC towards the major groove. This base has a relatively poor electron density indicating its high flexibility. Instead of completely flipping out, the N3-meC base stays partially intrahelical (Figure 8A), with a tilt angle around 18° (0.6° for B-form DNA) and a twist angle of 49° (36° for B-form DNA). The distances of P_T4 … P_N3-meC5 and P_N3-meC5 … P_T6 are stretched to 7.2 Å and 6.9 Å, respectively. In contrast, the complementary guanine of N3-meC remains intrahelical, stabilized by the stacking to A11′ and A9′, its two flanking bases. Water-mediated hydrogen bonds of N3_G10′… O1′_A11′ and O1′_G10′… N3_A9′ also contribute to the stabilization of the guanine. In addition, another water-mediated interaction between G10′ and T4, N1_G10′ to water (2.8 Å) and water to N1_T4 (3.1 Å), might also help to ‘lock’ the unpaired G10′ intrahelix. Although the neighboring base pair, T4:A11′, is not apparently influenced by this lesion, a marked change occurred to the base pair T6:A9′. This base pair is disrupted such that T6 adopts a syn conformation to form a Wobble base pair with A8′ (N3_T6 … N1_A8′, 2.8 Å; O2_T6 … N6_A8′, 3.1 Å), which is the base opposite the flipped out C7*. The disruption of the two base pairs around the N3-meA lesion site creates a pocket that is partly occupied by the unpaired N3-meC5 and A9′. Benefitting from potential stacking with the adjacent A8′ and G10′, nucleotide A9′ stays in this pocket in the absence of base pairing. These observed distortions may suggest that the introduction of N3-meC lesion to dsDNA results in a significant alternation to the duplex structure.
Figure 7.
Diagram of the DNA structure containing N3-meC:G. (A) Structure of dsDNA2 with N3-meC:G. N3-meC5 in magenta, opposite G10’ in blue. dsDNA 2: 5′-CTGT(3meC)TC*ATTGCG-3′ paired with 5′-TCGCAATAAGACA-3′. (B) Structure of dsDNA2 with N3-meC:G aligned with the unlesioned dsDNA in structure 3BTX. DNA- containing N3-meC in red, unlesioned DNA in green. dsDNA in 3BTX: 5′-CTGTATC*ATT GCG-3′ paired with 5′-TCGCAATAATACA-3′. Different packing pattern at the end of the dsDNA is observed.
Figure 8.
Structure of dsDNA2 with N3-meC lesion opposite with G and its flanking base pairs. (A) Side view of N3-meC5 lesion complementary with G10′ and neighboring base pairs. Atoms are colored as follows: carbon, green; nitrogen, blue; oxygen, red; phosphate, orange. A Wobble base pairing between T6 and A8′ is shown in place of the Watson–Crick base-pairing of T6 with A9′. Black dotted lines represent hydrogen bonds. (B) Interactions between G10′ with the neighboring nucleotides help unpaired G10′ and A9′ stay intrahelical. Atoms are in same color coding as (A) with water in purple.
DISCUSSION
Cytotoxic damages such as N1-meA and N3-meC can block Watson–Crick base pairing in duplex DNA, which leads to cytotoxic consequences to living cells. These base lesions are efficiently detected and repaired by the AlkB family proteins (24–31). To structurally characterize these methylation bases in duplex DNA, a disulfide cross-linked ABH2–dsDNA complex was employed as a host–guest-like system for DNA crystallization. To minimize potential influence from the bound protein, we placed the methylated bases away from the interface between the protein and DNA.
This protein–DNA complex, stabilized by covalent cross-linking between the protein and DNA, affords the first crystal structures of N1-meA and N3-meC in duplex DNA. We show that N1-meA forms a Hoogsteen base pair with the opposite T with the purine ring of N1-meA adopting a syn conformation, which is consistent with the previous NMR result (38). This crystal structure also provides detailed parameters on the N1-meA:T pair and its impact on the local DNA duplex structure. Due to steric clashes caused by the N1-methylated adenine, we observed a 3 Å shift of N1-meA base into the major groove in the structure, as well as significant changes in the values of the opening, tilt and twist angles. A shortening of C1′–C1′, RN9–YN1 and RC8–YC6 distances between N1-meA and the opposite T is observed, probably induced by the displacement of the N1-meA to the major groove and the movement of the complementary thymine toward the center of the helix. As a result of these conformational changes, the neighboring base pair G11:C4’ shifts to the 3′ direction, while the 5′ neighboring base pair T9:A6′ is left almost unaffected. A detailed investigation of ABH2–N1-meA also reveals that the protein residues, which normally establish contacts with the backbone in the middle of the lesion-containing strand and 5′-end of the complementary strand, lose most of the hydrogen-bonding interactions with the backbone of the complementary strand (Figure 1B). This observation suggests that: (i) these interactions to the complementary strand might be weak, perhaps non-existent when the ABH2 protein is scanning through the duplex DNA to detect potential base lesions; (ii) the covalent cross-linking is important for holding the complex together, thus providing necessary robustness to this host–guest-like system with a low binding affinity in the absence of cross-linking.
We also report the structure of the ABH2–dsDNA complex containing the N3-meC lesion. The electron density of N3-meC is poor due to the absence of any hydrogen-bonding interaction to the opposing G. However, N3-meC remains intrahelical with a movement of ∼2 Å to the major groove. The opposite guanine is also intrahelical and stacks to the neighboring A11′ and A9′. The disruption of the N3-meC5:G10′ base pair also yields an elongated back-bone P … P distance between C5 to T4 and T6. Furthermore, the flanking base pair between T6 and A9′ is broken, which leads to a Wobble base pairing between T6 and A8′. The melting of the DNA structure around the lesion site generates low thermodynamic stability and produces the distortion of the DNA backbone, an effect that may lead to the loss of interactions between the protein and the backbone around the A6′–C4′ stretch on the complementary strand. These changes in DNA structure may explain why this lesioned base cannot be crystallized through typical DNA-crystallization strategies.
Both N1-meA and N3-meC are efficiently detected and repaired by the AlkB family proteins, yet they show distinct structural features in duplex DNA. The only common property of the base pairs containing these two lesions is the low thermodynamic stability of the local duplex DNA structure induced by the methylation. Thus, the repair proteins could detect unstable regions in the genome to locate these base damages.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
FUNDING
National Institutes of Health (GM071440 to C.H.); Beamline 23ID-B (General Medicine and Cancer Institutes Collaborative Access Team ([GM/CA-CAT]) at the Advanced Photon Source at Argonne National Laboratory; National Institutes of Health and the United States Department of Energy. Funding for open access charge: GM071440.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the staff at beamline 23BM (General Medicine and Cancer Institute Collaborative Access Team [GM/CA-CAT]) at the Advanced Photon Source at Argonne National Laboratory for data collection, and Dr E. Duguid for data processing.
REFERENCES
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.De Bont R, van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19:169–185. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- 3.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz AR, Ellenberger T. Biological response to DNA damage. In: Friedberg EC, editor. DNA Repair and Mutagenesis. Washington DC: ASM Press; 2005. Chapter 1, pp. 2–7. [Google Scholar]
- 4.Lindahl T. Molecular biology: ensuring error-free repair. Nature. 2004;427:598. doi: 10.1038/427598a. [DOI] [PubMed] [Google Scholar]
- 5.Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science. 2001;291:1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- 6.Truglio JJ, Croteau DL, Van Houten B, Kisher C. Prokaryotic nucleotide excision repair: the UvrABC system. Chem. Rev. 2006;106:233–252. doi: 10.1021/cr040471u. [DOI] [PubMed] [Google Scholar]
- 7.Sancar A. DNA repair in humans. Annu. Rev. Genetics. 1995;29:69–105. doi: 10.1146/annurev.ge.29.120195.000441. [DOI] [PubMed] [Google Scholar]
- 8.Egli M. Nucleic acid crystallography: current progress. Curr. Opin. Chem. Biol. 2004;8:580–591. doi: 10.1016/j.cbpa.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 9.Drew HR, Wing RM, Takano T, Broka C, Tanaka S, Itakura K, Dickerson RE. Structure of a B-DNA dodecamer: conformation and dynamics. Proc. Natl. Acad. Sci. USA. 1981;78:2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hossain MT, Sunami T, Tsunoda M, Hikima T, Chatake T, Ueno Y, Matsudda A, Takénaka A. Crystallographic studies on damaged DNAs IV. N4-methoxycytosine shows a second face for Watson-Crick base-pairing, leading to purine transition mutagenesis. Nucleic Acid Res. 2001;29:3949–3954. doi: 10.1093/nar/29.19.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park H, Zhang K, Ren Y, Nadji S, Sinda N, Taylor J.-S. Crystal structure of a DNA decamer containing a cis-syn thymine dimer. Proc. Natl Acad. Sci. USA. 2002;99:15965–15970. doi: 10.1073/pnas.242422699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freisinger E, Fernandes A, Grollman AP, Kisker C. Crystallographic characterization of an exocyclic DNA adduct: 3,N4-etheno-2′-deoxycytidine in dodecamer 5′-CGCGAATTεCGCG-3′. J. Mol. Biol. 2003;329:685–697. doi: 10.1016/s0022-2836(03)00445-5. [DOI] [PubMed] [Google Scholar]
- 13.Cote ML, Yohannan SJ, Georgiadis MM. Use of an N-terminal fragment from Moloney murine leukemia virus reverse transcriptase to facilitate crystallization and analysis of a pseudo-16-mer DNA molecule containing G-A mispairs. Acta Crystallogr. D Biol. Crystallogr. 2000;56:1120–1131. doi: 10.1107/s0907444900008246. [DOI] [PubMed] [Google Scholar]
- 14.Najmudin S, Coté ML, Sun M, Yohannan S, Montano SP, Gu J, Georgiadis MM. Crystal structure of an N-terminal fragment from Moloney murine leukemia virus reverse transcriptase complexed with nucleic acid: functional implications for template-prime binding to the fingers domain. J. Mol. Biol. 2000;296:613–632. doi: 10.1006/jmbi.1999.3477. [DOI] [PubMed] [Google Scholar]
- 15.Goodwin KD, Long EC, Georgiadis MM. A host-guest approach for determining drug-DNA interactions: an example using netropsin. Nucleic Acid Res. 2005;33:4106–4116. doi: 10.1093/nar/gki717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee S, Bowman BR, Ueno Y, Wang S, Verdine GL. Synthesis and structure of duplex DNA containing the genotoxic nucleobase lesion N7-methylguanine. J. Am. Chem. Soc. 2008;130:11570–11571. doi: 10.1021/ja8025328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowman BR, Lee S, Wang S, Verdine GL. Structure of the E. coli DNA glycosylase AlkA bound to the ends of duplex DNA: A system for the structure determination of lesion-containing DNA. Structure. 2008;16:1166–1174. doi: 10.1016/j.str.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferre-D′Amare AR, Zhou K, Doudna JA. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 19.Ferre-D′Amare AR, Doudna JA. Crystallization and structure determination of a hepatitis delta virus ribozyme: use of the RNA-binding protein U1A as a crystallization module. J. Mol. Biol. 2000;295:541–556. doi: 10.1006/jmbi.1999.3398. [DOI] [PubMed] [Google Scholar]
- 20.Ye J-D, Tereshkov V, Frederiksen JK, Koide A, Fellouse FA, Sidhu SS, Kide S, Kossiakoff AA, Piccirilli JA. Synthetic antibodies for specific recognition and crystallization of structured RNA. Proc. Natl Acad. Sci. USA. 2008;105:82–87. doi: 10.1073/pnas.0709082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao H, Murakami H, Suga H, Ferre-D′Amare AR. Structural basis of specific tRNA aminoacylation by a small in vitro selected ribozyme. Nature. 2008;454:358–361. doi: 10.1038/nature07033. [DOI] [PubMed] [Google Scholar]
- 22.Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, Yoneda Y, Tsukihara T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science. 2009;326:1275–1279. doi: 10.1126/science.1178705. [DOI] [PubMed] [Google Scholar]
- 23.Sedgwick B, Bates PA, Paik J, Jacobs SC, Lindahl T. Repair of alkylated DNA: recent advances. DNA Repair. 2007;6:429–442. doi: 10.1016/j.dnarep.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 24.Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419:174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 25.Falnes PO, Johansen RF, Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002;419:178–182. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- 26.Delaney JC, Essigmann JM. Mutagenesis, genotoxicity, and repair of 1-methyadenine, 3-alkycytosine, 1-methyguanine, and 3-methylthymine in alkB Escherichia coli. Proc. Natl Acad. Sci. USA. 2004;101:14051–14056. doi: 10.1073/pnas.0403489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mishina Y, Yang C.-G, He C. Direct repair of the exocyclic DNA adduct 1, N6-ethenoadenine by the DNA repair AlkB proteins. J. Am. Chem. Soc. 2005;127:14594–14595. doi: 10.1021/ja055957m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dlaney JC, Smeester L, Wong C, Frick LE, Taghizadeh k, Wishnok JS, Drennan CL, Samson LD, Essigmann JM. AlkB reverses etheno DNA lesion caused by lipid oxidation in vitro and in vivo. Nat. Struct. Mol. Biol. 2005;12:855–860. doi: 10.1038/nsmb996. [DOI] [PubMed] [Google Scholar]
- 29.Aas PA, Otterlei M, Falnes P.Ø., Vågbø CB, Skorpen F, Akbari M, Sundheim O, Bjørås M, Slupphang G, Seeberg E, et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 30.Westbye MP, Feyzl E, Aas PA, Vägbø CB, Talstad VA, Kavll B, Hagen L, Sundhelm O, Akbari M, Llabakk N.-B, et al. Human AlkB homolog I is a mitochondrial protein that demethylates 3-methylcytosine in DNA and RNA. J. Biol. Chem. 2008;283:25046–25056. doi: 10.1074/jbc.M803776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ougland R, Hang C.-M, Lilv A, Johansem RF, Seeberg E, Hou Y.-M, Remme J, Falnes P.Ø. AlkB restores the biological function of mRNA and tRNA inactivated by chemical methylation. Mol. Cell. 2004;16:107–116. doi: 10.1016/j.molcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 32.Duncan T, Trewick SC, Koivisto P, Bates PA, Lindahl T, Sedgwick B. Reversal of DNA alkylation damage by two human dioxygenases. Proc. Natl Acad. Sci. USA. 2002;99:16660–16665. doi: 10.1073/pnas.262589799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ringvoll J, Nordstrand LM, Vågbø CB, Talstad V, Reite K, Aas PA, Lauritzen KH, Liabakk NB, Bjørk A, Doughty RW, et al. Repair deficient mice reveal mABH2 as the primary oxidative demethylase for repairing 1meA and 3meC lesions in DNA. EMBO J. 2006;25:2189–2198. doi: 10.1038/sj.emboj.7601109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang C-G, Yi C, Duguid EM, Sullivan CT, Jian X, Rice PA, He C. Crystal structure of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature. 2008;452:961–965. doi: 10.1038/nature06889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. D. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 36.Jones JW, Robins RK. Purine nucleosides. III. Methylatioh studies of certain naturally occurring purine nucleosides. J. Am. Chem. Soc. 1963;85:193–201. [Google Scholar]
- 37.Lu X-J, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang H, Zhan Y, Fenn D, Chi LM, Lam SL. Effect of 1-methyadenine on double-helical DNA structures. FEBS Lett. 2008;582:1629–1633. doi: 10.1016/j.febslet.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 39.Aishima J, Gitti RK, Noah JE, Gan HH, Schlick T, Wolberger C. A hoogsteen base pair embedded in undistorted B-DNA. Nucleic Acids Res. 2002;30:5244–5252. doi: 10.1093/nar/gkf661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abrescia NG, Thompson A, Huynh-Dinh T, Subirana JA. Crystal structure od an antiparallel DNA fragment with Hoogsteen base pairing. Proc. Natl Acad. Sci. USA. 2002;99:2806–2811. doi: 10.1073/pnas.052675499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.