Abstract
G-quadruplexes are nucleic acid secondary structures for which many biological roles have been proposed but whose existence in vivo has remained elusive. To assess their formation, highly specific G-quadruplex ligands are needed. Here, we tested Phen-DC3 and Phen-DC6, two recently released ligands of the bisquinolinium class. In vitro, both compounds exhibit high affinity for the G4 formed by the human minisatellite CEB1 and inhibit efficiently their unwinding by the yeast Pif1 helicase. In vivo, both compounds rapidly induced recombination-dependent rearrangements of CEB1 inserted in the Saccharomyces cerevisiae genome, but did not affect the stability of other tandem repeats lacking G-quadruplex forming sequences. The rearrangements yielded simple-deletion, double-deletion or complex reshuffling of the polymorphic motif units, mimicking the phenotype of the Pif1 inactivation. Treatment of Pif1-deficient cells with the Phen-DC compounds further increased CEB1 instability, revealing additional G4 formation per cell. In sharp contrast, the commonly used N-methyl-mesoporphyrin IX G-quadruplex ligand did not affect CEB1 stability. Altogether, these results demonstrate that the Phen-DC bisquinolinium compounds are potent molecular tools for probing the formation of G-quadruplexes in vivo, interfere with their processing and elucidate their biological roles.
INTRODUCTION
G-quadruplexes (G4) are four-stranded nucleic acid helical structures that spontaneously form in vitro within certain G-rich sequences. The unitary motif of this structure is composed of four guanines stabilized by non-canonical H-bonding in a coplanar arrangement (called a G-quartet) (1). The uninterrupted stacking of at least three G-quartets stabilized by monovalent cations (Na+ or K+) is sufficient to form a G4. Quadruplexes can result from intra-molecular folding of one DNA strand containing four triplets of G separated by few bases or from intermolecular association of several strands and adopt a large variety of conformations, depending on the size and sequence of intervening loops, and have been documented by numerous structural studies (2).
Evidences concerning the in vivo formation of G4 and their involvement in several biological pathways remain limited but are starting to emerge (3–5). Formation of G4 in transcribed human G-rich DNA arrays in bacteria was visualized by electron microscopy (5). In ciliates, the formation of G4 was detected by immunochemistry (6). In this organism, the formation of G4 is triggered by specific telomere end-binding proteins which in turn regulates telomere protection from degradation and cell-cycle-dependent accessibility to telomerase (6,7). Using genetic approaches, the formation of G4 was shown to participate in the instability of the human CEB1 minisatellite inserted on a yeast chromosome (8), in a gene conversion pathway resulting in pilin antigenic variation in the bacteria Neisseria gonorrhoeae (9), and in the instability of guanine-rich regions in the Caenorhabditis elegans genome in absence of the dog-1 helicase (10). Complementarily, computational studies have provided a wealth of information concerning the occurrence and location within the genome of sequences having potential to form intra-molecular G4, as inferred from their primary DNA sequence (11). These potential G4-forming sequences are statistically over-represented at several loci, including telomeres of most eukaryotic organisms, at the rDNA loci in yeast (12) and human (13), and are significantly enriched in promoters sequences in human (14), yeast (12) and in C. elegans (15). These studies suggest that G4-DNA structures could exert a regulatory effect in cis on gene expression, either by recruiting factors at promoters or helping maintain a chromatin organization to favor or repress transcription. Some of these hypotheses have started to be submitted to experimental challenge, but it is still unclear how much of these potential G4 really form in vivo and how they specifically affect, for example, replication, transcription or recombination of genomic regions surrounding them.
To address the biological roles of G4, another approach of general use is the stabilization of these structures in vivo using specific ligands. The presence of G4-forming sequences at human telomeres and the fact that the first generation of G4-binding ligands were able to inhibit telomerase (16) have largely contributed to make G4 the archetypal higher order nucleic acid structure for the design of selectively targeting ligands in the presence of duplex DNA. Among the large number of ligands produced to date (17), the porphyrin derivatives N-methyl-mesoporphyrin IX (NMM, Figure 1A) and TmPyP4, and the perylene dimide derivative PIPER (18) are commercially available and thus have been often used for biochemical and biological studies. However, these two porphyrins are not optimal tools for probing quadruplex functions in vivo since NMM is selective for G4 over duplex DNA but is a relatively low-affinity ligand (19) and TmPyP4 has a high affinity but a poor selectivity for quadruplex DNA (20,21). Equally, the propensity of PIPER to aggregate in aqueous media (22) and its binding to duplex DNA renders its biological use questionable. The natural product telomestatin, which fulfills both requirements of selectivity and affinity, was promising and thus has been used to probe quadruplex structures in vivo and in vitro (23,24). However, telomestatin suffers several disadvantages such as poor water-solubility, chemical instability, and at present is only accessible by an arduous multi-step synthetic pathway (25) that makes large-scale use difficult. In this context, we recently developed the bisquinolinium family of compounds that includes Phen-DC3 and Phen-DC6 [Figure 1A, referred as compounds 2a and 2b respectively in ref. (26)], two promising molecules that emerged for their strong quadruplex stabilizing ability and an exquisite selectivity for quadruplex over duplex DNA. Indeed, the G4 recognition properties of Phen-DC3 and Phen-DC6 rival or surpass those of the best G4-binders such as Braco-19, telomestatin and their pyridine analogues (360A, 307A), which all exhibit a high selectivity for G4 and an affinity in the nanomolar range (27–29). In addition, the chemical stability and ease of preparation of our Phen-DC compounds are important advantages (27).
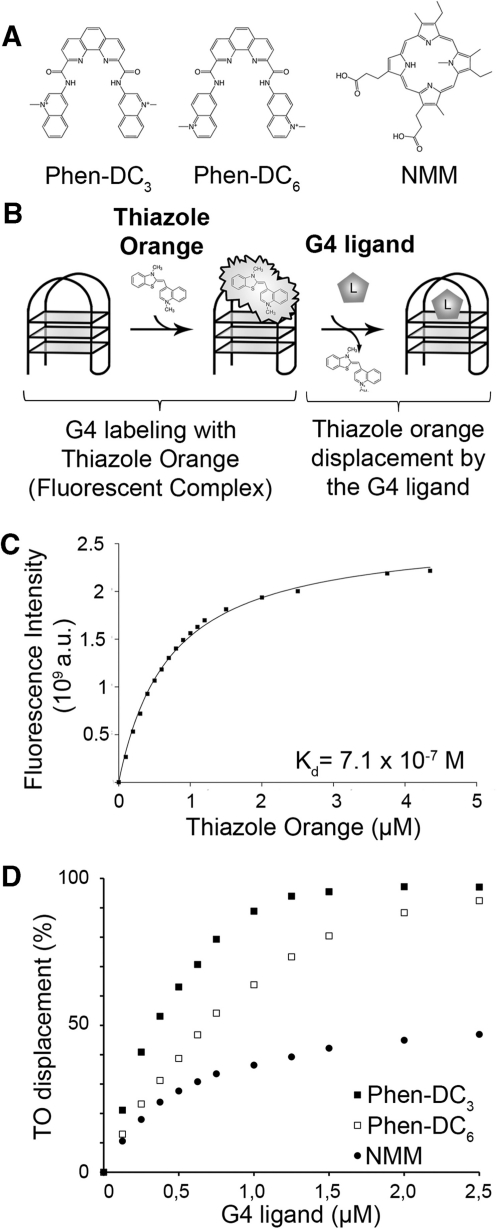
Figure 1.
Fluorescent intercalator displacement of the G-quadruplex formed by the CEB1 motif (G4-CEB1) with Phen-DC3, Phen-DC6 and NMM. (A) Phen-DC3, Phen-DC6 and NMM structures. (B) Schematic representation of the FID assay. (C) TO association curve with G4-CEB1. (D) TO displacement by (filled square) Phen-DC3, (open square) Phen-DC6 or (filled circle) NMM in presence of K+.
Although the Phen-DC compounds are very promising candidates to foster biological studies involving potential G4-forming sequences, their in vivo efficiency remains to be established (30). Therefore, we conducted in vitro and in vivo studies in the yeast Saccharomyces cerevisiae to assay the biological activity of the Phen-DC ligands with respect to the formation and the processing of G4, based on the ability of the human CEB1 minisatellite inserted in the yeast genome to efficiently form G4 structures and the ability of the yeast Pif1 helicase to resolve these structures (8). In budding yeast, the inactivation of Pif1, but not of other helicases, destabilizes the CEB1 tandem array and yields recombination-dependent size variants (8). We show that in vitro both Phen-DC compounds bind the G4 structure formed by CEB1 with high affinity, and inhibit G4 unwinding by Pif1 efficiently and selectively. In WT cells, Phen-DC compounds specifically induce the instability of G4-prone CEB1 arrays, a phenomenon further increased in Pif1-deficient mutants. These results demonstrate that the Phen-DC compounds are exquisite molecular tools to probe the in vivo formation of G4, to interfere with their processing, and thereof provide mechanistic insights.
MATERIALS AND METHODS
G-quadruplex fluorescence intercalator displacement assay
The fluorescence intercalator displacement (FID) assay was performed in lithium cacodylate buffer solution (10 mM LiAsO2Me2) containing NaCl or KCl (100 mM) and adjusted to pH 7.2 with HCl. The fluorescence spectra were recorded with a HORIBA Jobin-Yvon Fluoromax-3® spectrofluorimeter in the wavelength range from 510 to 750 nm, in a 3-ml quartz cell (path length 1 cm), using the following experimental parameters: excitation wavelength, 501 nm; increment, 1 nm; optical slit widths, 3.0/3.0 nm; integration time, 0.1 s. The fluorescence spectrum of the buffer solution was recorded and was systematically subtracted from all following spectra. The solutions of DNA (0.25 µM) were mixed with thiazole orange (TO) (final concentration 0.5 μM) and reference fluorescence spectra were recorded. Increasing concentrations (from 0 to 10 molar equivalents, i.e. from 0 to 2.5 µM) of ligands to be tested were then added. After a 3-min shaking and equilibration period, a fluorescence spectrum was recorded for each ligand addition step. The degree of TO displacement (percentage of ligand-induced decrease of TO fluorescence) is calculated from the fluorescence area FA as:
where FA0 is the fluorescence area of the reference spectrum. The percentage of displacement is then plotted as a function of the concentration of added compound.
Helicase assays
Helicase assays were carried out by incubating 100 nM of S. cerevisiae Pif1 and 2 nM of Cy5-labeled G4 DNA or 1 nM forked DNA substrate at 35°C, in a buffer containing 20 mM Tris pH 7.5, 50 mM NaCl, 100 µg/ml bovine serum albumin, 2 mM DTT, 5 mM Mg2+ and 4 mM ATP, and the indicated amount of the G4 binding ligands Phen-DC3, Phen-DC6 or NMM. Reactions were pre-incubated at 35°C for 10 min and started by addition of 4 mM ATP. Reactions (10 µl) with G4-DNA substrates were stopped by addition of 2 µl stop buffer (17% Ficoll, 50 mM EDTA, 3 mg/ml Proteinase K) and further incubated at 35°C for 10 min. Reaction products were loaded on a 8% polyacrylamide non-denaturing gel and resolved by electrophoresis at 4°C and 10V/cm in TBE 1X buffer containing 10 mM NaCl (for G4-DNA formed in NaCl) or 10 mM KCl buffer (for G4-DNA formed in KCl). Reactions containing the forked DNA substrate (10 µl) were stopped by addition of 2 µl of 17% Ficoll, 50 mM EDTA and 150 nM unlabelled fD20 oligonucleotide. Reaction products were loaded on a 10% polyacrylamide non-denaturing gel and resolved by electrophoresis at 4°C and 10 V/cm in TBE 1× buffer. Gels were scanned with a Storm PhosphorImager (Molecular Dynamics) at 635 nm and quantified using ImageQuant software (GE Healthcare). Two independent lots of NMM were purchased from Frontier Scientific and assayed with similar outcome.
Yeast strains and CEB1 minisatellites
The genotypes of the S. cerevisiae strains (S288C background) used here are reported in the Supplementary Table S1. The rad51Δ strain (AND1239-5C) containing CEB1-WT-1.7 was obtained after sporulation of the diploid obtained by crossing AND1202-11A (CEB1-WT-1.7 pif1::KanMX) with the BY4742 rad51::KanMX strain (31). The nucleotide sequences of the natural human CEB1-1.8 (see Supplementary Figure S6) and the synthetic CEB1-WT-1.7 and CEB1-Gmut-1.7 minisatellites, each containing 42 motifs, have been previously reported (8,32). The CEB1 arrays were inserted on the yeast chromosome VIII, upstream of the ARG4 promoter (32).
Induction of minisatellite instability
Cells taken from a fresh patch were suspended at a density of 2 × 105 cells/ml into 5 ml of rich Yeast–Peptone–Dextrose (YPD) medium containing the indicated concentration of Phen-DC3 and Phen-DC6 and 1% DMSO, or in YPD containing only 1% DMSO (control treatment). Cells were grown for eight generations at 30°C with agitation, plated as individual colonies on YPD plates and grown at 30°C. For the induction in synthetic complete (SC) medium with NMM and Phen-DC3, the final concentration of DMSO reached in the liquid culture and in the control treatment was 0.4%. For the NMM treatment in SC medium, cells were grown overnight in presence of the drug, diluted to 2 × 105 cells/ml in the same media and grown for an additional eight generations at 30°C, thus undergoing approximately four additional generations in presence of the drug compared to the control condition or the Phen-DC3 treatment.
Analysis of minisatellite instability
Colonies grown from treated cultures were inoculated in 96-well megaplaque in YPD for 24–48 h at 30°C. Pools of 4–16 colonies were made right before DNA extraction. DNA was digested with ApaI and SpeI to probe for the CEB1 minisatellite, or with AluI to probe for the DAN4 and FLO1 minisatellites (8). Digestion products were migrated in a 0.8% agarose–TBE 1× gel and analyzed by Southern blot using a radiolabeled probe corresponding to the minisatellite of interest. Blots were analyzed using a Storm PhosphorImager (Molecular Dynamics) and quantified using ImageQuant software (GE healthcare). To account for secondary rearrangements (occurring after plating) in the analysis, each visible band was quantified and normalized to the mean of the intensity of the parental bands. Bands with signal intensity two times lower than expected for a primary rearrangement were removed from the counting. In numerous instances pools of colonies were depooled for verification and sequencing.
Additional experimental protocols are reported in Supplementary Material and Methods.
RESULTS
Phen-DC compounds strongly bind to the G-quadruplex form of CEB1
Our screening and initial analysis of a large series of potential G4-binding ligands was performed with two quadruplex-forming oligonucleotides: 22AG [5′-AG3(T2AG3)3-3′] mimicking a human telomeric sequence and TBA [5′-G2T2G2TGTG2T2G2-3′], the thrombin binding aptamer sequence (27). The bisquinolinium-dicarboxamide derivatives Phen-DC3 and Phen-DC6 (illustrated in Figure 1A) were among the most selective G4 ligands.
Since G4 can fold differently depending on primary nucleotide sequence (33), we wished to evaluate the capacity of the Phen-DC compounds to bind to the quadruplex conformation adopted by the more complex human minisatellite CEB1 motif used in the present study. To this end, we folded a single-stranded oligonucleotide representing the prevalent 39 nt CEB1 repeat motif (G4-CEB1) (8) and assayed its interaction with the Phen-DC and NMM compounds using the G4-FID assay (34), whose principle is illustrated in Figure 1B. This method is based on the competitive displacement of the fluorescent intercalator probe TO by a putative G4 ligand. TO being highly fluorescent when bound to DNA and virtually non-fluorescent when free in solution, the decrease of TO fluorescence as function of increasing ligand concentration thus serves to evaluate binding affinity of the ligand for G-quadruplex DNA (expressed as DC50: concentration required to decrease the fluorescence signal of 50%). As shown in Figure 1C, fluorimetric titration of TO by the G4-CEB1 substrate showed a strong fluorescence increase, indicating that TO efficiently bound to the G4-CEB1 conformation. The titration curve adopted a regular shape that could be fitted with a 1/1 stoichiometry model, giving a Kd value in the micromolar range (Kd = 7.1 × 10−7 M). This behavior as well as the Kd value are similar to results obtained with the TBA and the human telomeric G4-DNA (27), thereby indicating that, in our experimental conditions, G4-CEB1 accommodates the probe at a single site corresponding to one of the two external G-quartets. The displacement of the TO by Phen-DC compounds was then examined in K+ (Figure 1D) and Na+ (Supplementary Figure S1) conditions. Both Phen-DC3 and Phen-DC6 isomers display a strong ability to displace TO (DC50 = 0.4–0.5 µM) with complete displacement (100% quenching) being reached at a low ligand/quadruplex ratio (5 and 10 for Phen-DC3 and Phen-DC6, respectively). Considering previous results obtained with the telomeric quadruplex and the low concentration of G4-DNA (0.25 µM) used in the test, these data unambiguously reveal a strong interaction between Phen-DC compounds and G4-CEB1 (Figure 1D, Supplementary Figure S1). In sharp contrast, the NMM was much less efficient in displacing TO since saturation was not reached even at high ligand/quadruplex ratio (>40). This result indicates that NMM exhibits a much lower affinity for G4-CEB1 than our Phen-DC compounds (Figure 1D).
Phen-DC compounds specifically inhibit G4-DNA unwinding by the Pif1 helicase in vitro
Recently, we showed that the yeast Pif1 helicase efficiently unwinds the G4-CEB1 substrate (8). To investigate the ability of the two Phen-DC derivatives to interfere with such G4 unwinding enzymatic activities, we tested their effect on G4-CEB1 unwinding by Pif1 in vitro. The G4-CEB1 substrates were made in two conditions: i.e. in presence of either Na+ or K+ as a monovalent cation (see Supplementary Materials and Methods) (8). This led to intermolecular G4-CEB1 structures with different gel migration properties, likely corresponding to a different folding of the G4-DNA. As shown in Figure 2A, Pif1 efficiently unwound the G4-CEB1 substrates formed in either conditions but was rapidly inhibited by low concentrations of Phen-DC6 [ki(K+) = 250 nM; ki(Na+) = 300 nM]. Phen-DC3 showed comparable activity (Supplementary Figure S2).
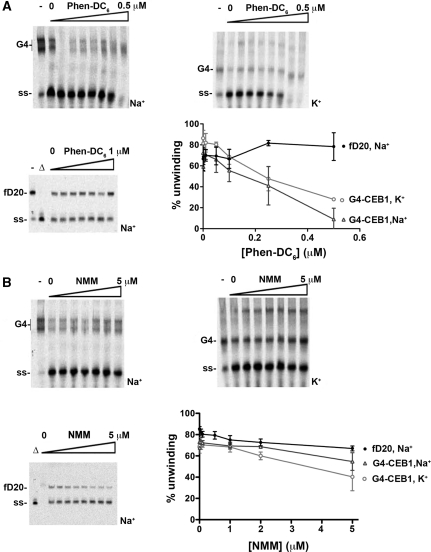
Figure 2.
Inhibition of G4-CEB1 unwinding by Phen-DC6 and NMM in vitro. (A) Pif1 unwinding of G4-CEB1 or forked DNA substrate (fD20) in presence of increasing concentrations of Phen-DC6. The G4-CEB1 substrates were formed in presence of Na+ or K+ as indicated. Quantifications (mean ± SD) from three independent experiments are shown. Open triangle indicates boiled substrate. Dash indicates absence of Pif1. Position of G4-CEB1 (G4) and unwound substrate (ss) are indicated. (B) Same as in (A), using NMM as a G4 ligand.
For comparison, we also tested the inhibition of G4-CEB1 unwinding by NMM, reported to be a potent inhibitor of G4-DNA unwinding by the Escherichia coli RecQ helicase and its eukaryotic homologues Sgs1 and BLM helicases (35). Strikingly, NMM was fairly inefficient in inhibiting G4 DNA unwinding by Pif1. At 5 µM concentration, the unwinding of G4-CEB1 by Pif1 was inhibited by ∼50% (Figure 2B). Therefore, Phen-DC compounds are more effective than NMM to inhibit G4-CEB1 unwinding by Pif1, a result consistent with the difference in their respective binding ability as evaluated by the G4-FID assay.
Next, to address the specificity of Phen-DC compounds for G4-DNA, we tested their effect on the unwinding of a forked 20-mer double-stranded DNA (dsDNA) substrate (fD20). Phen-DC6 had no effect on fD20 unwinding by Pif1 at concentrations up to 1 µM concentration (Figure 2A). We could not test higher concentrations of the drug in this assay because higher concentrations of Phen-DC6 resulted in non-specific aggregation of DNA substrates that were not resolved in the polyacrylamide gel, regardless of the substrate used (G4-DNA or dsDNA). This effect is reminiscent of DNA compaction by polyamines, a situation frequently encountered with cationic ligands (36,37). In comparison, NMM also exhibited specificity for G4-DNA substrates since unwinding of the forked substrate was not inhibited by high concentrations of NMM (5 µM) (Figure 2B). In conclusion, contrary to NMM, the Phen-DC compounds are active at low concentrations as potent and specific inhibitors of G4-CEB1 unwinding by Pif1 helicase in vitro.
The Phen-DC compounds induce genetic instability of the G4-prone CEB1 minisatellite
To test the effect of the Phen-DC compounds in vivo and their specificity towards G4-DNA substrates, we examined their capacity to trigger the instability of the G4-prone human CEB1 minisatellite inserted in the yeast genome (38) (Figure 3A). In WT yeast cells, the synthetic 1.7 kb CEB1 array (CEB1-WT-1.7) composed of 42 wild-type motifs of 39 nt (Figure 3B) is rather stable (8). It is highly destabilized in the absence of the Rad27/FEN1 endonuclease or of the Pif1 helicase, but in the latter case, only when the CEB1 repeats contains G4-forming sequences (8). Mutation of the G4-forming motifs in the synthetic CEB1 array (CEB1-Gmut-1.7, Figure 3B) stabilizes CEB1 in pif1Δ cells but not in rad27Δ cells (8). Thus, this system provides a well-defined genetic assay to monitor the formation of G4 structures in vivo.
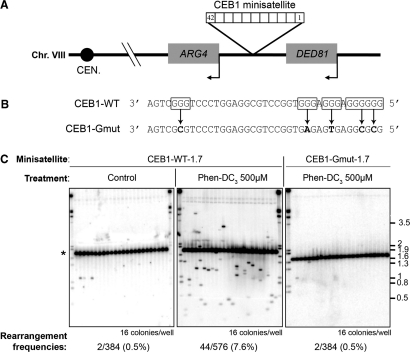
Figure 3.
Phen-DC compounds trigger G-quadruplex-dependent CEB1 instability. (A) Schematic representation of the CEB1 minisatellite insertion in the chromosome VIII. (B) Sequences of CEB1-WT and CEB1-Gmut motifs. G-runs involved in G-quadruplex formation are boxed. Point mutations introduced in CEB1-Gmut motif and preventing G-quadruplex formation in vitro are shown in bold. (C) Southern blot analysis of WT cells carrying CEB1-WT-1.7 (strain AND1212-10D) or CEB1-Gmut-1.7 (AND1227-5C) after control (1% DMSO) or 500 µM Phen-DC3 treatment. The number of colonies analyzed and the rearrangement frequencies are indicated below each gel. Size markers (kb) are indicated on the right. The position of the parental minisatellite alleles (42 repeats, 1.7 kb) is indicated by an asterisk.
We first treated WT cells carrying the synthetic CEB1-WT-1.7. The frequency of CEB1 instability was measured by the appearance of size variants (expansions or contractions) visualized by Southern blot analysis (Figure 3C). Treatment of cells grown in rich media (YPD) with increasing doses of Phen-DC3 or Phen-DC6 (up to 500 µM and 200 µM, respectively) had no effect on cell growth (Supplementary Figure S3A). In untreated cells or upon 1% DMSO control treatment (Phen-DC compounds are solubilized in pure DMSO), CEB1 rearrangements were rare: 2/708 and 2/384 respectively (Table 1). In contrast, upon treatment with 200 µM and 500 µM of Phen-DC3, the frequency of size variants increased to 4.2% (16/384 rearranged colonies) and 7.6% (44/576), respectively (Figure 3C, Table 1). This dose–response was statistically significant (P = 0.029). Similarly, treatment with 200 µM Phen-DC6 also stimulated significantly the formation of CEB1-WT-1.7 rearrangements to yield a frequency of 4.7% (18/384) (Table 1), indicating that both compounds, structurally close to each other, behave similarly and yielded variants of diverse size. Lower concentrations of Phen-DC were active in different media (see below). Again, treatment with 200 µM or 500 µM NMM did not stimulate CEB1-WT-1.7 instability (Table 1, Supplementary Figure S4). We concluded that the two Phen-DC isomers are equally able to enter the cells and to trigger a biological response in the nucleus, namely to induce an 8-15 fold stimulation of CEB1 rearrangements.
Table 1.
Frequencies of CEB1 rearrangements in WT cells
| Strain: | AND1212-10D |
AND1227-5C |
|||
|---|---|---|---|---|---|
| Minisatellite: | CEB1-WT-1.7 |
CEB1-Gmut-1.7 |
|||
| Rearrangt. Freq. | P-value versus control | Rearrangt. Freq. | P-value CEB1-WT-1.7 versus CEB1-Gmut-1.7 | ||
| YPD media | |||||
| Untreated | 2/708 (0.3%)a | NS | 0/192a | NS | |
| Treatment | |||||
| Control | 2/384 (0.5%) | NA | 2/384 (0.5%) | NS | |
| Phen-DC6 200 µM | 18/384 (4.7%) | 3.4e−4 | 1/384 (0.2%) | 6.4e−5 | |
| Phen-DC3 200 µM | 16/384 (4.2%) | 1.2e−3 | 0/384 | 2.6e−5 | |
| Phen-DC3 500 µM | 44/576 (7.6%) | 9.1e−8 | 2/384 (0.5%) | 9.1e−8 | |
| NMM 200 µM | 0/384 | NS | 3/368 (0.8%) | NS | |
| NMM 500 µM | 0/192 | NS | ND | NA | |
| SC media | |||||
| Treatment | |||||
| Control | 0/192 | NA | ND | NA | |
| Phen-DC3 2 µM | 2/384 (0.5%) | NS | ND | NA | |
| Phen-DC3 20 µM | 27/384 (7%) | 2.4e−5 | 3/384 (0.8%) | 5.8e−6 | |
| NMM 20 µM | 0/384 | NS | ND | NA | |
aData from ref. (8).
Rearrangement frequencies were compared using a two-tailed Fischer's exact test.
NA, not applicable; ND, not determined; NS, not significant.
Then, to test if the CEB1 instability triggered by the Phen-DC compounds relied specifically on the formation and/or stabilization of G4 structures, we carried out similar experiments with cells bearing a CEB1-Gmut-1.7 array. In contrast to the CEB1-WT-1.7 array, the CEB1-Gmut-1.7 array remained stable upon treatment with up to 500 µM Phen-DC3 or 200 µM Phen-DC6 (Figure 3C, Table 1). In these conditions and in both control experiments (untreated or incubated with 1% DMSO), the frequencies of CEB1 rearrangements are in the range of 0.5% or less, and are not significantly different from each other. Also, the treatment with 200 µM NMM had no effect (Table 1). To further test the specificity of the Phen-DC compounds, we also examined the behavior of two natural yeast minisatellites DAN4 and FLO1, which are devoided of potential G4-forming sequences. Neither minisatellite was destabilized upon treatment with Phen-DC3 or Phen-DC6 (0/384 rearrangement, at 200 µM in each case). Thus, CEB1 instability observed in WT cells upon treatment with Phen-DC compounds depends on the presence of G4-forming sequences, mimicking what is observed in pif1Δ cells (8).
Although consistent with its weak activity in our in vitro assays (Figure 2), the lack of effect of NMM on CEB1 stability is rather surprising since a lower dose of NMM (8 µM) was reported to significantly alter transcription of yeast genes bearing G4-forming sequences in their promoter regions (12). An important difference between this study and ours is that we used rich media (YPD) instead of SC media. Therefore, we re-examined the dose effect of NMM and Phen-DC compounds in cells grown in SC media. We found that NMM had a mild effect on growth at 40 µM. In contrast, 20 µM Phen-DC3 or 8 µM Phen-DC6 was sufficient to reduce growth up to 50% (Supplementary Figure S3B). These effects were observed whether or not the strains carried the CEB1 minisatellite (Supplementary Figure S3B). Consequently, although we preferred to pursue our study using YPD media and concentrations of drugs not affecting cell growth, we nevertheless examined CEB1 instability upon drug treatment in SC media. We observed that the treatment of cells with 20 µM NMM did not destabilized CEB1-WT-1.7 (0/384 rearrangement) (Table 1, Supplementary Figure S4). At 2 µM Phen-DC3, CEB1-WT-1.7 remained stable. In contrast, upon treatment with 20 µM Phen-DC3, we observed 7% instability (27/384) of CEB1-WT-1.7 and no instability of the CEB1-Gmut-1.7 allele (3/384 rearrangements, P = 5.8e−6) (Table 1, Supplementary Figure S5). Altogether, these results demonstrate that the Phen-DC molecules are active in both rich and synthetic complete medium.
The Phen-DC compounds increase CEB1 instability in Pif1-deficient strains
The occurrence of G4-dependent CEB1 rearrangements in treated wild-type cells revealed that some G4 stabilized by the Phen-DC compounds escape from the unwinding activity of Pif1. This is consistent with the observations that these compounds inhibit G4-CEB1 unwinding by the Pif1 helicase in vitro (Figure 2), but whether or not Pif1 remove all or a fraction of the G4 that form in CEB1 remained to be evaluated. To address this question, we examined the frequency of CEB1-WT-1.7, CEB1-1.8 and CEB1-Gmut-1.7 rearrangements triggered by Phen-DC3 or Phen-DC6 in the absence of Pif1 (pif1Δ cells) and in a strain carrying a helicase-dead allele of PIF1 [pif1-K264A (39)].
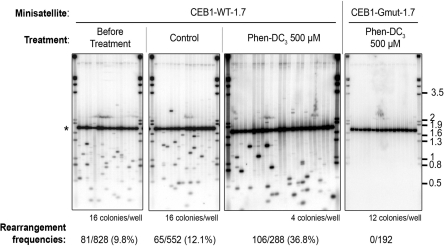
In our assays, the background of CEB1 rearrangements in WT cells is low (<1%) and roughly negligible compared to the rapid and strong effect of the Phen-DC compounds (Table 1), but this is not the case in PIF1 mutant cells, in which the G4-prone CEB1 minisatellite is substantially unstable [see ref. (8) and present study]. Thus, to take into account CEB1 rearrangements which may pre-exists before the treatment, we estimated the effect of the G4-ligands in two steps. First, we measured the frequency of pre-existing rearrangements in the starting cell patches and then, as in WT cells, divided the assay culture in two samples: one being treated by the Phen-DC compounds and the other incubated in 1% DMSO alone (control treatment). Thus, upon subtraction of the pre-existing events (see Supplementary Materials and Methods for details on the procedure used for correction), we could estimate the frequency of rearrangements generated during the drug treatment only (eight generations). The pif1Δ cultures treated with 200 µM Phen-DC6 or 500 µM Phen-DC3 exhibited a high frequency of CEB1-WT-1.7 rearrangements, 20.9% and 36.8%, respectively, instead of 12.1% in the control treatment (P-values versus control treatment are 4.1e−4 and 2.7e−16, respectively) (Figure 4, Table 2). Similarly, the frequency of CEB1-1.8 rearrangements raised from 5.2% to 12.2% (P = 1.1e−3) and 2.7% to 7.3%, (P = 5.5e−3) in the pif1Δ and pif1-K264A cells, respectively. Thus, taking into account the pre-existing events, estimated at 9.8% (CEB1-WT-1.7) and 2.1 % (CEB1-1.8) in pif1Δ cell cultures, and at 1.3% (CEB1-1.8) in the pif1-K264A cell cultures, the treatments yielded a 3- to 12-fold increase of CEB1 rearrangements. Finally, as in WT cells, the Phen-DC compounds did not stimulate instability of the CEB1-Gmut-1.7 minisatellite (Figure 4). Altogether, the above results demonstrate that the Phen-DC compounds are able to stimulate CEB1 rearrangements in the presence or absence of Pif1, and in this latter case, uncover the in vivo formation of a higher number of G4 than in WT cells.
Figure 4.
Treatment with Phen-DC3 increases the instability of CEB1-WT-1.7 in pif1Δ cells. Southern blot analysis of CEB1-WT-1.7 (AND1202-11A) or CEB1-Gmut-1.7 (AND1206-4C-D11P2) instability in pif1Δ cells, before and after control (1% DMSO) or 500 µM Phen-DC3 treatment. Other legends are as in Figure 3.
Table 2.
Frequency of CEB1 rearrangements in pif1Δ cells
| Strain | AND1202-11A | AND1206-4C-D11P2 | ORT4841 | ORT5087-5E |
|---|---|---|---|---|
| Minisatellite | CEB1-WT-1.7 | CEB1-Gmut-1.7 | CEB1-1.8 | CEB1-1.8 |
| Mutation | pif1Δ | pif1Δ | pif1Δ | pif1-K264A |
| Before treatment | 81/828 (9.8%) | 1/383 (0.2%)a | 4/192 (2.1%) | 5/375 (1.3%) |
| Treatment | ||||
| Control | 67/552 (12.1%) | ND | 20/384 (5.2%) | 10/375 (2.7%) |
| Phen-DC6 200 µM | 77/368 (20.9%) | 0/368 | ND | ND |
| Phen-DC3 500 µM | 106/288 (36.8%) | 0/192 | 41/336 (12.2%) | 26/358 (7.3%) |
aData from ref. (8).
ND: not determined.
Sequence of CEB1 rearrangements induced by the Phen-DC compounds in WT and Pif1 deficient cells
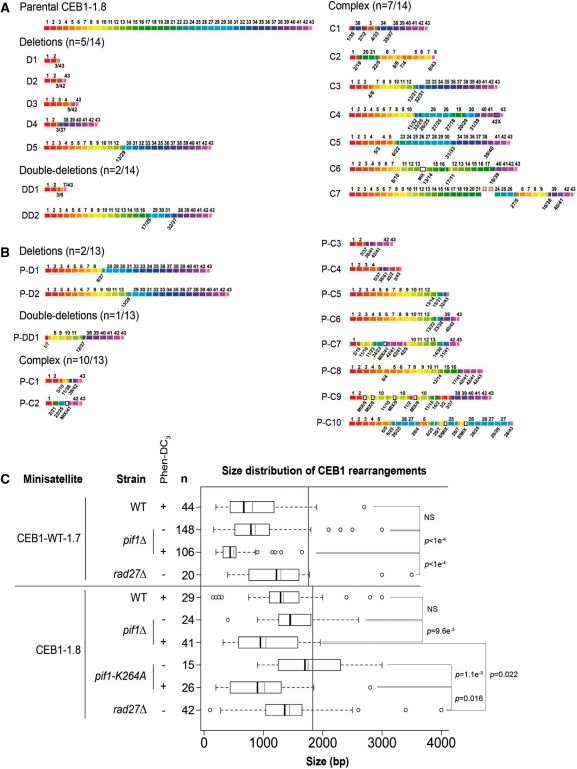
To determine the nature of the rearrangements induced upon Phen-DC treatment in WT and pif1Δ cells, we used the naturally polymorphic CEB1-1.8 minisatellite in which the presence of multiple base substitutions along the 42 motifs allows a precise analysis of the contribution of the parental motifs in the sequenced size variants (32). As for the synthetic CEB1-WT-1.7 minisatellite, the treatment of WT cells grown in YPD with 500 µM Phen-DC3 stimulated the rearrangements of the CEB1-1.8 allele ∼14-fold (29/768 versus 5/1824 in the untreated cells, P = 1.8e−11) and yielded variants of diverse size. Thus, the presence of base polymorphisms located outside the triplets of G involved in CEB1 quadruplex formation does not interfere with the drug target. The sequences of 14 and 13 CEB1-1.8 contractions obtained upon treatment of WT and pif1Δ cells are reported in Supplementary Figure S6. They were different from each other. In both strains, simple deletions with one chimerical motif, double deletion with two chimerical motifs, and complex events with multiple chimerical motifs and various internal reshuffling of the parental polymorphic markers occurred (Figure 5A and B), suggesting that the underlying mechanisms of rearrangements were similar in WT and pif1Δ cells. No de novo mutagenic events were observed. Thus, the nature of the rearrangements obtained after treatment with Phen-DC3 is diverse and reflects the same general pattern of events as in the untreated pif1Δ (8) and rad27Δ (32) mutants.
Figure 5.
Sequence and size of CEB1 rearrangements produced upon treatment with Phen-DC3. (A) Schematic representation of the natural CEB1-1.8 allele and of 14 rearrangements produced upon treatment of WT cells (ORT2914) with 500 µM Phen-DC3. Nucleotide sequences are reported in Supplementary Figure S6. Chimerical motifs combining the polymorphisms of more than three parental motifs (called ‘Mix’ in Supplementary Figure S6) are shown as a white box. Sequences of the two red motifs in C7 have not been determined. (B) Schematic representation of 13 rearrangements produced upon treatment of pif1Δ cells (ORT4841) with 500 µM Phen-DC3. (C) Size distribution of CEB1-WT-1.7 and CEB1-1.8 rearrangements in WT (AND1212-10D and ORT2914), pif1Δ (AND1202-11A and ORT4841), pif1-K264A (ORT5087-5E) or rad27Δ (AND1218-1A and ORD6713-8D) cells, with or without treatment with 500 µM Phen-DC3. Datasets are from this study, except the size distribution of rearranged CEB1-WT-1.7 alleles in rad27Δ cells, obtained from ref. (8) (strain AND1218-1A). The n indicates the number of variants analyzed. The vertical gray lines indicate the size of the parental alleles. For each distribution, dark bar: median; gray bar: mean; box: distance between the first and third quantiles; whiskers: extreme values; white circles: outliers. P-values were obtained by comparing the samples with a Mann–Whitney–Wilcoxon statistical test.
Phen-DC3 treatment of Pif1-deficient cells yields unusually short CEB1 rearrangements
In comparison to the rearrangements observed in the absence of Pif1 alone, we noted that the rearrangements produced upon treatment of Pif1-deficient cells yielded a larger number of CEB1 fragments of small size, corresponding to short variants of 15 or less repeat units (Figure 4). A comparative analysis of the size of the CEB1-WT-1.7 and CEB1-1.8 rearrangements obtained in pif1Δ or pif1-K264A cells with or without treatment is reported in Figure 5C. Strikingly, in all cases, despite the existence of pre-existing rearrangements, the mean size of the rearrangements was shorter in Phen-DC3-treated versus untreated Pif1-deficient cells (P ≤ 9.6e−3). We also examined the size of the CEB1 variants produced in the rad27Δ cells. This cell exhibits a high level of instability for CEB1-WT-1.7 (30%) (8) and CEB1-1.8 (41/158, 25.9%) that is comparable to the instability measured in pif1Δ-treated cells (Table 2). The mean size of the CEB1 rearrangements in rad27Δ is much higher than in both pif1Δ and pif1-K264A treated cells (P ≤ 2.2e−2) (Figure 5C). Hence, the increase in short alleles upon treatment of Pif1-deficient cells is not only a consequence of a high level of instability, but rather a specific effect of Phen-DC3 treatment.
CEB1 rearrangements induced by Phen-DC compounds depend on the homologous recombination pathway
In previous studies, we demonstrated that in rad27Δ and pif1Δ cells the formation of CEB1 rearrangements depends on the process of homologous recombination, since it was abolished in the absence of the RAD51 or RAD52 genes (8,32). Similarly, we examined the behaviour of the CEB1-WT-1.7 array in a rad51Δ strain upon treatment with 200 µM Phen-DC3 (Supplementary Figure S7). Compared to the WT strain (4.2% rearrangements, Table 1), we observed no rearrangements (0/384) in the absence of Rad51 (P = 2.6e−5). Therefore, Phen-DC treatment also leads to CEB1 rearrangements through the homologous recombination pathway.
DISCUSSION
In this study, we investigated the in vitro and in vivo activities of two very potent G4 ligands: Phen-DC3 and Phen-DC6. Based on in vitro biophysical and biochemical criteria, they are best ranked among current G4 ligands (26,27,29,30,40) and these promises clearly extend to their biological activity. We reported that the Phen-DC compounds: (i) exhibit a strong affinity for preformed G4-CEB1 structures in vitro, (ii) inhibit the unwinding by Pif1 of the resulting G4-ligand structure(s), (iii) manifest biological activity in intact yeast cells, (iv) destabilize the G4-prone but not the G4-mutated CEB1 array in WT cells, (v) further destabilize CEB1 in Pif1 deficient cells, (vi) induce recombination-dependent CEB1 size variants (expansions and contractions) similar to the spontaneous rearrangements produced in the absence of Pif1, and (vii) unexpectedly yield short size CEB1 variants. Thus, as hoped, the two Phen-DC compounds behave as potent biological drugs targeting G4 structures and interfering with their processing. To choose between the Phen-DC3 and Phen-DC6 molecules, the differences are subtle. Both are chemically stable, easy to prepare in large amounts (27), and qualitatively their in vitro and in vivo effects are similar as expected from their closely related chemical structures. The advantage of Phen-DC3 is a slightly higher solubility in buffered solutions, allowing the use of higher drug concentrations in culture media.
To be optimal, a ligand should exhibit high selectivity and affinity for its substrate. Convincingly, the two Phen-DC derivatives fulfill both requirements. They bind with high affinity to various types of G-quadruplexes (27,40) and to the human minisatellite G4-CEB1 (Figure 1). This binding behavior is attributed to the crescent shape and the size of the Phen-DC scaffold that fit with the dimensions of a G-quartet, both features being favorable to an optimal π-overlap between the two aromatic surfaces (26).
We extensively compared the effects of the Phen-DC compounds to NMM, a molecule previously reported to bind G4 (19,41,42), to inhibit their unwinding by RecQ (43), Sgs1, and BLM (35) helicases in vitro, and to exert G4-related biological effects in budding yeast (12) and N. gonorrhoeae (9). Thus, we expected to observe an effect of NMM in our assays. The Phen-DC compounds act at nanomolar concentrations (Ki = 250 nM in K+) as specific inhibitors of G4-CEB1 unwinding by Pif1 in vitro (Figure 2), which is a very active helicase on these substrates (8). In contrast, although our in vitro experiments confirmed that NMM is a selective G4 ligand (as assessed by FID), it is inefficient in inhibiting the Pif1 helicase over a wide range of concentrations (Figure 2) and does not induce any CEB1 instability in yeast (Table 1, Supplementary Figure S4). The low affinity of NMM for G4-CEB1 structures can explain the discrepancies observed between treatment with NMM and Phen-DCs. An alternative and not mutually exclusive interpretation is that these molecules could recognize different G4 topologies and/or creates distinct G4-ligand structures, which would be differently processed by Pif1 or other endogenous helicases. Along these lines, the examination of the sequence of CEB1 motifs as well as their organization in a direct tandem array (placing all G triplets on the same strand) suggest that the CEB1 minisatellite has the potential to form topologically distinct intra- and inter-motifs G4 structures with heterogeneous loop sizes (11). Structurally, these two quinolinium rings, which display a pronounced electron-acceptor character that contributes to the increased stacking interactions with guanines, have the potential to recognize various types of G4 via stacking on the external quartets terminating the quadruplex core. Thus, we envisage that the Phen-DC compounds may have a large spectrum of G4-structure targets (27,29,30), whereas NMM might be more specific of certain G4 structures (41).
The instability observed in treated wild type cells and its increase when Pif1 activity is compromised (revealing a higher number of G4) suggest that Pif1 greatly limits the effect of these G4 ligands in vivo. Mechanistically, this can be due to the unwinding of only a fraction of G4-CEB1 by Pif1 in vivo. In addition to the thermodynamic shift toward the folded state in presence of the ligand, which could trigger the formation and/or stabilization of G4 structures that would otherwise not stably form naturally, the persistency of G4 can be due to the inhibition of other potential G4-processing helicases. In S. cerevisiae, Sgs1 and Dna2 were reported to exhibit a G4-unwinding activity in vitro (44,45) but their in vivo activity on G4 substrates have not been demonstrated. We know that the absence of Sgs1 does not stimulate CEB1 instability, and that the deletion of the DNA2 gene in a pif1Δ strain has the same effect as the inactivation of Pif1 alone (8). Further studies will be undertaken to decipher redundant and overlapping G4 processing pathways in wild-type, single and multi-mutant contexts.
The structure of the CEB1 rearrangements produced in Phen-DC treated and untreated pif1Δ and rad27Δ cells are of similar types, including simple deletion, double deletions and a majority of complex events. They are interpreted to be the consequence of the repair of an initiating lesion through the homologous recombination pathway, in which multiple cycles of misaligned synthesis-dependent strand annealing (SDSA) (46) reactions generate the various reshuffling of the parental minisatellite (8,32) [see illustration Figure 6 in ref. (32)]. However, we noted that the CEB1-WT-1.7 rearrangements obtained in the pif1Δ-treated cells were unusually short, suggesting that the Phen-DC treatment was not exactly mimicking the untreated situations. To confront this unexpected observation, we made a comparative analysis of the size of the CEB1-WT-1.7 and CEB1-1.8 rearrangements obtained in the pif1Δ and the pif1-K264A mutants with or without treatment. Strikingly, in all pairwise comparisons of strains with the same genotype, the rearranged variants were shorter in the treated sample (Figure 5C). Why does the treatment of Pif1-deficient cells yield extensive contractions? An attractive possibility is that the concomitant formation (47) and the persistency of several G4 on the same strand may cause multiple distant lesions favoring large internal deletions. Mechanistically, these mutagenic events may be reminiscent of the deletions of G4-prone sequences reported in C. elegans mutated for the FANC-J helicase homolog dog-1 (10). The lesion initiating the CEB1 rearrangements could be a single-strand nick occurring within the G4-prone region as observed in N. gonorrhoeae (9), or a single-strand gap or a double-strand break resulting from the persistency of unprocessed intra-molecular G4 accumulating in CEB1 during replication, transcription or other biological processes generating single-stranded DNA.
To conclude, the present study demonstrates that the Phen-DC3 and Phen-DC6 bisquinolinium compounds are reliable and potent small molecules to specifically target and trigger a G-quadruplex-dependent biological process, namely tandem-repeat instability. The independent and combinatorial use of genetic and chemical interfering approaches provides new perspectives to probe the G4-forming sequences in yeast and other organisms (11).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Institut Curie; the Centre National de la Recherche Scientifique; and La Ligue Nationale contre le Cancer ‘Equipe Labellisée LIGUE 2007’ (to A.N.); by an E.U.; FP6 ‘MolCancerMed’ (LSHC-CT-2004-502943 to M.-P, T.-F.); graduate student fellowship from the Ministère de l’Education Nationale, de la Recherche et de la Technologie (to A.P.); a post-doctoral fellowship from the Association pour la Recherche sur le Cancer (to J.B.B.); Massachussets Institute of Technology - Institut Curie exchange program fellowship (to K.M.); a BDI graduate student fellowship from the Institut Curie and the Centre National de la Recherche Scientifique (to E.L.). Funding for open access charge: Institut Curie.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of our laboratories, A. Londono-Vallejo, J.-L. Mergny and E. Blackburn for helpful discussions; D. Monchaud for preparing the Phen-DC compounds; G. Millot for advices on statistical analyses.
REFERENCES
- 1.Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc. Natl Acad. Sci. USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phan AT, Kuryavyi V, Luu KN, Patel DJ. Structural diversity of G-quadruplex scaffolds. In: Neidle S, Balasubramanian S, editors. Quadruplex Nucleic Acids. UK: Ch. 3, RSC publishing, Cambridge; 2006. pp. 81–99. [Google Scholar]
- 3.Lipps HJ, Rhodes D. G-quadruplex structures: In vivo evidence and function. Trends Cell. Biol. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat. Struct. Mol. Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 5.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paeschke K, Juranek S, Simonsson T, Hempel A, Rhodes D, Lipps HJ. Telomerase recruitment by the telomere end binding protein-[beta] facilitates G-quadruplex DNA unfolding in ciliates. Nat. Struct. Mol. Biol. 2008;15:598–604. doi: 10.1038/nsmb.1422. [DOI] [PubMed] [Google Scholar]
- 7.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 8.Ribeyre C, Lopes J, Boule JB, Piazza A, Guedin A, Zakian VA, Mergny JL, Nicolas A. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009;5:e1000475. doi: 10.1371/journal.pgen.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cahoon LA, Seifert HS. An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science. 2009;325:764–767. doi: 10.1126/science.1175653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kruisselbrink E, Guryev V, Brouwer K, Pontier DB, Cuppen E, Tijsterman M. Mutagenic capacity of endogenous G4 DNA underlies genome instability in FANCJ-defective C. elegans. Curr. Biol. 2008;18:900–905. doi: 10.1016/j.cub.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 11.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hershman SG, Chen Q, Lee JY, Kozak ML, Yue P, Wang LS, Johnson FB. Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 2008;36:144–156. doi: 10.1093/nar/gkm986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanakahi LA, Sun H, Maizels N. High affinity interactions of nucleolin with G-G-paired rDNA. J. Biol. Chem. 1999;274:15908–15912. doi: 10.1074/jbc.274.22.15908. [DOI] [PubMed] [Google Scholar]
- 14.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong HM, Huppert JL. Stable G-quadruplexes are found outside nucleosome-bound regions. Mol. BioSyst. 2009;5:1713–1719. doi: 10.1039/b905848f. [DOI] [PubMed] [Google Scholar]
- 16.Sun D, Thompson B, Cathers BE, Salazar M, Kerwin SM, Trent JO, Jenkins TC, Neidle S, Hurley LH. Inhibition of human telomerase by a G-quadruplex-interactive compound. J. Med. Chem. 1997;40:2113–2116. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- 17.De Cian A, Lacroix L, Douarre C, Temime-Smaali N, Trentesaux C, Riou J.-F, Mergny J.-L. Targeting telomeres and telomerase. Biochimie. 2008;90:131–155. doi: 10.1016/j.biochi.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 18.Fedoroff OY, Salazar M, Han H, Chemeris VV, Kerwin SM, Hurley LH. NMR-based model of a telomerase-inhibiting compound bound to G-quadruplex DNA. Biochemistry. 1998;37:12367–12374. doi: 10.1021/bi981330n. [DOI] [PubMed] [Google Scholar]
- 19.Ren J, Chaires JB. Sequence and structural selectivity of nucleic acid binding ligands. Biochemistry. 1999;38:16067–16075. doi: 10.1021/bi992070s. [DOI] [PubMed] [Google Scholar]
- 20.Parkinson GN, Ghosh R, Neidle S. Structural basis for binding of porphyrin to human telomeres. Biochemistry. 2007;46:2390–2397. doi: 10.1021/bi062244n. [DOI] [PubMed] [Google Scholar]
- 21.Ping W, Lige R, Hanping H, Feng L, Xiang Z, Zheng T. A phenol quaternary ammonium porphyrin as a potent telomerase inhibitor by selective interaction with quadruplex DNA. ChemBioChem. 2006;7:1155–1159. doi: 10.1002/cbic.200600036. [DOI] [PubMed] [Google Scholar]
- 22.Kern JT, Thomas PW, Kerwin SM. The relationship between ligand aggregation and G-quadruplex DNA selectivity in a series of 3,4,9,10-perylenetetracarboxylic acid diimides. Biochemistry. 2002;41:11379–11389. doi: 10.1021/bi0263107. [DOI] [PubMed] [Google Scholar]
- 23.De Cian A, Guittat L, Shin-ya K, Riou J.-F, Mergny J.-L. Affinity and selectivity of G4 ligands measured by FRET. Nucleic Acids Symp. Ser. 2005;49:235–236. doi: 10.1093/nass/49.1.235. [DOI] [PubMed] [Google Scholar]
- 24.Temime-Smaali N, Guittat L, Sidibe A, Shin-ya K, Trentesaux C, Riou J.-FO. The G-quadruplex ligand telomestatin impairs binding of topoisomerase III to G-quadruplex-forming oligonucleotides and uncaps telomeres in alt cells. PLOS ONE. 2009;4:e6919. doi: 10.1371/journal.pone.0006919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doi T, Yoshida M, Shin-ya K, Takahashi T. Total synthesis of (r)-telomestatin. Org. Lett. 2006;8:4165–4167. doi: 10.1021/ol061793i. [DOI] [PubMed] [Google Scholar]
- 26.De Cian A, DeLemos E, Mergny JL, Teulade-Fichou MP, Monchaud D. Highly efficient G-quadruplex recognition by bisquinolinium compounds. J. Am. Chem. Soc. 2007;129:1856–1857. doi: 10.1021/ja067352b. [DOI] [PubMed] [Google Scholar]
- 27.Monchaud D, Allain C, Bertrand H, Smargiasso N, Rosu F, Gabelica V, De Cian A, Mergny JL, Teulade-Fichou MP. Ligands playing musical chairs with G-quadruplex DNA: A rapid and simple displacement assay for identifying selective G-quadruplex binders. Biochimie. 2008;90:1207–1223. doi: 10.1016/j.biochi.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 28.De Cian A, Cristofari G, Reichenbach P, De Lemos E, Monchaud D, Teulade-Fichou MP, Shin-Ya K, Lacroix L, Lingner J, Mergny JL. Reevaluation of telomerase inhibition by quadruplex ligands and their mechanisms of action. Proc. Natl Acad. Sci. USA. 2007;104:17347–17352. doi: 10.1073/pnas.0707365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Cian A, Grellier P, Mouray E, Depoix D, Bertrand H, Monchaud D, Teulade-Fichou MP, Mergny JL, Alberti P. Plasmodium telomeric sequences: structure, stability and quadruplex targeting by small compounds. ChemBiochem. 2008;9:2730–2739. doi: 10.1002/cbic.200800330. [DOI] [PubMed] [Google Scholar]
- 30.Henn A, Joachimi A, Goncalves DP, Monchaud D, Teulade-Fichou MP, Sanders JK, Hartig JS. Inhibition of dicing of guanosine-rich shRNAs by quadruplex-binding compounds. ChemBiochem. 2008;9:2722–2729. doi: 10.1002/cbic.200800271. [DOI] [PubMed] [Google Scholar]
- 31.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 32.Lopes J, Ribeyre C, Nicolas A. Complex minisatellite rearrangements generated in the total or partial absence of Rad27/hFEN1 activity occur in a single generation and are Rad51 and Rad52 dependent. Mol. Cell. Biol. 2006;26:6675–6689. doi: 10.1128/MCB.00649-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neidle S. The structures of quadruplex nucleic acids and their drug complexes. Curr. Op. Struct. Biol. 2009;19:239–250. doi: 10.1016/j.sbi.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 34.Monchaud D, Allain C, Teulade-Fichou M.-P. Development of a fluorescent intercalator displacement assay (G4-FID) for establishing quadruplex-DNA affinity and selectivity of putative ligands. Bioorg. Med. Chem. Let. 2006;16:4842–4845. doi: 10.1016/j.bmcl.2006.06.067. [DOI] [PubMed] [Google Scholar]
- 35.Huber MD, Lee DC, Maizels N. G4 DNA unwinding by BLM and Sgs1p: Substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 2002;30:3954–3961. doi: 10.1093/nar/gkf530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raspaud E, Olvera de la Cruz M, Sikorav JL, Livolant F. Precipitation of DNA by polyamines: A polyelectrolyte behavior. Biophys. J. 1998;74:381–393. doi: 10.1016/S0006-3495(98)77795-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slama-Schwok A, Peronnet F, Hantz-Brachet E, Taillandier E, Teulade-Fichou MP, Vigneron JP, Best-Belpomme M, Lehn JM. A macrocyclic bis-acridine shifts the equilibrium from duplexes towards DNA hairpins. Nucleic Acids Res. 1997;25:2574–2581. doi: 10.1093/nar/25.13.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopes J, Debrauwere H, Buard J, Nicolas A. Instability of the human minisatellite CEB1 in rad27Δ and dna2-1 replication-deficient yeast cells. EMBO J. 2002;21:3201–3211. doi: 10.1093/emboj/cdf310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou J, Monson EK, Teng S, Schulz VP, Zakian VA. Pif1p helicase, a catalytic inhibitor of telomerase in yeast. Science. 2000;289:771–774. doi: 10.1126/science.289.5480.771. [DOI] [PubMed] [Google Scholar]
- 40.De Cian A. Paris: Université Pierre et Marie Curie - Paris VI; 2007. Identification and characterization of G-quadruplex ligands: to target telomeres and/or telomerase? [Google Scholar]
- 41.Arthanari H, Basu S, Kawano TL, Bolton PH. Fluorescent dyes specific for quadruplex DNA. Nucleic Acids Res. 1998;26:3724–3728. doi: 10.1093/nar/26.16.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y, Geyer CR, Sen D. Recognition of anionic porphyrins by DNA aptamers. Biochemistry. 1996;35:6911–6922. doi: 10.1021/bi960038h. [DOI] [PubMed] [Google Scholar]
- 43.Wu X, Maizels N. Substrate-specific inhibition of RecQ helicase. Nucleic Acids Res. 2001;29:1765–1771. doi: 10.1093/nar/29.8.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun H, Bennett RJ, Maizels N. The Saccharomyces cerevisiae Sgs1 helicase efficiently unwinds G-G-paired DNAs. Nucleic Acids Res. 1999;27:1978–1984. doi: 10.1093/nar/27.9.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Masuda-Sasa T, Polaczek P, Peng XP, Chen L, Campbell JL. Processing of G4 DNA by Dna2 helicase/nuclease and RPA provides insights into the mechanism of Dna2/RPA substrate recognition. J. Biol. Chem. 2008;36:24359–24373. doi: 10.1074/jbc.M802244200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pâques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Renciuk D, Kejnovska I, Skolakova P, Bednarova K, Motlova J, Vorlickova M. Arrangements of human telomere DNA quadruplex in physiologically relevant K+ solutions. Nucleic Acids Res. 2009;37:6625–6634. doi: 10.1093/nar/gkp701. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.