Abstract
During meiosis there is an imperative to create sufficient crossovers for homologue segregation. This can be achieved during repair of programmed DNA double-strand breaks (DSBs), which are biased towards using a homologue rather than sister chromatid as a repair template. Various proteins contribute to this bias, one of which is a meiosis specific kinase Mek1. It has been proposed that Mek1 establishes the bias by creating a barrier to sister chromatid repair, as distinct from enforcing strand invasion with the homologue. We looked for evidence that Mek1 positively stimulates strand invasion of the homologue. This was done by analysing repair of DSBs induced by the VMA1-derived endonuclease (VDE) and flanked by directly repeated sequences that can be used for intrachromatid single-strand annealing (SSA). SSA competes with interhomologue strand invasion significantly more successfully when Mek1 function is lost. We suggest the increase in intrachromosomal SSA reflects an opportunistic default repair pathway due to loss of a MEK1 stimulated bias for strand invasion of the homologous chromosome. Making use of an inhibitor sensitive mek1-as1 allele, we found that Mek1 function influences the repair pathway throughout the first4–5 h of meiosis. Perhaps reflecting a particular need to create bias for successful interhomologue events before chromosome pairing is complete.
INTRODUCTION
During meiosis programmed DNA double-strand breaks (DSBs) are created in order that sexually reproducing organisms can benefit from the outcome of their repair by homologous recombination. In most organisms a major benefit of crossovers is the creation of stable connections between homologues before the first meiotic nuclear division. These stable connections are made because non-sister chromatids of homologous chromosomes become covalently linked, and sister chromatids are tightly associated by sister chromatid cohesion until first anaphase (1). In most species studied, the ordered segregation of homologous chromosomes to opposite poles is dependent upon the interhomologue joints mediated by crossovers and sister chromatid cohesion. Without crossovers or sister chromatid cohesion homologue segregation becomes randomized, causing first meiotic non-disjunction.
In some organisms the mechanisms ensuring that sufficient DSBs are repaired using crossovers could involve stochastic processes. For example, in Schizosaccharomyces pombe there is evidence that recombination intermediates [joint molecules (JMs)] are more abundant among sister chromatids compared with homologous chromosomes (2). Since S. pombe has only three pairs of chromosomes, random distribution of interhomologue repair may be sufficient to guarantee the required one crossover per homologous pair. In budding yeast meiosis there is a strong bias towards using the homologue as template in preference to the sister chromatid (3,4). Notionally, there are two distinct ways to enforce a bias of using the homologue versus sister chromatid as repair template. One method could to be to positively promote a search for, and strand invasion of, the homologous chromosome. Another method of creating bias towards the homologue could be a barrier to sister chromatid repair (BSCR) preventing strand invasion of the sister chromatid so that the homologue is used by default.
Mutating DMC1, a meiosis-specific RecA orthologue (5) prevents cells from repairing meiotic DSBs even though Rad51 is still present and the sister chromatid could in theory be used as a template (5). Repair and interhomologue crossovers can be rescued by over-expressing either RAD51 or RAD54, or by deleting the Rad51 inhibitor HED1 (6–9). Rad51 in vegetative cells can bring about strand invasion with the sister chromatid, but this is inhibited during meiosis unless the activity of Rad51 is experimentally increased.
Further information on how the bias of repair is created comes from the study of a complex containing the proteins Mek1 (also known as Mre4), Hop1 and Red1. This complex is required for a diverse range of meiotic functions required for creating viable spores; these include creating meiosis specific chromosome structures [axial elements and synaptonemal complex (SC)], heteroduplex formation and establishing a pachytene checkpoint or surveillance system to prevent exit from pachytene before all DSBs are repaired (10–14). Alleles of all three genes have been isolated in screens for decreased interhomologue repair or increased intersister recombination during meiosis (15,16). Deleting RED1 has been seen to reduce the steady-state population of interhomologue JMs with little impact on the intersister JM frequency, suggesting that the Mek1–Hop1–Red1 complex may specifically stimulate an interhomologue repair pathway (3). More recent results indicate that the population of intersister JMs in red1 cells does account for all DSBs created (N. Kleckner, personal communication).
Evidence of there being a BSCR comes from analysis of Mek1 function. MEK1 is a meiosis-specific kinase required for interhomologue DSB repair (13,17). Activation of wild-type Mek1 protein requires autophosphorylation, dimerization and probably recruitment to the chromosomes (18,19). These activation events are dependent on Hop1 and Red1 (19,20). Either deleting MEK1 or preventing activation of Mek1 kinase activity prevents the accumulation of DSBs in dmc1 cells (14,18,20). Repair of DSBs in mek1 dmc1 double mutants is dependent on RAD54, and it is thought that Rad54-mediated MEK1-independent repair uses the sister chromatid as template (18). These observations have been used to support the view that the Mek1 complex is required for interhomologue bias because it enforces a BSCR.
Recently, it has been suggested that in MEK1 cells Rad51-mediated strand invasion of the sister chromatid is prevented by poor interaction of Rad54 and Rad51 due to Mek1 phosphorylation of Rad54 (21). Thus, in mek1 cells Rad51 strand invasion activity is increased as inhibition of the Rad54–Rad51 interaction is ameliorated (21).
While a BSCR is plausible, published data are also consistent with the idea that Mek1 activity positively commits the cells to strand invade the homologue. Based on this model DSBs accumulate in dmc1 cells because they are committed to interhomologue repair but are unable to achieve it because there is insufficient RecA activity [Rad51 activity being inhibited by both Mek1 phosphorylation of Rad54 and Hed1 binding to Rad51 (7,8,21)]. Then in mek1 dmc1 cells, sufficient Rad51 activity is released because Rad54 is not phosphorylated, and repair is intersister because there is no positive enforcement of the interhomologue strand invasion route.
We set out to gather evidence on the possibility that Mek1 does positively promote interhomologue strand invasion. Using two different reporter cassettes that contain the recognition sequence for the VMA1-derived homing endonuclease (VDE), we have previously reported on various factors that influence DSB repair during meiosis (22,23). Here we make use of the VDE-induced DSBs (VDE–DSBs) to remove the likelihood of intersister repair. This allows us to study the impact of losing Mek1 function on competition between interhomologue gene conversion and intrachromatid repair by single-strand annealing (SSA) of flanking directly repeated sequences. Equal intersister repair by gene conversion is unlikely because both chromatids containing the VDE recognition sequence receive a VDE–DSB during meiosis. This view is evidenced by our observation that VDE–DSB repair is rare (<1%) when the homologue is absent and there are no flanking repeated sequences (Tittcomb and Goldman, unpublished data). Furthermore, we have established that VDE–DSB repair using flanking repeated sequences in meiosis is independent of Rad54, and is unlikely therefore to represent an unequal repair between repeated sequences on sister chromatids (22). These properties of the VDE–DSB reporter provide unique approach to look at repair partner bias. Decreases in interhomologue repair can be assessed by observing increases in intrachromatid repair by SSA, which is easy to measure and is unlikely to require (or reflect) the lowering of any BSCR.
Using various mutant forms of mek1, our data show that as for Spo11–DSBs (14), the lifespan of VDE–DSBs is reduced when Mek1 function is absent. When provided with a choice to repair a VDE–DSB by interchromosomal gene conversion or by intrachromatid SSA, mek1 cells have a strong bias towards the latter.
We provide evidence that this influence of Mek1 on repair at the VDE–DSB is dependent on its kinase function. Also the time during which Mek1 activity can influence VDE–DSB repair is coincident with the period during which it influences Spo11–DSB repair and spore viability. The data are consistent with a role for Mek1 in either promoting or stabilizing interhomologue strand invasion.
MATERIALS AND METHODS
Yeast strains and media
Strain genotypes not previously published are listed in Table 1. All strains are derived from the SK1 background in which we have developed the VDE assays (22,23). Published wild-type strain numbers for VDE–DSB1 and VDE–DSB2 are dAG206 and dAG639, respectively. Yeast strains were maintained using standard laboratory techniques. All incubations were performed at 30°C.
Table 1.
Unpublished diploid strains
| Strain | Relevant genotype |
|---|---|
| dAG702 | mek1Δ::LEU2 |
| dAG732 | mek1Δ::LEU2 TFP1::VDE ade2/ade2::URA3- [arg4-VDE,ura3] ARG4/arg4-nsp ura3 NUC1/nuc1Δ::LEU2 TRP1/trp1::hisG |
| dAG1542 | ade2::mek1-as1 mek1Δ::LEU2 |
| dAG1543 | ade2::mek1-T327A mek1Δ::LEU2 |
All diploid strains are SK1, MATa/α TFP1/TFP1::VDE ura3::URA3-[arg4-vde]/ura3::URA3-[arg4-bgl] SPO11/spo11(Y135F)-HA3His6::KanMX and homozygous for arg4-nsp,bgl, leu2, lys2 ho::LYS2 and homozygous for all other loci reported in the table unless otherwise indicated.
The ura3::arg4-vde (VDE–DSB1) and ade2::arg4-vde (VDE–DSB2) reporter cassettes were made as described previously (22,23).
To obtain mek1Δ strains, the mek1::LEU2 allele in the SK1 strain S2683 (gift of N. Hollingsworth) was crossed into our ura3::arg4-vde, ade2::arg4-vde and TFP1::VDE (VDE expressing) haploids by mating and dissection. Haploids of the appropriate genotypes were mated to create diploids dAG702 and dAG732, which are homozygous for mek1::LEU2 and containing, respectively, VDE–DSB1 and VDE–DSB2. Full details of all intermediate haploid strains and mating strategies are available on request.
Strains homozygous for mek1-as1 (with the mutation Q241G) or mek1-T327A (with GST tag) were made from mating haploids transformed with pLW21 and pTS32-4, respectively. Both plasmids include the native MEK1 promoter (20). These integrating plasmids (gift of Nancy M. Hollingsworth) are based on the pRS402 vector with ADE2 for integration and as a selective marker (20). The plasmids were transformed in linear form (StuI restriction endonuclease) into mek1::LEU2 haploid strains hAG793 (containing the arg4-vde allele) and hAG794 (expressing VDE) and mated to create dAG1542 and dAG1543.
Time courses
Diploid strains were grown on YPG plates and then struck onto YEPD plates to isolate single colonies. A single colony was inoculated into 10 ml YEPD, cultivated overnight, diluted into PSP2 (1/500, 1/250, 1/125) (0.67% yeast nitrogen base, 0.1–0.2% yeast extract, 1% KAc, 1.02% potassium phthalate) and grown overnight again to OD = 1.6–1.7. Cells from PSP2 cultures were harvested, washed with warm 1% KAc and resuspended into the same volume of sporulation medium (1% KAc 250 ml). Sporulation media were supplemented according to the requirements of the strains.
The inhibitor 4-amino-1-(tert-butyl)-3-(1′-naphthylmethyl)pyrazolo[3,4-d]pyrimidine (1-NM-PP1; Toronto Research Chemicals inc.) was dissolved in DMSO (99.9%) and kept at –20°C at 10 mM. 1-NM-PP1 was added once to each culture to a final concentration of 2 µM at specific time points as described in the ‘Results’ section. Viability of the spores was assessed by dissection on YEPD after 24 h of sporulation.
DNA isolation and Southern blot analysis
Sporulating cells (25 ml) were collected at hourly intervals and processed for storage and DNA isolation according to (24), hexamine cobalt (III) chloride was excluded from solutions. Restriction endonuclease digestion and Southern hybridization was performed as previously described (22,23). In brief, analysis of VDE cutting in VDE–DSB1 was done using probe specific for chromosome V coordinates 117126–117992 following digest of genomic DNA with EcoRV and BglII. For analysis of VDE–DSB1 appearance and repair, genomic DNA was digested with SpeI and probed with DNA specific for chromosome V coordinates 117126–117992. Repair at VDE–DSB2 was monitored by digestion of genomic DNA with SpeI and the probe specific for chromosome XV coordinates 566120–566811. In all cases, the digested genomic DNA was separated under native conditions and blotted to Zetaprobe membrane (Bio-Rad) with Vacugene-XL system (PharmaciaBiotech). Quantification of hybridization signal was performed using Kodak phosphor screens that were scanned on a Personal-FX phosphorimager (BioRad) and QuantityOne software.
Calculations
To determine the proportion of VDE–DSB1 alleles in the population that had been cut, the quantity of signal present in the EcoRV and BglII band representing two chromatids with the arg4-vde allele was divided by half of the signal found in the band representing four chromatids of the natural ARG4 locus on chromosome VIII, and normalized to the 0 h time point. The proportion of arg4-vde chromatids that were visible as DSBs and repaired by SSA or gene conversion was calculated using the SpeI digest from which three bands are visible. The 11.5-kb fragment contains DNA from each of parental arg4-vde chromatids (P), arg4-vde chromatids that have been gene converted to either arg4-bgl or ARG4 (GC) and the donor homologue arg4-bgl allele (D), this band is referred to as (P+GC+D). The 7.8-kb fragment contains broken arg4-vde chromatids (VDE–DSB1). The 2.3-kb fragment contains the SSA deletion product (SSAΔ):
Total signal in lane; TL = (P + GC + D) + VDE–DSB1 + SSAΔ
Signal attributable to arg4-vde alleles; Tv = TL/2
Signal attributable to donor arg4-bgl alleles on homologue; D = TL/2
Proportion of arg4-vde alleles in DSB state = DSB1/Tv
Proportion of arg4-vde alleles in SSAΔ = SSAΔ/Tv
Proportion of arg4-vde alleles that are either unbroken or gene converted = [(P + GC + D) – D]/Tv
RESULTS
We set out to test the idea that Mek1 positively adds drive or stabilization to strand invasion of the homologous chromosome. We used an assay in which intersister repair is inhibited independently of Mek1, and there is an alternative repair route that needs neither a sister chromatid nor a homologous chromosome. Thus, any reduction in interhomologue repair in mek1 mutant cells could not be explained by removal of an intersister repair barrier and ‘passive’ competitive bias for invading the nearer homologous duplex.
The assay depends upon a site-specific DSB created by the meiosis-specific homing endonuclease VDE. This DSB site (referred to as VDE–DSB1) is located inside an allele of ARG4 (arg4-vde), and flanked by directly repeated sequences of URA3 on chromosome V. A similar insert on the opposite homologue contains an arg4-bgl allele that can act as donor for VDE–DSB1 repair, but cannot be cut by VDE. The repeated sequences create an opportunity for repair by SSA (an intrachromatid event). SSA is not blocked by the meiotic BSCR, as in wild-type meiosis it can compete effectively with interhomologue strand invasion repair (23). Repair between sister chromatids is rendered unlikely because VDE cuts both chromatids in the majority of cells. This view is supported by tetrad analysis, which has shown that very few arg4-vde alleles are present in the offspring and that the arg4-vde sister chromatids repair independently of each other (23). Repair of the VDE–DSB1 by an unequal strand invasion event between repeated sequences on sister chromatids is also unlikely. This is evidenced by the fact that VDE–DSB repair using flanking repeats is RAD54-independent (22). In wild-type cells there is a close to equal competition in this assay for VDE–DSB1 to repair by interhomologue strand invasion and gene conversion or SSA [(23); described further below]. Thus, if MEK1 only creates a BSCR, mutating it should have no impact on the competition between gene conversion and SSA at VDE–DSB1. Alternatively, if MEK1 directly promotes strand invasion with the homologue, then removing such function should cause a shift to favour repair by SSA.
To determine if Mek1 positively promotes interhomologue strand invasion, we compared the proportion of VDE–DSB1 repair by SSA in wild-type cells, mek1Δ cells and cells expressing different mek1 alleles that have lost kinase activity.
The proportion of cells receiving VDE–DSB1 is independent of MEK1
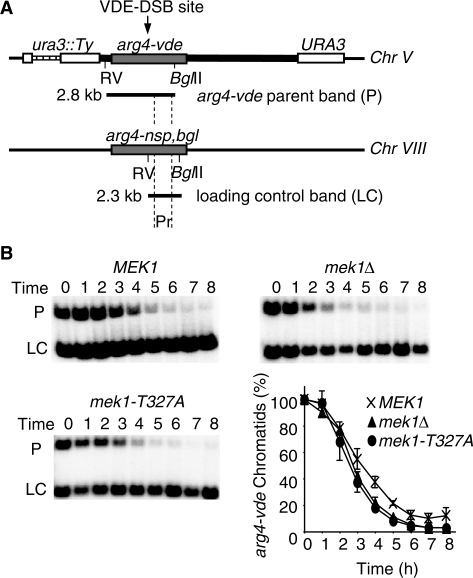
Before determining what proportion of VDE–DSB1s are repaired by SSA in mek1 cells, it was important to establish that Mek1 does not influence the rate at which the arg4-vde allele is cut. This can be done by restriction endonuclease digestion with EcoRV and BglII, followed by probing close to the VDE–DSB1 site (Figure 1; ‘Materials and Methods’ section). The EcoRV/BglII digest, followed by native agarose gel electrophoresis isolates the parental arg4-vde allele from all other related species (VDE–DSB1s, repaired molecules and arg4-bgl on the homologue). Throughout the time course and regardless of repair mechanisms it is possible to monitor the proportion of arg4-vde chromatids that have suffered a DSB (23). The parental arg4-vde containing band was diminished at similar rates in all strains tested (Figure 1B). Most importantly, by the final time point assessed (after 8 h of meiosis) there was no difference in the total proportion of arg4-vde chromatids that had been cut by VDE. From this we conclude that direct comparisons of the amount of SSA product formed are legitimate and not a reflection of different frequencies of VDE–DSB1 formation.
Figure 1.
The arg4-vde allele receives DSBs independently of MEK1 status. (A) The chromosome V ura3::arg4-vde reporter cassette containing the VDE–DSB1 and the chromosome VIII region containing arg4-nsp,bgl site as described previously (22,23). Following restriction endonuclease digestion with EcoRV (RV) and BglII and probing a Southern blot with probe (Pr), (B) the chromosome V arg4-vde parental fragment (P) and chromosome VIII arg4-nsp,bgl fragment (LC) are isolated from other species (not shown). Using the band LC as a loading control it possible to determine the fraction of arg4-vde alleles that have received a DSB, regardless of repair status (repaired molecules move into bands not shown). Neither of the MEK1 alleles tested influence the rate at which the arg4-vde allele receives a DSB.
VDE–DSB1 repair is influenced by MEK1
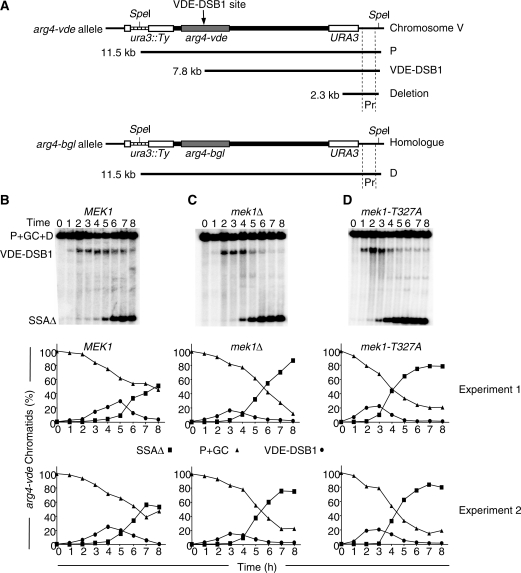
Analysis of VDE–DSB1 repair was undertaken using restriction endonuclease digestion with SpeI and Southern analysis using a probe distal to the furthest flanking repeated sequence. Three bands are visible from this analysis, a high molecular weight band (11.5 kb), a mid molecular weight band (7.8 kb) and a low molecular weight band (2.3 kb; Figure 2). The 11.5-kb band contains a fragment that could be derived from uncut parental arg4-vde chromatids (P), arg4-vde chromatids that have been gene converted to either arg4-bgl or ARG4 (GC) and the donor homologue arg4-bgl allele (D). The 7.8-kb band contains broken arg4-vde chromatids that have not yet been repaired (VDE–DSB1). The 2.3-kb band contains the SSA deletion product (SSAΔ).
Figure 2.
MEK1 influences the repair of VDE–DSB1. (A) Restriction endonclease digestion with SpeI and probing a Southern blot as indicated (Pr) produces three bands. An 11.5-kb band containing DNA from three sources; parental arg4-vde (P), gene conversion products from using the arg4-bgl allele on the homologous template as donor (GC), parental arg4-bgl allele from the donor homologue (D). Two other bands are present, one at 7.8 kb contains the broken arg4-bgl alleles (VDE–DSB1) and one at 2.3 kb contains the deletion product of SSA (SSAΔ). Thick black line represents plasmid DNA, stippled line represents a natural Ty element. (B–D) Representative Southern blots with quantification from strains homozygous for the indicated MEK1 alleles. The graphs show duplicate experiments. The proportion of arg4-vde chromatids with a VDE–DSB1 (circles) or repaired to SSAΔ (squares) are calculated and plotted as described in ‘Materials and Methods’ section. The proportion of DNA in the P+GC+D band that is attributable to only P+GC was calculated as described and displayed for comparison (triangles). Removing Mek1 function by either deletion or using a site-specific mutation in the kinase domain increases the proportion of VDE–DSB1s repaired by SSA. By 8 h when almost all arg4-vde alleles have been cut (Figure 1B) and there is almost no unrepaired VDE–DSB1 detected, the proportion of repair by GC can be deduced (triangle at 8 h). GC is significantly reduced when Mek1 function is absent.
In wild-type cells the VDE–DSB1 appears and disappears with time; reminiscent of what is seen at Spo11-induced DSBs (Figure 2B). By 8 h of meiosis (when few VDE–DSBs remain) the deletion product indicating SSA repair events represents ∼50% of the original arg4-vde alleles. The highest molecular weight band containing P+GC+D chromatids remains dense throughout the time course as 50% of the total probe target, D, remains in this band. Because 50% of the total signal in the lane is from D, it is possible to calculate the proportion of chromatids that are P+GC. As P becomes ∼0 by 8 h the proportion of VDE–DSB1 repair by gene conversion is revealed to be ∼50% in wild-type cells (Figure 2B).
Deleting MEK1 causes significant changes in VDE–DSB1 repair (Figure 2C). The VDE–DSB1 band peaks earlier than in wild-type cells indicating more rapid repair. The balance of repair is shifted towards SSA such that by 8 h of meiosis ∼80% of arg4-vde alleles have suffered a deletion, leaving ∼20% of them gene converted. This increase in propensity to repair VDE–DSB1 by SSA at the cost of gene conversion implies that Mek1 contributes to the ability for meiotic DSBs to repair by interhomologue strand invasion.
Loss of Mek1 kinase activity is sufficient to increase repair by SSA
To determine if this influence of Mek1 resulted from its activity as a kinase, we also examined VDE–DSB1 repair in cells expressing mek1-T327A. This allele has significantly reduced kinase activity, which is normally activated in part by phosphorylation of T327 (19,20). Cells expressing this kinase-deficient allele behaved almost identically to mek1Δ cells (Figure 2D). Similar results were also obtained from cells expressing mek1-K199R a kinase dead protein (25) (data not shown). Thus, the influence of Mek1 on the lifespan of VDE–DSBs and balance of VDE–DSB1 repair by SSA is likely dependent on Mek1 regulating a repair protein by phosphorylation.
The timing of Mek1’s influence on VDE–DSB1 repair coincides with the timing of its influence on spore viability
Cells lacking Mek1 function produce few viable spores (13,17). By using two mek1 alleles, which can be inactivated by exposure to the inhibitor 1-NM-PP1, it has been established that the kinase activity of Mek1 is essential for spore viability and that this function is required during the first 4 h of meiosis (20).
The mek1-as1 allele and inhibitor was used to check if the influence of Mek1 on VDE–DSB1 coincides temporally with its influence on spore viability. The inhibitor was added to sporulating cells expressing mek1-as1 as the only source of Mek1 protein.
Confirming that inhibitor was working, we found the pattern of spore viability to be very similar to that previously reported (20) (data not shown). In brief, when inhibitor was added at the start of meiosis spore viability was 0% (20 tetrads), when inhibitor was added at or after 5 h of meiosis spore viability was 85% of wild-type.
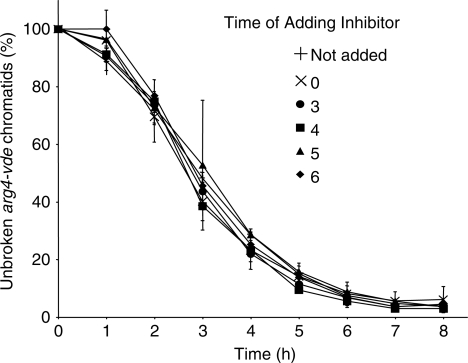
Meiotic time courses were run adding no inhibitor or adding inhibitor once at 0 h, or on the hour from 3 to 6 h. For all time points tested, and regardless of addition of inhibitor, the mek1-as1 allele had no significant impact on VDE–DSB1 formation (Figure 3).
Figure 3.
The rate of cutting the arg4-vde allele is unaffected by the mek1-as1 expression or the addition of inhibitor. Southern analysis was undertaken as described in Figure 1 for cells expressing the mek1-as1 allele with inhibitor added at the times shown. The proportion of arg4-vde containing chromatids that have received a VDE–DSB1 is similar for all time courses tested.
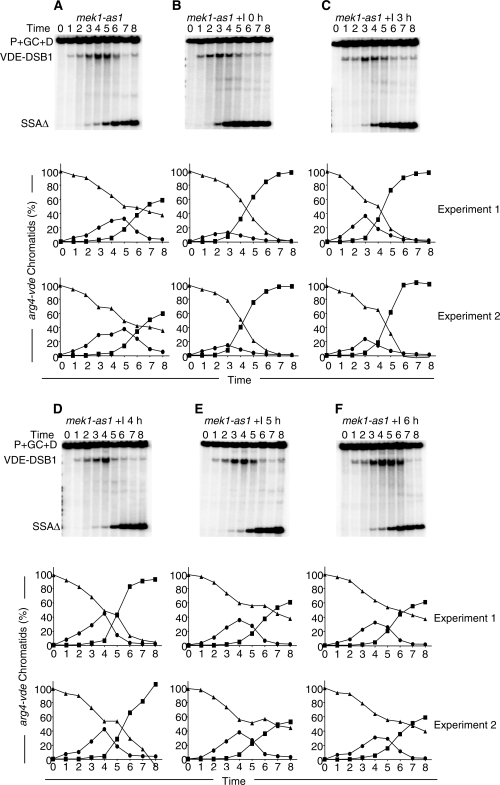
Repair of the VDE–DSB1 was first monitored in mek1-as1 cells without inhibitor added. In mek-as1 cells VDE–DSB1 repair was essentially the same as wild-type (Figure 4A). When inhibitor was added at the start of the meiotic time course the expectation was that the VDE–DSB1 would be repaired in a manner similar to mek1Δ cells, with increased SSA reflecting a reduced gene conversion frequency. This was confirmed with the VDE–DSB1 having a shorter life span than in wild-type cells and the VDE–DSB1 was repaired by SSA in virtually all cells; indicating an almost complete shift away from gene conversion and towards SSA (Figure 4B). Near identical results to these were obtained when inhibiter was added after 3 or 4 h of meiosis (Figure 4C and D).
Figure 4.
The influence of Mek1 function on VDE–DSB1 repair is confined to the first 4 h of meiosis. (A) DNA from sporulating cells expressing mek1-as1 in place of MEK1 was subject to Southern analysis as described for Figure 2. The inhibitor was added where indicated by +I at the times shown. (B–F) The Southern blots and graphs of duplicate experiments are as described in Figure 2B–D. The mek1-as1 allele behaved like wild-type (compare Figure 2). When inhibitor was added at any of 0, 3 or 4 h of meiosis there was shift in repair to bias SSA (squares) rather than gene conversion (triangles at 8 h). Adding inhibitor at 0 and 3 h increased the turnover rate of VDE-DSB1, reducing the peak levels (circles). Adding inhibitor later than 4 h had no discernable impact on VDE–DSB1 repair.
In contrast, adding inhibitor at either 5 or 6 h of meiosis had no discernable impact on VDE–DSB1 repair (Figure 4E and F). The balance of VDE–DSB1 repair by gene conversion and SSA returned to be more like wild-type, with the same levels of SSA seen for mek1-as1 cells without inhibitor. It is worth noting that ∼25% of the arg4-vde population were detected as broken when inhibitor was added at 5 h (Figures 4E). Therefore, had loss of Mek1 kinase activity been an important issue even after 5 h of meiosis, this would have been detectable by an increase in SSA.
These data indicate that kinase activity of Mek1 up to the first 5 h of meiosis contributes to the normal timing of VDE–DSB1 repair, and encourages repair by interhomologue gene conversion at the expense of repair using flanking repeated sequences.
Return to growth experiments confirm that interchromosomal recombination is reduced when Mek1 is inactive and the VDE–DSB1 can repair by SSA
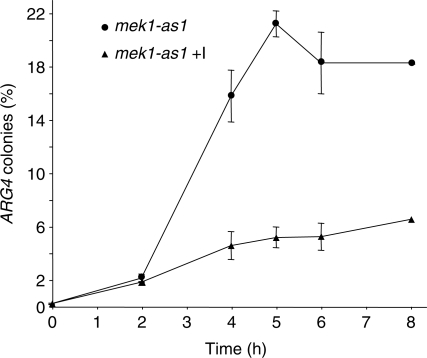
To confirm that gene conversion is reduced in line with the increased SSA when inhibitor is added to mek1-as1 cells, we assessed the proportion of colony forming units that are Arg+ in return to growth (RTG) experiments. Using mek1-as1 cells either with or without inhibitor added at the start of meiosis, cells were withdrawn from meiotic culture and plated on rich medium and synthetic medium lacking arginine. The latter selects cells for growth if there has been at least one interchromosomal gene conversion of arg4-vde to ARG4. For the mek1-as1 cells not exposed to inhibitor the frequency of cells with at least one chromatid repaired to ARG4 increased from less than 1% at 0 h to between 18 and 20% over 8 h of meiosis (Figure 5). For the mek1-as1 cells exposed to inhibitor the frequency of ARG4 colonies reached ∼7% by 8 h of meiosis.
Figure 5.
Mek1 function is required for interchromosomal gene conversion independent of competition for repair with a sister chromatid. Sporulating mek1-as1 cells were withdrawn from meiotic culture at the time indicated, either following addition of inhibitor at 0 h, or without inhibitor. Diluted cells were plated on both rich medium and medium selective for cells expressing ARG4. The calculated frequency of ARG4 colony forming units is significantly higher when Mek1 is active, and the low frequency when Mek1 is inactive could account for the high proportion of repair by SSA (Figure 4). The data are averages from triplicate experiments with standard error indicated.
Using RTG assays the difference between Mek1 active and Mek1 inhibited will be underestimated for two reasons. First, this method does not distinguish between gene conversion on one versus both arg4-vde chromatids because the colonies tested are still diploid (where as the Southern analysis reports on the proportion of sister chromatids repaired by SSA). Thus, the proportion of repair by gene conversion may be relatively underestimated in Mek1 active cells. Secondly, a proportion of cells will not repair the VDE–DSB1 until after RTG, when Mek1 function may not be relevant. In this case the proportion of repair by gene conversion may be overestimated to a greater extent in Mek1 inactive cells. RTG therefore provides a conservative estimate of the differences between meiotic gene conversion frequencies. Even so, the low level of gene conversion found when inhibitor was added is consistent with the high proportion of SSA. This result supports the view that Mek1 is required to stimulate interchromosomal repair independently of raising any BSCR.
MEK1 only influences VDE–DSB repair when gene conversion is an option
A possible explanation for the increased SSA in mek1 cells is that it results from a reduced regulation of resection independently of what is happening at theVDE–DSB1 ends. For successful SSA, resection must uncover the flanking repeated sequences and a mutation causing hyperessection might therefore give SSA a competitive advantage over interhomologue gene conversion. Such hyperresection could arise due to pleiotropic effects or a direct role for MEK1 in regulating resection. We have found previously that repair of the VDE–DSB1 is sensitive to the availability of proteins required for DSB repair (22). For example, in dmc1 cells (which accumulate many kilobasepairs of ssDNA) VDE–DSB1 repair is severely impaired. The failure to repair VDE–DSB1 in dmc1 cells can be completely rescued by preventing Spo11–DSBs formation i.e. in spo11 dmc1 cells (22). Therefore, it is plausible that the fast turnover of Spo11–DSBs in mek1 cells (13,14,17) increases the availability of repair proteins to VDE–DSB1, leading to hyperresection and rapid repair by SSA.
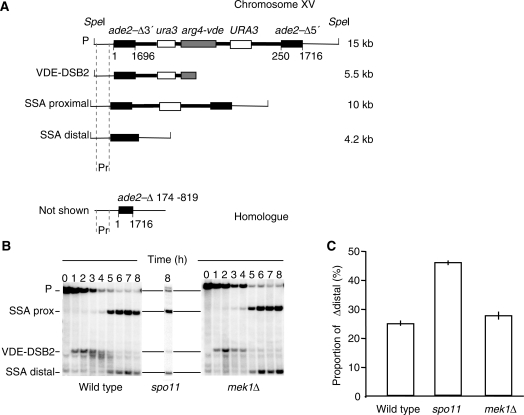
If mutating MEK1 causes hyperresection for either direct or pleiotropic reasons then removing its function should influence resection tract length independently of a competition between gene conversion and SSA. To test for this we assessed repair of VDE–DSB2, which is created in the context of a hemizygous insert that contains nested repeated DNA sequences flanking the break site (22). In this chromosomal insert, repair by SSA can arise after resection of 3.0 kb using the proximal URA3 repeated sequences or after up to 10.0 kb of resection using the distal ADE2 repeated sequences (Figure 6A). Increase in the relative use of the distal repeated sequences compared with proximal repeated sequences is an indicator for hyperresection.
Figure 6.
Mek1 function does not influence the use of proximal versus distal flanking repeats when strand invasion is not an option for VDE–DSB2. DNA from sporulating cells was subject to Southern analysis as previously described, where the wild-type data has been published (22). (A) The bands in the representative Southern blots are; P = parental fragment, SSA prox = product of repair by SSA using proximal repeats (3-kb resection), VDE–DSB2 = the broken molecules, SSA distal = product of repair by SSA using distal repeats (10-kb resection). (B and C) Southern blots and proportion of molecules repaired using distal repeats at 8 h. Loss of Spo11 function causes a hyper resection phenotype at VDE–DSB2 as indicated by the increased proportion of repair to SSA distal, consistent with results seen for VDE–DSB1 (23). Mutating MEK1 has no impact on the proportion of molecules repaired with the longer resection tract.
As a positive control we compared proximal versus distal repeat SSA using VDE–DSB2 in cells completely lacking Spo11–DSBs due to a mutation in SPO11. We have previously shown that spo11-Y135F cells have an increased propensity to repair VDE–DSB1 with long resection tracts, even though gene conversion atVDE–DSB1 is still possible (23). Here we report that atVDE–DSB2 the chance of spo11-Y135F cells repairing with the distal flanking repeats is twice that seen for wild-type cells, confirming that the assay has the resolution to detect hyperresection (Figure 6B and C).
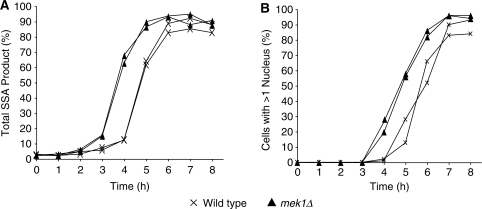
In mek1 cells we found the chance of repairingVDE–DSB2 with the distal flanking repeats was indistinguishable from wild type (Figure 5B and C). In other words, deleting MEK1 does not cause hyperresection at VDE–DSB2. Another possibility is that regardless of resection length appearing to be normal in mek1 cells, MEK1 might directly inhibit SSA itself. Indeed SSA products do appear 1 h earlier in mek1Δ cells (Figure 7A). Counting DAPI stained nuclei through the time courses revealed the mek1 culture was 1 h more advanced in meiosis than wild-type cells (Figure 7B). Rather than SSA being more efficient in mek1 cells due to a direct effect, the earlier appearance of product could reflect either earlier entry of the population into meiosis, or more rapid progress through meiosis I.
Figure 7.
SSA products appear earlier in mek1Δ, reflecting earlier entry into meiosis. (A) The total amount of SSA product (as proportion of arg4-vde containing chromatids) detected in the same duplicate time courses used in Figure 6. SSA product appears earlier in mek1Δ cells. (B) The number of nuclei in each of 100 cells was counted after staining with DAPI, the proportion of cells that had passed through first meiosis is displayed. The population of mek1Δ cells entered meiosis 1 h ahead of wild-type cells, coincident with the earlier appearance of SSA product.
These results provide evidence against the idea that the mek1 phenotype at VDE–DSB1 is due either to an indirect effect of increasing recombination protein supply or a direct effect of losing negative regulation of either long resection or SSA. Rather, the data are consistent with the view that the increased repair by SSA at VDE–DSB1 is due to a failure of interhomologue strand invasion in mek1 cells.
DISCUSSION
The question of how in meiosis DSBs are directed to be repaired using the homologous chromosome as template is fundamental to understanding how DNA repair is regulated in meiosis. This in turn may shed light more broadly on how vegetative cells choose different repair pathways. Conceptually there are two mechanisms that could contribute to the bias in choice of repair partner. There could be a BSCR, so use of the homologue is a default solution. Alternatively, there could be a biochemical activity that specifically enhances the ability of single-stranded intermediates to invade the homologous chromosome. There is no reason why these two possibilities should be mutually exclusive, in fact, both positive choice of template and inhibition of donor template are well-established mechanisms in both budding yeast and fission yeast mating type switching (26,27). However, it is important to note that the main argument for there being a BSCR comes from the fact that rescue of dmc1 arrested DSBs in mek1 cells is RAD54-dependent (18). RAD54 is implicated during RTG and meiosis in specifically raising intersister repair in dmc1 cells (6,28). But in dmc1 cells Rad54 is likely to be important to promote any strand invasion activity as it stimulates the only remaining RecA orthologue, Rad51 (reviewed in (29)). Currently available data does not refute the possibility that DSB repair in mek1 dmc1 cells is via intersister JMs because MEK1 is required to stimulate the interhomologue route. Our data add weight to the argument that Mek1 promotes or stabilizes interhomologue strand invasion.
The repair choice in our system is between interhomologue strand invasion and intrachromosomal SSA. The latter is increased to account for virtually all VDE–DSB1 cells lacking Mek1 function. In theory this could be due to a direct impact on MEK1 normally inhibiting SSA. If the current model of how MEK1 creates a BSCR (via influencing Rad51–Rad54 strand invasion) is correct, then a direct impact of MEK1 on SSA seems unlikely as neither of these proteins contribute to SSA (22,30). On the other hand, it has previously been observed that SSA increases when strand invasion events are diminished by mutation (30). We therefore interpret the increase of SSA in mek1 cells to reflect a failure to strand invade the homologue rather than the loss of a general barrier to intrachromosomal repair. Since turnover of VDE–DSBs is faster in mek1 cells, it is possible that the normal timing of events somehow creates the bias for interhomologue repair, and the faster meiosis in mek1 cells creates a competitive advantage for SSA.
The importance of timing
An important feature of the data presented using the mek1-as1 allele plus inhibitor is the all or none effect between adding inhibitor at 4 h versus 5 h. At 5 h around 10% of the VDE–DSB1s that will be made are still to arise, and ∼25% of arg4-vde chromatids are in a broken state, yet adding inhibitor had no discernible influence. One possible reason for this is that all VDE–DSB1 repair destined to be by strand invasion of the homologue is over by 5 h. In other words, the only molecules available for SSA after 5 h come from the proportion of VDE–DSB1s that in a wild-type situation (no inhibitor) will repair by SSA anyway. This is consistent with the observation that in wild-type cells or mek1-as1 cells without inhibitor, most SSA product appears after 5 h of meiosis. In contrast, adding inhibitor at 0 or 3 h causes more than half the SSA product to appear before 5 h, presumably because molecules normally destined to gene convert before 5 h have been diverted to the SSA pathway.
Following addition of inhibitor after 4 h of meiosis SSA repairs nearly all chromatids by 8 h, even though a substantial proportion of VDE–DSB1 are formed up to 2 h before inhibitor was added. What were these broken molecules doing between 2 and 4 h of meiosis? At a Spo11–DSB, in the SK1 strain, there is arguably a ∼1-h gap between DSB formation and JM appearance (31). JMs must be preceded by strand invasion events. If VDE–DSBs behave as do Spo11–DSBs, a proportion of VDE–DSB1 molecules appearing before 4 h and normally destined to gene convert should have strand invaded before inhibitor was added at 4 h. It is worth considering then, that MEK1 is required to stabilize strand invasion events to ensure commitment to interhomologue recombination. In this case when inhibitor was added at 4 h, strand invasion events destabilized at VDE–DSB1s were shifted onto the SSA pathway.
Is there a link between Mek1 phosphorylation of Rad54 and a bias for strand invading the homologue during meiosis?
One plausible route to Mek1 positively enforcing strand invasion between homologous chromosomes is through modification of RAD52 epitasis group proteins. Niu et al. (21) have reported that Mek1 phosphorylation of Rad54 destabilizes its interaction of Rad51, presumably reducing Rad51s strand invasion activity during meiosis. The Rad51–Rad54 interaction is also inhibited by the binding of Hed1 to Rad51 (7,8). Even though both Mek1 and Hed1 might inhibit strand invasion activity of Rad51, RAD51 is required for efficient meiotic DSB repair and promotion of the bias for interhomologue repair (3,32). To explain these apparently contradictory observations, it has been suggested that a major role of Rad51 in meiosis is structural to promote Dmc1 function (33). Could it be that in mek1 cells additional Rad51 activated for strand invasion disrupts the Dmc1 pathway, inadvertently destabilizing interhomologue strand invasion? This seems unlikely since either mutating the target threonine to be non-phosphorylatable (RAD54-T132A) or deleting HED1 have no obvious meiotic phenotype in otherwise wild-type cells (8,21). It is therefore more likely that Mek1 promotes or stabilizes interhomologue strand invasion through as yet unidentified substrate(s).
How might Mek1 distinguish a sister chromatid from a homologue?
A clear difference between paired homologous chromosomes and sister chromatids is the repertoire of proteins that connects them together. Homologous chromosomes are connected by a proteinaceous structure, the SC. This tripartite structure is made of lateral elements running the length of each homologue and a central region that connects the homologues together (34). In budding yeast the SC is not formed until after strand invasion and other recombination intermediates are made. But, the chromosome axial element that will form part of the SC (and is shared by sister chromatids), are present as DSBs are created (35). Sister chromatids are connected by cohesins, including a specialized meiotic protein Rec8 (36). Either or both of axial elements and cohesins could be used to distinguish a non-sister chromatid (homologue) from a sister chromatid.
Sister chromatid cohesion is partially compromised in mek1 kinase mutants and red1Δ cells (10), and therefore it is plausible that the Mek1–Red1–Hop1 complex interacts with cohesins and uses them to identify the sister chromatid. Also, DSB repair in meiosis is very inefficient in rec8 mutant cells, implying that sister chromatid cohesion may be needed to help direct repair towards the homologue (36). Further evidence for this comes from the fact that another protein, Mnd1, is normally required for DSB repair but this requirement is dependent on the presence of Rec8 (37).
There is also evidence that axial element proteins are important to create the interhomologue bias (3,33,38). Red1 and Hop1 are structural components of the axial elements while Mek1 is required for full length SC (10,11,13,39,40). Thus, Mek1 is physically located in the right place to interact with proteins that will form the SC, and could therefore identify proteins specific to paired homologous chromosomes. Interestingly, ectopic recombination between heterologous chromosomes (which do not pair with axial elements) is significantly less sensitive to hop1 mutation than is allelic interhomologue recombination (41). This is consistent with the view that influence of Mek1–Hop1–Red1 on interchromosomal recombination could be via axial element structures.
Finally, it is also worth asking whether or not the perceived MEK1-dependent BSCR and MEK1-dependent positive enforcement of interhomologue strand invasion need to be separable processes. It is important to point out that the proposed BSCR is not insurmountable even in wild-type cells, as intersister JMs do appear at a significant frequency (3,4). One possibility is that MEK1 functions to tip the balance of equilibrium strongly in favour of interhomologue strand invasion. Interhomologue repair may otherwise be kinetically disfavoured because the sister chromatid is so much nearer than the homologue, especially when early DSBs are made before chromosome pairing is complete.
FUNDING
Biotechnology and Biological Sciences Research Council [50/G20296 to A.S.H.G.]. Funding for open access charge: The open access charges for this paper were partially waived by Oxford University Press.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
Thanks to Nancy Hollingsworth and Michael Lichten for strains and plasmids. The authors are grateful to Nancy Hollingsworth, Michael Lichten, Enrique (Fadri) Martinez-Perez, Helen Bryant, Lydia Hulme and Andrew Cosgrove for discussions and comments on an earlier version of the manuscript and to Nancy Hollingsworth, Michael Lichten and Nancy Kleckner for discussing their unpublished results.
REFERENCES
- 1.Watanabe Y. Modifying sister chromatid cohesion for meiosis. J. Cell. Sci. 2004;117:4017–4023. doi: 10.1242/jcs.01352. [DOI] [PubMed] [Google Scholar]
- 2.Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, Smith GR. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127:1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwacha A, Kleckner N. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997;90:1123–1135. doi: 10.1016/s0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- 4.Schwacha A, Kleckner N. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell. 1994;76:51–63. doi: 10.1016/0092-8674(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 5.Bishop DK, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 6.Bishop DK, Nikolski Y, Oshiro J, Chon J, Shinohara M, Chen X. High copy number suppression of the meiotic arrest caused by a dmc1 mutation: REC114 imposes an early recombination block and RAD54 promotes a DMC1-independent DSB repair pathway. Genes Cells. 1999;4:425–444. doi: 10.1046/j.1365-2443.1999.00273.x. [DOI] [PubMed] [Google Scholar]
- 7.Busygina V, Sehorn MG, Shi IY, Tsubouchi H, Roeder GS, Sung P. Hed1 regulates Rad51-mediated recombination via a novel mechanism. Genes Dev. 2008;22:786–795. doi: 10.1101/gad.1638708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsubouchi H, Roeder GS. Budding yeast Hed1 down-regulates the mitotic recombination machinery when meiotic recombination is impaired. Genes Dev. 2006;20:1766–1775. doi: 10.1101/gad.1422506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsubouchi H, Roeder GS. The importance of genetic recombination for fidelity of chromosome pairing in meiosis. Dev. Cell. 2003;5:915–925. doi: 10.1016/s1534-5807(03)00357-5. [DOI] [PubMed] [Google Scholar]
- 10.Bailis JM, Roeder GS. Synaptonemal complex morphogenesis and sister-chromatid cohesion require Mek1-dependent phosphorylation of a meiotic chromosomal protein. Genes Dev. 1998;12:3551–3563. doi: 10.1101/gad.12.22.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hollingsworth NM, Ponte L. Genetic interactions between HOP1, RED1 and MEK1 suggest that MEK1 regulates assembly of axial element components during meiosis in the yeast Saccharomyces cerevisiae. Genetics. 1997;147:33–42. doi: 10.1093/genetics/147.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nag DK, Scherthan H, Rockmill B, Bhargava J, Roeder GS. Heteroduplex DNA formation and homolog pairing in yeast meiotic mutants. Genetics. 1995;141:75–86. doi: 10.1093/genetics/141.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rockmill B, Roeder GS. A meiosis-specific protein kinase homolog required for chromosome synapsis and recombination. Genes Dev. 1991;5:2392–2404. doi: 10.1101/gad.5.12b.2392. [DOI] [PubMed] [Google Scholar]
- 14.Xu L, Weiner BM, Kleckner N. Meiotic cells monitor the status of the interhomolog recombination complex. Genes Dev. 1997;11:106–118. doi: 10.1101/gad.11.1.106. [DOI] [PubMed] [Google Scholar]
- 15.Thompson DA, Stahl FW. Genetic control of recombination partner preference in yeast meiosis. Isolation and characterization of mutants elevated for meiotic unequal sister-chromatid recombination. Genetics. 1999;153:621–641. doi: 10.1093/genetics/153.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hollingsworth NM, Ponte L, Halsey C. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 1995;9:1728–1739. doi: 10.1101/gad.9.14.1728. [DOI] [PubMed] [Google Scholar]
- 17.Leem SH, Ogawa H. The MRE4 gene encodes a novel protein kinase homologue required for meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 1992;20:449–457. doi: 10.1093/nar/20.3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niu H, Wan L, Baumgartner B, Schaefer D, Loidl J, Hollingsworth NM. Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol. Biol. Cell. 2005;16:5804–5818. doi: 10.1091/mbc.E05-05-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niu H, Li X, Job E, Park C, Moazed D, Gygi SP, Hollingsworth NM. Mek1 kinase is regulated to suppress double-strand break repair between sister chromatids during budding yeast meiosis. Mol. Cell. Biol. 2007;27:5456–5467. doi: 10.1128/MCB.00416-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wan L, de los Santos T, Zhang C, Shokat K, Hollingsworth NM. Mek1 kinase activity functions downstream of RED1 in the regulation of meiotic double strand break repair in budding yeast. Mol. Biol. Cell. 2004;15:11–23. doi: 10.1091/mbc.E03-07-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niu H, Wan L, Busygina V, Kwon Y, Allen JA, Li X, Kunz RC, Kubota K, Wang B, Sung P, et al. Regulation of meiotic recombination via Mek1-mediated Rad54 phosphorylation. Mol. Cell. 2009;36:393–404. doi: 10.1016/j.molcel.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson R, Borde V, Neale MJ, Bishop-Bailey A, North M, Harris S, Nicolas A, Goldman ASH. Excess single-stranded DNA inhibits meiotic double-strand break repair. PLoS Genet. 2007;3:e223. doi: 10.1371/journal.pgen.0030223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neale MJ, Ramachandran M, Trelles-Sticken E, Scherthan H, Goldman ASH. Wild-type levels of Spo11-Induced DSBs are required for normal single-strand resection during meiosis. Mol. Cell. 2002;9:835–846. doi: 10.1016/s1097-2765(02)00498-7. [DOI] [PubMed] [Google Scholar]
- 24.Allers T, Lichten M. A method for preparing genomic DNA that restrains branch migration of Holliday junctions. Nucleic Acids Res. 2000;28:e6. doi: 10.1093/nar/28.2.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de los Santos T, Hollingsworth NM. Red1p, a MEK1-dependent phosphoprotein that physically interacts with Hop1p during meiosis in yeast. J. Biol. Chem. 1999;274:1783–1790. doi: 10.1074/jbc.274.3.1783. [DOI] [PubMed] [Google Scholar]
- 26.Egel R. Fission yeast mating-type switching: programmed damage and repair. DNA Repair. 2005;4:525–536. doi: 10.1016/j.dnarep.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 27.Haber JE. Mating-type gene switching in Saccharomyces cerevisiae. Annu. Rev. Genet. 1998;32:561–599. doi: 10.1146/annurev.genet.32.1.561. [DOI] [PubMed] [Google Scholar]
- 28.Arbel A, Zenvirth D, Simchen G. Sister chromatid-based DNA repair is mediated by RAD54, not by DMC1 or TID1. EMBO J. 1999;18:2648–2658. doi: 10.1093/emboj/18.9.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heyer WD, Li X, Rolfsmeier M, Zhang XP. Rad54: the Swiss Army knife of homologous recombination? Nucleic Acids Res. 2006;34:4115–4125. doi: 10.1093/nar/gkl481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ivanov EL, Sugawara N, Fishman-Lobell J, Haber JE. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allers T, Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106:47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- 32.Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 33.Sheridan S, Bishop DK. Red-Hed regulation: recombinase Rad51, though capable of playing the leading role, may be relegated to supporting Dmc1 in budding yeast meiosis. Genes Dev. 2006;20:1685–1691. doi: 10.1101/gad.1447606. [DOI] [PubMed] [Google Scholar]
- 34.Heyting C. Synaptonemal complexes: structure and function. Curr. Opin. Cell. Biol. 1996;8:389–396. doi: 10.1016/s0955-0674(96)80015-9. [DOI] [PubMed] [Google Scholar]
- 35.Zickler D, Kleckner N. The leptotene-zygotene transition of meiosis. Annu. Rev. Genet. 1998;32:619–697. doi: 10.1146/annurev.genet.32.1.619. [DOI] [PubMed] [Google Scholar]
- 36.Klein F, Mahr P, Galova M, Buonomo SB, Michaelis C, Nairz K, Nasmyth K. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98:91–103. doi: 10.1016/S0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- 37.Zierhut C, Berlinger M, Rupp C, Shinohara A, Klein F. Mnd1 is required for meiotic interhomolog repair. Curr. Biol. 2004;14:752–762. doi: 10.1016/j.cub.2004.04.030. [DOI] [PubMed] [Google Scholar]
- 38.Storlazzi A, Xu L, Schwacha A, Kleckner N. Synaptonemal complex (SC) component Zip1 plays a role in meiotic recombination independent of SC polymerization along the chromosomes. Proc. Natl Acad. Sci. USA. 1996;93:9043–9048. doi: 10.1073/pnas.93.17.9043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith AV, Roeder GS. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J. Cell. Biol. 1997;136:957–967. doi: 10.1083/jcb.136.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hollingsworth NM, Byers B. HOP1: a yeast meiotic pairing gene. Genetics. 1989;121:445–462. doi: 10.1093/genetics/121.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steele DF, Morris ME, Jinks-Robertson S. Allelic and ectopic interactions in recombination-defective yeast strains. Genetics. 1991;127:53–60. doi: 10.1093/genetics/127.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]