Abstract
Genes, like organisms, struggle for existence, and the most successful genes persist and widely disseminate in nature. The unbiased determination of the most successful genes requires access to sequence data from a wide range of phylogenetic taxa and ecosystems, which has finally become achievable thanks to the deluge of genomic and metagenomic sequences. Here, we analyzed 10 million protein-encoding genes and gene tags in sequenced bacterial, archaeal, eukaryotic and viral genomes and metagenomes, and our analysis demonstrates that genes encoding transposases are the most prevalent genes in nature. The finding that these genes, classically considered as selfish genes, outnumber essential or housekeeping genes suggests that they offer selective advantage to the genomes and ecosystems they inhabit, a hypothesis in agreement with an emerging body of literature. Their mobile nature not only promotes dissemination of transposable elements within and between genomes but also leads to mutations and rearrangements that can accelerate biological diversification and—consequently—evolution. By securing their own replication and dissemination, transposases guarantee to thrive so long as nucleic acid-based life forms exist.
INTRODUCTION
Since life first emerged, organisms have been struggling for survival and competing over the finite resources within their ecosystems (1,2). This struggle for survival not only is confined to the organism level but it also applies to individual genes (3) and even non-coding DNA segments (4,5). As a corollary, a gene’s success can be determined by its ability to persist in nature and to be spread throughout genomes and biomes (6). For this to take place, genes need some sequence plasticity to adapt to different environments while retaining enough sequence conservation to preserve the structure of their encoded proteins and the identity of their encoded biological functions (7,8).
Every time a new genome is sequenced, many genes are identified and annotated based on their homology to sequences available in databases, but new genes with novel functions are also identified, adding to the universal gene pool. To date, no study has systematically and directly surveyed the millions of protein-encoding genes (PEGs) deposited in sequence databases to identify their relative prevalence. There have been several challenges to such an endeavor: (i) the absence of numerical parameters to assess a gene’s prevalence; (ii) the lack of fair representation of the tree of life within available sequence data (9,10) and (iii) the difficulty of defining what is meant by ‘same gene’ in different organisms and ecosystems.
To overcome these difficulties, (i) we calculated both the abundance and ubiquity of all known biological functions encoded in genomes and ecosystems to estimate their prevalence, with the assumption that these values will be correlated with gene fitness; (ii) we surveyed both genomic and metagenomic data sets to reduce bias caused by the uneven sampling of the tree of life in genomic data sets; and (iii) we defined similar genes as those encoding proteins with similar specific biological functions. In some instances, this definition could be regarded as an oversimplification, notably in cases of convergent evolution or homoplasy, where multiple genes of different origin evolve to perform similar biological functions. However, the majority of current gene annotations are specific enough to distinguish many instances of paralogous genes or different classes within gene/protein families. It is also understandable that different genes are under different selection pressures, as some are forced to endure mutations and tolerate sequence variability to escape pressure (e.g. bacterial genes encoding immunogenic proteins that are under pressure of the host immune system and genes encoding surface proteins that are easily recognized by predators) while others are under strict sequence conservation pressure (e.g. genes encoding housekeeping enzymes and essential biological functions).
Importantly, in determining gene prevalence we distinguished between ubiquity and abundance. Ubiquity is one of the indicators of essentiality, while abundance without ubiquity is an indicator of adaptive, organism-specific or habitat-specific functionality. In other words, ubiquitous genes are assumed to be those that carry essential functions and are thus indispensable in every genome (elements of core genomes) or every ecosystem (eco-essential genes). On the other hand, genes that are overly abundant in few ecosystems and absent in others are likely to play essential habitat-specific roles (e.g. photosynthesis, anaerobic metabolism, detoxification, etc.).
Contenders for the ‘fittest gene’ title include the gene encoding ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO), an enzyme that plays a critical role in the fixation of carbon dioxide via the Calvin cycle and that has been touted as the single most successful, most abundant enzyme on the planet (11). Genes encoding ribosomal proteins are also plausible candidates. However, those are largely limited to cellular life forms, are not essential and almost absent in viruses (12), and are divergent between eukaryotes and prokaryotes. Additionally, DNA polymerase genes and other genes involved in DNA synthesis and nucleotide/nucleoside metabolism (e.g. ribonucleotide reductases, RNR) are essential for DNA-based life and are not restricted to cellular organisms, being found in viral genomes as well. Their essentiality favors them as strong competitors; yet, they are often present at one or few copies per genome. To our surprise, none of the previous candidate genes topped the list of the most abundant, most ubiquitous genes. Instead, our analysis singled out genes encoding transposases as the most abundant genes in genomes and metagenomes, and the most ubiquitous in metagenomes.
ANALYSIS OF GENOMES AND METAGENOMES
To determine the most abundant non-hypothetical PEG, we examined almost 10 million annotated genes or gene tags: over 3.2 million PEGs in fully sequenced viral, bacterial, archaeal and eukaryotic genomes (2137 genomes on 1 May 2009) and over 6.7 million environmental gene tags (EGT)—with significant matches to known proteins—in 187 random community genomes (metagenomes). For functional assignments, we mostly relied on the annotations available in the SEED database (13) because it uses subsystems-based controlled vocabulary curated by human experts and automatically propagated among genomes (14). For consistency, the same SEED subsystems were used for the annotation of all metagenomic data sets described in this study (15).
Analysis of complete genome sequences
We screened 2137 complete genomes (47 archaeal, 725 bacterial, 29 eukaryotic and 1336 viral genomes at the time this study was performed) available in the SEED database (URL: http://seed-viewer.theseed.org) and identified 37 258 PEGs (1.163% of all PEGs) annotated as transposase-related. Out of these, 26 625 (0.825% of all PEGs) were explicitly annotated as ‘transposases’, 360 were annotated as ‘degenerate transposases’, and then there were a variety of insertion sequence-related transposases, which may or may not be functional. Even when these ambiguous annotations were excluded from the final counts, transposases remained the most abundant PEGs in the completely sequenced genomes (Figure 1).
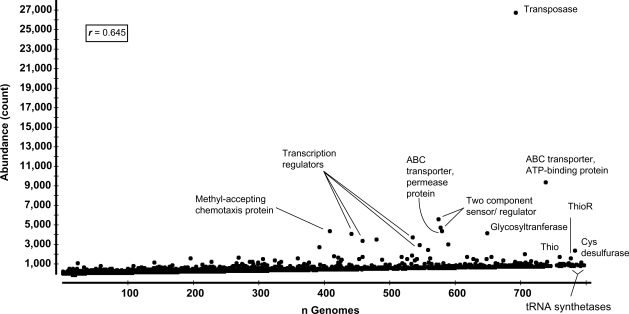
Figure 1.
Abundance of different functional roles in 2137 genomes plotted against the ubiquity of these functional roles (defined as the number of genomes in which the functional role is represented at least once). r, Pearson’s product moment correlation between abundance and ubiquity; Cys, cysteine; Thio, thioredoxin; ThioR, thioredoxin reductase. Proteins annotated solely based on their location or posttranslational modification but not their biological functions (e.g. membrane proteins, cytoplasmic proteins, secreted proteins, transmembrane proteins and generic lipoproteins) were excluded; an exception was the ‘outer membrane protein’ annotation as it describes specific bacterial proteins rather than protein localization.
These data imply that out of a set of 2000 randomly sampled genes (the average number of genes in a typical bacterial genome), 22 genes are expected to encode transposases, at least 16 of which are likely functional. Obviously, genomes that have transposase genes tend to have them in multiple copies; this explains why although two-thirds of sequenced genomes (mostly viral) lack known functional transposases, the average number of transposases—when present—is 38.42 per genome (Table 1 and Supplementary Table S1). This observation is also in agreement with reports that transposases are unequally distributed among bacterial genomes, with higher abundance in facultative pathogens and free-living bacteria than in obligate pathogens and endosymbionts (16), and with extraordinarily high numbers in some species, e.g. Crocosphaera watsonii (17,18).
Table 1.
The 20 most abundant non-hypothetical protein-encoding genes in all sequenced genomes
| Rank | Functional role | nG Count | V | A | B | E | C/n | % |
|---|---|---|---|---|---|---|---|---|
| 1 | Transposase | 693 | 15 (1.1%) | 31 (66%) | 630 (86.9%) | 17 (58.6%) | 38.42 | 0.83 |
| 26 625 | 21 | 736 | 25 226 | 642 | ||||
| 2 | ABC transporter, ATP-binding protein | 738 | 1 (<1%) | 39 (83%) | 682 (94.1%) | 16 (55.2%) | 12.71 | 0.29 |
| 9382 | 1 | 264 | 8998 | 119 | ||||
| 3 | Sensor histidine kinase | 574 | – | 22 (46.8%) | 550 (75.9%) | 2 (6.9%) | 9.71 | 0.17 |
| 5575 | 294 | 5276 | 5 | |||||
| 4 | DNA-binding response regulator | 578 | – | 13 (27.7%) | 562 (77.5%) | 3 (10.3%) | 8.20 | 0.15 |
| 4708 | 33 | 4669 | 6 | |||||
| 5 | Methyl-accepting chemotaxis protein | 408 | 1 (<1%) | 15 (31.9%) | 391 (53.9%) | 1 (3.4%) | 10.76 | 0.14 |
| 4389 | 1 | 64 | 4318 | 6 | ||||
| 6 | ABC transporter, permease protein | 580 | – | 33 (70.2%) | 545 (75.2%) | 2 (6.9%) | 7.55 | 0.14 |
| 4,377 | 137 | 4238 | 2 | |||||
| 7 | Glycosyltransferase (EC 2.4.1.-) | 649 | – | 41 (87.2%) | 598 (82.5%) | 10 (34.5%) | 6.43 | 0.13 |
| 4172 | 287 | 3863 | 22 | |||||
| 8 | Transcriptional regulator, LysR family | 441 | – | 8 (17%) | 430 (59.3%) | 3 (10.3%) | 9.15 | 0.13 |
| 4037 | 10 | 4017 | 10 | |||||
| 9 | Transcriptional regulator, TetR family | 535 | – | 14 (29.8%) | 521 (71.9%) | – | 6.93 | 0.12 |
| 3709 | 45 | 3664 | ||||||
| 10 | Acetyltransferase, GNAT family | 480 | – | 19 (40.4%) | 458 (63.2%) | 3 (10.3%) | 7.33 | 0.11 |
| 3516 | 53 | 3453 | 10 | |||||
| 11 | Transcriptional regulator, AraC family | 459 | 1 (<1%) | 7 (14.9%) | 450 (62.1%) | 1 (3.4%) | 7.37 | 0.11 |
| 3382 | 1 | 7 | 3373 | 1 | ||||
| 12 | Long-chain-fatty-acid–CoA ligase (EC 6.2.1.3) | 589 | – | 31 (66%) | 533 (73.5%) | 25 (86.2%) | 5.08 | 0.09 |
| 2995 | 68 | 2728 | 199 | |||||
| 13 | Transcriptional regulator, MarR family | 546 | – | 23 (48.9%) | 522 (72%) | 1 (3.4%) | 5.32 | 0.09 |
| 2905 | 71 | 2831 | 3 | |||||
| 14 | Permeases of the major facilitator superfamily | 393 | 1 (<1%) | 12 (25.5%) | 375 (51.7%) | 5 (17.2%) | 6.95 | 0.09 |
| 2733 | 1 | 22 | 2701 | 9 | ||||
| 15 | Acetyltransferase (EC 2.3.1.-) | 559 | 4a (<1%) | 22 (46.8%) | 532 (73.4%) | 5 (17.2%) | 4.36 | 0.08 |
| 2436 | 4 | 57 | 2374 | 5 | ||||
| 16 | Cysteine desulfurase (EC 2.8.1.7) | 783 | – | 36 (76.6%) | 722 (99.6%) | 25 (86.2%) | 3.02 | 0.07 |
| 2362 | 66 | 2239 | 57 | |||||
| 17 | 3-oxoacyl-[acyl-carrier protein] reductase (EC 1.1.1.100) | 706 | – | 27 (57.4%) | 665 (91.7%) | 14 (48.3%) | 2.80 | 0.06 |
| 1975 | 68 | 1863 | 44 | |||||
| 18 | Integrase | 534 | 70 (5.2%) | 11 (23.4%) | 448 (61.8%) | 5 (17.2%) | 3.43 | 0.06 |
| 1829 | 70 | 19 | 1729 | 11 | ||||
| 19 | Outer membrane protein | 415 | 1 (<1%) | 10 (21.3%) | 402 (55.4%) | 2 (6.9%) | 4.34 | 0.06 |
| 1803 | 1 | 12 | 1786 | 4 | ||||
| 20 | Permease of the drug/metabolite transporter (DMT) superfamily | 518 | – | 28 (59.6%) | 486 (67%) | 4 (13.8%) | 3.37 | 0.05 |
| 1746 | 53 | 1688 | 5 |
nG: number of genomes in which the functional role is present at least once; Count: number of genes in all sequenced genomes; V, A, B, E: viruses, archaea, bacteria, eukarya, respectively; C/n: average number of genes per positive genome; %: percentage of genes to the total number of genes in all genomes (n = 3 204 918).
aAcetyltransferase-like proteins that were missed in the automated analysis.
While the abundance of transposase genes in microbial genomes has been recognized for long time, only recently has it been exploited for inferring microbial cohabitation patterns and lateral gene transfer (19). Next to transposases, the most abundant functional roles in all sequenced genomes include ABC transporters, transcriptional regulators of different families, signal transduction kinases, chemotaxis proteins, acetyl- and glycosyl- transferases and cysteine desulfurase (Table 1 and Supplementary Table S1). On the other hand, the most ubiquitous functional roles in sequenced genomes are encoded by low-copy-number genes that consequently have a low overall abundance. Only four out of the 100 most ubiquitous functional genes have a mean copy number >2 per genome. These are genes encoding thioredoxin reductase; thioredoxin; cysteine desulfurase and the ABC transporter, ATP-binding protein (Supplementary Table S2). The list of most ubiquitous functional roles in genomes was topped by tRNA synthetases (Figure 1), and other genes associated with protein synthesis and post-translational protein sorting (e.g. translation elongation factor and preprotein translocase, Supplementary Table S2).
Analysis of metagenomic sequences
In spite of the striking prevalence and high copy numbers of transposase genes in fully sequenced genomes, the use of those data sets is prone to biases. The available genomes unevenly represent the tree of life as they mostly correspond to cultured organisms from just four bacterial phyla (9). Moreover, there is an over-representation of microbes of interest to humans (20), such as bacterial pathogens and microbes used in agriculture or industry (21). Finally, while viruses are at least 10 times as abundant as bacteria in nature (22,23), sequenced viral genomes are lagging behind both in terms of numbers (∼2:1 viral to bacterial genome ratio) and annotation quality (most encoded proteins are of unknown functions). In contrast, analysis of community genomes (metagenomes) offers a less-biased representation of life forms and biological functions in various habitats.
The term ‘metagenome’ describes the collective genomes found in a particular ecosystem (24,25). Since the first uncultured viral community genomic sequences were published in 2002 (26), metagenomics has emerged as a rapid and efficient method of identifying not only the species present in a given ecosystem but also the ecosystem-associated metabolic signatures or patterns (27–31). The emergence of low-cost, high-throughput next-generation sequencing technologies (32–37) has enabled the quick implementation of metagenomics in the analysis of different environments, allowing an unprecedented view of biodiversity (25,38–42). Over the past few years, metagenomic sequencing has been used to explore a wide range of environments, encompassing various marine ecosystems (28,43–47), hydrothermal vents (48,49), corals (50–52), salterns (53,54), soil (55–57), sludge (58), mines (59), human and animal guts (60–64) and lungs (65), microbialites (66,67) and even mosquitoes (31).
Metagenomic analysis is shifting the paradigm from organism/genome-centric to gene-centric and pathway-centric approaches to understanding biodiversity (68,69). Several bioinformatics and statistical tools allow the metabolic reconstruction of a particular ecosystem by enumerating EGTs in metagenomes and binning them either phylogenetically or biochemically (15,68,70–73), as well as the comparison of multiple metagenomes (59,74,75).
In this study, we followed a gene-centric approach by enumerating EGTs, and estimating the abundance and ubiquity of their different functional roles in 187 different metagenomic samples representing a broad range of environments. Assessing EGT abundance in metagenomic data is slightly different from determining PEG frequency in fully sequenced genomes. In genomes, a single, full-length copy of a gene reflects a single occurrence of that gene in one cell of an organism. In metagenomic data, multiple occurrence of an EGT can be attributed to either multiple copies of the same gene, multiple orthologs (from different genomes), multiple paralogs or just multiple sequences covering different parts of the exact same DNA segment. Moreover, the coding sequence length is a potential confounding factor: longer genes are more likely to be sampled by random sequencing (unless the sample is large enough to provide 100% coverage). For those reasons, the frequency of each EGT was normalized to the mean length of the most similar proteins [from BLASTX (76) results] to generate an abundance index, which was further divided by the number of informative sequence reads (those sequence tags matching annotated proteins in known databases) to generate a normalized abundance index (see the legend of Table 2 for more details).
Table 2.
The 20 most abundant functional roles in metagenomes
| Rank | Functional role | nMG | nCAI |
|---|---|---|---|
| 1 | Transposase | 178 | 4026.17 |
| 2 | Retrotransposon-related p150 protein | 69 | 3412.12 |
| 3 | Viral structural protein | 126 | 1909.75 |
| 4 | ABC transporter, ATP-binding protein | 170 | 1528.03 |
| 5 | Replication-associated protein | 32 | 1481.67 |
| 6 | Photosystem II CP43 protein (PsbC) | 47 | 1429.44 |
| 7 | Photosystem II protein D2 (PsbD) | 71 | 1224.89 |
| 8 | Replication protein Rep | 66 | 1213.18 |
| 9 | Photosystem II protein D1 (PsbA) | 83 | 930.2 |
| 10 | Cytochrome b6-f complex subunit, cytochrome b6 | 51 | 925.32 |
| 11 | Viral nonstructural protein | 39 | 847.57 |
| 12 | ATP synthase alpha chain (EC 3.6.3.14) | 157 | 804.47 |
| 13 | Ribonucleotide reductase of class Ia (aerobic), alpha subunit (EC 1.17.4.1) | 165 | 776.57 |
| 14 | Thymidylate synthase thyX (EC 2.1.1.-) | 140 | 771.16 |
| 15 | Single-stranded DNA-binding protein | 151 | 769.41 |
| 16 | Major capsid protein | 100 | 745.51 |
| 17 | ATP synthase beta chain (EC 3.6.3.14) | 156 | 661.21 |
| 18 | UDP-glucose 4-epimerase (EC 5.1.3.2) | 169 | 657.36 |
| 19 | Ribonucleotide reductase of class Ia (aerobic), beta subunit (EC 1.17.4.1) | 150 | 652.32 |
| 20 | Integrase | 164 | 633.18 |
nMG: number of metagenomes in which the functional role is present at least once; nCAI: normalized cumulative abundance index. For each metagenome, a normalized abundance index (nAI) was calculated as the relative, length-normalized number of functional roles per million EGTs, and the nAI values for each functional role were added up to generate the normalized cumulative abundance index (nCAI).
The metagenomic data sets, which have been sequenced by different research groups, have been analyzed, consistently annotated and made publicly available through the metagenomics RAST server [http://metagenomics.theseed.org (15)]. They include both free-living and metazoan-associated viral, bacterial and eukaryotic sequences from autotrophic and heterotrophic communities from a wide variety of environments. In the analyzed metagenomes, the two most abundant functional genes are related to transposable elements [transposase and the retrotransposon-related p150 protein (77)]. Next to these, a set of photosynthesis-related genes; genes encoding viral structural, nonstructural, capsid and integrase proteins; genes associated with DNA replication; and genes involved in DNA synthesis are all among the most abundant biological functions in environmental metagenomes (Table 2 and Supplementary Table S3).
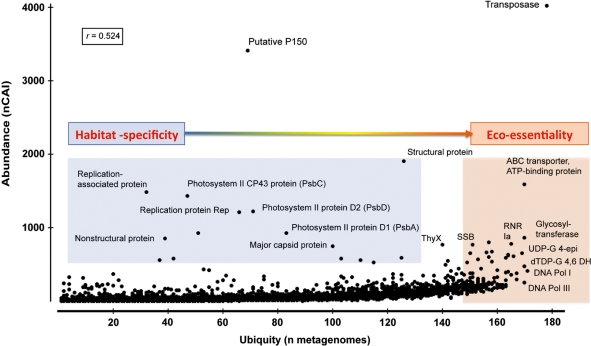
Since gene abundance in metagenomes is sensitive to sampling bias and sequencing depth, we also combined ubiquity with abundance data. The combined analysis confirmed the prevalence of transposases (abundant in 95% of the analyzed metagenomes) over the retrotransposon-related p150 genes (overly abundant in only 36% of these metagenomes) and other replication and DNA metabolism-related genes that are equally ubiquitous but less abundant than transposases (Figure 2). The abundance of all analyzed non-hypothetical functions does not necessarily correlate with their ubiquity (Pearson correlation index = 0.524, Figure 2), i.e. many EGTs were pervasive in some ecosystems but absent in others (e.g. photosystem II proteins, p150 and viral structural genes; Table 2). Ubiquitous EGTs, on the other hand, include those matching transposases, DNA polymerases and enzymes involved in nucleotide metabolism (e.g. dTDP-glucose 4,6-dehydratase, UDP-glucose 4-epimerase and RNR; see Table 3 and Supplementary Table S4). Most of the ubiquitous EGTs are likely to be ‘housekeeping’ and essential for life, rather than habitat-specific (Figure 2). Additionally, many of these EGTs (e.g. DNA polymerases and RNRs) are found in all cellular and non-cellular biological entities, including viruses. As with genome sequence data, transposases are unequally distributed in ecosystems. This unequal distribution is in accordance with studies of ocean community genomics that showed a depth-dependent abundance of transposase genes (30) and a recent study that reported an unusually high abundance of transposase and retroviral integrase genes in a hydrothermal chimney biofilm (49).
Figure 2.
The normalized cumulative abundance indices (nCAI) of different functional roles in 187 metagenomes plotted against the ubiquity of these functional roles (defined as the number of metagenomes in which the functional role is represented at least once). r, Pearson’s product moment correlation between abundance and ubiquity; DNA Pol, DNA polymerase; dTDP-G 4,6 DH, dTDP-glucose 4,6 dehydratase; Rep, replication-associated protein; RNR, ribonuleotide reductase; SSB, single-stranded DNA-binding protein; ThyX, thymidylate synthase thyX (EC 2.1.1.-); UDP-G 4-epi, UDP-glucose 4-epimerase.
Table 3.
The 20 most ubiquitous functional roles in metagenomes
| Rank | Functional role | nMG | % |
|---|---|---|---|
| 1 | Transposase | 178 | 95.19 |
| 2 | DNA polymerase I (EC 2.7.7.7) | 171 | 91.44 |
| 3 | dTDP-glucose 4,6-dehydratase (EC 4.2.1.46) | 170 | 90.91 |
| 4 | DNA polymerase III alpha subunit (EC 2.7.7.7) | 170 | 90.91 |
| 5 | ABC transporter, ATP-binding protein | 170 | 90.91 |
| 6 | UDP-glucose 4-epimerase (EC 5.1.3.2) | 169 | 90.37 |
| 7 | Heat shock protein 60 family chaperone GroEL | 167 | 89.30 |
| 8 | Chaperone protein DnaK | 167 | 89.30 |
| 9 | Ribonucleotide reductase of class II (coenzyme B12-dependent) (EC 1.17.4.1) | 166 | 88.77 |
| 10 | Ribonucleotide reductase of class Ia (aerobic), alpha subunit (EC 1.17.4.1) | 165 | 88.24 |
| 11 | Replicative DNA helicase (EC 3.6.1.-) | 165 | 88.24 |
| 12 | Integrase | 164 | 87.70 |
| 13 | Long-chain-fatty-acid–CoA ligase (EC 6.2.1.3) | 164 | 87.70 |
| 14 | Phosphate starvation-inducible protein PhoH, predicted ATPase | 163 | 87.17 |
| 15 | Carbamoyl-phosphate synthase large chain (EC 6.3.5.5) | 163 | 87.17 |
| 16 | DNA primase (EC 2.7.7.-) | 163 | 87.17 |
| 17 | Glycosyltransferase | 163 | 87.17 |
| 18 | Valyl-tRNA synthetase (EC 6.1.1.9) | 163 | 87.17 |
| 19 | Thymidylate synthase (EC 2.1.1.45) | 163 | 87.17 |
| 20 | ATP-dependent Clp protease ATP-binding subunit clpX | 162 | 86.63 |
nMG: number of metagenomes in which the functional role is present at least once; %: percentage of nMG to the total number of metagenomes analyzed (187).
Other than the predominance of transposases, ABC transporter ATP-binding proteins and phage integrases (Table 2), there is little agreement in the gene abundance data between genomes and metagenomes (Tables 1 and 2). In genomic data, the most abundant functional roles reflect the over-representation of bacterial proteins in currently available fully sequenced genomes (2.5 million bacterial proteins versus 560 000 eukaryotic, 100 000 archaeal and 40 000 viral proteins). This bias may decrease when more viral genomes are sequenced and better annotated to reflect their actual distribution in nature. In metagenomic data, abundance indices reflect an overrepresentation of bacterial, archaeal and viral over eukaryotic sequences in currently available data sets; however, this overrepresentation is in agreement with reports that bacteria and archaea dominate the cellular world (78) while viruses are the most abundant biological entities (22,23).
DISCUSSION
The main assumption of this study is that the most successful genes are likely to be prevalent in genomes and ecosystems. We defined the most prevalent gene as the one ‘spreading its DNA around’ and not the one expressing the most protein molecules. Thus, while RuBisCO, for example, is claimed as the most abundant enzyme on Earth (11) based on the estimated number of its protein molecules, its genes are neither the most abundant nor most widely distributed (Supplementary Table S5). In addition, we focused on PEGs and did not include genes encoding ribosomal RNA in the analysis; those are absent in viruses and usually present in multiple copies in cellular genomes [1–15, mean = 4, (79)], which would place them at the 12th rank in gene abundance in all sequenced genomes (compare with Table 1).
This study demonstrates that transposases are the most abundant genes in both completely sequenced genomes and environmental metagenomes, and are also the most ubiquitous in metagenomes. Transposase genes encode DNA-binding enzymes, members of the polynucleotidyl transferase superfamily, that catalyze ‘cut-and-paste’ or ‘copy-and-paste’ reactions promoting the movement of DNA segments to new sites (80). The term transposase is often used to describe what are classically known as DNA transposases or type II transposases. These move double-stranded DNA directly by excision and insertion, and are sometimes associated with insertion sequences, but often just catalyze their own mobilization (81,82). The major group of dsDNA transposases is known as DDE transposases due to their possession of a non-contiguous, highly conserved catalytic triad of two aspartate (D) and one glutamate (E) residues (83). Other protein families that essentially use transposition but lack the DDE motif include tyrosine and serine recombinases, and rolling-circle transposases (82). In addition, within these transposase subclasses, several protein family domains [PFam domains (84)] have been described (49,83), yet a large fraction of transposases identified in genomes and metagenomes remain unclassified.
There are two other classes of transposable elements (Types I and III) that are distinguished as separate categories and were not as abundant or ubiquitous as Type II transposases in our analyzed data sets. Type I includes retrotransposons, which use the enzyme retrotransposase to move DNA by reverse transcription of an RNA intermediate (85). Retrotransposases (Type I transposases) are suggested to be responsible for the majority of ‘junk’ repeats, which make up >40% of the human genome and seem to code for no other genes (86–88). Type III transposable elements are associated with miniature inverted-repeat transposable elements (MITEs) (89,90). Transposases, in general, and Type II transposases, in particular, constitute a highly diverse group of enzymes. It is difficult to provide a robust, consistent scheme for classifying transposase sequences in ecosystems; however, structure-based classification schemes are being developed (83).
The prevalence of transposons (Type II) and retrotransposons (Type I) in eukaryotic genomes has been well documented, but in these genomes they are mostly associated with non-coding, repetitive DNA (91–93). Moreover, Type II transposases are continuously being detected in bacterial, archaeal and, to a lesser extent, bacteriophage genomes. In this work, we demonstrate that these jumping genes are also almost omnipresent in every ecosystem that contains nucleic acid-based life forms.
OUTLOOK
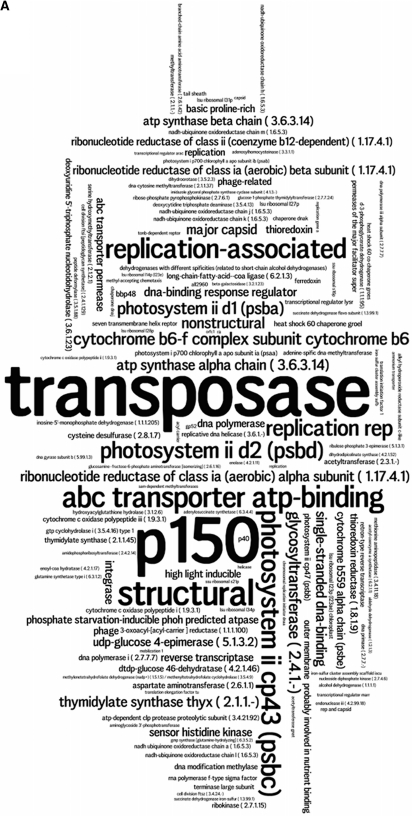
Transposase genes have been classically considered as ‘selfish genes’ with no other purpose than spreading themselves and are thus expected to be universal DNA parasites (6,85). If this were their only raison-d’être, they have certainly fulfilled it by surviving, persisting and prevailing in all ecosystems. An open question is whether their ubiquity is also an indication of eco-essentiality. The finding that transposases are as ubiquitous as housekeeping DNA-processing enzymes but that they outnumber all essential genes (Figure 3) supports the idea that these mobile, self-replicating genes strive to inhabit and multiply in as many genomes as possible.
Figure 3.
Word clouds (created on http://www.wordle.net) representing (A) the 100 most abundant functional roles (Supplementary Table S3) and (B) the 100 most ubiquitous functional roles (Supplementary Table S4) in metagenomes. The font size of each functional role is proportional to its (A) abundance index or (B) number of metagenomes in which it is present.
Besides the obvious detrimental effect that transposition can cause to host genomes—by inactivating housekeeping genes or impairing the chromosome’s structure—transposases also play beneficial roles (92). For example, transposases may mobilize or activate genes that enhance their hosts’ fitness (94,95), induce advantageous rearrangements (96) or enrich the host’s gene pool (97–100). There are accruing documented examples of transposase genes co-opted by the host to encode transcription factors (99), centromere-binding proteins (100) or generators of diversity in the immune system (97,98), a process described as exaptation [or domestication, from a host-centric view (94)]. Such cases can involve one or a few transposases per genome or, as more recently shown, thousands of transposases (95).
Despite their ubiquity and abundance, there is neither evidence nor reason to believe that transposases encode conserved essential cellular functions. In our opinion, the role of transposases as diversifying agents (94,101) is beneficial enough to be selected for; however, the cost of transposon-induced mutations also puts pressure on the cells to inactivate or delete their transposases (16,91,93,101).
In conclusion, the prevalence of transposases in metagenomes and completely sequenced genomes from bacteria, archaea, eukaryotes and viruses is in accordance with suggestions that they may offer a selective advantage to the genomes and ecosystems that they ‘parasitize’ (17,94,101). The diversification they induce in these genomes and ecosystems is arguably an essential way of maintaining, diversifying and evolving life on our planet.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation, Division of Biological Infrastructure (DBI-0850356 to R.A.E. and DBI-0850206 to M.B.); the NMPDR project was supported by National Institutes of Health (HHSN266200400042C). Funding for open access charge: National Science Foundation, Division of Biological Infrastructure (DBI-0850356 to R.A.E.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Anca Segall, Elizabeth Dinsdale, Forest Rohwer, Peter Salamon, Jim Nulton and Ben Felts for stimulating discussions and helpful suggestions, and Moselio Schaechter and Stanley Maloy for valuable suggestions to improve the manuscript.
REFERENCES
- 1.Darwin C. On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life. London: John Murray; 1859. [PMC free article] [PubMed] [Google Scholar]
- 2.Huxley JS. Evolution: The Modern Synthesis. 1st edn. London: Harper; 1942. [Google Scholar]
- 3.Dawkins R. The Selfish Gene. Oxford: Oxford University Press; 1976. [Google Scholar]
- 4.Edgell DR, Fast NM, Doolittle WF. Selfish DNA: the best defense is a good offense. Curr. Biol. 1996;6:385–388. doi: 10.1016/s0960-9822(02)00502-x. [DOI] [PubMed] [Google Scholar]
- 5.Doolittle WF, Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980;284:601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- 6.Orgel LE, Crick FH. Selfish DNA: the ultimate parasite. Nature. 1980;284:604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- 7.Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koonin EV. Darwinian evolution in the light of genomics. Nucleic Acids Res. 2009;37:1011–1034. doi: 10.1093/nar/gkp089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hugenholtz P. Exploring prokaryotic diversity in the genomic era. Genome Biol. 2002;3:REVIEWS0003. doi: 10.1186/gb-2002-3-2-reviews0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, Kunin V, Goodwin L, Wu M, Tindall BJ, et al. A phylogeny-driven genomic encyclopaedia of bacteria and archaea. Nature. 2009;462:1056–1060. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dhingra A, Portis A.R., Jr, Daniell H. Enhanced translation of a chloroplast-expressed RbcS gene restores small subunit levels and photosynthesis in nuclear RbcS antisense plants. Proc. Natl Acad. Sci. USA. 2004;101:6315–6320. doi: 10.1073/pnas.0400981101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kristensen DM, Mushegian AR, Dolja VV, Koonin EV. New dimensions of the virus world discovered through metagenomics. Trends Microbiol. 2010;18:11–19. doi: 10.1016/j.tim.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crecy-Lagard V, Diaz N, Disz T, Edwards R, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, et al. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ochman H, Davalos LM. The nature and dynamics of bacterial genomes. Science. 2006;311:1730–1733. doi: 10.1126/science.1119966. [DOI] [PubMed] [Google Scholar]
- 17.Mes TH, Doeleman M. Positive selection on transposase genes of insertion sequences in the Crocosphaera watsonii genome. J. Bacteriol. 2006;188:7176–7185. doi: 10.1128/JB.01021-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zehr JP, Bench SR, Mondragon EA, McCarren J, DeLong EF. Low genomic diversity in tropical oceanic N2-fixing cyanobacteria. Proc. Natl Acad. Sci. USA. 2007;104:17807–17812. doi: 10.1073/pnas.0701017104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hooper SD, Mavromatis K, Kyrpides NC. Microbial co-habitation and lateral gene transfer: what transposases can tell us. Genome Biol. 2009;10:R45. doi: 10.1186/gb-2009-10-4-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aziz RK. The case for biocentric microbiology. Gut. Pathogen. 2009;1:16. doi: 10.1186/1757-4749-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmed N. A flood of microbial genomes-do we need more? PLoS ONE. 2009;4:e5831. doi: 10.1371/journal.pone.0005831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furuse K, Osawa S, Kawashiro J, Tanaka R, Ozawa A, Sawamura S, Yanagawa Y, Nagao T, Watanabe I. Bacteriophage distribution in human faeces: continuous survey of healthy subjects and patients with internal and leukaemic diseases. J. Gen. Virol. 1983;64(Pt 9):2039–2043. doi: 10.1099/0022-1317-64-9-2039. [DOI] [PubMed] [Google Scholar]
- 23.Breitbart M, Rohwer F. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 2005;13:278–284. doi: 10.1016/j.tim.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 24.Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem. Biol. 1998;5:R245–R249. doi: 10.1016/s1074-5521(98)90108-9. [DOI] [PubMed] [Google Scholar]
- 25.Riesenfeld CS, Schloss PD, Handelsman J. Metagenomics: genomic analysis of microbial communities. Annu. Rev. Genet. 2004;38:525–552. doi: 10.1146/annurev.genet.38.072902.091216. [DOI] [PubMed] [Google Scholar]
- 26.Breitbart M, Salamon P, Andresen B, Mahaffy JM, Segall AM, Mead D, Azam F, Rohwer F. Genomic analysis of uncultured marine viral communities. Proc. Natl Acad. Sci. USA. 2002;99:14250–14255. doi: 10.1073/pnas.202488399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 28.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 29.Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, Podar M, Short JM, Mathur EJ, Detter JC, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 30.DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU, Martinez A, Sullivan MB, Edwards R, Brito BR, et al. Community genomics among stratified microbial assemblages in the ocean’s interior. Science. 2006;311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- 31.Dinsdale EA, Edwards RA, Hall D, Angly F, Breitbart M, Brulc JM, Furlan M, Desnues C, Haynes M, Li L, et al. Functional metagenomic profiling of nine biomes. Nature. 2008;452:629–632. doi: 10.1038/nature06810. [DOI] [PubMed] [Google Scholar]
- 32.Ronaghi M, Karamohamed S, Pettersson B, Uhlen M, Nyren P. Real-time DNA sequencing using detection of pyrophosphate release. Anal. Biochem. 1996;242:84–89. doi: 10.1006/abio.1996.0432. [DOI] [PubMed] [Google Scholar]
- 33.Ronaghi M. Pyrosequencing sheds light on DNA sequencing. Genome Res. 2001;11:3–11. doi: 10.1101/gr.11.1.3. [DOI] [PubMed] [Google Scholar]
- 34.Bennett S. Solexa Ltd. Pharmacogenomics. 2004;5:433–438. doi: 10.1517/14622416.5.4.433. [DOI] [PubMed] [Google Scholar]
- 35.Kartalov EP, Quake SR. Microfluidic device reads up to four consecutive base pairs in DNA sequencing-by-synthesis. Nucleic Acids Res. 2004;32:2873–2879. doi: 10.1093/nar/gkh613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schuster SC. Next-generation sequencing transforms today’s biology. Nat. Methods. 2008;5:16–18. doi: 10.1038/nmeth1156. [DOI] [PubMed] [Google Scholar]
- 38.Schloss PD, Handelsman J. Biotechnological prospects from metagenomics. Curr. Opin. Biotechnol. 2003;14:303–310. doi: 10.1016/s0958-1669(03)00067-3. [DOI] [PubMed] [Google Scholar]
- 39.Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 2004;68:669–685. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edwards RA, Rohwer F. Viral metagenomics. Nat. Rev. Microbiol. 2005;3:504–510. doi: 10.1038/nrmicro1163. [DOI] [PubMed] [Google Scholar]
- 41.Xu J. Microbial ecology in the age of genomics and metagenomics: concepts, tools, and recent advances. Mol. Ecol. 2006;15:1713–1731. doi: 10.1111/j.1365-294X.2006.02882.x. [DOI] [PubMed] [Google Scholar]
- 42.Casas V, Rohwer F. Phage metagenomics. Methods Enzymol. 2007;421:259–268. doi: 10.1016/S0076-6879(06)21020-6. [DOI] [PubMed] [Google Scholar]
- 43.Angly FE, Felts B, Breitbart M, Salamon P, Edwards RA, Carlson C, Chan AM, Haynes M, Kelley S, Liu H, et al. The marine viromes of four oceanic regions. PLoS Biol. 2006;4:e368. doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yooseph S, Sutton G, Rusch DB, Halpern AL, Williamson SJ, Remington K, Eisen JA, Heidelberg KB, Manning G, Li W, et al. The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McDaniel L, Breitbart M, Mobberley J, Long A, Haynes M, Rohwer F, Paul JH. Metagenomic analysis of lysogeny in Tampa Bay: implications for prophage gene expression. PLoS ONE. 2008;3:e3263. doi: 10.1371/journal.pone.0003263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Persson OP, Pinhassi J, Riemann L, Marklund BI, Rhen M, Normark S, Gonzalez JM, Hagstrom A. High abundance of virulence gene homologues in marine bacteria. Environ. Microbiol. 2009;11:1348–1357. doi: 10.1111/j.1462-2920.2008.01861.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grzymski JJ, Murray AE, Campbell BJ, Kaplarevic M, Gao GR, Lee C, Daniel R, Ghadiri A, Feldman RA, Cary SC. Metagenome analysis of an extreme microbial symbiosis reveals eurythermal adaptation and metabolic flexibility. Proc. Natl Acad. Sci. USA. 2008;105:17516–17521. doi: 10.1073/pnas.0802782105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brazelton WJ, Baross JA. Abundant transposases encoded by the metagenome of a hydrothermal chimney biofilm. ISME J. 2009;3:1420–1424. doi: 10.1038/ismej.2009.79. [DOI] [PubMed] [Google Scholar]
- 50.Dinsdale EA, Pantos O, Smriga S, Edwards RA, Angly F, Wegley L, Hatay M, Hall D, Brown E, Haynes M, et al. Microbial ecology of four coral atolls in the Northern Line Islands. PLoS ONE. 2008;3:e1584. doi: 10.1371/journal.pone.0001584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vega Thurber RL, Barott KL, Hall D, Liu H, Rodriguez-Mueller B, Desnues C, Edwards RA, Haynes M, Angly FE, Wegley L, et al. Metagenomic analysis indicates that stressors induce production of herpes-like viruses in the coral Porites compressa. Proc. Natl Acad. Sci. USA. 2008;105:18413–18418. doi: 10.1073/pnas.0808985105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vega Thurber R, Willner-Hall D, Rodriguez-Mueller B, Desnues C, Edwards RA, Angly F, Dinsdale E, Kelly L, Rohwer F. Metagenomic analysis of stressed coral holobionts. Environ. Microbiol. 2009;11:2148–2163. doi: 10.1111/j.1462-2920.2009.01935.x. [DOI] [PubMed] [Google Scholar]
- 53.Santos F, Meyerdierks A, Pena A, Rossello-Mora R, Amann R, Anton J. Metagenomic approach to the study of halophages: the environmental halophage 1. Environ. Microbiol. 2007;9:1711–1723. doi: 10.1111/j.1462-2920.2007.01289.x. [DOI] [PubMed] [Google Scholar]
- 54.Rodriguez-Brito B, Li L, Wegley L, Furlan M, Angly F, Breitbart M, Buchanan J, Desnues C, Dinsdale E, Edwards R, et al. Viral and microbial community dynamics in four aquatic environments. ISME J. 2010 doi: 10.1038/ismej.2010.1. [12 February 2010, Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 55.Kim KH, Chang HW, Nam YD, Roh SW, Kim MS, Sung Y, Jeon CO, Oh HM, Bae JW. Amplification of uncultured single-stranded DNA viruses from rice paddy soil. Appl. Environ. Microbiol. 2008;74:5975–5985. doi: 10.1128/AEM.01275-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pang H, Zhang P, Duan CJ, Mo XC, Tang JL, Feng JX. Identification of cellulase genes from the metagenomes of compost soils and functional characterization of one novel endoglucanase. Curr. Microbiol. 2009;58:404–408. doi: 10.1007/s00284-008-9346-y. [DOI] [PubMed] [Google Scholar]
- 57.Zhang K, He J, Yang M, Yen M, Yin J. Identifying natural product biosynthetic genes from a soil metagenome by using T7 phage selection. Chembiochem. 2009;10:2599–2606. doi: 10.1002/cbic.200900297. [DOI] [PubMed] [Google Scholar]
- 58.Kunin V, He S, Warnecke F, Peterson SB, Garcia Martin H, Haynes M, Ivanova N, Blackall LL, Breitbart M, Rohwer F, et al. A bacterial metapopulation adapts locally to phage predation despite global dispersal. Genome Res. 2008;18:293–297. doi: 10.1101/gr.6835308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edwards RA, Rodriguez-Brito B, Wegley L, Haynes M, Breitbart M, Peterson DM, Saar MO, Alexander S, Alexander E.C., Jr, Rohwer F. Using pyrosequencing to shed light on deep mine microbial ecology. BMC Genomics. 2006;7:57. doi: 10.1186/1471-2164-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Breitbart M, Hewson I, Felts B, Mahaffy JM, Nulton J, Salamon P, Rohwer F. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 2003;185:6220–6223. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frank DN, Pace NR. Gastrointestinal microbiology enters the metagenomics era. Curr. Opin. Gastroenterol. 2008;24:4–10. doi: 10.1097/MOG.0b013e3282f2b0e8. [DOI] [PubMed] [Google Scholar]
- 63.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tuohy KM, Gougoulias C, Shen Q, Walton G, Fava F, Ramnani P. Studying the human gut microbiota in the trans-omics era–focus on metagenomics and metabonomics. Curr. Pharm. Des. 2009;15:1415–1427. doi: 10.2174/138161209788168182. [DOI] [PubMed] [Google Scholar]
- 65.Willner D, Furlan M, Haynes M, Schmieder R, Angly FE, Silva J, Tammadoni S, Nosrat B, Conrad D, Rohwer F. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS ONE. 2009;4:e7370. doi: 10.1371/journal.pone.0007370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Desnues C, Rodriguez-Brito B, Rayhawk S, Kelley S, Tran T, Haynes M, Liu H, Furlan M, Wegley L, Chau B, et al. Biodiversity and biogeography of phages in modern stromatolites and thrombolites. Nature. 2008;452:340–343. doi: 10.1038/nature06735. [DOI] [PubMed] [Google Scholar]
- 67.Breitbart M, Hoare A, Nitti A, Siefert J, Haynes M, Dinsdale E, Edwards R, Souza V, Rohwer F, Hollander D. Metagenomic and stable isotopic analyses of modern freshwater microbialites in Cuatro Cienegas, Mexico. Environ. Microbiol. 2009;11:16–34. doi: 10.1111/j.1462-2920.2008.01725.x. [DOI] [PubMed] [Google Scholar]
- 68.Eisen JA. Environmental shotgun sequencing: its potential and challenges for studying the hidden world of microbes. PLoS Biol. 2007;5:e82. doi: 10.1371/journal.pbio.0050082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hugenholtz P, Tyson GW. Microbiology: metagenomics. Nature. 2008;455:481–483. doi: 10.1038/455481a. [DOI] [PubMed] [Google Scholar]
- 70.Angly F, Rodriguez-Brito B, Bangor D, McNairnie P, Breitbart M, Salamon P, Felts B, Nulton J, Mahaffy J, Rohwer F. PHACCS, an online tool for estimating the structure and diversity of uncultured viral communities using metagenomic information. BMC Bioinformatics. 2005;6:41. doi: 10.1186/1471-2105-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krause L, Diaz NN, Goesmann A, Kelley S, Nattkemper TW, Rohwer F, Edwards RA, Stoye J. Phylogenetic classification of short environmental DNA fragments. Nucleic Acids Res. 2008;36:2230–2239. doi: 10.1093/nar/gkn038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kunin V, Copeland A, Lapidus A, Mavromatis K, Hugenholtz P. A bioinformatician’s guide to metagenomics. Microbiol. Mol. Biol. Rev. 2008;72:557–578. doi: 10.1128/MMBR.00009-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schloss PD, Handelsman J. A statistical toolbox for metagenomics: assessing functional diversity in microbial communities. BMC Bioinformatics. 2008;9:34. doi: 10.1186/1471-2105-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rodriguez-Brito B, Rohwer F, Edwards RA. An application of statistics to comparative metagenomics. BMC Bioinformatics. 2006;7:162. doi: 10.1186/1471-2105-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huson DH, Richter DC, Mitra S, Auch AF, Schuster SC. Methods for comparative metagenomics. BMC Bioinformatics. 2009;10(Suppl. 1):S12. doi: 10.1186/1471-2105-10-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sassaman DM, Dombroski BA, Moran JV, Kimberland ML, Naas TP, DeBerardinis RJ, Gabriel A, Swergold GD, Kazazian H.H., Jr Many human L1 elements are capable of retrotransposition. Nat. Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- 78.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc. Natl Acad. Sci. USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee ZM, Bussema C, 3rd, Schmidt TM. rrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea. Nucleic Acids Res. 2009;37:D489–D493. doi: 10.1093/nar/gkn689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rice PA, Baker TA. Comparative architecture of transposase and integrase complexes. Nat. Struct. Biol. 2001;8:302–307. doi: 10.1038/86166. [DOI] [PubMed] [Google Scholar]
- 81.Craig NL, Craigie R, Gellert M, Lambowitz AM. Mobile DNA II. Washington, DC: ASM press; 2002. [Google Scholar]
- 82.Curcio MJ, Derbyshire KM. The outs and ins of transposition: from mu to kangaroo. Nat. Rev. Mol. Cell. Biol. 2003;4:865–877. doi: 10.1038/nrm1241. [DOI] [PubMed] [Google Scholar]
- 83.Hickman AB, Chandler M, Dyda F. Integrating prokaryotes and eukaryotes: DNA transposases in light of structure. Crit. Rev. Biochem. Mol. Biol. 2010;45:50–69. doi: 10.3109/10409230903505596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wright S, Finnegan D. Genome evolution: sex and the transposable element. Curr. Biol. 2001;11:R296–R299. doi: 10.1016/s0960-9822(01)00168-3. [DOI] [PubMed] [Google Scholar]
- 86.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 87.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 88.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wessler SR, Bureau TE, White SE. LTR-retrotransposons and MITEs: important players in the evolution of plant genomes. Curr. Opin. Genet. Dev. 1995;5:814–821. doi: 10.1016/0959-437x(95)80016-x. [DOI] [PubMed] [Google Scholar]
- 90.Feschotte C, Mouches C. Evidence that a family of miniature inverted-repeat transposable elements (MITEs) from the Arabidopsis thaliana genome has arisen from a pogo-like DNA transposon. Mol. Biol. Evol. 2000;17:730–737. doi: 10.1093/oxfordjournals.molbev.a026351. [DOI] [PubMed] [Google Scholar]
- 91.Ohshima K, Hattori M, Yada T, Gojobori T, Sakaki Y, Okada N. Whole-genome screening indicates a possible burst of formation of processed pseudogenes and Alu repeats by particular L1 subfamilies in ancestral primates. Genome Biol. 2003;4:R74. doi: 10.1186/gb-2003-4-11-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mills RE, Bennett EA, Iskow RC, Devine SE. Which transposable elements are active in the human genome? Trends Genet. 2007;23:183–191. doi: 10.1016/j.tig.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 93.Pace J.K., 2nd, Feschotte C. The evolutionary history of human DNA transposons: evidence for intense activity in the primate lineage. Genome Res. 2007;17:422–432. doi: 10.1101/gr.5826307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Benjak A, Forneck A, Casacuberta JM. Genome-wide analysis of the “cut-and-paste” transposons of grapevine. PLoS ONE. 2008;3:e3107. doi: 10.1371/journal.pone.0003107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nowacki M, Higgins BP, Maquilan GM, Swart EC, Doak TG, Landweber LF. A functional role for transposases in a large eukaryotic genome. Science. 2009;324:935–938. doi: 10.1126/science.1170023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mendiola MV, Bernales I, de la Cruz F. Differential roles of the transposon termini in IS91 transposition. Proc. Natl Acad. Sci. USA. 1994;91:1922–1926. doi: 10.1073/pnas.91.5.1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 98.Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 99.Lin R, Ding L, Casola C, Ripoll DR, Feschotte C, Wang H. Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science. 2007;318:1302–1305. doi: 10.1126/science.1146281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Casola C, Hucks D, Feschotte C. Convergent domestication of pogo-like transposases into centromere-binding proteins in fission yeast and mammals. Mol. Biol. Evol. 2008;25:29–41. doi: 10.1093/molbev/msm221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Condit R, Stewart FM, Levin BR. The population biology of bacterial transposons: A priori conditions for maintenance as parasitic DNA. Am. Nat. 1988;132:129–147. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.