Abstract
Using peptide arrays and binding to native histone proteins, we show that the ADD domain of Dnmt3a specifically interacts with the H3 histone 1–19 tail. Binding is disrupted by di- and trimethylation of K4, phosphorylation of T3, S10 or T11 and acetylation of K4. We did not observe binding to the H4 1–19 tail. The ADD domain of Dnmt3b shows the same binding specificity, suggesting that the distinct biological functions of both enzymes are not related to their ADD domains. To establish a functional role of the ADD domain binding to unmodified H3 tails, we analyzed the DNA methylation of in vitro reconstituted chromatin with Dnmt3a2, the Dnmt3a2/Dnmt3L complex, and the catalytic domain of Dnmt3a. All Dnmt3a complexes preferentially methylated linker DNA regions. Chromatin substrates with unmodified H3 tail or with H3K9me3 modification were methylated more efficiently by full-length Dnmt3a and full-length Dnmt3a/3L complexes than chromatin trimethylated at H3K4. In contrast, the catalytic domain of Dnmt3a was not affected by the H3K4me3 modification. These results demonstrate that the binding of the ADD domain to H3 tails unmethylated at K4 leads to the preferential methylation of DNA bound to chromatin with this modification state. Our in vitro results recapitulate DNA methylation patterns observed in genome-wide DNA methylation studies.
INTRODUCTION
DNA methylation is a major form of epigenetic modification and plays essential roles in gene expression regulation and chromatin structure remodeling (1–3). The methylation state of DNA is closely connected to other epigenetic signals including histone modifications, such as methylation or acetylation, which are known to activate or silence gene expression (4). The methylation of CpG dinucleotides (CpG) in mammalian cells is catalyzed by DNA methyltransferases (Dnmts), comprising Dnmt3a and 3b, which establish DNA methylation patterns during embryonic development and Dnmt1, which maintains the methylation pattern after DNA replication (1,2,5). Dnmt3a and 3b contain large N-terminal parts including a PWWP domain and a PHD-like ADD domain, which interact with other proteins, and a C-terminal domain harboring the catalytic center. The isolated catalytic domains of Dnmt3a and 3b are enzymatically active (6). Another member of the Dnmt3 family, Dnmt3L (Dnmt3-like), is homologous to the Dnmt3 enzymes, but lacks catalytic activity. It acts as a regulatory factor and can stimulate the catalytic activity of Dnmt3a and 3b (7–10).
The ADD domain of Dnmt3L was shown to interact specifically with histone H3 tails that are unmethylated at lysine 4 (11). The Dnmt3a/3L complex forms a heterotetramer (12), suggesting that the interaction of the Dnmt3L ADD domain with the H3 tail could direct DNA methylation by Dnmt3a (11). The ADD domains of Dnmt3a and 3b share considerable homology with Dnmt3L and recently binding of the Dnmt3a ADD domain to H3 tails unmodified at K4 has been shown and the structure of this complex solved (13). In addition, an interaction of the Dnmt3a ADD with H4R3me2s peptides has been reported as well (14). Here, we studied the interaction of the Dnmt3a and 3b ADD domains with modified histone tails by applying a hypothesis free peptide array binding approach.
Independent experimental evidence suggesting an influence of histone tail modification on DNA methylation has been provided through several epigenomic studies. Genome-wide DNA methylation and histone modification studies revealed a strong anti-correlation of DNA methylation and histone H3 lysine 4 trimethylation (H3K4me3) (15–18), and a correlation of H3K9me3 with DNA methylation (16). A functional connection of these two silencing marks (DNA methylation and H3K9 methylation) has been observed before in Neurospora crassa (19,20), plants (21) and mammalian cells (22), where disruption of the H3K9me3 signal led to a loss of DNA methylation. As described above, biochemical and structural data suggest a direct role of the ADD domain of Dnmt3L in the targeting of Dnmt3a to chromatin unmethylated at H3K4 (11). However, so far experimental evidence for preferential methylation of DNA bound to chromatin, which carries a particular modification pattern, has not been provided. In order to establish the molecular mechanism of DNA methylation guidance by histone modification states, we set up a complete in vitro system that allowed us to study the influence of chromatin modifications on the activity of purified Dnmt3a or Dnmt3a/3L. To this end, histones were generated by native peptide ligation to contain specifically H3K4 and H3K9 methylation. The modified histones were assembled into octamers, bound to DNA and the reconstituted oligonucleosomes were used as substrates for methylation with Dnmt3a and Dnmt3a/3L.
MATERIALS AND METHODS
For details of ‘Materials and Methods’ section; see Supplementary Data.
Recombinant chromatin preparation
Expression and purification of Xenopus laevis histones were performed as described (23). H3K4me3 and H3K9me3 were generated by native protein ligation. Ligation of the activated H3 peptide to the truncated H3 histone and purification of the ligation product was performed as described (24). Assembly of histone octamers containing H3unmod, H3K4me3 and H3K9me3, as well as reconstitution of recombinant oligonucleosomes was performed by salt dialysis as described, using the 12× 200 × 601 template (23,25).
Purification of Dnmt enzymes
The Dnmt3a and Dnmt3b ADD domains (amino acids 472–610 of murine Dnmt3a and 422–560 of murine Dnmt3b1) were cloned as GST fusion proteins and purified by standard procedures. The expression and purification of Dnmt3a2, Dnmt3L and Dnmt3a-C were carried out as described (10,12). The purity of the proteins was determined on 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel to be better than 90% (Supplementary Figure S4).
Binding of protein domains to peptides arrays
CelluSpots arrays were provided by Intavis AG (Köln, Germany). The array was blocked, incubated with purified GST-tagged ADD domains of Dnmt3a or 3b (1 µM) and the binding detected by anti-GST antibody. We confirmed the correct synthesis of the spots containing T3P, K4me2, K4me3, S10P, T11P and H4R3me2s by incubation with modification specific antibodies (Supplementary Figure S5), methylation of the array with different histone lysine methyltransferases and by high-performance liquid chromatography (HPLC) and mass spectrometric analysis of peptides synthesized in parallel and cleaved off from the matrix for characterization (data not shown).
Bisulfite conversion, subcloning and sequencing
The modified chromatin was methylated with Dnmt3a complexes and the DNA was purified and treated with bisulfite as described (26).
RESULTS
Binding of the Dnmt3a ADD domain to modified histone tail peptides
We used an array comprising 384 peptide spots prepared by the CelluSpots method (27) to study the interaction of the purified Dnmt3a and 3b ADD domains with modified histone tails (Figure 1). The array contains peptides from eight different regions of the N-terminal tails of histones, namely H3 1–19, 7–26, 16–35 and 26–45, H4 1–19 and 11–30, H2A 1–19 and H2B 1–19 (Figure 1 and Supplementary Data S1), featuring 59 post-translational modifications in many different combinations. Each array was presenting the same peptides in duplicate for quality control. Binding to the internal duplicates was highly reproducible (Supplementary Figure S1). In addition, the results were confirmed by binding of the ADD domain to peptide arrays originating from an independent peptide synthesis (data not shown).
Figure 1.
Binding of the Dnmt3a and Dnmt3b ADD domains to peptide arrays and native histones. (A) Binding of Dnmt3a ADD domain to peptide arrays comprising 384 different peptides. The enlargement shows the binding to the H3 1–19 peptides. Peptides containing H3K4me2 or me3, H3T3P, H3S10P or H3T11P are shaded green, red and blue, respectively. The positions of the unmodified H3 1–19 as well as the peptides di- and trimethylated at K4 are annotated. (B) Design of the CelluSpots histone tail peptide arrays. For a detailed annotation of all spots cf. Supplementary Data S1. (C) Binding of Dnmt3b ADD domain to peptide arrays comprising 384 different peptides. (D) Binding of ADD domains to native histones isolated from human cells. Histones were separated by polyacrylamide gel electrophoresis and blotted to Nitrocellulose membrane. The membrane was stained with Ponceau S (PS). Then, membranes were incubated with GST tagged ADD domains, washed and ADD binding detected with anti-GST antibody staining (AS).
Peptide binding of the ADD domains from Dnmt3a and 3b was very similar (Figure 1). In general, we observed highly specific binding of the Dnmt3a and 3b ADD domains to differentially modified H3 tails (Figure 1A). Binding to H2A and H2B tails was not observed. Binding at H4 tails was only observed at H4K16ac, if they contained additional modifications in different combinations. Binding of the ADD domain to H4R3me2s peptides that has been reported recently (14) could not be detected (Supplementary Figure S2). We tested binding of the ADD domain to native histones isolated from human cells by western blotting and detected strong binding to the H3 tail, but no binding to H4, H2A or H2B tails (Figure 1D). This result suggests that the special combinations of modifications on H4 leading to ADD binding on the peptide array are not frequent in chromatin isolated from human cells. Based on these results, we focused on the H3 tail interaction for the further functional studies.
Binding of H3 tails was disrupted by modification of H3K4 other than monomethylation. In addition, the phosphorylation of T3, S10 or T11 inhibited peptide binding. As shown clearly in Figure 1A, binding of the ADD domain to H3 1–19 peptides is only possible if none of these inhibiting modifications is present. Monomethylation of K4 reduced binding, but did not completely prevent the interaction. In addition, non-natural acetylation of the N-terminus completely disrupted binding, indicating that the free amino terminus is an important contact point as well (Supplementary Figure S3). These results agree with equilibrium peptide binding results for the Dnmt3a ADD domain reported by Otani et al. (13), who showed that H3K4me2, H3K4me3 and N-acetylation inhibit H3 tail binding, while trimethylation at K9 has no influence on binding. Also in their experiments, binding to H4 1–19 tails and H4 1–19 modified by R3me2s was very weak or undetectable. Other modifications were not tested in that study.
We conclude that similar to the ADD domain of Dnmt3L, the Dnmt3a and Dnmt3b ADD domains bind to the H3 tail. With Dnmt3L, mono-, di- and trimethylation of K4 was shown to inhibit binding, but the effect of monomethylation was weaker than that of di- or trimethylation (11). Since we have available active full-length Dnmt3a2 [which is an isoform of Dnmt3a (28) containing the ADD domain] and its catalytic domain (not containing the ADD) (6), we focused our functional studies on the targeting of DNA methylation activity of Dnmt3a2 by its ADD domain.
Reconstitution of specifically modified oligonucleosomes
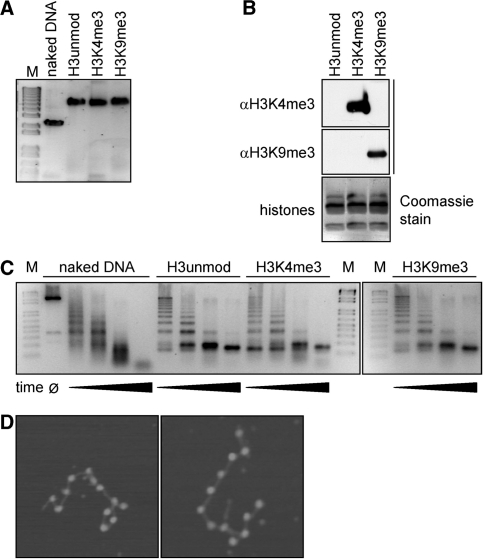
With the aim to study the interaction of DNA methylation and histone modifications in nucleosomes, we generated recombinant unmodified chromatin and chromatin containing H3K4me3 and H3K9me3 modifications. Wild-type Xenopus laevis histones were expressed and purified. Histone H3 proteins modified at H3K4me3 and H3K9me3 were generated by native protein ligation (24). The assembly of histone octamers containing unmodified H3, H3K4me3 and H3K9me3 and the reconstitution of recombinant oligonucleosomes was performed using the 12 × 200 × 601 template (12 tandem repeats of a 200 bp 601 DNA sequence). The nucleosome positioning sequence locates at the center of each monomeric 200 bp of the 601 repeat, and the 12 repeats were arranged on the array in a head to tail manner (25,29). The successful assembly of chromatin was verified by native gel electrophoresis (Figure 2A) and analytical ultracentrifugation (data not shown). Identity and purity of the histone proteins was confirmed by SDS-PAGE (Figure 2B) and by mass spectrometry. To further control the saturation level of the oligonucleosomal reconstitution, assembled material was analyzed after digestion with micrococcal nuclease (Figure 2C). In addition, samples were analyzed by scanning force microscopy after mild fixation with glutaraldehyde and deposition on mica support (Figure 2D) (30). At a 1.1 to 1 octamer:DNA ratio, all samples showed the MNase digestion pattern characteristic for nucleosome bound DNA; 11 ± 1 nucleosomes on arrays were observed in SFM, confirming the successful reconstitution of recombinant chromatin.
Figure 2.
Analysis of reconstituted oligonucleosomes. (A) Native agarose gel (0.5%, 0.2× TBE) of free DNA or oligonucleosomes after reconstitution on 12 × 200 × 601 sequence containing uniformly the indicated H3 species. Gel was stained post-running with ethidium bromide (M, molecular size markers). (B) Reconstituted oligonucleosomes containing the indicated H3 species were run on SDS PAGE gel and analyzed by western blotting using the indicated antibodies. A separately run gel was stained with Coomassie blue. (C) DNA used for reconstitution or assembled oligonucleosomes containing different H3 species were digested with MNase for varying times. Samples were run on agarose gels (1.5%, 1× TBE) and stained with ethidium bromide (M, molecular size markers). (D) Scanning force microscopy images of reconstituted oligonucleosomes. At a 1.1 : 1 octamer : DNA ratio the number of visible nucleosomes on one DNA molecule typically ranged from 10 to 12.
Methylation of DNA bound to oligonucleosomes
In the next step, the recombinant chromatin was methylated by full-length Dnmt3a2, the C-terminal catalytic domain of Dnmt3a, and the complex of full-length Dnmt3a2 and Dnmt3L. For methylation analysis, the DNA was isolated and treated with bisulfite. By using primers specific for the DNA sequence of the monomeric 601 repeat (Figure 3), we amplified the PCR products, which represent a mixture of the DNA repeat in all 12 nucleosomes, and analyzed the DNA methylation pattern by sequencing of individual clones. The analyzed region spanned the nucleosome and included 16 out of the 18 CpG sites of each 601 repeat. DNA methylation was investigated in both DNA strands. Control methylation reactions were performed with naked DNA from the 12 × 200 × 601 template, which was used for the chromatin reconstitution. The Dnmt3a2/3L complex methylated DNA with about 1.5-fold enhanced rate as compared to Dnmt3a2 alone, which is a similar level of stimulation of Dnmt3a by Dnmt3L on long DNA substrates as reported in the literature (31). We observed in all reactions that nucleosome bound DNA was methylated with a strong preference at the first three CpG sites, which are located in the linker DNA region immediately upstream of the nucleosomal bound DNA (Figure 3). The activities of all enzymes were reduced on chromatin when compared to naked DNA, with Dnmt3a-C showing stronger reduction than full-length enzymes (Figure 3). In addition, the naked DNA showed a totally different methylation pattern, indicating that the preference for methylation of the first three sites was not related to the DNA sequence.
Figure 3.
DNA methylation of unmodified chromatin and naked DNA by full-length Dnmt3a2, full-length Dnmt3a2/Dnmt3L complex and Dnmt3a-C. DNA methylation analysis was performed for both strands of DNA in chromatin substrates and DNA. Results shown are the average of three independent experiments. Altogether about 250 clones were sequenced for each strand and each data point. (A) Methylation levels of single CpG sites in the upper and lower strand of the DNA. (B) DNA sequence of the 601 monomeric unit (200 bp). The grey color shades the sequences of the forward and reverse primers. The red-color-labeled CpG sites were analyzed in this study. The nucleosome positioning sequence (147 bp) is underlined.
Methylation of DNA bound to specifically modified oligonucleosomes
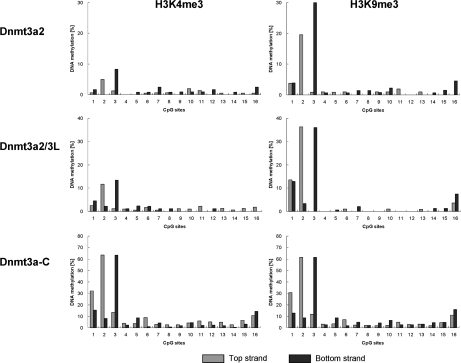
Next, we investigated the methylation of the oligonucleosomes carrying H3K4me3 or H3K9me3 modifications following the same approach (Figure 4). Since we observed the strongest methylation in the first three CpG sites, we restricted the analysis to these sites (Figure 5). Introduction of the H3K9me3 modification did not significantly change the methylation levels—an observation that is in agreement with previous results obtained using mononucleosomes (32) and with the lack of any effect of H3K9me3 on ADD binding to H3 tails. In contrast, the DNA methylation level introduced into the H3K4me3 modified chromatin by full-length Dnmt3a2 or the complex of full-length Dnmt3a2 with Dnmt3L was strongly decreased when compared to the unmodified chromatin. To analyze whether this reduction of methylation of H3K4me3 modified chromatin was related to the ADD domain of Dnmt3a, we methylated the recombinant chromatin using only the catalytic domain of Dnmt3a (Dnmt3a-C). The results showed that without the ADD domain there was no significant methylation difference between the unmodified chromatin and the chromatin containing H3K4me3, suggesting that it is the K4 methylation specific H3 tail binding of the ADD domain that guides methylation of full-length Dnmt3a in this experiment. This observation also confirms that the different modified chromatin substrates used here are of comparable quality.
Figure 4.
DNA methylation levels of single CpG sites in chromatin with H3K4me3 or H3K9me3 modifications by full-length Dnmt3a2, full-length Dnmt3a2/Dnmt3L and Dnmt3a-C. Results shown are the averages of three independent experiments. Altogether about 250 clones were sequenced for each strand and each data point.
Figure 5.
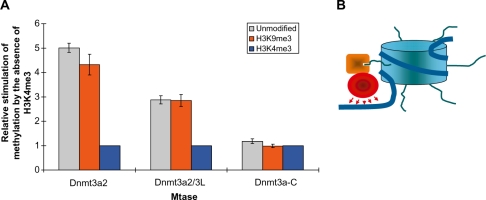
Methylation of DNA on modified oligonucleosomes. (A) Methylation of the first three CpG sites by full-length Dnmt3a2, full-length Dnmt3a2/Dnmt3L, but not Dnmt3a-C is stimulated by the absence of H3K4me3 (data taken from Figures 3 and 4). For each comparison, the methylation levels were normalized to H3K4me3, which showed the lowest methylation. Results shown are the average of three independent experiments; error bars indicate the standard deviations. (B) Schematic picture showing how the H3 tail interaction of Dnmt3a’s ADD domain (orange) anchors the catalytic domain (red) to methylate the linker DNA regions of nucleosomes.
DISCUSSION
It is an open question how DNA methylation patterns are generated by DNA methyltransferases. For the catalytic domain of Dnmt3a it has been observed that it shows some flanking sequence preferences (33,34) and it favors the methylation of CpG sites in a distance equal to one helical turn of DNA (12,35). Both these properties were shown to influence the DNA methylation in cells, but they cannot explain the generation of a specific DNA methylation pattern during embryogenesis and germ cell development. In addition, being an epigenetic signal, the DNA methylation pattern cannot be determined only by the primary DNA sequence alone.
We have shown that the ADD domains of Dnmt3a and Dnmt3b interact with the H3-tail unmethylated at K4 similar as the ADD domain of Dnmt3L (11). Thereby, these domains assist the binding of full-length Dnmt3a (or the Dnmt3a/Dnmt3L complex) to chromatin. Using designer nucleosomes containing defined modifications, we investigated whether H3K4me3 or H3K9me3 modifications influence the methylation of the chromatin bound DNA by Dnmt3a2 and its catalytic domain (which lacks the N-terminal part including the ADD domain). Nucleosome bound DNA was methylated with a strong preference for the linker DNA region. This finding confirms a previous results obtained with full-length Dnmt3a and Dnmt3a-C using mononucleosomes reconstituted with a different DNA template and a different method to study methylation of chromatin bound DNA (32). We conclude that the core nucleosome is protected against DNA methylation by Dnmt3a, but the linker regions of the DNA are accessible. The finding that histone bound DNA is protected from methylation is consistent with results obtained with chromatin isolated from mammalian cells (36) also showing that addition of H1 further inhibits DNA methylation. We observed that the catalytic domain of Dnmt3a showed a higher activity than Dnmt3a2. This result was reproducible over many purifications and many years. It may suggest that the N-terminal part of Dnmt3a2 has a repressive regulatory function on the catalytic domain, as it is often seen in other regulated enzymes like proteases or kinases.
The preferential methylation of linker DNA is observed for Dnmt3a2 and its isolated catalytic domain, indicating that it is not mediated by the ADD domain. However, with Dnmt3a2 the DNA methylation levels of chromatin with unmodified H3 tail or with the H3K9me3 modification is four to five times higher than methylation of H3K4me3 containing chromatin. This difference was not seen with the isolated catalytic domain of Dnmt3a. This result indicates that the disruption of the ADD-H3 tail interaction by di- or tri-methylation of H3K4 interferes with the chromatin recruitment of Dnmt3a2 and methylation of DNA. With Dnmt3a2/3L, methylation of unmodified and H3K9me3 methylated chromatin was about three times higher than with H3K4me3 chromatin. The reduced stimulation might be explained by the fact that in the Dnmt3a/3L tetramer the Dnmt3L ADD domains also interact with the H3 tails, but Dnmt3L does not contribute a catalytic domain. In contrast, in case of Dnmt3a2, each ADD-H3 tail interaction leads to the recruitment of one catalytic domain to the chromatin bound DNA.
Using peptide array containing several hundred peptides in different modification states, we identified additional modifications of the H3 tail that interfere with binding, namely acetylation of K4 and phosphorylation of T3, S10 or T11. The effects at T3 and K4 can be readily explained in the light of the Dnmt3a ADD structure (13): T3 is in 4.4 Å distance to Glu545, an interaction that would be disrupted by phosphorylation of the threonine. K4 is inserted into a very tight acidic pocket formed by Asp529 and Asp531 that does not allow for its acetylation. The effects in the C-terminal part of the H3 peptide cannot be easily interpreted, but in the structure the peptide was covalently bound to the N-terminus of the ADD domain, such that the positioning of the C-terminal part of the H3 peptide might not be fully reliable.
Our data provide the first evidence that preferential binding of a DNA methyltransferase to histone tails carrying specific post-translational modification patterns directly leads to the favored methylation of DNA bound to the modified chromatin (Figure 5B). Here, we demonstrate that this effect can be achieved by the ADD domain in Dnmt3a, which binds to chromatin with the same specificity as the Dnmt3L ADD domain. However, other domains and factors may further contribute to the targeting of Dnmt3a. For example, the PWWP domain of Dnmt3a has been shown to be involved in heterochromatin targeting of the enzyme (37,38). Our unpublished data suggest that this domain is also directly engaged in an interaction with histone tails (manuscript in preparation). Together with the DNA binding of the catalytic domains, these interactions may be responsible for the strong and direct interaction of Dnmt3a with chromatin (39). Another silencing mark that is associated with DNA methylation is H3K9me3 (19–22). Our results make a direct interaction of Dnmt3a with H3K9me3 (via the ADD domain or any other part of the enzyme) unlikely, suggesting that this correlation might be mediated by additional proteins, like HP1, which reads H3K9me3 and has been shown to directly interact with Dnmt3a (40).
It is interesting that the ADD domains of Dnmt3L (11), Dnmt3a (this work and Ref. 13) and Dnmt3b (this work) all basically have the same preference for binding to H3 tails unmodified at K4. Dnmt3a and 3b show clearly distinct biological functions with Dnmt3a being involved in the setting of parental imprints (41,42) and Dnmt3b in the methylation of pericentromeric repeats (41,43). Our results suggest that the ADD domains of both proteins are not responsible for this difference in function. The finding that the ADD domains from Dnmt3a and Dnmt3L bind to H3 tails with the same specificity suggests that the recruitment of Dnmt3a to chromatin with unmethylated H3 tail may not be the primary function of Dnmt3L.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The DFG (JE 252/6 and JE 252/7); NIH grant DK08267; and by the Max Planck Society (to W.F. and R.R.). S. S. was recipient of a Boehringer Ingelheim predoctoral fellowship. Funding for open access charge: DFG.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 2.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 3.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 4.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Hermann A, Gowher H, Jeltsch A. Biochemistry and biology of mammalian DNA methyltransferases. Cell. Mol. Life Sci. 2004;61:2571–2587. doi: 10.1007/s00018-004-4201-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gowher H, Jeltsch A. Molecular enzymology of the catalytic domains of the Dnmt3a and Dnmt3b DNA methyltransferases. J. Biol. Chem. 2002;277:20409–20414. doi: 10.1074/jbc.M202148200. [DOI] [PubMed] [Google Scholar]
- 7.Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 8.Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- 9.Chedin F, Lieber MR, Hsieh CL. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc. Natl Acad. Sci. USA. 2002;99:16916–16921. doi: 10.1073/pnas.262443999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gowher H, Liebert K, Hermann A, Xu G, Jeltsch A. Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J. Biol. Chem. 2005;280:13341–13348. doi: 10.1074/jbc.M413412200. [DOI] [PubMed] [Google Scholar]
- 11.Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otani J, Nankumo T, Arita K, Inamoto S, Ariyoshi M, Shirakawa M. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep. 2009;10:1235–1241. doi: 10.1038/embor.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Q, Rank G, Tan YT, Li H, Moritz RL, Simpson RJ, Cerruti L, Curtis DJ, Patel DJ, Allis CD, et al. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat. Struct. Mol. Biol. 2009;16:304–311. doi: 10.1038/nsmb.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 16.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Rohde C, Tierling S, Jurkowski TP, Bock C, Santacruz D, Ragozin S, Reinhardt R, Groth M, Walter J, et al. DNA methylation analysis of chromosome 21 gene promoters at single base pair and single allele resolution. PLoS Genet. 2009;5:e1000438. doi: 10.1371/journal.pgen.1000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hodges E, Smith AD, Kendall J, Xuan Z, Ravi K, Rooks M, Zhang MQ, Ye K, Bhattacharjee A, Brizuela L, et al. High definition profiling of mammalian DNA methylation by array capture and single molecule bisulfite sequencing. Genome Res. 2009;19:1593–1605. doi: 10.1101/gr.095190.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- 20.Tamaru H, Zhang X, McMillen D, Singh PB, Nakayama J, Grewal SI, Allis CD, Cheng X, Selker EU. Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nature Genet. 2003;34:75–79. doi: 10.1038/ng1143. [DOI] [PubMed] [Google Scholar]
- 21.Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- 22.Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 23.Luger K, Rechsteiner TJ, Flaus AJ, Waye MM, Richmond TJ. Characterization of nucleosome core particles containing histone proteins made in bacteria. J. Mol. Biol. 1997;272:301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- 24.Shogren-Knaak MA, Peterson CL. Creating designer histones by native chemical ligation. Methods Enzymol. 2004;375:62–76. doi: 10.1016/s0076-6879(03)75004-6. [DOI] [PubMed] [Google Scholar]
- 25.Huynh VA, Robinson PJ, Rhodes D. A method for the in vitro reconstitution of a defined “30 nm” chromatin fibre containing stoichiometric amounts of the linker histone. J. zzMol. Biol. 2005;345:957–968. doi: 10.1016/j.jmb.2004.10.075. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Rohde C, Tierling S, Stamerjohanns H, Reinhardt R, Walter J, Jeltsch A. DNA methylation analysis by bisulfite conversion, cloning, and sequencing of individual clones. Methods Mol. Biol. 2009;507:177–187. doi: 10.1007/978-1-59745-522-0_14. [DOI] [PubMed] [Google Scholar]
- 27.Winkler DF, Hilpert K, Brandt O, Hancock RE. Synthesis of peptide arrays using SPOT-technology and the CelluSpots method. Methods Mol. Biol. 2009;570:157–174. doi: 10.1007/978-1-60327-394-7_5. [DOI] [PubMed] [Google Scholar]
- 28.Chen T, Ueda Y, Xie S, Li E. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J. Biol. Chem. 2002;277:38746–38754. doi: 10.1074/jbc.M205312200. [DOI] [PubMed] [Google Scholar]
- 29.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 30.Leuba SH, Bustamante C. Analysis of chromatin by scanning force microscopy. Methods Mol. Biol. 1999;119:143–160. doi: 10.1385/1-59259-681-9:143. [DOI] [PubMed] [Google Scholar]
- 31.Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J. Biol. Chem. 2004;279:27816–27823. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- 32.Gowher H, Stockdale CJ, Goyal R, Ferreira H, Owen-Hughes T, Jeltsch A. De novo methylation of nucleosomal DNA by the mammalian Dnmt1 and Dnmt3A DNA methyltransferases. Biochemistry. 2005;44:9899–9904. doi: 10.1021/bi047634t. [DOI] [PubMed] [Google Scholar]
- 33.Handa V, Jeltsch A. Profound flanking sequence preference of Dnmt3a and Dnmt3b mammalian DNA methyltransferases shape the human epigenome. J. Mol. Biol. 2005;348:1103–1112. doi: 10.1016/j.jmb.2005.02.044. [DOI] [PubMed] [Google Scholar]
- 34.Oka M, Rodic N, Graddy J, Chang LJ, Terada N. CpG sites preferentially methylated by Dnmt3a in vivo. J. Biol. Chem. 2006;281:9901–9908. doi: 10.1074/jbc.M511100200. [DOI] [PubMed] [Google Scholar]
- 35.Jurkowska RZ, Anspach N, Urbanke C, Jia D, Reinhardt R, Nellen W, Cheng X, Jeltsch A. Formation of nucleoprotein filaments by mammalian DNA methyltransferase Dnmt3a in complex with regulator Dnmt3L. Nucleic Acids Res. 2008;36:6656–6663. doi: 10.1093/nar/gkn747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takeshima H, Suetake I, Tajima S. Mouse Dnmt3a preferentially methylates linker DNA and is inhibited by histone H1. J. Mol. Biol. 2008;383:810–821. doi: 10.1016/j.jmb.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 37.Ge YZ, Pu MT, Gowher H, Wu HP, Ding JP, Jeltsch A, Xu GL. Chromatin targeting of de novo DNA methyltransferases by the PWWP domain. J. Biol. Chem. 2004;279:25447–25454. doi: 10.1074/jbc.M312296200. [DOI] [PubMed] [Google Scholar]
- 38.Chen T, Tsujimoto N, Li E. The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol. Cell. Biol. 2004;24:9048–9058. doi: 10.1128/MCB.24.20.9048-9058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeong S, Liang G, Sharma S, Lin JC, Choi SH, Han H, Yoo CB, Egger G, Yang AS, Jones PA. Selective anchoring of DNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA. Mol. Cell. Biol. 2009;29:5366–5376. doi: 10.1128/MCB.00484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31:2305–2312. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 42.Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 43.Xu GL, Bestor TH, Bourc'his D, Hsieh CL, Tommerup N, Bugge M, Hulten M, Qu X, Russo JJ, Viegas-Pequignot E. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.