Abstract
A strong, negative cis-element located at the first intron +502/+835 (I300) of zebrafish myf5 has been reported. To elucidate the molecular mechanism underlying this repression network, we microinjected zebrafish single-cell embryos with I300 RNA, resulting in the dramatic reduction of luciferase activity driven by the myf5 promoter. Within this I300 segment, we identified an intronic microRNA (miR-In300) located at +609/+632 and found that it was more highly expressed in the older mature somites than those newly formed, which negatively correlated with the distribution of zebrafish myf5 transcripts. We proved that miR-In300 suppressed the transcription of myf5 through abolishing myf5 promoter activity, and we subsequently identified the long isoform of the Dickkopf-3 gene (dkk3) as the target gene of miR-In300. We further found that injection of the dkk3-morpholinos (MOs) resulted in downregulation of myf5 transcripts in somites, whereas co-injection of myf5 mRNA with dkk3-MO1 enabled rescue of the defects induced by dkk3-MO1 alone. Finally, injection of miR-In300-MO enhanced both myf5 transcripts in somites and the level of Dkk3 protein in zebrafish embryos. Based on these findings, we concluded that miR-In300 binds to its target gene dkk3, which inhibits the translation of dkk3 mRNA and, in turn, suppresses zebrafish myf5 promoter activity.
INTRODUCTION
In vertebrates, the determination and differentiation of trunk skeletal muscle is controlled by the basic helix–loop–helix family of transcription factors, such as Myf5, Myod, Myogenin and MRF4 (1). Myf5 is the first myogenic regulatory factor which is expressed in mammals (2), birds (3,4) and fish (5) during early embryogenesis. In zebrafish, myf5 is primarily detectable in the somites and segmental plates (5,6). The transcription level of myf5 elevates substantially until 16 h post-fertilization (hpf), gradually declines to undetectable levels by 33 hpf, and is strictly repressed after somitogenesis (5). The cis-element –82/–62 segment enables direct somite-specific expression of zebrafish myf5 (7), which is directly bound by FoxD3 (8). Thus, the cell lineage-specific expression of myf5 is delicately regulated. More importantly, Lin et al. (9) had previously reported that a strong, negative cis-regulatory motif located at the first intron of zebrafish myf5, +502/+835 (I300), specifically represses the promoter activity of myf5. Interestingly, they found that the sense strand of I300 is capable of repressing GFP expression driven by the upstream region of zebrafish myf5, but that the I300 with antisense strand substantially loses its repressive ability. Furthermore, when the repressive segment is placed in the upstream 3 kb of myf5, repression is nearly abolished. Thus, the I300 repressive function appears to behave in an orientation-, position- and myf5-dependent manner. However, the molecular mechanism underlying this I300-mediated repression of zebrafish myf5 transcription is totally unknown. Based on these lines of evidence, we hypothesized that I300 could function as a self-regulatory, microRNA-triggered modulator whereby negative feedback regulation is initiated by RNA-mediated silencing.
MicroRNAs are single-stranded RNA molecules, about 19–30 nt in length, which repress gene expression through sequence-specific base-pairing with target mRNA. Mature microRNA molecules are partially complementary to one or more mRNA molecules, and their main function is to downregulate gene expression. MicroRNAs were first described by Lee and colleagues (10) in Caenorhabditis elegans in lin-4. Subsequently, hundreds of microRNAs have been identified in other organisms, such as Drosophila, zebrafish, Xenopus, mammals and plants, by using molecular cloning and bioinformatics (11–16). MicroRNAs are generated either from specific RNA genes or from intronic splicing. While microRNAs derived from intronic splicing require the presence of RNA polymerase II and spliceosomal components (17), this is not the case for microRNAs derived from small RNA genes. An estimated one-fourth of all microRNAs are intronic, and, although several of these have been identified in worm, mouse and in human cells (18), the target gene and function of intronic microRNAs remain largely unknown. One exception is miR-208 whose target gene, THARP-1, regulates cardiac growth (18).
In this study, we demonstrated that a novel intronic microRNA, miR-In300, which is derived from I300 of the first intron of zebrafish myf5, enables significant repression of myf5 promoter activity through silencing the long isoform of the Dickkopfs-3 gene (dkk3), indicating that miR-In300 is an effective negative regulator of myf5 expression.
MATERIALS AND METHODS
Knockdown experiments
The morpholinos (MOs) designed specifically for knocking down myf5 and dkk3 were TACCTGCATAAGAGGTGTAGGGTCT (myf5-MO) and GAGGCTGAATCCGAGCAGAAACATG (dkk3-MO1), respectively. A negative control MO designed for dkk3 was GACGCTCAATCCGACCACAAAGATG (dkk3-control-MO), in which the mutated-mismatched nucleotides are underlined. In order to study the efficacy and specificity of dkk3-MO in blocking the translation of the dkk3 mRNA, we designed dkk3-MO2, (ATGATGCAAGACTCTCGTACCTTTA), in which the matching nucleotide sequences were located at the boundary between exon 2 and intron 2. In addition, we also constructed a plasmid containing a dkk3-GFP, in which the 27th nucleotide after ATG of dkk3 mRNA was fused with GFP cDNA. The MO designed for knocking down miR-In300 was AAAATCTGCATTCAAAATGCTTTTATCTACC (miR-In300-MO), in which miR-In300-MO bound to the miR-In300 motif within the pre-miR-In300 sequence to avoid forming a mature miR-In300 by the lack of hairpin structure. All MOs were prepared at a stock concentration of 1 mM and diluted to the desired concentration for microinjection into each embryo.
Northern blot analysis of small RNAs by polyacrylamide gel electrophoresis
Total RNAs at an amount of 70 µg were separated on an 6% and 15% polyacrylamide/8M urea gel (Amersham Pharmacia) to analyze I300 RNA and miR-In300, respectively, by a Protein III Apparatus (BioRad). Gel was preheated to 55°C by electric current and water bath circulation before loading sample. The separated RNA on gel was electro-blotted onto a Hybond-N+ membrane (Amersham) by Trans-Blot SD semi-dry electrophoretic transfer cell (BioRad). After ultraviolet (UV) cross-linking and air drying, the blotted membrane was prehybridized with hybridization buffer (0.02% SDS, 5× SSC, 50% Fromamide deionized, 0.1% N-lauroylsarcosine, 2% Blocking solution and 20 μg/ml salmon sperm DNA) at 37–42°C for 60 min, hybridized with radiolabeled I300 or miR-In300 and incubated at 37–42°C for 3–4 h or overnight. The I300 was 32P-labeled, and miR-In300 was DIG-labeled and LNA-incorporated (Exiqon). The membrane was washed two to four times at 40°C with 2× SSC and 0.5% SDS for 15 min and exposed to an X-ray film (Kodak, NY) at −80°C.
Searching for the target gene of miR-In300
To determine the target gene of miR-In300, we used our novel labeled microRNA pull-down (LAMP) assay system in which the pre-microRNA was labeled with digoxigenin (DIG) and then mixed with cell extracts as described previously (19). Briefly, DIG-labeled pre-miRNA was synthesized by using the DIG RNA labeling kit (Roche). After cell extracts were incubated with 70 μg of digoxigeninylated RNA at 4°C for 30 min, the total volume was adjusted to 1 ml with binding buffer (25 mM Tris-base pH 7.4, 60 mM KCl, 2.5 mM EDTA, 0.2% Triton X-100, 80 U of rRnasin), and the mixture was incubated at 30°C for 60 min. The purified RNA was collected when DNase I (20 U) was added and incubated at 37°C for 30 min before the phenol/chloroform extraction was performed. The putative target genes were precipitated by anti-DIG antiserum, cloned out by reverse transcriptase-polymerase chain reaction (RT-PCR) or collected by a pull-down assay. Then, we analyzed these cDNAs or putative clones for miR-In300 targeting.
(Other experimental procedures are available in Supplementary Data).
RESULTS
I300 segment of zebrafish myf5 specifically suppresses myf5 promoter activity
A plasmid containing –6300/–1 of zebrafish myf5 fused with a luciferase gene and a segment of I300 (+502/+835 of myf5) DNA were either transfected into the cell lines or injected into the zebrafish embryos. Results showed that the expression of luciferase reporter gene was dramatically reduced. Luciferase activity was decreased by 50% and 80% in the cell lines and zebrafish embryos, respectively (Supplementary Figure S1), which enabled the sense strand of I300 DNA to repress the myf5 promoter activity.
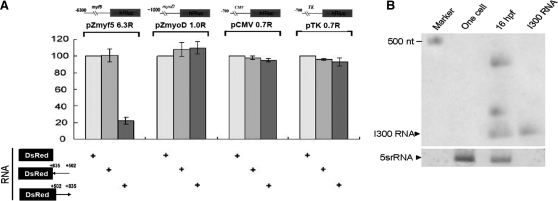
To accomplish this, RNA encoding I300 fused with red fluorescence protein (RFP, DsRed) was synthesized in vitro (DsRed +502/+835) and co-injected with four plasmids consisting of –6300/–1 of myf5 (pZmyf5 6.3), −1014/–1 of myod (pZmyoD 1.0), cytomegalovirus promoter (pCMV), and thymidine kinase promoter (pTK). Results showed that I300 RNA did, in fact, enable the downregulation of luciferase activity driven by the myf5 promoter in the RFP-expressing embryos by 20% from that of control (Figure 1A). These results are consistent with those obtained from co-injection with sense I300 DNA (+502/+835). However, the decrease of luciferase activity was neither from promoters of myod, CMV or TK, nor from injection of antisense I300 RNA (DsRed +835/+502), indicating that the I300 RNA-mediated repression is myf5 promoter- and sense strand-specific. As expected, since the I300 RNA is spliced from intron 1 of myf5, Northern blot analysis showed that I300 RNA was predominantly detected in the somites of embryos at the 16 hpf stage when myf5 was highly expressed, whereas I300 RNA was not detected at the one-celled stage when myf5 was not expressed (Figure 1B). As this evidence demonstrated that I300 RNA is endogenous in zebrafish embryos, we were motivated to investigate whether RNA-mediated regulation is involved in myf5 promoter activity.
Figure 1.
Intron 1 RNA is a repressive element for myf5-specific expression. (A) Plasmids used for the transient luciferase assay were microinjected into the one-cell stage of fertilized embryos. The luciferase gene was under-controlled in different promoters, including the myf5 promoter (pZmyf5 6.3R), myod promoter (pZmyoD 1.0R), CMV promoter (pCMV 0.7R) and TK promoter (pTK 0.7R). RNAs used for microinjection into zebrafish embryos were mRNA encoding red fluorescence protein (DsRed), sense strand of the first intron +502/+835 (I300) of zebrafish myf5 followed by DsRed mRNA (DsRed +502-> +835), and antisense strand of I300 followed by DsRed mRNA (DsRed +835-> +502). Luciferase activity was reduced only in the embryos co-injected with DsRed fused with sense I300 and the plasmid containing myf5 promoter. (B) Detection of the existence of primary transcript of I300 RNA by northern blot analysis. Using I300 RNA as the positive control, a positive signal (333 nt) was shown in the 16-hpf embryos, but not in the one-cell stage (0 hpf) embryos. The 5S rRNA (5srRNA) was served as a loading control.
The first intron of zebrafish myf5 contains miR-In300
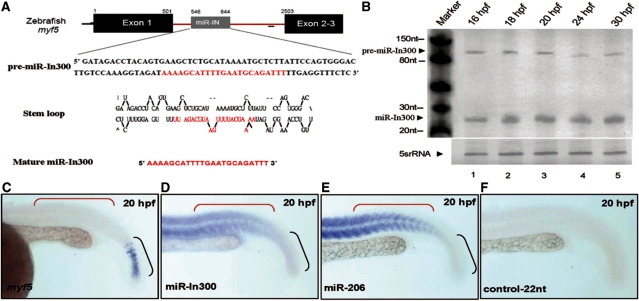
I300 RNA is derived from the first intron of the primary mRNA of zebrafish myf5. Using computer analysis, we predicted that the pre-miR-In300 sequence may locate at +546/+644 and that the mature miR-In300 may locate at +609/+632 (Figure 2A). In order to establish the soundness of these predictions, we first used northern blot to confirm the ability of pre-miR-In300 to process normally to a mature miR-In300 in the presence of endogenous Dicer of cell extracts (lanes 1 and 3, Supplementary Figure S2A). However, when Dicer was completely depleted from the cell extracts (Supplementary Figure S2B), the pre-miR-In300 was not processed to mature miR-In300 (lanes 2 and 4, Supplementary Figure S2A). Therefore, we next used anti-biotin beads to perform immunoprecipitation, and the complex of biotin-labeled miR-In300 and miRNP was obtained from cell extracts. When we used anti-AGO antiserum to perform western blot analysis, we found that AGO was detected in that complex (Supplementary Figure S2C). This result was consistent with the control group when miR-1 was used in parallel (Supplementary Figure S2C). Since miR-In300 was picked up by AGO, we are confident that it had indeed been processed correctly from pre-miR-In300 by Dicer. Furthermore, northern blot analysis also demonstrated the existence of a 22-nt miR-In300 in the somites of embryos during 16–30 hpf when the myf5 transcripts were expressed (Figure 2B). However, among these stages, the miR-In300 signal appeared at its highest levels after 20 hpf (lane 3 of Figure 2B).
Figure 2.
Expression patterns of zebrafish miR-In300. (A) miR-In300 is generated from intron 1 (+502/+2502) of the zebrafish myf5 gene. Pre-miR-In300 (+546/+644) and mature microRNA sequences (indicated in red; +609/+632) are presented. The predicted secondary structure of pre-miR-In300 is also illustrated. (B) Detection of the existence of miR-In300 transcript by northern blot analysis. The total RNAs extracted from the various stages of zebrafish embryos. The RNA level of miR-In300 was gradually increased in the embryos from 16 hpf to 20 hpf. The 5S rRNA (5srRNA) was served as a loading control. (C–F) Expression patterns of myf5, miR-In300 and miR-206 in somites at 20 hpf were detected by whole-mount in situ hybridization. myf5 was expressed only in the newly formed somites and in the presomitic mesoderm (PSM) at 20 hpf (C), whereas miR-In300 was predominant in the older formed somites, but only mildly present in the newly formed somites at 20 hpf (D). A muscle-specific microRNA in zebrafish, miR-206, was used as a positive control and was detected in mature muscle, but not in PSM at 20 hpf (E). In contrast, the antisense strand of myf5 intron 1 (A, red line) served as negative control and did not present any signal (F).
Based on whole-mount in situ hybridization, we found that miR-In300 was highly expressed in the older mature somites, but much less so in the newly formed somites (Figure 2D). This asymmetrical distribution stood in exact contrast to the distribution of the expression of myf5 mRNA (Figure 2C and D), which is substantially greater at the early somite stages. This did not result from hybridization to the genomic DNA because no signal appeared when the control probe, which was complementary to the first intron at +1341/+1361 of myf5, was used (Figure 2F). Moreover, the positive control, miR-206, was shown in mature somites (Figure 2E).
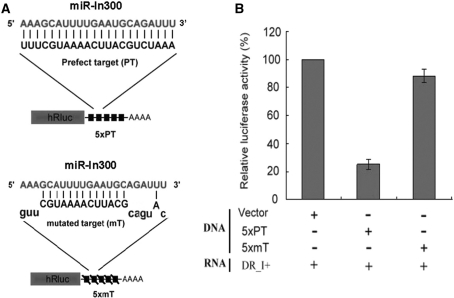
In order to further confirm that the zebrafish miR-In300 motif functions as an RNAi, we next designed a plasmid construct, TK-Luciferase-5 X PT, in which a perfectly matched complementary sequence of miR-In300 in five copies (5 X PT) was inserted in the 3′ untranslated region (UTR) of luciferase cDNA and driven by the TK promoter (Figure 3A). Zebrafish embryos were co-injected with I300 RNA and TK-Luciferase-5 X PT. Results showed that the luciferase activity of these embryos was dramatically reduced, down to 25% of that observed in the embryos co-injected with I300 RNA and a control construct, TK-Luciferase, which does not contain a 5 X PT-inserted motif at the 3′ UTR of luciferase cDNA (Figure 3B). This result was consistent with embryos co-injected with I300 RNA and another control construct, TK-Luciferase-5 X mT, in which a mismatched complementary sequence of miR-In300 in five copies (5 X mT) was inserted at the 3′ UTR (Figure 3A). This evidence clearly demonstrated that miR-In300 does function as a microRNA by its repression of reporter gene expression through sequence-specific base-pairing with target mRNA at the 3′ UTR.
Figure 3.
Injection of the sense strand of I300 RNA containing miR-In300 repressed the target mRNA expression in embryos. (A) Two plasmids containing a luciferase reporter gene (hRluc) fused with five copies (5 X) of either a perfectly matched target sequence (PT) or a mutated-mismatched target sequence (mT) for miR-In300 were constructed. (B) Relative luciferase activities of zebrafish embryos microinjected with the materials as indicated. Co-injection of the sense strand of I300 RNA (DR_I+) with a vector DNA containing hRluc gene served as a standard for comparison to the luciferase activities driven by co-injection of DR_I+ with a plasmid DNA containing either 5 X PT or 5 X mT. In vivo transgenesis enabled the sense strand of I300 RNA to repress the gene expression of luciferase in the 5 X PT construct, but not in the 5 X mT construct.
Either overexpression or knockdown of miR-In300 causes misexpression of myf5 in somites
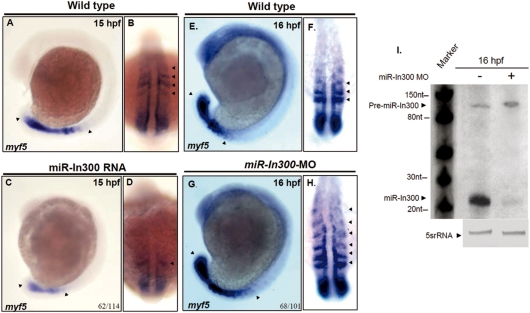
To further confirm whether miR-In300-mediated repression affects zebrafish myf5 promoter activity in vivo, we microinjected 300 pg of miR-In300 dsRNA into single-cell fertilized eggs. Results showed that myf5 expression was decreased, both in the newly formed somites of S-II, S-I, S0 and S1, as well as the presomitic mesoderm (PSM) at 15 hpf (Figures 4A and B versus 4C and D). On the other hand, we employed an antisense MOs which specifically blocks miR-In300 (miR-In300-MO) to examine myf5 promoter activity at 16 hpf. First, we used northern blot analysis to prove that miR-In300-MO caused a decrease in the amount of miR-In300 (Figure 4I). Second, when miR-In300-MO was injected, we found that the expression of myf5 was enhanced not only in the somites at S-II, S-I, S0 and S1, but also extended to S3 in the embryos (Figures 4E and F versus 4G and H). This evidence suggests that miR-In300 may suppress the transcription of myf5 mRNA through abolishing the myf5 promoter activity in the older formed somites at 16 hpf.
Figure 4.
Change of miR-In300 level resulted in abnormal expression patterns of myf5. Expression patterns of myf5 (A and B) in somites at the 12–14 somite stage, as indicated. However, when miR-In300 dsRNA was injected, the expression of myf5 was greatly reduced in the newly formed somites and in the presomitic mesoderm (PSM), which became smaller (C and D). In wild-type embryos, myf5 was expressed in S-II to S1 somites (E and F), while in the miR-In300-MO-injected embryos, myf5 was expressed in S-II to S3 somites (G and H). Detection of the existence of miR-In300 transcripts by northern blot analysis. Results showed that miR-In300 was decreased in the miR-In300-MO-injected embryos at 16 hpf (I). The 5S rRNA (5srRNA) was served as a loading control.
miR-In300 targets the long isoform of dkk3 mRNA
To determine the target gene of miR-In300, we used our novel LAMP assay system (19). Accordingly, we obtained around 3000 putative clones for miR-In300 targeting. Among them, 667 clones were screened, and 20 putative clones potentially containing sequences complementary to miR-In300 were selected. Nine out of 20 contained the EST sequences, while the remaining 11 clones contained an undefined DNA sequence. Seven of the nine clones contained the same EST sequences. The full length of cDNA corresponding with this EST was cloned and identified as a zebrafish long-isoform dkk3 gene (NM_001159283.1), also named dkk3-related gene, encoding a 293-amino-acid polypeptide with Cys-rich domain located at amino acid positions 134–182 and 170–275 (Figure S3A). The EST sequence was complementarily matched with three miR-In300-binding sites at 3′ UTR by miRanda v.1.0b software analysis (Figure S3B). Using RT-PCR, we revealed that the dkk3 transcript was maternally inherited and expressed in the somites from 16 to 30 hpf (Supplementary Figure S4). The remaining two clones contained another EST sequence, which was part of a sequence located at the 3′ UTR of another undefined target gene. When we used it as a probe to detect the expression pattern, results showed that it was also co-localized with miR-In300 in trunk somites (data not shown). Although similar, this clone was not the focus of this study.
Therefore, based on computer analysis, the putative miR-In300 binding site in the 3′ UTR of the dkk3 mRNA was highly complementary with the miR-In300. However, in order to confirm that the miR-In300 target sequence was located at the 3′ UTR of dkk3 and enabled mediation of translational repression, we monitored the luciferase activity in embryos injected with a construct in which the 3′ UTR of dkk3 was inserted downstream of the luciferase cDNA driven by TK promoter (TK-Rluc-dkk3 3′ UTR) (Figure 5A). Results showed that luciferase activity was greatly decreased in the embryos injected with I300 RNA and TK-Rluc-dkk3 3′ UTR, compared to that of embryos injected with I300 RNA with a plasmid in which the luciferase cDNA did not contain 3′ UTR of dkk3 (TK-Rluc) (Figure 5C). If embryos received injection with I300 RNA using a plasmid in which the luciferase cDNA contained mutated sequences of dkk3 3′ UTR (TK-Rluc-mdkk3 3′ UTR) (Figure 5B), the luciferase activity was not responsive to miR-In300 (Figure 5C). Furthermore, if miR-In300-MO was used to reduce the endogenous miR-In300 and co-injected with TK-Rluc-dkk3 3′ UTR to embryos, the luciferase activity was initially increased, but then reduced when embryos did not receive miR-In300-MO (Figure 5C). This evidence strongly supports the implication that the recognition sequence of zebrafish miR-In300 is specifically bound at the 3′ UTR of dkk3 mRNA in a canonical binding assay.
Figure 5.
miR-In300 represses the translation of dkk3 mRNA. Schematic illustration of plasmid constructs used for microinjection. (A) Plasmid Tk-Rluc-dkk3 3′ UTR, in which the dkk3 3′ UTR was fused with the 3′ UTR of the luciferase gene and driven by TK promoter. Three putative miR-In300 binding sites, which are located at dkk3 3′ UTR, are indicated by empty boxes, and their possible targeted sequences are also presented. (B) Plasmid TK-Rluc-mdkk3 3′ UTR, which was the same as plasmid Tk-Rluc-dkk3 3′ UTR, except that the mutated target sequences (M1, M2 and M3) of dkk3 3′ UTR were included, are indicated by crossed boxes and the lowercase alphabet. (C) Luciferase activities in the microinjected embryos. TK-Rluc was constructed without ligating any sequence into the 3′ UTR region of the luciferase gene, which served as basal control. DsRed-I300 (+) was a sense strand RNA of I300. Compared to the embryos injected with TK-Rluc and I300 RNA, the luciferase activity of embryos injected with Tk-Rluc-dkk3 3′ UTR and I300 RNA decreased greatly, which was only 1/4th of the luciferase activity induced by injection of TK-Rluc and I300 RNA. However, the luciferase activities of embryos injected with I300 RNA and TK-Rluc-mdkk3 3′ UTR, which contained mutated sequences, remained unchanged, compared to the luciferase activity driven by I300 RNA and TK-Rluc. Co-injection of miR-In300-MO with Tk-Rluc-dkk3 3′ UTR showed that the luciferase activity was increased 3-fold over that of embryos injected with Tk-Rluc-dkk3 3′ UTR alone, without co-injection of miR-In300-MO.
The specific defects induced by the injected MOs
Two different MOs were specifically designed to inhibit the translation of dkk3 mRNA. One was complementary to 25 bp after AUG (dkk3-MO1), and another, dkk3-MO1, contained five mutated nucleotides (dkk3-control-MO). Injection of 5 ng dkk3-MO1 resulted in defective embryos characterized by reduced head size and wide, U-shaped somites with irregular boundaries and the expression of myf5 was downregulated in the somites when the myf5 probe was used to perform whole-mount in situ hybridization and the embryos were observed at 13 hpf and 16 hpf (Supplementary Figure S5). However, injection of dkk3-control-MO did not result in any defective embryos, even when the dkk3-control-MO was injected as high as 8 ng per embryo (Supplementary Figure S5).To confirm that the phenotypes of morphants were specific as a result Dkk3 loss of function, we constructed a dkk3-GFP and microinjected 0.1 ng of dkk3-GFP RNA in the presence of 1.5 ng dkk3-MO1, which resulted in the absence of GFP expression (Supplementary Figure S6). In contrast, there was no effect on the expression of GFP in embryos co-injected with 0.1 ng of dkk3-GFP RNA and 1.5 ng dkk3-control-MO (Supplementary Figure S6). Furthermore, we co-injected synthetic dkk3 mRNA and dkk3-MO1, which enabled synthetic dkk3 mRNA to rescue the morphological defects induced by 5 ng dkk3-MO1 (Figure 6E versus 6F). In contrast, by injection of dkk3 mRNA alone, no defective phenotype was observed. To further study whether the phenotypes induced by dkk3-MO1 were specific, we designed dkk3-MO2, whose corresponding sequences were located at the boundary between exon 2 and intron 2. Results showed that the defective phenotypes caused by injection of 4 ng dkk3-MO2 were identical to those caused by dkk3-MO1 (Supplementary Figure S5). Therefore, we concluded that the phenotypes of dkk3-MO1-injected embryos were specific.
Figure 6.
The myf5 promoter activity is controlled by the dkk3 gene. (A) Plasmid and MO, as indicated by +, were co-microinjected into the one-celled stage of fertilized embryos to carry out the transient luciferase assay. The luciferase activity driven by the upstream 6.3 kb of zebrafish myf5 promoter (pZmyf5 6.3R) and the co-injected dkk3-control-MO was measured in average (n = 6) and served as 100%. Compared to the embryos injected with pZmyf5 6.3R and dkk3-control-MO, the luciferase activity was greatly reduced in the embryos injected with pZmyf5 6.3R and dkk3-MO1. Meanwhile, the average of luciferase activity driven by pZmyf5 6.3R was also measured and served as 100%. Compared to the embryos injected with pZmyf5 6.3R, the luciferase activity was dramatically increased for the embryos in which the endogenous miR-In300 production had been inhibited by injection of miR-In300-MO. Zebrafish embryos derived from the transgenic line Tg (myf5 (80K): GFP), whose somites display GFP reporter, were used. (B) When dkk3-control-MO was injected, the GFP expression in somites remained unchanged at 16 hpf, whereas (C) the GFP was greatly reduced in somites when dkk3-MO1 was injected. (D–H) Whole-mount in situ hybridization of myf5 transcripts in zebrafish embryos at 16 hpf. Compared to the control (D), the myf5 signal in the somites of either dkk3-MO1-injected embryos (E) or miR-In300-dsRNA-injected embryos (G) was decreased. Co-injection of either dkk3-MO1 with dkk3 mRNA (F) or excess miR-In300-dsRNA with dkk3 mRNA (H) enabled embryos to rescue the defects induced by injecting either dkk3-MO1 alone (E) or miR-In300-dsRNA alone (G). The numbers shown in the lower-right corner of panels B, C, E, F, G and H indicate the number of phenotypes out of the number of embryos examined.
The promoter activity of myf5 is modulated by dkk3 expression
To study whether dkk3 affects zebrafish myf5 expression, we first co-injected pZmyf5 6.3R with dkk3-MO1, which resulted in the repression of luciferase activity (Figure 6A). However, co-injection of pZmyf5 6.3R with dkk3-control-MO had no effect on the luciferase activity in embryos (Figure 6A), indicating that the absence of dkk3 reduces the expression of zebrafish myf5 in vivo. Next, we injected either dkk3-MO1 or dkk3-control-MO into embryos derived from the zebrafish transgenic line Tg (myf5 (80K): GFP) in which the upstream 80 kb segment of myf5 fused with the GFP reporter (20). Results showed that the expression of GFP was repressed dramatically in transgenic embryos injected with dkk3-MO1 (Figure 6C), but not in embryos injected with dkk3-control-MO (Figure 6B). Therefore, our data suggest that dkk3-MO1 specifically inhibits the expression of myf5 promoter activity in zebrafish embryos. Furthermore, the myf5 signal in the somites of miR-In300-dsRNA-injected embryos was decreased (Figure 6G). Co-injection of excess miR-In300-dsRNA with dkk3 mRNA enabled embryos to rescue the defects induced by injecting miR-In300-dsRNA alone (Figure 6H). Interestingly, when pZmyf5 6.3R and miR-In300-MO were co-injected into embryos, the luciferase activity was increased, suggesting that the myf5 promoter activity was enhanced by the inhibition of endogenous miR-In300 (Figure 6A).
To further validate in vivo the correlation between dkk3 and miR-In300, the expressions of Dkk3 protein and mRNA, as well as miR-In300, during embryonic development of zebrafish, were investigated. Cell extracts of embryos from different developmental stages were prepared for western blot analysis by using anti-Dkk3 antibody. Results showed that the protein level of Dkk3 was greatly reduced in the dkk3-MO1-injected embryos (lane 7, Figure 7A), suggesting that the anti-Dkk3 antibody could recognize the Dkk3 protein. Furthermore, we found that Dkk3 protein was highly produced at the 16 hpf stage (lane 1, Figure 7A) when, at the same time, myf5 was highly expressed, but miR-In300 was weakly expressed. We found that the level of Dkk3 protein was greatly decreased at 18 hpf (lane 2, Figure 7A) and undetectable at 20 hpf, as well as later stages examined, such as 24 hpf and 30 hpf (lane 4 and lane 5,Figure 7A). On the other hand, miR-In300 was highly expressed after 20 hpf (Figure 2B). Interestingly, when the protein lysates were extracted from wild-type embryos at 16 hpf and from the miR-In300-MO-injected embryos at 20 hpf, the protein level of Dkk3 in the miR-In300-MO-injected embryos at 20 hpf was as high as the protein level of Dkk3 in the wild-type embryos at 16 hpf (lane 8, Figure 7A). At 20 hpf, however, the protein level of Dkk3 became undetectable in the wild type (lane 3, Figure 7A). Taken together, these data therefore suggested that 1) the miR-In300 level is negatively correlated with the protein level of Dkk3 protein and 2) the absence of endogenous miR-In300 causes an increase of Dkk3 protein in zebrafish embryos. From these lines of evidence, we can, in turn, conclude that miR-In300 specifically binds to its target gene dkk3, which leads to three interacting consequences: (i) inhibiting the translation of dkk3 mRNA, thus (ii) decreasing the Dkk3 protein level, an event ultimately (iii) suppressing the promoter activity of zebrafish myf5.
Figure 7.
Western blot analysis of Dkk3 protein during embryogenesis. (A) Total protein lysates were extracted from zebrafish embryos at 16, 18, 20, 24 or 30 hpf, as indicated. The molecular weight of protein markers and the positions of positive reactive bands for antiserum against Dkk3 and alpha-tubulin (A-tubulin) are also indicated. The protein level of Dkk3 was greatly reduced in the protein lysates extracted from embryos at 18 hpf. The Dkk3 protein became undetectable at 20, 24 and 30 hpf. The Dkk3 protein was reduced in the lysates extracted from the dkk3-MO1-injected embryos (lane 7), whereas the Dkk3 protein was greatly enhanced in the lysates extracted from the miR-In300-MO-injected embryos at 20 hpf (lane 8). The intensity of positive bands for antiserum against A-tubulin served as a loading control. (B) Possible model that illustrates the modulation of myf5 expression through miR-In300 mediation during somitogenesis in zebrafish embryos. The miR-In300 is an intronic microRNA within the first intron of zebrafish myf5, and dkk3 is the target gene of miR-In300. The miR-In300 blocks the target sequences located at the 3′ UTR of dkk3 mRNA, which results in repressing the translation of dkk3 mRNA. The absence of Dkk3 protein in zebrafish embryos then causes the downregulation of myf5 promoter activity through an unknown signal transduction pathway. Consequently, myf5 mRNA expression is gradually decreased at the later stages of zebrafish embryogenesis.
DISCUSSION
Around three-fourths of identified microRNAs are intergenic, whereas the remainder belongs to the class known as intronic microRNAs. To date, about 90 intronic microRNAs have been predicted among humans, mice, and C. elegans, but most of them have originated from bioinformatics BLASTing and have not yet been validated (21). Recently, an intronic microRNA known as miR-208 was confirmed as a cardiac-specific microRNA, which is encoded by an intron of the α-myosin heavy chain (αMHC) gene and is reported to be required for cardiomyocyte hypertrophy, fibrosis and expression of βMHC in response to stress and hypothyroidism (22). In this study, we are the first to report a de novo intronic microRNA, miR-In300, to isolate its cognate target gene, dkk3, and to provide evidence that it functions as a negative modulator to reduce myf5 promoter activity in somites of zebrafish by silencing dkk3 during embryonic stage.
As development progresses, we observed that the expression of myf5 is decreased in the somites at 24 hpf and is not detectable at 30 hpf. Interestingly, it is during the same developmental stages that the expression level of miR-In300 is lower in the newly formed somites where myf5 starts its transcription, but gradually appears in somites where myf5 had previously been expressed. As a result, up to 30 hpf, almost all somites have the same expression level of miR-In300, but after 30 hpf, we noticed that miR-In300 remains highly expressed. Therefore, we hypothesize that only pre-miR-In300, but not mature miR-In300, is generated when myf5 is transcribed initially in the newly formed somites. After mature miR-In300 is processed, the miR-In300 is accumulated in the older mature somites by the long half-life of microRNA, a phenomenon which is in agreement with Rooij et al. (18) who reported that the half-life of miR-208 is around 21 days. This hypothesis is also supported by Giraldez et al. (22) who reported that mature miR-430 of zebrafish is not shown until 4 hpf, but pre-miR-430 starts to present at 2.5 hpf.
In this study, we discovered that dkk3 mRNA is the target of miR-In300. The Dkk family is composed of four main members (Dkk1, 2, 3, 4 and Soggy). Many reports have described that all family members, except Dkk3, typically regulate Wnt/β-catenin signaling (23,24). Recently, however, Yue et al. (25) demonstrated that Dkk3 is involved in Wnt signaling. Whether Dkk3 plays the same role in the simitogenesis of zebrafish embryos is open to further investigation; however, this study has presented evidence suggesting that it does modulate zebrafish myf5 promoter activity. This is noteworthy in view of the fact that dkk3 is known as a tumor suppressor gene in higher vertebrates (26) Specifically, evidence clarifying the roles and relationships between miR-In300 and myf5 promoter activity was provided by our two luciferase assays, which first indicated that either I300 DNA or I300 RNA is capable of silencing zebrafish myf5 promoter activity (Supplementary Figure S1 and Figure 1A). We next demonstrated that the recognition sequence of zebrafish miR-In300 is specifically located at the 3′ UTR of dkk3 mRNA and inhibits luciferase activity (Figure 5), suggesting that miR-In300 binds to the dkk3 3′ UTR, thereby validating dkk3 as a target gene of miR-In300. Although the detailed regulatory pathway involved in Dkk3-mediated myf5 expression requires further study, we have conclusively shown that the following sequence of events leads to the gradual disappearance of myf5 mRNA expression in late-stage somites. First, during myf5 mRNA transcription, an I300 segment +502/+835 is generated from the first intron of primary zebrafish myf5 mRNA transcripts. Second, within this I300 segment, an intronic microRNA, miR-In300, is processed to maturity from +609/+632. Third, miR-In300 inhibits myf5 promoter activity by silencing the dkk3 gene and blocking the expression of myf5 mRNA at mature somite regions.
The decrease of Dkk3 protein level, which becomes undetectable after 20 hpf (Figure 7A), seems to be correlated to the down-regulation of myf5 mRNA after 16 hpf, whereas the level of dkk3 mRNA remains unchanged (Supplementary Figure S4F and G). Importantly, this is also the period when, as noted above, miR-In300 gradually appears in somites in which myf5 had previously been expressed. This evidence suggests that the distribution of miR-In300 is negatively correlated with the protein level of Dkk3 during zebrafish embryogenesis. Furthermore, as shown in Figure 7A, inactivation of Dkk3 expression by miR-In300 is further demonstrated by up-regulation of dkk3 in the miR-In300-MO-injected embryos. For example, those 20-hpf embryos injected with miR-In300-MO to abolish the endogenous miR-In300 displayed a high protein level of Dkk3, which is similar to the level of Dkk3 protein observed in the 16 hpf embryos (Figure 7A). Taken together, these lines of evidence support our hypotheses that dkk3 is a target gene of miR-In300, that microRNA is indeed responsible for the post-transcriptional silencing of dkk3 expression, and, finally, that miR-In300 is a strong negative modulator of zebrafish myf5 transcription (Figure 7B).
In summary, we have demonstrated that (i) miR-In300 is derived from the first intron of myf5; (ii) the miR-In300-mediated myf5 promoter activity is gene-specific and regulates dkk3 mRNA translation; and (iii) the transfection of exogenous miR-In300 can significantly reduce myf5 transcription through the silencing of dkk3 gene translation, an event which most likely occurs with optimum efficiency during myf5 transcription.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Council of Republic of China under the grant of NSC 97-2313-B-002-036-MY3, ROC.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Pownall ME, Gustafsson MK, Emerson C.P., Jr Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annu. Rev. Cell. Dev. Biol. 2002;18:747–783. doi: 10.1146/annurev.cellbio.18.012502.105758. [DOI] [PubMed] [Google Scholar]
- 2.Buckingham M. Making muscle in mammals. Trends Genet. 1992;8:144–148. doi: 10.1016/0168-9525(92)90373-C. [DOI] [PubMed] [Google Scholar]
- 3.Hacker A, Guthrie S. A distinct developmental programme for the cranial paraxial mesoderm in the chick embryo. Development. 1998;125:3461–3472. doi: 10.1242/dev.125.17.3461. [DOI] [PubMed] [Google Scholar]
- 4.Hirsinger E. Notch signalling acts in postmitotic avian myogenic cells to control MyoD activation. Development. 2001;128:107–116. doi: 10.1242/dev.128.1.107. [DOI] [PubMed] [Google Scholar]
- 5.Chen YH, Lee WC, Liu CF, Tsai HJ. Molecular structure, dynamic expression, and promoter analysis of zebrafish (Danio rerio) myf-5 gene. Genesis. 2001;29:22–35. doi: 10.1002/1526-968x(200101)29:1<22::aid-gene1002>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 6.Coutelle O, Blagden CS, Hampson R, Halai C, Rigby PW, Hughes SM. Hedgehog signalling is required for maintenance of myf5 and myoD expression and timely terminal differentiation in zebrafish adaxial myogenesis. Dev. Biol. 2001;236:136–150. doi: 10.1006/dbio.2001.0193. [DOI] [PubMed] [Google Scholar]
- 7.Chen YH, Lee HC, Liu CF, Lin CY, Tsai HJ. Novel regulatory sequence -82/-62 functions as a key element to drive the somite-specificity of zebrafish myf5. Dev. Dyn. 2003;228:41–50. doi: 10.1002/dvdy.10357. [DOI] [PubMed] [Google Scholar]
- 8.Lee HC, Huang HY, Lin CY, Chen YH, Tsai HJ. Foxd3 mediates zebrafish myf5 expression during early somitogenesis. Dev. Biol. 2006;290:359–372. doi: 10.1016/j.ydbio.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 9.Lin CY, Chen YH, Lee HC, Tsai HJ. Novel cis-element in intron 1 represses somite expression of zebrafish myf5. Gene. 2004;334:63–72. doi: 10.1016/j.gene.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 10.Lee RC, Feinbaum RL, Ambros V. C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–848. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 11.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;312:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 12.Ambros V, Lee RC. Identification of microRNAs and other tiny noncoding RNAs by cDNA cloning. Methods Mol. Biol. 2004;265:131–158. doi: 10.1385/1-59259-775-0:131. [DOI] [PubMed] [Google Scholar]
- 13.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 14.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 15.Reinhart BJ, Bartel DP. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297:1831. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- 16.Watanabe T, Takeda A, Mise K, Okuno T, Suzuki T, Minami N, Imai H. Stage-specific expression of microRNAs during Xenopus development. FEBS Lett. 2005;579:318–324. doi: 10.1016/j.febslet.2004.11.067. [DOI] [PubMed] [Google Scholar]
- 17.Lin SL, Miller JD, Ying SY. Intronic microRNA (miRNA) J. Biomed. Biotechnol. 2006;2006:26818. doi: 10.1155/JBB/2006/26818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rooij VanE, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 19.Hsu RJ, Yang HJ, Tsai HJ. Labeled microRNA pull-down assay system: an experimental approach for high-throughput identification of microRNA-target mRNAs. Nucleic Acids Res. 2009;37:e77. doi: 10.1093/nar/gkp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen YH, Wang YH, Chang MY, Lin CY, Weng CW, Westerfield M, Tsai HJ. Multiple upstream modules regulate zebrafish myf5 expression. BMC Dev. Biol. 2007;3:7:1. doi: 10.1186/1471-213X-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 23.Krupnik VE, Sharp JD, Jiang C, Robison K, Chickering TW, Amaravadi L, Brown DE, Guyot D, Mays G, Leiby K, et al. Functional and structural diversity of the human Dickkopf gene family. Gene. 1999;238:301–313. doi: 10.1016/s0378-1119(99)00365-0. [DOI] [PubMed] [Google Scholar]
- 24.Mao B, Wu W, Li Y, Hoppe D, Stannek P, Glinka A, Niehrs C. LDL-receptor-related protein 6 is a receptor for Dickkopf proteins. Nature. 2001;411:321–325. doi: 10.1038/35077108. [DOI] [PubMed] [Google Scholar]
- 25.Yue W, Sun Q, Dacic S, Landreneau RJ, Siegfried JM, Yu J, Zhang L. Downregulation of Dkk3 activates beta-catenin/TCF-4 signaling in lung cancer. Carcinogenesis. 2008;29:84–92. doi: 10.1093/carcin/bgm267. [DOI] [PubMed] [Google Scholar]
- 26.Kuphal S, Lodermeyer S, Bataille F, Schuierer M, Hoang BH, Bosserhoff AK. Expression of Dickkopf genes is strongly reduced in malignant melanoma. Oncogene. 2006;25:5027–5036. doi: 10.1038/sj.onc.1209508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.