Abstract
MicroRNAs (miRNAs) play an important role in diverse physiological processes and are potential therapeutic agents. Synthetic oligonucleotides (ONs) of different chemistries have proven successful for blocking miRNA expression. However, their specificity and efficiency have not been fully evaluated. Here, we show that peptide nucleic acids (PNAs) efficiently block a key inducible miRNA expressed in the haematopoietic system, miR-155, in cultured B cells as well as in mice. Remarkably, miR-155 inhibition by PNA in primary B cells was achieved in the absence of any transfection agent. In mice, the high efficiency of the treatment was demonstrated by a strong overlap in global gene expression between B cells isolated from anti-miR-155 PNA-treated and miR-155-deficient mice. Interestingly, PNA also induced additional changes in gene expression. Our analysis provides a useful platform to aid the design of efficient and specific anti-miRNA ONs for in vivo use.
INTRODUCTION
The discovery of small regulatory RNAs such as microRNAs (miRNAs) has added a new layer of control to the mechanisms directing gene expression. To date, the number of known human miRNA loci is 721 (1). At least 30% of protein-coding genes are estimated to be regulated by miRNAs, establishing them as one of the largest classes of regulatory molecules. By binding to complementary regions on their target mRNAs, they mediate gene silencing by translational repression, mRNA degradation or both (2). MiRNAs regulate a wide variety of biological processes such as proliferation, differentiation, cellular migration, cell fate determination and apoptosis. Not surprisingly, deregulation of miRNA expression has been linked to human diseases such as cancer and autoimmune dysfunction. One of the first miRNAs identified with oncogenic potential was miR-155. Encoded within an exon of the non-coding RNA BIC, miR-155 is deregulated in a number of different human cancers, most of which are of B-cell origin (3–6). Moreover, miR-155 over-expression during B-cell development is sufficient to trigger B-cell transformation (7). High levels of miR-155 are not restricted to transformed cells. In the course of an immune response, lymphoid cells up-regulate miR-155 (8,9). Induction of miR-155 profoundly affects gene expression in T and B cells, resulting in reduced expression of dozens of target genes (8,10).
The emergence of miRNAs as regulators of malignant transformation or autoimmunity is likely to have an impact on gene therapies designed to block tumour progression or inflammation. Further, the potential for miRNA antagonists as therapeutic agents in vivo has been illustrated for miR-122 (11,12), a liver-specific miRNA that has direct control over cholesterol biosynthesis and which is required for hepatitis C infection (13). An LNA/DNA anti-miR-122 oligonucleotide (ON) has been shown to be effective in suppressing hepatitis C viremia and is currently in phase I clinical trials (14).
A central goal in efforts to define miRNA antagonists with potential use in the clinic is the ability to synthesize stable and specific miRNA antagonists on a scale suitable for in vivo studies. Peptide nucleic acid (PNA; Figure 1a) is an uncharged ON analogue in which the sugar–phosphodiester backbone of DNA/RNA has been replaced by an achiral structure consisting of N-(2-aminoethyl)-glycine units. PNA ONs show high affinity and sequence specificity for complementary RNA and DNA, and also bear high chemical and metabolic stability (15). PNA has been exploited as an antisense agent for various applications in diagnostics and as potential therapeutics, often with attached peptides (16). PNA has also been shown to possess antisense biological activities in vivo with little or no toxicity (17–20). We showed previously that a 23-mer PNA, containing just four lys residues, was able to inhibit miR-122 in human liver cells and primary rat hepatocytes in culture without need for a transfection agent or attachment of a cell-penetrating peptide (21). However, PNAs have not been used hitherto for inhibiting miRNAs in vivo. Indeed, the widespread usage of PNA in vivo has been limited by the unavailability of an automated laboratory synthesis method that allows efficient synthesis of PNA oligomers of sufficient length and in sufficient quantities for in vivo studies from commercially available monomers.
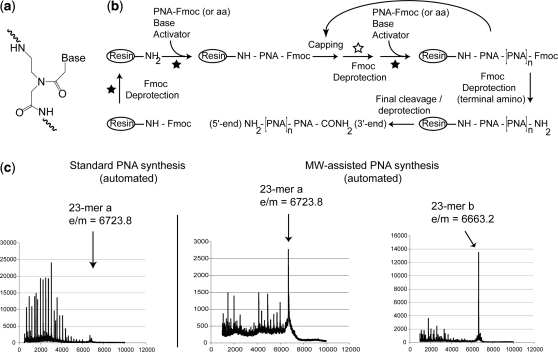
Figure 1.
(a) Schematic representation of a PNA monomer unit. (b) Schematic representation of the cycle of microwave-assisted PNA synthesis on solid support. Fmoc/Bhoc are orthologous protecting groups for primary and nucleobase amino groups, respectively. The applied microwave pulse is indicated by a filled star symbol. The empty star represents an applied microwave pulse when the growing chain consists of natural amino acids only, or no microwave if the synthesized chain contains at least one PNA monomer. Activator: PyBOP; base: DIPEA for amino acids or DIPEA/Lutidine for PNA; final cleavage/deprotection: TFA/TIS/H2O 95:2.5:2.5. (c) Typical MALDI-TOF mass spectra of crude 23-mer PNA (sequence a) synthesized by standard automated non-microwave method on a PAL-PEG-PS solid support (left panel) or by microwave-assisted synthesis on a PAL-PEG-PS support (sequence a, centre panel) or on a ChemMatrix solid support (sequence b, right panel).
In this article, we describe an automated microwave-assisted method for medium-scale (50 µmol) synthesis of PNA, using commercially available Fmoc/Bhoc PNA monomers and a CEM Liberty microwave peptide synthesizer, which results in rapid assembly and synthesis of high-purity PNA in good yields. We then illustrate the usefulness of PNA for miRNA inhibition by showing its ability to inhibit miR-155 expression and function in primary murine B cells both in culture and in vivo. By comparing the gene expression profiles of B cells isolated from the spleens of mice treated with anti-miR-155 PNA, untreated B cells and miR-155-deficient B cells, we find that PNA-treated mice showed alterations in gene expression similar to those from miR-155-deficient mice, thus validating anti-miR PNA targeting in vivo. In addition, numerous additional genes are also altered in expression levels upon in vivo PNA treatment.
MATERIALS AND METHODS
Synthesis of K-PNA-K3 ONs
The microwave-assisted method for PNA assembly was developed using a commercial microwave peptide synthesizer (Liberty, CEM Corporation) and carried out using commercially available Fmoc/Bhoc (9-fluorenylmethoxycarbonyl/benzhydryloxycarbonyl) PNA monomers (Link Technologies, Scotland). PNA oligomers containing a C-terminal amide were synthesized with Rink-Amide ChemMatrix (Matrix Innovation) solid support, and PyBOP (benzotriazol-1-yltris-pyrrolidinophosphonium hexafluorophosphate) as coupling agent. Detailed methods of synthesis are supplied in Supplementary Material. PNA ONs are 23-mer sequence a, Cys-K-5′-ACAAACACCATTGTCACACTCCA-3′-KKK (21); 23-mer anti-miR-155 K-PNA-K3 (sequence b), K-5′-ACCCCTATCACAATTAGCATTAA-3′-KKK; 23-mer scrambled-anti-miR-155 K-PNA-K3, K-5′-ACCCAATCGTCAAATTCCATATA-3′-KKK; 18-mer non-targeting K-PNA-K3-1, dK-5′-CACCATTGTCACACTCCA-3′-dKdKdK; 18-mer non-targeting K-PNA-K3-2, Cys-K-5′-CACCATTGTCACACTCCA-3′-KKK; and 18-mer non-targeting K-PNA-K3-3, dK-5′-ACCTCCAACTGCCTATAC-3′-dKdKdK. Sequences contained lys residues in either all l or all d form, for cellular and in vivo studies, respectively. PNA ONs were purified by reversed-phase high-pressure liquid chromatography (RP-HPLC) using a C18 Waters XBridge BEH130 10-µm column at 36°C in trifluoroacetic acid buffers (buffer A: 0.1% trifluoroacetic acid (TFA) in water; buffer B: 90% acetonitrile/10% water/0.1% TFA). Collected fractions were analysed by matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF) on a Voyager DE Pro BioSpectrometry workstation by mixing eluate:matrix in the ratio 1:1 (in microliters). Matrix consisted of α-cyano-4-hydroxynnamic acid (10 mg/ml) in acetonitrile/3% TFA (1:1, vol/vol). Fractions containing pure material were combined, lyophilized and subsequently dissolved in water. Oligomers were dissolved in de-ionized water and stored at −80°C after filtering through a 0.22- µm cellulose acetate membrane.
Primary B-cell purification, cell transfections and RNA extractions
Splenic primary B cells were purified from adult C57BL/6J mice or miR-155-deficient mice backcrossed six times to C57BL/6J background (8). For cell culture, splenocytes were T-cell depleted using Thy1.2 and complement. Cells were subsequently stimulated using 10–20 µg/ml lipopolysaccharide (LPS; Sigma) and 25 ng/ml IL-4. Electroporation of primary B cells with K-PNA-K3 ONs was carried out with an Amaxa system (Lonza) using the corresponding mouse nucleofector buffer solution and following manufacturer’s instructions. After electroporation, cells were cultured in the presence of LPS and IL-4 for 24–48 h before RNA extraction and analysis. RNA was extracted with Trizol following manufacturer’s instructions. For free-delivery experiments, K-PNA-K3 ONs were incubated for 4–8 h with 2 × 106 B cells, previously activated with LPS and IL-4 for 12–16 h, in 500-µl PNA-transfection buffer (Optimem; 20 µg/ml LPS; 25 ng/ml IL-4). Subsequently, cells were cultured in FCS-containing media in the presence of LPS and IL-4. RNAs were purified and analysed as described earlier.
In vivo experiments
All mice used in this study were maintained in a 12-h light/dark cycle, and experiments were conducted according to the Home Office guidelines. ONs were dissolved in saline and administered to mice based on body weight in the indicated doses by the intraperitoneal route. At study termination, mice were sacrificed and tissues were collected for further analysis.
Polyacrylamide gel electrophoresis and northern blots
Polyacrylamide gel electrophoresis (PAGE) and northern blots were carried out as described previously (21). RNA band size was estimated by running a 32P-labelled RNA ladder (Decade-Markers, Applied Biosystems), following manufacturer’s instructions.
Real-time reverse-transcription PCR (qRT-PCR)
Messenger RNAs of selected genes were quantified relative to the indicated house-keeping genes using a 7900 HT Fast Real-Time PCR System (Applied Biosystems) and a one-step RT–PCR approach (QuantiTect Probe RT–PCR, Qiagen) (21).
Microarray hybridization and analysis
For microarray experiments, splenic B cells were purified by positive selection using MACS B220-microbeads according to the manufacturer’s instructions (Miltenyi) and purity was >95%. Following extraction of RNA using Trizol, integrity was assessed by capillary electrophoresis using a BioAnalyser 2100 (Agilent Technologies), and its concentration was determined using a NanoDrop ND-1000 spectrophotometer. Aliquots of total RNA (200 ng) were then labelled using Affymetrix’s WT Sense Target labelling kit, and hybridized to Mouse Gene 1.0 ST arrays (Affymetrix) following the manufacturer’s instructions. After washing, arrays were scanned using a GS 3000 scanner (Affymetrix).
Microarray CEL files were processed and normalized with Affymetrix Power Tools (APTs), using the RMA-sketch method and obtaining summarized expression values for each transcript. Quality checks and all further analysis were carried out in Bioconductor (22). Probe-sets, not from the ‘main’ category and those with cross-hybridization potential, were removed according to the Affymetrix NetAffx release 29 annotation. In order to assess differential expression, limma was used to fit linear models, using an empirical Bayes approach to shrink the estimated variance (23). After this step, non-specific filtering was carried out, by calculation of the interquartile range and by excluding 50% of the probe-sets with lowest variability. Differentially expressed genes were then selected that passed a fold-change cut-off of 1.2 with a false discovery rate (FDR) of 10%, as estimated using the Benjamini–Hochberg method (24). These criteria were selected to highlight the differences and similarities between differentially expressed genes in PNA-treated and miR-155-knockout samples. All microarray data has been deposited in the ArrayExpress database (accession number: E-MEXP-2587).
RESULTS
Efficient synthesis of PNAs using a microwave-assisted method
PNA oligomers (Figure 1a) have been synthesized previously on solid support using Boc/benzyl chemistry (15) by Fmoc/DDE chemistry (25) and by a novel PNA synthesis method using a benzothiazole-2-sulfonyl group as amino-protecting group (26). More commonly Fmoc/Bhoc chemistry is now used, because such protected PNA monomers are commercially available (27–31). Synthesis of a 23-mer anti-miR PNA using a conventional peptide synthesizer was reported earlier (21,31). At this length, we found that isolated yields of PNA were quite low (just a few percent). Further, there was a need for double-coupling reactions of 30 min each (31), because the efficiency of PNA-coupling reactions was poor.
The synthesis of peptides and peptidomimetics has been revolutionized using microwave-assisted solid-phase coupling methods that rely on the application of a microwave pulse to drive coupling and deprotection reactions to completion. Microwave assistance is particularly valuable for difficult peptide couplings to reduce chain aggregation and to improve coupling efficiencies (32–34). A method has been described recently for the orthogonal synthesis of a short PNA peptide using DDE/Mmt-protected PNA monomers (35), but such monomers are not commercially available. Here, we present the implementation of microwave-assisted synthesis of PNA using a commercial microwave peptide synthesizer (Liberty, CEM Corporation) and carried out using commercially available Fmoc/Bhoc PNA monomers.
A cycle of PNA synthesis is analogous to that of standard Fmoc solid-phase peptide synthesis and consists of PNA monomer coupling, capping of unreacted amino groups and terminal Fmoc deprotection (Figure 1b). A microwave pulse is applied in all amino acid- or PNA-coupling steps. For PNA residues, a single coupling of 30 min duration is sufficient for optimal oligomer assembly. Microwave energy is also applied during Fmoc-deprotection steps, but only for those involving amino acids when no PNA is present on the support. Microwave pulsing was not used for Fmoc-deprotection reactions of terminal PNA residues or for amino acids when PNA was present on the support, as side reactions on the PNA were observed (36). In all cases, power, temperature and duration of the microwave pulse were adjusted depending on the particular step (Supplementary Data). Compared with our previously reported method for PNA synthesis carried out at room temperature on a conventional robotic synthesizer (21,31), the assembly of a 23-mer PNA oligomer (including 4 × lys residues) by the microwave-assisted method is overall 2.5-fold faster and was completed within 24 h.
With the conventional synthesis method on a synthesis scale of 5–10 µmol with the PAL-PEG-PS solid support (31), the MALDI-TOF mass spectrum of the crude synthesis product, before purification of a 23-mer PNA plus 5 amino acids (sequence a, Cys-K-PNA-K3), showed only a low yield of the desired product (Figure 1c, left panel, and data not shown). In contrast, the mass spectrum of the same Cys-K-PNA-K3 23-mer PNA synthesized using the microwave-assisted method and the same PAL-PEG-PS support showed a clearly improved amount of 23-mer product (sequence a, Figure 1c, middle panel). The overall isolated yield after HPLC purification was 5% (Table 1). We then investigated the alternative ChemMatrix solid support for the synthesis of a 23-mer containing four lys residues (K-PNA-K3, sequence b, anti-miR-155), and the mass spectrum of the crude synthesis product showed a further improved result (Figure 1c, right panel). The overall isolated yield after HPLC was 10% (Table 1). Shorter 18-mer oligomers were also synthesized with final overall isolated yields of ∼15% in the case of the PAL-PEG-PS support and ∼20% in the case of the ChemMatrix support (Table 1). The yields obtained provide milligram quantities of PNA sufficient for mouse studies.
Table 1.
Examples of PNA ONs synthesized by microwave-assisted solid-phase synthesis
| Peptide sequence | Length | Solid support | Yield (%) | Name |
|---|---|---|---|---|
| Cys-K-5′-ACAAACACCATTGTCACACTCCA-3′-KKK | 23 + 5aa | PAL-PEG-PS | 5.0 | Cys-K-PNA-K3 23-mer (sequence a) |
| dK-5′-CACCATTGTCACACTCCA-3′-dKdKdK | 18 + 4aa | PAL-PEG-PS | 15.7 | dK-PNA-dK3-1 18-mer |
| Cys-K-5′-CACCATTGTCACACTCCA-3′-KKK | 18 + 5aa | PAL-PEG-PS | 14.9 | Cys-K-PNA-K3-2 18-mer |
| dK-5′-ACCCCTATCACAATTAGCATTAA-3′-dKdKdK | 23 + 4aa | ChemMatrix | 10.4 | Cys-K-PNA-K3 23-mer (sequence b) |
| dK-5′-CACCATTGTCACACTCCA-3′-dKdKdK | 18 + 4aa | ChemMatrix | 22.3 | dK-PNA-dK3-1 18-mer |
| dK-5′-ACCTCCAACTGCCTATAC-3′-dKdKdK | 18 + 4aa | ChemMatrix | 19.8 | dK-PNA-dK3-3 18-mer |
PNA ONs contained an N-terminal Cys for post-synthetic modification and four lys residues for improved solubility and cell uptake. Yield refers to the percentage of product after HPLC purification relative to the amount of starting solid support. Sequences a and b refer to ONs shown in Figure 1c.
PNA ONs inhibit miR-155 expression in cultured B cells
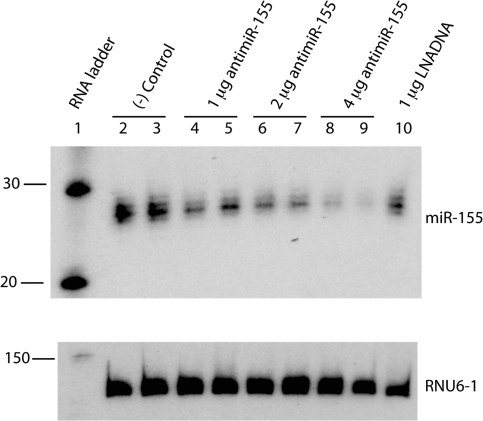
We focused our analysis of inhibition of miR-155 on B cells due to the functional relevance of miR-155 in this cell type and previous identification of target genes (8–10). It is known that bacterial LPS triggers miR-155 expression in cultured B cells (9,10), and therefore we first determined the kinetics of miR-155 expression by incubation of mouse primary B cells with LPS (Supplementary Figure S1). miR-155 expression was induced very rapidly and reached a plateau at 24-h post-initiation of treatment, which was sustained for at least 3 days, in agreement with previous data (10,37). Thus, we tested the activity of a 23-mer anti-miR-155 K-PNA-K3 ON by electroporation into LPS-activated primary B cells (Figure 2). Northern blot analysis showed the expected dose–response on miR-155 inhibition, with a 60% knockdown of miR-155 with 4-µg PNA.
Figure 2.
A total of 2 × 106 primary B cells were electroporated with the indicated amounts of anti-miR-155 PNA or control anti-miR-155 LNA/DNA (Exiqon). RNA was extracted and analysed by northern blot 24 h post-transfection. Cells were cultivated with LPS supplemented media overnight prior to PNA transfection. The indicated masses correspond to 1.47, 2.94 and 5.88 µM PNA, or 1.39 µM LNA ON.
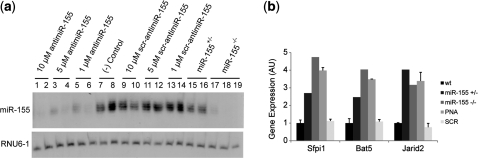
The K-PNA-K3 was evaluated further for its ability to be internalized by primary B cells and to block miR-155 expression in the absence of any additional transfection agent or other internalization procedure, as previously obtained for miR-122 inhibition in hepatocytes (21). Primary B cells were incubated with different concentrations of anti-miR-155 K-PNA-K3 or its scrambled sequence, scr-anti-miR-155, in the presence of LPS. Figure 3a shows that anti-miR-155 K-PNA-K3 was efficiently internalized by activated primary B cells unaided and substantially reduced the levels of endogenous miR-155 at concentrations of 1–10 µM. The miRNA inhibition effect was sequence specific, as the scrambled control (scr-anti-miR-155) did not show this ability. As expected, in B cells obtained from miR-155-deficient mice (miR-155−/−), miR-155 expression was not detected, while B cells from heterozygous mice for miR-155 contained intermediate levels of the miRNA (Figure 3a).
Figure 3.
miR-155 inhibition in LPS-activated primary B cells with K-PNA-K3 ONs by free delivery. (a) Northern blot of total RNA from LPS-activated B cells treated with the corresponding K-PNA-K3 ON in media without antibiotics for 4–6 h, washed and further incubated for 18–20 h in full media. RNA from miR-155+/− and miR-155−/− B cells were included as controls. Each lane corresponds to an individual experimental replicate. (b) qRT-PCR of miR-155 validated target genes, Bat5, Jarid2 and Sfpi1 (Pu.1) in miR-155+/−, miR-155−/− or wild-type primary B cells, incubated with 10-µM anti-miR-155 PNA. B2m expression was used as endogenous control. Primary B-cell cultures were pre-activated with LPS overnight before incubation with PNA.
To evaluate further the functional activity of anti-miR PNA ONs, we measured the extent of de-repression of endogenous targets of miR-155. It has been shown previously that in activated B cells, miR-155 regulates the abundance of numerous mRNAs, including Bat5, Jarid2 and Sfpi1 (10). Cells treated with the anti-miR-155 K-PNA-K3 ON at 10 µM showed a strong 3- to 4-fold de-repression of these target genes (Figure 3b), partially mimicking the effect of deletion of miR-155−/−. Interestingly, deletion of only one miR-155 allele (miR-155+/−) caused strong target de-repression, indicating a miR-155 gene dosage effect. The lack of activity seen for the scrambled control ON by northern blot is corroborated by the lack of a measurable effect on miR-155-regulated genes in cells incubated with this ON.
Efficient inhibition of miR-155 in vivo by a PNA anti-miR
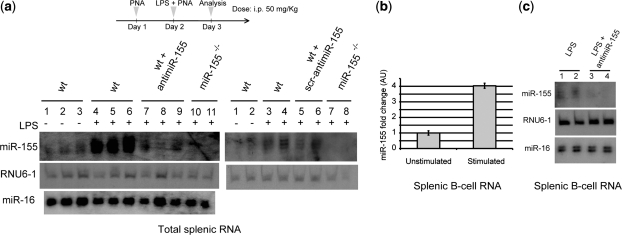
Next, we examined the ability of anti-miR-155 dK-PNA-dK3 ON (sequence b, Table 1, line 4; d-lys residues were used to ensure proteolytic stability) to block miR-155 expression in vivo. Because naïve cells express low amounts of miR-155, we examined whether LPS could promote acute induction of miR-155 in splenocytes, as is known to occur in bone marrow cells (38). We found that intraperitoneal injection of 100 µg of LPS was the optimal sub-lethal dose for inducing miR-155 expression in the spleen. Moreover, miR-155 expression peaked at 24-h post-administration and returned to steady state levels by 72 h (Supplementary Figure S2). Based on these observations, mice were dosed systemically at 50 mg PNA/kg/day for 2 days and analysed 24 h after the last injection (at which time miR-155 expression is maximal). As shown in Figure 4a, RNA isolated from the spleens of LPS-treated mice showed a strong induction of miR-155, which was completely abolished by administration of the anti-miR-155 PNA. The effect appeared to be sequence specific as evidenced by the lack of activity of the control scrambled PNA sequence, and the unaltered levels of the unrelated miRNA, miR-16. Because miR-155 is up-regulated in splenic B cells upon LPS treatment, northern blot analysis showed that the observed miR-155 inhibition by anti-miR-155 K-PNA-K3 was also seen in this splenic B-cell population (Figure 4b and c).
Figure 4.
In vivo miR-155 inhibition with dK-PNA-dK3 ONs. (a) Schematic representation of a dosage regimen and northern blot analysis of total RNA (whole spleen, Day 3) from animals treated with the indicated dK-PNA-dK3 anti-miR. Each lane represents an individual mouse sample. RNAs from miR-155−/− or wild-type mice with no LPS administration were included as controls. (b) miR-155 levels in splenic B cells purified from mice receiving either LPS (i.p., 100 µg) or vehicle control (PBS). Measurement by qRT-PCR using RNU6-2 RNA as endogenous control. Error bars indicate the standard deviation between experimental replicates (duplicates). (c) Ten micrograms of RNA from isolated splenic B cells from the indicated mice group were subjected to northern blot analysis for miR-155, miR-16 and RNU6-1 detection. Each lane corresponds to an individual mouse sample.
Genome-wide analysis of gene expression shows enrichment of miR-155-binding sites in mRNAs up-regulated in B cells from mice deficient in miR-155−/− or treated with anti-miR-155
To evaluate the efficacy and specificity of the PNA treatment in mice, we compared gene expression profiles of splenic B cells from either LPS administered miR-155-deficient animals receiving vehicle control (phosphate buffered saline, PBS), or LPS administered wild-type animals receiving either PBS or anti-miR-155 dK-PNA-dK3 with the dosing regimen shown in Figure 4a.
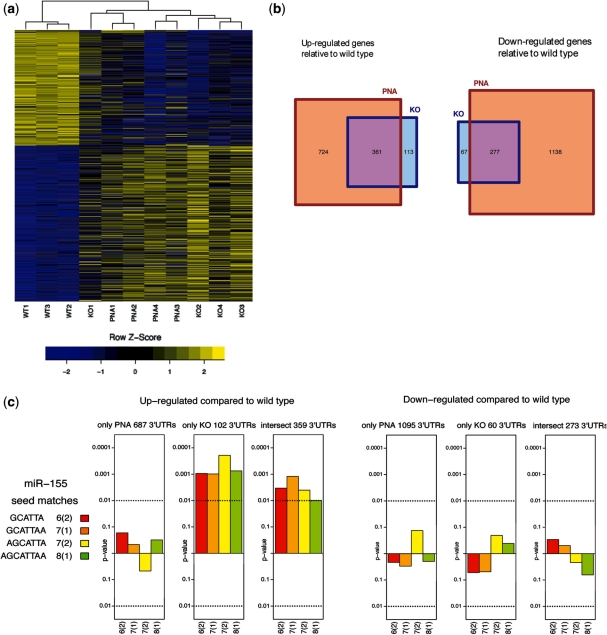
In all, 474 transcripts were significantly up-regulated and 344 were down-regulated in miR-155-deficient B cells relative to their wild-type counterparts (differential expression more than 1.2-fold and 10% FDR). Among the deregulated genes are transcription factors, adhesion molecules, chemotaxis regulators and signalling molecules. In support with our previous observation that anti-miR-155 PNA efficiently blocks miR-155 both in cell culture and in vivo, the ON treatment induced changes in mouse B-cell gene expression that recapitulated deficiency of miR-155 (Figure 5a). In particular, 76% of the up-regulated and 80% of the down-regulated genes in the miR-155-deficient samples passed the same fold-change and FDR cut-offs in the PNA-treated samples (Figure 5b and Supplementary Table S1). However, when all differentially expressed genes are considered, a substantially higher number of genes are found in the PNA-treated versus miR-155−/− group, as shown by the Venn diagram in Figure 5b. Gene ontology analysis revealed enrichment in pathways related to protein catabolism and general cellular metabolism (data not shown).
Figure 5.
Affymetrix gene expression profiling of B cells from wild-type mice treated with PBS and of mice treated with dK-PNA-dK3 (3 × 50 mg/kg) and of B cells from miR-155−/− mice. (a) Heat-map profile showing relative expression across all samples of the differentially expressed transcripts between miR-155−/− and wild-type B cells from mice spleens. (b) Venn diagrams showing the overlap of differentially expressed genes in PNA-treated and miR-155−/− groups, relative to wild-type B cells. (c) Seed-enrichment analysis for the groups indicated in the Venn diagram, considering only genes with an annotated 3′-UTR sequence. In all cases, gene lists were selected with a fold-change >1.2 and FDR <10%.
We next focused our analysis on predicted miR-155 mRNA targets. This was based on computational and experimental results indicating that miRNA targets contain a perfect Watson–Crick matching sequence of 6–8 nt between the 5′-end of an miRNA (referred to as the ‘seed’ region) and the 3′-unstranslated region (3′-UTR) of the mRNAs (39). Moreover, we have previously shown how the enrichment in 3′-UTR sequences complementary to the miR-155 seed region reflects direct targeting by this miRNA (8,10,40). As expected, miR-155 seed matches are statistically over-represented in the 3′-UTR sequences of the transcripts up-regulated in miR-155-deficient B cells (Figure 5c). This effect is observed both for transcripts consistently up-regulated (‘intersect’) as well as those exclusively up-regulated in the miR-155 knockout. The 724 transcripts that were only up-regulated in the PNA samples and all the down-regulated groups showed no seed complement enrichment (Figure 5c). We also found that several experimentally validated mRNA targets of miRNA-155 were up-regulated in both miR-155−/− and PNA-treated groups (Supplementary Table S1): Inpp5d (also known as Ship1) (41), Agtrap (42), Arid2 and Zfp652 (43).
Several other up-regulated mRNAs, not all containing miR-155 seed sequences, were identified in miR-155−/− and PNA-treated groups that had already been found in our previous microarray studies of miR-155-deficient B and T cells activated in culture: Sgk3, Mt1, Cxcl10 (10), Ccl5, Map3k7ip1 and Il7r (8). These results demonstrate that PNA inhibition of miR-155 in vivo recapitulates to a great extent the effect of genetic deletion of miR-155. Additional changes in gene expression may have resulted from cascade effects of genes up-regulated by miR-155. For example, the transcription factor c/ebp was shown by LNA/DNA anti-miR treatment to be a target of miR-155, and its up-regulation caused subsequent down-regulation of G-CSF in myeloid cells (44).
In addition, we found novel targets up-regulated in both, miR-155−/− and PNA-treated groups, which contained one or more miR-155 seed sequences, for example: Tifa (TRAF-interacting protein with forkhead-associated domain), Pik3ip1 (phosphoinositide-3-kinase interacting protein 1), Map3k14 (mitogen-activated protein kinase kinase kinase 14), Il6ra (interleukin 6 receptor α) and Apobec1 (apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1).
As suggested by the seed enrichment analysis (Figure 5c), it is likely that up-regulation of the 724 transcripts found only in the PNA-treated group is not due to direct miR-155 inhibition, instead due to a combination of other indirect effects of in vivo PNA anti-miR treatment, including off-target effects and PNA chemistry-specific effects. Enrichment was observed neither in the 3′-UTRs of these transcripts for seed matches of miR-155* (the passenger strand of the mature miR-155) nor for sequences complementary to the first eight residues at the 5′-end of the PNA (equivalent to the seed sequence of an miRNA) (Supplementary Figure S3). We also did not find any propensity for complementary matching of the full-length PNA to any regions in the whole transcripts (data not shown).
DISCUSSION
The intrinsic role of miRNAs as regulators of gene expression, especially those miRNAs relevant to human diseases, has opened up the possibility of modulating miRNA expression for therapeutic purposes. ONs of different chemistries such as LNA/DNA mixmers, MOEs and ‘antagomirs’ have proven successful for blocking miRNA expression in vivo, as exemplified by targeting of miR-122 and miR-155 (11,12,14,44,45). We have now shown that PNA ONs are also efficient in blocking miRNA function in vivo. We chose to investigate PNA ONs as inhibitors of miR-155, because we previously characterized the function of this miRNA in primary B cells (10). The in vivo studies reported here required the production and purification of milligram quantities of PNA ONs, which was achieved using microwave-assisted PNA assembly. This methodology allowed rapid synthesis and a significant increase in the quantity and quality of the crude PNA product (Figure 1 and Table 1).
The efficient unaided delivery of a 23-mer anti-miR-155 K-PNA-K3 in cultured primary B cells in the presence of LPS is unprecedented (Figure 3a). Such cells are generally known to be difficult to transfect (46,47). By measurement of the expression of miR-155 target genes that we have previously identified in cultured B cells (10), we were able to demonstrate a significant effect of the PNA treatment on the regulation of miR-155 target genes (Figure 3b). This finding strengthens the general utility of PNA for miRNA targeting in primary cell culture, as we had previously shown efficient inhibition of miR-122 in primary hepatocytes by a K-PNA-K3 (21).
When evaluated in vivo, anti-miR-155 dK-PNA-dK3 led to a dramatic and sequence-specific decrease in LPS-induced miR-155 levels in whole spleen or in the purified splenic B-cell fraction (Figure 4). Hitherto, only a few demonstrations of PNA effectiveness in vivo have been shown. Applications include PNA modified with a peptide nuclear localization signal for splicing redirection, PNA targeting of mRNAs or exon skipping (16,19,47–49). We have, therefore, shown for the first time that lysine-bearing PNAs are also suitable for inhibiting miRNAs in whole animals.
Experiments examining the ability of synthetic ONs to block miRNA function have uncovered the potential of such molecules for therapeutics. Our results extend these studies showing the usefulness of PNAs for miRNA targeting in animals and highlight the utility of comparing the genetic and chemical approaches for the identification of specific and unspecific effects. With this approach, we have been able to show for the first time the extensive overlap in the expression profiles of B cells from PNA-treated and miR-155−/− mice (Figure 5a). Interestingly, we found the expression levels of a significant number of other genes also affected, including 724 genes up-regulated and 1138 genes that were down-regulated only in the PNA-treated group (Figure 5b). The mechanisms involved are not clear and this will require a separate detailed study, but are likely to derive from a combination of indirect effects that might include off-target effects of the PNA and PNA chemistry-specific effects. Further gene expression studies of this type will help to determine whether synthetic ONs of different backbone chemistry might confer particular advantages over others in terms of reduced unrelated gene expression alterations or a stronger direct ‘knockout’ recapitulation. It should be noted that we did not observe any signs of toxicity in the mice treated with PNA ONs (data not shown), in agreement with previous results for PNA in vivo treatment (17,20), and a great deal of detailed analysis will be needed to relate any particular gene expression changes to particular unwanted side effects. However, such a genomics approach could provide a useful platform as one criterion in the design and assessment of ONs as blockers of RNA function and/or as therapeutics.
B lymphocytes play an important role in immune responses against Gram-negative bacteria and their activation by LPS leads to enhanced antigen presentation, proliferation and differentiation. Similar to myeloid cells, systemic administration of LPS induced expression of miR-155 in splenic B cells, which peaked at 24 h (Supplementary Figure S2). The response to LPS by B cells is the activation of the nuclear factor kappa B (NF-KB) and mitogen-activated protein kinase (MAPK) signalling pathways, and miR-155 itself seems to be at least transiently under the control of NF-kB (9,48). Our microarray analysis of up-regulated genes in miR-155-deficient or PNA-treated B cells in the presence of LPS identified several genes that regulate these pathways, for example, Map3k5/8/14 (mitogen-activated protein 3-kinase 5, 8 and 14, respectively), Peli1 (Pellino 1; only found in miR-155-deficient cells) and Tifa. Interestingly, Map3k8/14, Peli1 and Tifa are known activators of the NF-kB signalling pathway and both contain at least one miRNA binding site in their 3′-UTRs, suggesting they may be direct miR-155 targets.
Among other up-regulated genes containing miR-155-binding sites, we found Sgk3 (Serum/Glucocorticoid Regulated Kinase 3), Pik3ip1 (Phosphoinositide-3-kinase Interacting Protein 1) and Inpp5d (Inositol polyphosphate-5-phosphatase D, Ship1), all components of the PI3K pathway, a key modulator of B-cell development and signalling (49). Inpp5d has already been validated as a miR-155 target gene (41).
The contribution of all these genes to the regulation by miR-155 of the B-cell response by LPS remains to be understood, but it is likely to reflect deregulation of multiple cellular processes, involving the induction of both pro- and anti-inflammatory mediators. Thus, our results have identified potential new targets under miR-155 regulation in activated B cells in vivo, as well as confirmed previously known targets of miR-155. These observations can guide future studies to unveil how the network of miR-155-regulated genes contributes to the LPS response in vivo.
Noteworthy also is the relationship between miR-155 and Inpp5d, which has interesting implications concerning the possible therapeutic use of anti-miR-155 ONs. Diffuse large B-cell lymphoma (DLBCL) has been characterized with low levels of Inpp5d (50). Non-germinal centre-DLBCL patients showed increased expression of miR-155 and a concomitant decrease in Inpp5d levels, which also correlated with poorer survival (50). It is, therefore, possible that therapies aimed at increasing Inpp5d levels by reducing miR-155 expression could be beneficial in the treatment of specific types of lymphoma.
The combination of genetic approaches and synthetic ON targeting miRNAs can be instrumental to dissect in vivo responses. Improving the design of synthetic ONs to block miRNA function to increase efficiency of silencing but avoiding non-specific effects will provide new tools for uncovering the function of miRNAs and better therapies for the treatment of human diseases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
CDA-MRC fellowship (to E.V.); Biotechnology and Biological Sciences Research Council. A.G.T. is a Milstein Student of the Darwin Trust, Edinburgh, Scotland. Funding for the open access charge: Medical Research Council.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Small Animal Barrier Unit Staff for technical assistance and Martin Turner and Mariann Bienz for critical reading of the manuscript.
REFERENCES
- 1.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pillai R, Bhattacharyya S, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Calin G, Croce C. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 4.Eis P, Tam W, Sun L, Chadburn A, Li Z, Gomez M, Lund E, Dahlberg J. Accumulation of miR-155 and Bic RNA in human B cell lymphomas. Proc. Natl Acad. Sci. USA. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van den Berg A, Kroesen B.-J, Kooistra K, De Jong D, Briggs J, Blokzijl T, Jacobs S, Kluiver J, Diepstra A, Maggio E, et al. High expression of B-cell receptor inducible gene BIC in all subtypes of Hodgkin lymphoma. Genes Chromosomes Cancer. 2003;37:20–28. doi: 10.1002/gcc.10186. [DOI] [PubMed] [Google Scholar]
- 6.Wang M, Tan L, Dijkstra M, van Lom K, Robertus J.-L, Harms G, Blokzijl T, Kooistra K, van t’Veer M, Rosati S, et al. miRNA analysis in B-cell chronic lymphocytic leukaemia: proliferation centres characterized by low miR-150 and high BIC/miR-155 expression. J. Pathol. 2008;215:13–20. doi: 10.1002/path.2333. [DOI] [PubMed] [Google Scholar]
- 7.Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce C. Pre-B cell proliferation and lymphoblastic leukaemia/high grade lymphoma in Eµ-MiR155 transgenic mice. Proc. Natl Acad. Sci. USA. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thai T-H, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–607. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 10.Vigorito E, Perks K, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das P, Miska E, Rodriguez A, Bradley A, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:1–13. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjärn M, Hansen HF, Berger U, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 12.Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 13.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 14.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Ørum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2009;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egholm M, Buchardt O, Nielsen PE, Berg RH. Peptide nucleic acids (PNA). Oligonucleotide analogues with an achiral backbone. J. Am. Chem. Soc. 1992;114:1895–1897. [Google Scholar]
- 16.Lebleu B, Moulton HM, Abes R, Ivanova GD, Abes S, Stein DA, Iversen PL, Arzumanov A, Gait MJ. Cell penetrating peptide conjugates of steric block oligonucleotides. Adv. Drug Delivery Rev. 2008;60:517–529. doi: 10.1016/j.addr.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sazani P, Gemignani F, Kang S.-H, Maier MA, Manoharan M, Persmark M, Bortner D, Kole R. Systemically delivered antisense oligomers upregulate gene expression in mouse tissues. Nat. Biotech. 2002;20:1228–1233. doi: 10.1038/nbt759. [DOI] [PubMed] [Google Scholar]
- 18.Boffa LC, Cutrona G, Cilli M, Mariani MR, Matis S, Pastorini M, Damonte G, Millo E, Roncella S, Ferrarini M. Therapeutically promising PNA complementary to a regulatory sequence for c-myc: pharmacokinetics in an animal model of human Burkitt’s lymphoma. Oligonucleotides. 2005;15:85–93. doi: 10.1089/oli.2005.15.85. [DOI] [PubMed] [Google Scholar]
- 19.Chaubey B, Tripathi S, Pandey VN. Single acute-dose and repeat-doses toxicity anti-HIV-1 PNATAR-penetratin conjugates after intraperitoneal administration to mice. Oligonucleotides. 2008;18:9–20. doi: 10.1089/oli.2007.0088. [DOI] [PubMed] [Google Scholar]
- 20.Yin H, Lu Q, Wood M. Effective exon skipping and restoration of dystrophin expression by peptide nucleic acid antisense oligonucleotides in mdx mice. Mol. Therapy. 2008;16:38–45. doi: 10.1038/sj.mt.6300329. [DOI] [PubMed] [Google Scholar]
- 21.Fabani M, Gait MJ. miR-122 targeting with LNA/2′-O-methyl oligonucleotide mixmers, peptide nucleic acids (PNA), and PNA-peptide conjugates. RNA. 2008;14:336–346. doi: 10.1261/rna.844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 24.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. Ser. B Methodol. 1995;57:289–300. [Google Scholar]
- 25.Bialy L, Diaz-Mochon JJ, Specker E, Keinicke L, Bradley M. Dde-protected PNA monomers, orthogonal to Fmoc, for the synthesis of PNA-peptide conjugates. Tetrahedron. 2005;61:8295–8305. [Google Scholar]
- 26.Lee H, Jeon JH, Lim JC, Choi H, Yoon Y, Kim SK. Peptide nucleic acid synthesis by novel amide formation. Org. Lett. 2007;9:3291–3293. doi: 10.1021/ol071215h. [DOI] [PubMed] [Google Scholar]
- 27.Thomson SA, Josey JA, Cadilla R, Gaul MD, Hassman CF, Luzzio MJ, Pipe AJ, Reed KL, Ricca DJ, Wiethe RW, et al. Fmoc mediated synthesis of peptide nucleic acids. Tetrahedron. 1995;51:6179–6194. [Google Scholar]
- 28.Mayfield LD, Corey DR. Automated synthesis of peptide nucleic acids and peptide nucleic-acid-peptide conjugates. Anal. Biochem. 1999;268:401–404. doi: 10.1006/abio.1998.3052. [DOI] [PubMed] [Google Scholar]
- 29.Braasch DA, Nulf CJ, Corey DR. Synthesis and purification of peptide nucleic acids. Curr. Protocols Nucleic Acids Chem. 2002;2:4.11.1–4.11.18. doi: 10.1002/0471142700.nc0411s09. [DOI] [PubMed] [Google Scholar]
- 30.Turner JJ, Ivanova GD, Verbeure B, Williams D, Arzumanov A, Abes S, Lebleu B, Gait MJ. Cell-penetrating peptide conjugates of peptide nucleic acids (PNA) as inhibitors of HIV-1 Tat-dependent trans-activation in cells. Nucleic Acids Res. 2005;33:6837–6849. doi: 10.1093/nar/gki991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turner JJ, Williams D, Owen D, Gait MJ. Disulfide conjugation of peptides to oligonucleotides and their analogues. Curr. Protocols Nucleic Acids Chem. 2005;2:4.28.1–4.28.21. doi: 10.1002/0471142700.nc0428s24. [DOI] [PubMed] [Google Scholar]
- 32.Bacsa B, Desai B, Dibo G, Kappe CO. Rapid solid-phase synthesis using thermal and controlled micrpwave irradiation. J. Peptide Sci. 2006;12:633–638. doi: 10.1002/psc.771. [DOI] [PubMed] [Google Scholar]
- 33.Fara MA, Diaz-Mochon JJ, Bradley M. Microwave-assisted coupling with DIC/HOBt for the synthesis of difficult peptoids and fluorescently labelled peptides – a gentle heat goes a long way. Tetrahedron Lett. 2006;47:1011–1014. [Google Scholar]
- 34.Coantic S, Subra G, Martinez J. Microwave-assisted solid phase peptide synthesis on high loaded resins. Int. J. Pep. Res. Ther. 2008;14:143–147. [Google Scholar]
- 35.Svenson N, Diaz-Mochon JJ, Bradley M. Microwave-assisted orthogonal synthesis of PNA-peptide conjugates. Tetrahedron Lett. 2008;49:6498–6500. [Google Scholar]
- 36.Uhlmann E, Peyman A, Breipohl G, Will DW. PNA: synthetic polyamide nucleic acids with unusual binding properties. Angewandte Chem. Int. Ed. 1998;37:2796–2823. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2796::AID-ANIE2796>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 37.Teng G, Hakimpour P, Landgraf P, Rice A, Tuschl T, Casellas R, Papavasiliou FN. MicroRNA-155 is a negative reulator of activation-induced cytidine deaminase. Immunity. 2008;28:621–629. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, Paquette RL, Baltimore D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J. Exp. Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 40.van Dongen S, Abreu-Goodger C, Enright AJ. Detecting microRNA binding and siRNA off-target effects from expression data. Nat. Method. 2008;5:1023–1025. doi: 10.1038/nmeth.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Connell R, Chaudhuri A, Rao D, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc. Natl Acad. Sci. USA. 2009;106:7113–7118. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin MM, Lee EJ, Buckenberger JA, Schmittgen TD, Elton TS. MicroRNA-155 regulates human angiotensin II type 1 receptor expression in fibroblasts. J. Biol. Chem. 2006;281:18277–18284. doi: 10.1074/jbc.M601496200. [DOI] [PubMed] [Google Scholar]
- 43.Yin Q, McBride J, Fewell C, Lacey M, Wang X, Lin Z, Cameron J, Flemington EK. MicroRNA-155 is an Epstein–Barr virus-induced gene that modulates Epstein–Barr virus-regulated gene expression pathways. J. Virol. 2008;82:5295–5306. doi: 10.1128/JVI.02380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Worm J, Stenvang J, Petri A, Frederiksen K, Obad S, Elmen J, Hedtjarn M, Straarup E, Hanse J, Kauppinen S. Silencing of microRNA-155 in mice during acute inflammatory response leads to derepression of c/ebp Beta and down-regulation of G-CSF. Nucleic Acids Res. 2009;37:1–9. doi: 10.1093/nar/gkp577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, Mckay R, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metabolism. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 46.Buschle M, Brenner MK, Chen ISY, Drexler HG, Gignac SM, Rooney CM. Transfection and gene expression in normal and malignant primary B lymphocytes. J. Immunol. Methods. 1990;133:77–85. doi: 10.1016/0022-1759(90)90321-l. [DOI] [PubMed] [Google Scholar]
- 47.Seiffert M, Stilgenbauer S, Döhner H, Lichter P. Efficient nucleofection of primary human B cells and B-CLL cells induces apoptosis, which depends on the microenvironment and on the structure of transfected nucleic acids. Leukemia. 2007;21:1977–1983. doi: 10.1038/sj.leu.2404863. [DOI] [PubMed] [Google Scholar]
- 48.Tili E, Michaille J.-J, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-α stimulation and their possible roles in regulating the response to endotoxin shock. J. Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 49.Okkenhaug K, Vanhaesbroeck B. PI3K in lymphocyte development, differentiation and activation. Nat. Rev. Immunol. 2003;3:317–330. doi: 10.1038/nri1056. [DOI] [PubMed] [Google Scholar]
- 50.Pedersen IM, Otero D, Kao E, Miletic AV, Hother C, Ralfkiaer E, Rickert RC, Gronbaek K, David M. Onco-miR-155 targets SHIP1 to promote TNF-dependent growth of B cell lymphomas. EMBO Mol. Med. 2009;1:288–295. doi: 10.1002/emmm.200900028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.