Abstract
Accurate DNA segregation is essential for genome transmission. Segregation of the prototypical F plasmid requires the centromere-binding protein SopB, the NTPase SopA and the sopC centromere. SopB displays an intriguing range of DNA-binding properties essential for partition; it binds sopC to form a partition complex, which recruits SopA, and it also coats DNA to prevent non-specific SopA–DNA interactions, which inhibits SopA polymerization. To understand the myriad functions of SopB, we determined a series of SopB–DNA crystal structures. SopB does not distort its DNA site and our data suggest that SopB–sopC forms an extended rather than wrapped partition complex with the SopA-interacting domains aligned on one face. SopB is a multidomain protein, which like P1 ParB contains an all-helical DNA-binding domain that is flexibly attached to a compact (β3–α)2 dimer-domain. Unlike P1 ParB, the SopB dimer-domain does not bind DNA. Moreover, SopB contains a unique secondary dimerization motif that bridges between DNA duplexes. Both specific and non-specific SopB–DNA bridging structures were observed. This DNA-linking function suggests a novel mechanism for in trans DNA spreading by SopB, explaining how it might mask DNA to prevent DNA-mediated inhibition of SopA polymerization.
INTRODUCTION
Partition or segregation, is an essential process that ensures the maintenance of genomic DNA during cell division. The partition machinery of low copy number plasmids represent excellent model systems to study DNA segregation at a detailed atomic level because they are composed of just three components: a centromere DNA site, a partition NTPase and a centromere-binding protein (CBP) (1–3). These partition (par) systems, which are encoded on a cassette on the respective plasmid, are of two main types: types I and II (4). This useful categorization is based primarily on the kind of NTPase present. Type I systems are the most abundant and contain NTPase proteins that harbor deviant Walker A type ATPase folds, while the type II systems utilize actin-like NTPases (1–5). The type I par systems can be further subdivided into types Ia and b based on size and limited sequence homologies of the Par proteins. The type Ia NTPase and CBPs are ∼30–45 kDa while the type Ib homologs are smaller, containing 192–308 and 46–131 residues, respectively. More recently, type III par systems employing tubulin-like GTPases and type IV systems that use a single protein with a predicted coiled-coil domain for partition, have been described (6–9).
Unlike the NTPases, the CBPs show little sequence homology even within a given family. Despite this, a crucial and shared function of these proteins is the formation of a so-called partition complex by binding to their centromere DNA, which typically consists of multiple tandem repeats. Structures have shown that despite a lack of sequence homology, both the small type II and Ib CBPs contain ribbon–helix–helix (RHH) DNA-binding folds (10–13). Less is known about the more complex and larger type Ia CBPs. Indeed, structural information is only available for two type Ia plasmid CBPs, P1 ParB and RP4 KorB, and their complexes with DNA (14–17). These data combined with biochemical studies indicate that these proteins have several domains, including a flexible N-terminal NTPase-binding domain, a central HTH-domain, which is the only conserved domain among these proteins and a C-terminal dimer-domain (3). Structures were determined separately for the KorB HTH-containing DNA-binding domain and the C-terminal dimer-domain, which has an SH3-like fold (15,16). The P1 centromere is arguably the most complicated centromere as in addition to two different repeat elements, called A- and B-boxes, it also contains a central IHF binding site (14,17). Structures of P1 ParB(142–333) bound to the minimal centromere, consisting of two A-boxes and one B-box, revealed that ParB recognizes the two repeat elements using two DNA-binding motifs. Its HTH-domain binds the A-box, while its C-terminal dimer-domain, which is connected to the HTH-domain by a short flexible linker, interacts with the B-box (5,14,17). Notably, the P1 ParB dimer-domain has a completely different structure than the KorB SH3-like C-terminal dimer-domain. Despite the insight revealed by these structures, the higher order structures that are formed by type I partition complexes are not known.
After formation of the partition complex, the next step is recruitment of the NTPase, which mediates the actual separation of replicated plasmids. Recent data indicates that both the ParM and Walker NTPases form polymers to drive partition (18–23). These polymers are metastable structures that are stabilized by their interactions with their partner CBPs when found in the context of the partition complex. Thus, the CBPs themselves are not alone sufficient to regulate and promote NTPase polymer dynamics and some aspect of the partition complex structure appears critical for this function. Recent insight into the role of the partition complex in type II partition comes from the pSK41 ParR-centromere structure. This structure revealed that the ParR-centromere partition complex adopts a specific superhelical conformation that is optimal for engulfing and stabilizing the ParM NTPase filaments (13). Whether the more complicated type Ia CBPs also form superhelices or other superstructures to stabilize their metastable NTPase polymers is not known. One of the goals of this study was to gain insight into this question by examining DNA binding by the type Ia CBP SopB, of the F plasmid par system. The sopABC system was one of the first partition cassettes identified and represents perhaps the most studied par system from a genetic, cellular and biochemical standpoint. However, to date no structures have been available for the Sop proteins.
The F par system consists of the CBP SopB, the NTPase SopA and the centromere sopC. Compared to P1 parS, sopC is a much simpler centromere as it consists of 12 43-bp repeat elements and one repeat can support F plasmid segregation (24,25). Within each DNA element is a short 16-bp inverted repeat, which is bound by a single SopB dimer. Recent studies, which have provided important insight into F plasmid partition, have shown that SopB displays an intriguing range of DNA-binding properties that are essential for the partition process (19,26–28). Specifically, SopB not only forms a SopB–sopC partition complex that recruits SopA but SopB also stabilizes SopA filaments in an unusual manner; by coating DNA and preventing non-specific SopA–DNA interactions that inhibit polymer formation (19). Indeed, SopA is localized to the nucleoid of the cell and its non-specific interactions with the nucleoid DNA prevent unwanted polymer formation until partition is desired (19,29,30). Remarkably, SopB is able to reverse this inhibition by spreading in trans or between different DNA molecules, creating a DNA-depleted zone near the partition complex allowing SopA polymers to stably form. To gain insight into the striking DNA binding properties exhibited by SopB and thus its myriad functions in partition, we biochemically and structurally characterized the interactions between SopB and DNA.
MATERIALS AND METHODS
Crystallization of I23 crystal forms of full-length SopB, SopB(155–323) and SopB(155–272) bound to the 18mer sopC repeat
An artificial sopB gene, encoding the full-length (FL) SopB protein, was codon optimized for Escherichia coli expression and purchased from Genscript Corporation, Piscatway, NJ, USA (www.genscript.com). The gene contained NdeI and XhoI sites. Following digestion with these enzymes, the gene was ligated into the pET15b vector predigested with NdeI and XhoI. BL21(DE3) cells were transformed with the expression vector. The expressed protein contains an N-terminal hexa-histidine tag (his-tag) and the protein was purified in one step using Ni–NTA chromatography. Purified FL SopB was mixed at a molar ratio of 1:1 with 18mer duplex DNA containing the SopB consensus binding site (CTGGGACCATGGTCCCAG) and crystallized by hanging drop vapor diffusion using 20% PEG 3000, 0.1 M phosphate/citrate pH 4.2, 0.2 M lithium sulfate. The crystals grew within 4–8 h and continued to grow for 2–3 days. When data were collected on crystals at room temperature or cryo-preserved with the cryo-solution, which consisted of the crystallization solution mixed with 30% glycerol, the best diffraction that was observed was 5.0 Å. However, the diffraction was dramatically improved to 3.35 Å by an annealing procedure involving successive re-immersion of a frozen crystal in the cryo-solution. The crystals take the cubic space group I23.
The coding regions for sopB(155–323) and sopB(155–272) were ligated into pET15b (using NdeI and XhoI sites) and the expressed protein purified in one step via Ni–NTA chromatography. Proteins were mixed at a molar ration of 1:1 protein dimer: DNA duplex, and multiple crystallization screens were carried out because the condition used to obtain crystals of the FL SopB-18mer complex did not result in crystals using the smaller SopB fragments. SopB(155–323)-18mer crystals were grown using 1.3 M ammonium sulfate, 0.1 M Tris pH 8.5, 25% glycerol. SopB(155–272)-18mer crystals were obtained with a crystallizaton solution consisting of 25.5% PEG 4000, 0.17 M lithium sulfate, 0.1 M Tris pH 8.0, 15% glycerol. SopB(155–323)-18mer crystals were cryo-preserved directly from the drop while SopB(155–272)-18mer crystals were cryo-preserved by dipping the crystals in solutions consisting of the crystallization reagent plus 20% additional glycerol. Both SopB(155–323)-18mer and SopB(155–272)-18mer crystals were cubic with space group I23 (Table 1).
Table 1.
Selected crystallographic data for SopB–DNA structures
| Complex | FL sopB-18 | SopB(155–323)-18 | SopB(155–272)-18 | SopB(155–272)-18 | SopB(155–272)-18 |
|---|---|---|---|---|---|
| Space group | I23 | I23 | I23 | P3121 | P21 |
| Cell dimensions (Å) | a = b = c = 203.7 | a = b = c = 200.8 | a = b = c = 200.5 | a = b = 119.8; c = 181.5 | a = 111.4; b = 47.1 c = 118.2; β = 115.9° |
| Resolution (Å) | 142.9–3.36 | 141.0–2.86 | 100.0–2.99 | 103.7–3.99 | 99.0–2.98 |
| Overall Rsym (%)a | 7.3 (55.6)b | 4.4 (37.1) | 5.1 (44.8) | 12.4 (48.0) | 6.7 (39.9) |
| Overall I/σ(I) | 6.8 (1.8) | 12.7 (2.2) | 10.6 (2.0) | 6.1 (1.9) | 7.3 (2.1) |
| No. of total reflections | 76 369 | 88 722 | 110 540 | 65 544 | 44 792 |
| No. of unique reflections | 19 203 | 30 569 | 26 809 | 20 020 | 22 277 |
| Refinement statistics | |||||
| Resolution (Å) | 142.9–3.35 | 141.0–2.86 | 100.0–2.99 | N/A | 99.0–2.98 |
| Rwork/Rfree(%)c | 29.9/33.3 | 24.5/26.9 | 24.2/26.6 | N/A | 21.2/26.4 |
| RMSD | |||||
| Bond angles (°) | 1.52 | 1.18 | 1.10 | N/A | 1.43 |
| Bond lengths (Å) | 0.010 | 0.006 | 0.006 | N/A | 0.010 |
| Ramachandran analysis | |||||
| Most favored (%/n) | 86.3/182 | 87.7/185 | 89.2/189 | N/A | 82.8/338 |
| Add. allowed (%/n) | 12.8/27 | 11.8/25 | 9.9/21 | N/A | 15.0/61 |
| Gen. allowed (%/n) | 0.5/1 | 0.5/1 | 0.9/2 | N/A | 2.0/8 |
| Disallowed (%/n) | 0.5/1 | 0.0/0 | 0.0/0 | N/A | 0.2/1 |
N/A, not applicable as due to the low resolution, the structure was not refined after the molecular replacement solution was obtained.
aRsym = ΣΣ|Ihkl − Ihkl(j)|/ΣIhkl, where Ihkl(j) is observed intensity and Ihkl is the final average value of intensity.
bValues in parentheses are for the highest resolution shell.
cRwork = Σ||Fobs| − |Fcalc||/Σ|Fobs| and Rfree = Σ||Fobs| − |Fcalc||/Σ|Fobs|; where all reflections belong to a test set of 5% randomly selected data.
Structure determination of FL SopB-18mer, SopB(155–323)-18mer and SopB(155–272)-18mer I23 crystal forms
The structure of the FLSopB-18mer complex was solved to 3.5 Å by multiple wavelength anomalous diffraction (MAD) phasing using a crystal in which thymines were substituted with 5-bromouracil (CXGGGACCAXGGTCCCAG, where X corresponds to the locations of the 5-bromouracil substitutions). MAD data were collected at beamline 8.2.1 and processed with MOSFLM. The bromine sites were located using SOLVE (31). Model building was carried out using O and the structure was minimally refined using CNS (32,33). The SopB-18mer structure contains two SopB subunits (residues 157–271 of one subunit and 157– 270 of the second subunit) and all the nucleotides in the 18mer DNA duplex and has Rwork/Rfree values of 29.9%/33.3% to 3.35 Å resolution.
MAD data were also collected for a SopB(155–323)-18mer crystal containing the same 5-bromouracil substitutions as for the FL SopB-18mer crystal. The data were collected to 3.3 Å resolution and the sites were again located using SOLVE (31). The constructed model was minimally refined using CNS and used as a starting model for refinement with the high resolution 2.86 Å data. The final refined SopB(155–323)-18mer structure contains two SopB subunits (residues 157–271 of one subunit and 157–270 of the second subunit), all the nucleotides in the 18mer DNA duplex and 10 sulfate ions and has Rwork/Rfree values of 24.5%/26.9% to 2.86 Å resolution. The refined SopB(155–323)-18mer structure was solved by molecular replacement (MR) and the structure was refined to 2.99 Å resolution using CNS (32). The model includes two SopB subunits (residues 157–271 of one subunit and 157–270 of the second subunit), all the nucleotides in the 18mer DNA duplex and nine sulfate ions. The Rwork/Rfree values are 24.2%/26.6%.
Crystallization and structure determination of P3121 SopB(155–272)-18mer and P21 SopB(155–272)-18mer complexes
A second crystal form of SopB(155–272)-18mer grew in the same drops as the I23 form, took the trigonal space group P3121 and diffracted to 3.9 Å. A data set was collected and used to solve the structure by MR using the SopB(155–272) dimer as a search model. The structure contained a complicated asymmetric unit (ASU) consisting of three SopB dimers and four 18mer DNA complexes. Due to low resolution, the structure was not refined. A third crystal form was obtained using SopB(155–272) in which the his-tag was removed. The his-tag free protein in complex with the 18mer produced a monoclinic P21 crystal form using 30% PEG 400, 0.1 M HEPES pH 7.5, 0.2 CaCl2. The structure was solved by MR using the refined SopB(155–272)-18mer complex as a search model. The ASU consists of two DNA duplexes, one SopB dimer and two SopB monomers. The final model includes residues 157–268, 157–267, 157–267 and 157–266 of the four subunits in the ASU and all the nucleotides of each of the 18-bp duplexes as well as 16 solvent molecules. The final model has Rwork/Rfree values of 21.2%/26.4% to 2.98 Å resolution.
Crystallization and structure determination of SopB(275–323)
The region encoding sopB(275–323) was ligated into pET15b as for the gene expressing the FL protein, such that the N-terminal his-tag was expressed for ease of purification. After purification, the his-tag was cleaved from the protein with thrombin. After cleavage the protein was further purified via size exclusion chromatography and the protein was then concentrated to 30 mg/ml for crystallization. Crystals were grown using 30% PEG 4000, 0.2 M sodium acetate, 0.1 M Tris pH 8.5 as a crystallization solution. Native data and derivative data were collected in house using a Rigaku FR-E SuperBright microfocus rotating anode (Rigaku USA) equipped with an R-Axis HTC and processed with CrystalClear. An initial native data set and two derivatives (mercuric chloride and potassium hexachloroplatinate) were collected at room temperature. Heavy atom sites were located using SOLVE (31). The structure was traced using O and refined in CNS (32–33). A high-resolution data set was collected at beamline 8.2.1 and used for final refinement (Table 2).
Table 2.
Selected crystallographic data for SopB(275–323)
| Crystal | Native | HgCl2 | KAuCl4 |
|---|---|---|---|
| Space group | C2 | C2 | C2 |
| Cell dimensions (Å) | a = 89.7, b = 46.2, c = 36.4 | a = 89.3, b = 46.1, c = 36.4 | a = 89.1, b = 46.2, c = 36.5 |
| β = 100.8° | β = 100.3° | β = 100.4° | |
| Resolution (Å) | 30.73–1.99 | 28.11–2.40 | 35.89–2.40 |
| Overall Rsym (%)a | 4.1 (14.3)b | 9.1 (29.7) | 5.2 (11.2) |
| Overall I/σ(I) | 12.1 (4.8) | 9.7 (2.1) | 8.9 (3.0) |
| No. of total reflections | 19 383 | 9852 | 7018 |
| No. of unique reflections | 9905 | 5425 | 4968 |
| Refinement statistics | |||
| Space group | C2 | ||
| Cell constants (Å) | a = 89.1, b = 45.7, c = 36.5 | ||
| β = 100.8° | |||
| Resolution (Å) | 43.85–1.58 | ||
| Overall Rsym (%)a | 5.4 (30.2)b | ||
| Overall I/σ(I) | 10.1 (2.1) | ||
| No. of total reflections | 29 836 | ||
| No. of unique reflections | 18 124 | ||
| Rwork/Rfree (%)c | 19.8/23.1 | ||
| RMSD | |||
| Bond angles (°) | 1.61 | ||
| Bond lengths (Å) | 0.012 | ||
| Ramachandran analysis | |||
| Most favored (%/n) | 91.9/113 | ||
| Add. allowed (%/n) | 8.1/10 | ||
| Gen. allowed (%/n) | 0/0 | ||
| Disallowed (%/n) | 0/0 |
aRsym = ΣΣ|Ihkl − Ihkl(j)|/ ΣIhkl, where Ihkl(j) is observed intensity and Ihkl is the final average value of intensity.
bValues in parentheses are for the highest resolution shell.
cRwork = Σ||Fobs| − |Fcalc||/Σ|Fobs| and Rfree = Σ||Fobs| − |Fcalc||/Σ|Fobs|; where all reflections belong to a test set of 5% randomly selected data.
Fluorescence polarization assays
Fluorescence polarization (FP) assays were performed using a Panvera Beacon Fluorescence Polarization system (34). The 43mer (top strand:GGTCTGATTATTAGTCTGGGACCACGGTCCCACTCGTATCGTC) and 18mer (CTGGGACCATGGTCCCAG) used in the assays were 5′-fluorescein labeled. For each assay, increasing concentrations of SopB were titrated into the binding mixture containing 2 nM DNA in 20 mM Tris pH 7.5, 100 mM NaCl. The excitation and emission wavelengths were 490 and 530 nm, respectively. All data were processed in Kaleidagraph and fit with the equation P = {(Pbound – Pfree)[Protein]/(Kd+[Protein])}+Pfree, where P is the polarization magnitude at a given protein concentration, Pfree is the initial polarization of the free oligonucleotide and Pbound is the maximum polarization when the oligonucleotide is saturated by SopB. Non-linear least squares analysis was applied to determine Pbound and Kd.
Circular dichroism (CD) spectroscopy experiments
CD experiments were carried out on FL SopB in 50 mM HEPES pH 7.5, 100 mM NaCl at a concentration of 1.2 mg/ml and SopB(155–272) in the same buffer at 0.4 mg/ml on a JASCO 700 CD Spectrophotomer. The CD spectra were deconvoluted for secondary-structure content using the K2D2 program (35).
RESULTS AND DISCUSSION
Structure determination of SopB-centromere complexes
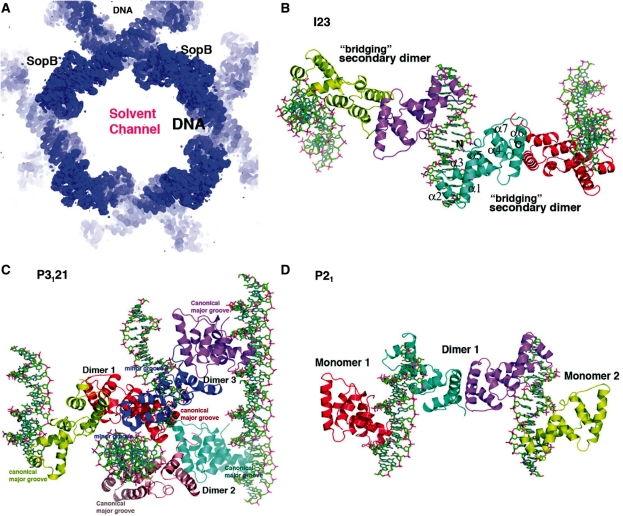
Crystals of the 323 residue FL SopB were obtained in complex with an 18 bp sopC centromere repeat, CTGGGACCATGGTCCCAG. The crystals, which harbor one SopB dimer and one DNA duplex in the crystallographic ASU, contained a high solvent content of ∼73%. The structure was solved to 3.35 Å resolution by MAD (Materials and Methods). Despite the relatively weak diffraction, solvent leveling produced a map of excellent quality permitting the structure to be traced (Figure 1A and B). All nucleotides of the 18-bp DNA duplex could be built. However, density was only observed for SopB residues 157–271 from one subunit and 157–270 of the second subunit. These residues contain all the determinants required for sequence specific binding to the sopC consensus site (36). The absence of density for the SopB N- and C-terminal regions suggested that either these regions were highly flexible or were proteolysed during crystallization. Consistent with the former possibility, the first and last residues observed, 157 and 271, each face into large solvent channels, which could easily accommodate the disordered or flexibly attached domains (Figure 1A). This was supported by SDS–PAGE analysis of washed crystals, which revealed that the FL protein was indeed present in the crystals (Supplementary Figure S1).
Figure 1.
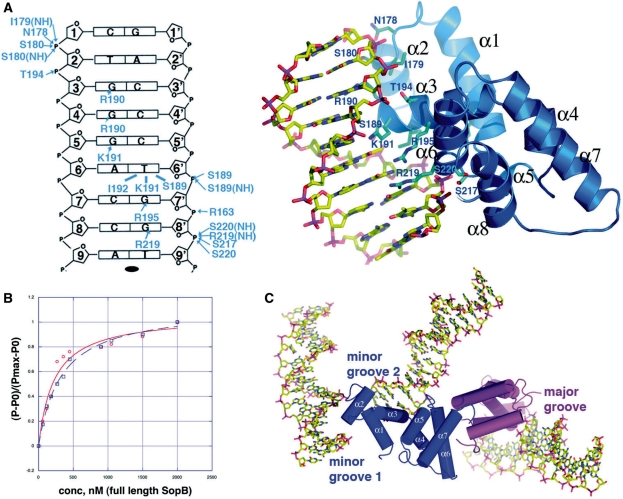
Crystal structures of SopB-18mer complexes. (A) A section of the experimental MAD electron density map (shown as a blue mesh) for the FL SopB-18mer complex, contoured at 1.5σ and calculated to 3.5 Å resolution. Labeled are the location of the pseudocontinuous DNA and one of the SopB intermolecular dimers, which bridge between DNA duplexes to form the crystal lattice. Also labeled is one of the large solvent channels. (B) Ribbon diagram of the I23 SopB-18mer complex. The crystallographic asymmetric unit (ASU) consists of two subunits (cyan and magenta) and one 18-mer DNA duplex. Shown also are subunits involved in secondary dimer/bridging interactions, generated in the crystals. For the cyan subunit, the secondary structural elements are labeled and the first and last residues observed in the structure are labeled N and C, respectively. This figure (C and D) and Figures 2A–D, 3, 4, 5A and C, and 6A and B were made with PyMOL (55). (C) Ribbon diagram of the P3121 SopB(155–272)-18mer complex. The molecules in the ASU are all shown consisting of three SopB secondary dimers and four 18mer duplexes, which all pack pseudocontinuously in the crystal. The specific HTH-major groove interacting subunits are labeled canonical major groove and the one subunit that interacts non-specifically with two minor grooves (colored blue) is also shown and its minor groove contacts labeled. (D) Ribbon diagram of the P21 SopB(155–272)-18mer complex. The ASU consists of one secondary dimer and two SopB subunits and two 18-mer DNA duplexes.
The structure indicates that SopB consists of three main regions: 1–155, 155–272 and 272–323 that are flexibly linked. Support for this supposition comes from studies showing that the N-terminal region of SopB, which contains the SopA interacting region, is highly flexible and that SopB residues 272–280 can be cleaved by multiple proteases (36,37). It appears that the reason only the SopB DNA-binding domain is visible in the crystal is because it is bound to the DNA, which is central in the formation of the crystal lattice via its pseudocontinuous packing. Indeed, examination of the crystal packing shows that the pseudocontinuously packed DNA constructs one direction of the lattice while the SopB DNA-binding domain assembles another direction via its contacts between DNA. These protein and DNA arrays are separated by enormous solvent channels (Figure 1A). The presence of a significant disordered, and thus invisible, portion of a protein within a crystal is not unprecedented. For instance, when the structure of the 780 residue FL zebra fish β-catenin was solved, 125 residues from its N-terminus and 98 residues from its C-terminus were not observed in electron density maps (38). The authors found that the N- and C-terminal domains of β-catenin are flexibly attached to and do not interact with the armadillo repeat domain, which was the only visible part of the structure. Similarly, when the structure of the 150 residue intact Max transcription factor was solved bound to DNA, its C-terminal region, accounting for 45% of the total protein, was disordered (39). Thus, a protein with a large number of disordered residues or flexibly attached domains can still form crystals if there are adequate solvent channels to accommodate them. However, only the regions involved in the formation of stable lattice contacts will be visible.
The fact that the FL SopB-18mer crystals contained a large percentage of disordered macromolecule led us to postulate that the utilization of shorter SopB fragments might result in better diffracting crystals. Thus, SopB(155–323) and SopB(155–272) were produced and used in crystallization screens with the 18-mer sopC palindrome. Cubic crystals, similar to the FL SopB-18mer crystals, were obtained with both fragments. Interestingly, however, different conditions were required to produce each of the crystals and the crystals obtained with the truncated proteins diffracted better than those grown with FL SopB (‘Materials and Methods’ section). The SopB(155–323)-18mer structure was solved by MAD and like the FL SopB-18mer complex, only SopB residues 157–270 were visible. SDS–PAGE analyses, however, confirmed that the entire SopB(157–323) fragment was present in the crystals (Supplementary Figure S1). The SopB(155–272)-18mer structure was subsequently solved by molecular replacement (MR). The final structures of the FL SopB-18mer, SopB(155–323)-18mer and SopB(155–272)-18mer complexes have Rwork/Rfree values of 29.9%/33.3%, 24.5%/26.9% and 24.2%/26.6% to 3.35, 2.86 and 2.99 Å resolution, respectively (Material and Methods).
P3121 SopB(155–272)-18mer and P21 SopB(155–272)-18mer structures
Two additional SopB–DNA crystal forms were obtained of the SopB(155–272)-18mer complex. One grew in the same drops as the cubic crystals and were trigonal, space group P3121 and diffracted to 3.9 Å resolution. The other crystal form, grown under different conditions, was monoclinic, P21, and required removal of the N-terminal his-tag from SopB(155–272). The structures were both solved by MR and both provide multiple views of the SopB–DNA complex; the P3121 crystals contain three SopB(155–272) dimers and four 18-mer DNA duplexes in the ASU while the P21 crystals have one SopB(155–272) dimer, two SopB(155–272) monomers and two 18-mer DNA duplexes in the ASU (Figure 1C and D).
Overall structure of SopB-18mer complexes
The region of SopB that is visible in all the SopB-18mer structures (residues 155–270) is essentially identical between the structures [root mean squared deviations (RMSDs) for comparisons of corresponding Cα atoms range from 0.3 to 0.8 Å]. The region is comprised of seven helices (α1; residues 159–172, α2; 179–186, α3; 190–201, α4; 204–209, α5; 218–227, α6; 232–248, α7; 254–265) and can be divided into two functional regions; an N-terminal HTH-containing DNA-binding element (residues 155–227) and a C-terminal motif consisting of α6 and α7, which, dimerize with α6′ and α7′ (where ′ indicates other subunit of a ‘dimer’) from a SopB subunit bound on a different DNA duplex. Thus, strikingly these ‘dimer’ contacts link together or bridge SopB molecules bound to different DNA molecules (Figure 1B). Database searches revealed that SopB residues 155–272 show structural similarity to the DNA-binding domains of P1 ParB and KorB (14,16–17). SopB(155–272) can be superimposed on the DNA-binding domains of KorB and P1 ParB with RMSDs of 3.0 and 3.1 Å for 93 and 98 corresponding Cα atoms, respectively (Supplementary Figure S2). Notably, this structural homology includes α6 and α7 of SopB. However, unlike SopB, the corresponding regions in KorB and ParB do not participate in oligomerization (Supplementary Figure S2). Helix 6 in KorB is too short to be involved in dimer formation while helix 6 in ParB is bent and not properly oriented to make the type of dimer contacts observed in the SopB structures.
The fact that the N- and C-terminal regions are not visible in the FL SopB-18mer structure indicates that these three principal regions of SopB are flexibly attached. To further test this theory, we carried out CD studies on SopB(155–272) and FL SopB. The SopB(155–272) fragment was found to contain 41% helix by CD, which compares favorably to the 43%, determined from the structure (35). If the DNA-binding domain is the only folded region of SopB then the FL protein should contain 17% helix and no β-strands. However, our CD analysis showed that it contained 30% helix and 13% β-strand, indicating that the N- and/or C-terminal regions contain folded structures or domains and therefore, must be flexibly attached to the central DNA-binding domain.
Structure determination of the C-terminal SopB(275–323) dimerization domain
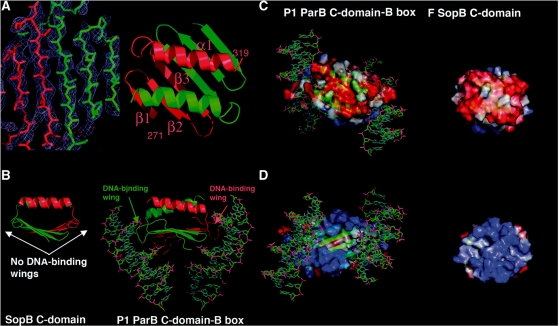
Previous studies suggest that SopB residues 275–323 are involved in dimer formation and therefore must form a folded domain, consistent with our CD studies (36). Thus, we next produced and crystallized SopB(275–323). The structure was solved by multiple isomorphous replacement (MIR) methods (Figure 2A). The structure contains three subunits in the ASU (one dimer and one subunit from which the crystal symmetry generates the dimer) and has been refined to an Rwork/Rfree of 19.8%/23.1% to 1.58 Å resolution and includes residues 271–319, 272–319 and 275–319 of the three subunits and 156 water molecules. The structure revealed that, indeed, residues 275–323 form a highly intertwined dimer composed of 3 β-strands and 1 α-helix with topology (β1; 276–281, β2; 284–289, β3; 292–298 α1; 306–318) (Figure 2A–D). The dimer is stabilized almost entirely by backbone, β-strand hydrogen bonds. The interface of the dimer buries an extensive 5990 Å2 of protein surface from solvent, consistent with it forming the primary dimerization domain of SopB, which anchors the SopB subunits on a palindromic DNA site (see Figure 3). That the C-domain is a physiologically relevant oligomer is also supported by the finding that the identical dimer is observed twice in the crystal (the dimers overlay with an RMSD of 0.6 Å) and size exclusion chromatography data providing a MW of 13 kDa for SopB(275–323) [compared to a calculated MW of 11.2 kDa for a SopB(275–323) dimer].
Figure 2.
Structure of SopB(275–323) dimer-domain. (A) Left, a section of the experimental MIR electron density map for the SopB(275–323) structure (blue mesh) contoured at 1σ. Right, ribbon diagram of the SopB(275–323) structure with one subunit colored red and labeled, and the other subunit colored green. (B) Comparison of the P1 ParB dimer-domain–DNA complex (right) with the corresponding SopB dimer-domain (left). Note that although the overall structure and topology are similar, P1 ParB has extended loops between its β1–β2 and β2–β3 units that are responsible for DNA binding that are not present in SopB. (C) Comparison of the electrostatic surfaces of the P1 ParB and SopB. Shown is the helical containing face of each dimer-domain, which is strongly electronegative (red) in both structures. (D) Electrostatic surface of the face opposite to that shown in (C). This face, which is involved in DNA-binding in the P1 ParB protein, is electropositive (blue) in each structure.
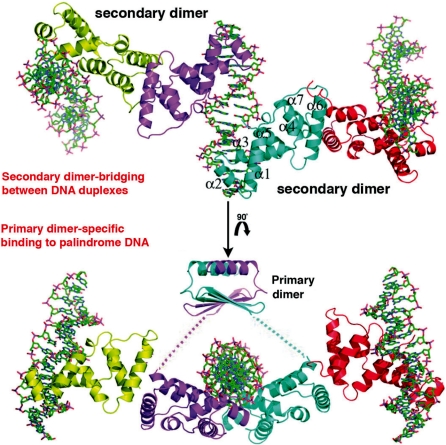
Figure 3.
SopB primary and secondary (bridging) dimers. Relationship of the SopB primary and secondary dimers. The structure of the SopB primary dimer bound to the palindromic DNA site was produced by combining the DNA-binding and dimer-domains (the flexible linkage is indicated by dashed lines). Above is shown the secondary dimer contacts that permit SopB to bridge or spread between multiple, adjacent DNA sites (generated by a 90° rotation).
In the SopB(275–323) dimer, the β3 from each subunit combine to form a continuous six stranded β-sheet and the helices interact to form an antiparallel coiled-coil. Although this structure shows no homology to the SH3-like dimer-domain from the KorB protein, it is similar in overall topology to the dimer-domain of P1 ParB, despite the lack of sequence homology between the two domains (Figure 2B–D). Specifically, both proteins have the same (β3α)2 topology, which has not been observed in other proteins. This was surprising because the dimer-domain of P1 ParB functions as a DNA-binding motif that specifically recognizes B-box DNA, while the SopB C-terminal dimer-domain displays no DNA-binding capacity (14,17). Interestingly, the P1 ParB and SopB C-domains are not only topologically similar but they share analogous electrostatic properties whereby one face of each domain is electronegative and the other, strongly electropositive (Figure 2C and D). Although the electropositive face of P1 ParB is involved in DNA-binding, all the specificity determining contacts from the ParB dimer-domain to the B-box are provided by residues on its extended loop between β1–β2 and, to a lesser extent, the loop between β2 and β3 (14,17). These loops are absent in the SopB dimer-domain, explaining why it does not participate in DNA binding (Figure 2B). The correlation between the structures, topology and electrostatics of the P1 ParB and SopB dimer-domains suggest that they may be evolutionarily related and that either P1 ParB gained DNA binding potential by acquiring the critical DNA-binding wings or SopB lost this potential by deletion of these loops.
SopB α6/α7 dimerization and possible functions in partition
Our structures reveal that the SopB C-domain composed of residues 275–323 is the primary dimerization motif of SopB. However, previous homologue specificity scanning (HSS) studies, which used the high degree of sequence similarity between the F and N15 Sop proteins (49 and 78% sequence identities between the SopB and SopA proteins, respectively), to localize functional regions of the Sop proteins mapped the self-association domain of SopB to the C-terminal 79 residues, 245–323 (36,37,40). Interestingly, this region includes most of α6 and all of α7, which our SopB–DNA structures show forms a ‘secondary’ dimer interaction (Figure 3). Thus, the data suggest that both oligomer interactions might be important in SopB function. That the secondary dimer is functionally relevant is supported by the significant buried surface area, 1400 Å2, in this interface, which is on par with interfaces in oligomeric proteins (41,42). In addition, our gel filtration studies demonstrated that the SopB(155–272) fragment is almost entirely (∼80%) dimeric. Thus, the data indicate that SopB contains multiple folded domains including a central DNA binding domain, which also functions in secondary dimerization (residues 155–272) and a C-terminal, primary dimer-domain from residues 273 to 323. Whether residues 1–154 fold into one or several domains remains to be determined (Supplementary Figure S3A–C).
SopB secondary dimer contacts
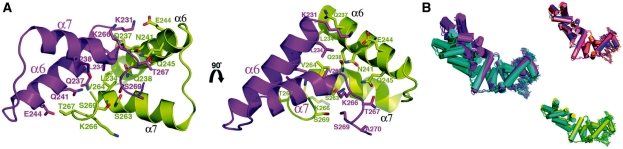
The α6/α7 interface requires the asymmetric arrangement of α6/α7 with α6′/α7′, unlike typical dimers in which contacts from each subunit to the other are identical (Figure 4A and B). This is revealed by superimposition of each subunit of all the secondary dimers observed in our three different SopB-DNA crystal forms (see Figures 1B–D and 4B). These overlays show that sets of contacts made by one subunit are different from those made by the other subunit. However, this specific asymmetric dimer arrangement is strikingly conserved in all dimers (as revealed by these overlays) suggesting that the two specific types of interactions permit optimal interface formation. While the contacts involved in the formation of the SopB secondary dimer interface are similar to those in other oligomer interfaces in that both polar and non-polar contacts are utilized for its formation, a distinctive feature of the SopB interface is the abundance of Gln and Asn residues. These residues play key roles in dimer stabilization by forming hydrogen bonds with the protein backbone atoms as well as polar side chains (Figure 4A). Residues involved in these interactions include Qln237, Qln238, Asn241 and Qln245. Hydrophobic contacts are also important and those between Leu234, Leu234′, Val264 and Val264′ serve to tie together one end of the dimer. Additional polar contacts include hydrogen bonds between the side chains of Glu244 and Thr267′, the side chains of Ser269 and Ser269′, and a salt bridge between Glu244 and Lys231′ (Figure 4A). Gln, Asn and Glu residues, which are abundant in the SopB secondary interface, are all known to interact with and coordinate calcium (43). Gel filtration studies carried out on SopB(155–272) in the presence of 50 mM CaCl2 resulted in a reduction of the dimer fraction from 80 to 40%, further supporting the secondary dimer formation observed in all the crystal forms. Moreover, these findings also explain why two out of the four subunits in the SopB(155–272)-18mer P21 crystal structure, which was obtained using 200 mM CaCl2, were monomeric. By contrast, all eight subunits in the I23 and P3121 crystals formed an α6/α7–α6′/α7′ secondary dimer with an adjacent DNA-bound SopB subunit (Figure 1B).
Figure 4.
The SopB α6–α7 secondary dimer interactions. (A) Ribbon diagram showing the residues that are involved in the formation of the secondary dimer interaction. Two views are included that are related by a ∼90° rotation. Residues that contribute to the interface are shown as sticks and labeled. Also labeled are α6 and α7. (B) Superimposition of subunits of all the secondary dimers reveals that the dimerization is not symmetric but mediated by one subunit making one set of contacts and the other making a different set of contacts. This asymmetric arrangement is observed in all dimers.
The α6/α7 dimer interaction is particularly intriguing as it provides a molecular mechanism to explain the myriad partition functions carried out by SopB including plasmid pairing, plasmid multimerization and, importantly, in trans spreading between DNA elements (19,44,45). Indeed, pairing could be mediated by a secondary dimer involving SopB molecules bound on adjacent plasmids. Studies in the Lane laboratory have shown that the SopB spreading function is essential for partition because it counteracts the inhibition of SopA polymerization. In particular, SopA localizes to the nucleoid and in the presence of ATP, binds to DNA in a non-sequence specific manner. When this occurs SopA polymerization is thwarted. SopB spreading onto nucleoid DNA, however, creates a DNA depleted or masked zone, which permits stable SopA polymer growth. The secondary dimerization allows SopB to interact with several DNA sites, in trans, both specifically and non-specifically (see below), thus suggesting an explanation for this unusual spreading phenomenon.
Specific and non-specific SopB–DNA complexes
Both specific and non-specific DNA binding by SopB is important for its partition functions. Its specific DNA-binding function is necessary for it to interact with each of its 12 centromere repeats to form a partition complex, which recruits SopA. Its non-sequence specific DNA-binding activity is needed to allow it to spread onto and around its centromere site to mask DNA and prevent SopA depolymerization. The multiple views of SopB-18mer complexes present in the combined crystal structures reveal how it can accomplish both forms of DNA binding (Figure 5A–C). The structures show that SopB achieves high specificity in binding its GC-rich consensus site primarily through the utilization of multiple arginine residues that contact guanine nucleobases. The majority of these base interactions are provided by residues from the recognition helix, α3, of the HTH. At the 5′ most end of each half site, Arg190 reads guanine 3 and makes weak interactions with guanine 4. Guanine 5 is contacted by Lys191, which also makes hydrophobic contacts to thymine 6′ (Figure 5A). In addition to Lys191, the side chain of Ile192 and the Cβ of Ser189 also contact thymine 6′ and Arg195 reads guanine 7′ (Figure 5A). One base specific contact, provided by Arg219 to guanine 8′, is made by a residue outside the HTH. The two central nucleotides, adenine 9 and thymine 9′, are not contacted by SopB explaining their lack of conservation in sopC centromere elements. Thus, these interactions effectively read every nucleotide of the SopB consensus sequence, G3G4G5A6C7C8a9t9′G8′G7′T6′C5′C4′C3′ (the consensus nucleobases are shown in capital letters). In addition to the large number of base specific contacts, there are a slew of interactions from SopB residues to phosphate groups at the 5′-end of each half site (Figure 5A). Importantly, these contacts show that while SopB specifies or reads only 14 of the 18 nt in its consensus site, it requires 18-bp for high affinity binding as the extra up- and down-stream nucleotides at the 5′-end of each half site provide phosphate contacts that are crucial for proper docking of the HTH element on to the DNA (Figure 5A and B). At one 5′-end, the contacts include hydrogen bonds and electrostatic interactions from α1 residue Arg163, α3 residues Ser189, Ser217, Arg219 and Ser220. At the other 5′-end of the half site, phosphate contacts are provided from α2 residues Asn178, Ile(NH)179 and Ser180, and α3 residue Thr194.
Figure 5.
SopB–DNA interactions. (A) Left, schematic representation of SopB–DNA interactions. Only one half site of the 18-mer duplex is shown as the identical contacts are made to each half site. The strands are labeled 1–9 and 1′–9′ (where ′ indicates other strand of the duplex). Bases are represented as rectangles and labeled according to sequence. The ribose groups are shown as pentagons. Hydrophobic contacts are indicated by lines and hydrogen bonds, by arrows. Right, close up of the SopB–DNA specific major groove interactions that are made to each half site and indicated schematically. Interacting residues are shown as blue sticks and the secondary-structural elements are labeled. (B) Fluorescence polarization DNA-binding isotherms comparing SopB binding to the 43- and 18-mer centromere sites. Each data set was normalized, and normalized polarizations were plotted along the y-axis against the protein concentrations, which are plotted along the x-axis. (C) Close up of the triple bridging interaction that is comprised of specific and non-specific SopB–DNA found in the P3121 crystal form. Specifically, this dimer is the one that is also colored blue and magenta in Figure 1C. The blue subunit makes non-specific minor groove contacts to two DNA duplexes while the magenta subunit makes the canonical major groove contacts indicated in Figure 5A and found in all the subunits of the crystals except the P3121 blue subunit.
The position of the F SopB HTH within the protein, i.e. residues 179–202, is consistent with HSS studies, which demonstrated that residues 177–198 and 179–200 of F SopB and N15 SopB, respectively, contain all the determinants required for sequence specific DNA binding (36). Indeed, swapping these residues from F SopB to N15 SopB and vice versa resulted in switching the DNA binding specificities of the proteins despite the fact that N15 SopB and F SopB recognize centromeres that differ in only one nucleotide (at position 4 and 4′, per half site) (Supplementary Figure S3C). The F SopB–DNA structure shows that the guanines at these positions, guanine 4, are weakly contacted by Arg190 (∼4.2 Å). This arginine is conserved in the N15 SopB HTH. However, residues adjacent to Arg190 on α3 are not conserved between F and N15 SopB and modeling indicates that these different residues could make distinct contacts to nucleotide 4 in the N15 SopB–DNA complex (Supplementary Figure S3C). However, the precise contacts cannot be predicted without structural information. The remainder of the N15 SopB–DNA contacts can be determined from the F SopB–DNA structure as residues Arg190, Arg195 and Arg219, which contact guanine 3, guanine 7′ and guanine 8′, are conserved in both SopB proteins.
SopB interacts directly with 18-bp of its centromere repeat. However, the FL centromere repeat consists of 43-bp, suggesting that the extra nucleotides between the 18-bp elements might play some role in DNA binding. However, FP studies, which revealed Kds of 223 ± 44 and 324 ± 43 nM for FL SopB binding the 18 and 43mer, respectively, demonstrated that the intervening nucleotides do not contribute to the high affinity binding of SopB to its centromere DNA sites [notably, higher concentrations of SopB are required to observe non-specific binding (27)] (Figure 5B). However, it is possible that the intervening sequences not bound by SopB might be intrinsically bent. However, these DNA sequences are not conserved and do not contain nucleotide stretches that have strong propensities for deformation or bending. Therefore, the role of these intervening sequences remains to be determined.
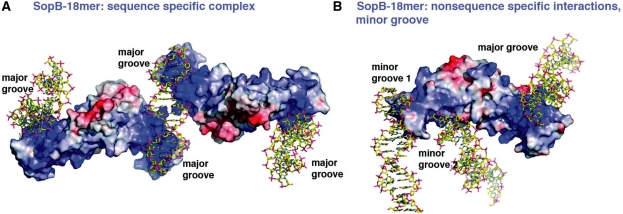
SopB-minor groove complex: a mechanism for non-specific DNA spreading
The abundance of basic residues in the SopB DNA-binding unit suggests that it might mediate non-specific interactions with DNA through the formation of phosphate contacts (Figure 6A and B). In fact, one secondary SopB dimer in the P3121 SopB(155–272)-18mer structure was captured binding both non-specifically and specifically to DNA (Figures 5C and 6B). Remarkably, this interaction links or bridges together three different DNA duplexes; one SopB subunit bridges between two DNA duplexes non-specifically while the HTH of its ‘dimer’ partner makes sequence specific contacts with the major groove of the third DNA duplex (Figures 5C and 6B). In the SopB-non-specific complexes, one SopB subunit engages residues from α1 to bind the minor groove of one DNA duplex and residues from α1, α3 and α6 to bind the minor groove of a second, adjacent DNA duplex. Several basic residues are involved in these interactions. In the SopB-minor groove 1 interaction these residues are Arg163, Arg166 and Arg170. Contacts to minor groove 2 are much more extensive and involve residues Ser169, Gln165, Gln172, α3 amide nitrogens, Lys201, Lys204, Lys231 and Lys236. Thus, the structure shows that SopB utilizes multiple basic and polar residues and its electropositive surface to bind DNA non-sequence specifically. Importantly, this structure suggests that the secondary dimerization motif plays a vital role in the formation of non-specific SopB–DNA complexes and non-specific DNA spreading as it allows the SopB to link or connect, via minor and major grooves contacts, multiple DNA duplexes.
Figure 6.
Electrostatic surface representations of SopB-18mer specific and non-specific complexes. (A) SopB dimers are shown as surface representations with electropositive regions colored blue and electronegative regions red. The DNA is shown as sticks. Shown is the specific complex in which the DNA major grooves are contacted by the basic HTH motifs. (B) Electrostatic surface representation of the non-specific SopB–DNA complex. In this complex, the SopB secondary dimer bridges three different DNA duplexes by making non-specific contacts to the minor grooves of two DNA duplexes (from one subunit) and specific contacts to the major groove by the HTH of the second subunit.
The conformation of the SopB bound DNA
The DNA sites bound by SopB are essentially B-DNA in conformation with an average twist of 33.7 Å and rise of 3.4 Å compared to 34.3 and 3.4 Å, respectively, for B-DNA. Moreover, contrary to previous suggestions that SopB may distort DNA, the measured bend angle in the various SopB bound DNA sites range from only 6–8° (Figure 5A) (46). The issue of higher order superstructure formation by SopB–sopC complexes has been controversial. Studies carried out >10 year ago suggested that SopB–sopC complexes form wrapped structures. This conclusion was based largely on the supercoiling deficit caused by SopB–sopC complex formation (47–49). However, more recent experiments from the Lane laboratory indicated that SopB does not cause DNA distortion and explained the supercoiling deficit by SopB DNA coating (27). Our finding that SopB does not bend or unwind DNA is consistent with the more recent data and suggests the possibility that the partition complex formed when multiple SopB proteins bind the sopC centromere may be an extended, not highly wrapped, complex. Moreover, our data revealing the mechanism of non-specific DNA binding by SopB as well as its ability to form a secondary dimer that bridges between DNA, unveils a mechanism by which SopB is able to coat DNA, thus supporting DNA coating as an explanation for the previously observed supercoiling deficit.
Model for F plasmid partition
Our combined data provide key insight into the multiple and complex roles played by SopB in the F plasmid segregation process. With the assertion that SopB does not distort its centromere DNA, we modeled the F plasmid partition complex as an extended complex (Supplementary Figure S4). Although the in vivo DNA centromere site would clearly exhibit some degree of flexibility, the model indicates that a more extended and unwrapped partition complex model would align the N-terminal SopA interacting regions of the SopB molecules primarily on one face, which is 90° away from the α6/α7 dimerization regions. This would therefore position the secondary dimer interaction motifs favorably to interact with SopB bound on adjacent DNA. The primary role of the partition complex is to recruit the SopA NTPase. SopA polymers have been visualized by electron microscopy and found to form bundles of large polymers up to 40 nm wide (19). The capture of these large SopA polymers would be best executed by an extended partition complex structure that casts out a large number of SopB N-terminal region ‘nets’ such as suggested by our structural data. Indeed, because the role of partition complexes is to recruit their partner NTPase polymers, it seems logical that the nature, structure and size of partition complexes are designed to provide optimal interaction with the NTPase polymer. Consistent with this idea, data from type Ib partition systems hint that they might similarly employ large NTPase polymers to interact with extended partition complexes. Specifically, the ParF NTPase from the E. coli TP228 type Ib partition system forms very large polymers (>70 nm) and while the nature of the TP228 partition complex is not known, studies on the type Ib Streptococcus pyogenes pSM19035 partition complex, formed by ω-DNA interactions, indicate that it is extended in nature (11,18). By contrast, type II partition systems have been shown to form highly wrapped ParR-centromere superhelical partition complexes with central pore regions 18 nm in diameter. The formation of these wrapped complexes lead to the creation, within the pore, of a high concentration of NTPase-interacting domains of ParR (13). The small pore of the ParR–parC superhelix is an optimal size to engulf and stabilize the ends of the smaller 6 nm actin-like ParM NTPase filaments (13,50–54).
Thus, we propose a model for F plasmid partition in which SopB first binds to the sopC centromere to form a more extended partition complex, which casts out many SopA-interacting nets primarily on one face, allowing it to effectively capture large SopA polymers. However, SopA polymers must be stably formed to drive plasmid separation and this is not possible due to the inhibitory nature of SopA–DNA contacts. SopB reverses this inhibition, stabilizing SopA polymer formation, by coating DNA near its centromere site, both in cis and in trans. Our structures suggest a novel mechanism for this coating whereby SopB α6/α7 dimerization bridges between two or more DNA sites. We show that SopB can bind non-specifically to the DNA minor groove revealing how non-specific spreading is mediated. Once the DNA is adequately coated or masked in this manner by SopB, nucleated SopA polymers are stabilized and can grow, thus driving plasmid segregation.
ACCESSION NUMBERS
Coordinates for the I23 SopB(155–323)-18mer, SopB(155–272)-18mer, P21 SopB(155–272)-18mer complexes and SopB(275–323) structures have been deposited with the Protein Data Bank under the Accession codes 3MKY, 3MKW, 3MKZ and 3KZ5 respectively.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
M.D. Anderson Trust Fellowship, Burroughs Wellcome Career Development Award and the National Institutes of Health (GM074815 to M.A.S.). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Advanced Light Source (ALS) and their support staff. The ALS is supported by the Director, Office of Science, Office of Basic Energy Sciences, and Material Science Division of the US Department of Energy at the Lawrence Berkeley National Laboratory.
REFERENCES
- 1.Hayes F, Barillà D. Assembling the bacterial segrosome. Trends Biochem. Sci. 2006;31:247–250. doi: 10.1016/j.tibs.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Hayes F, Barillà D. The bacterial segrosome: a dynamic nucleoprotein machine for DNA trafficking and segregation. Nat. Rev. Microbiol. 2006;4:133–143. doi: 10.1038/nrmicro1342. [DOI] [PubMed] [Google Scholar]
- 3.Schumacher MA. Structural biology of plasmid partition: uncovering the molecular mechanisms of DNA segregation. Biochem. J. 2008;412:1–18. doi: 10.1042/BJ20080359. [DOI] [PubMed] [Google Scholar]
- 4.Gerdes K, Møller-Jensen J, Bugge Jensen R. Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol. 2000;37:455–466. doi: 10.1046/j.1365-2958.2000.01975.x. [DOI] [PubMed] [Google Scholar]
- 5.Surtees JA, Funnell BE. The DNA binding domains of P1 ParB and the architecture of the P1 plasmid partition complex. J. Biol. Chem. 2001;276:12385–12394. doi: 10.1074/jbc.M009370200. [DOI] [PubMed] [Google Scholar]
- 6.Anand SP, Akhtar P, Tinsley E, Watkins SC, Khan SA. GTP-dependent polymerization of the tubulin-like RepX replication protein encoded by the pXO1 plasmid of Bacillus anthracis. Mol. Microbiol. 2008;67:881–890. doi: 10.1111/j.1365-2958.2007.06100.x. [DOI] [PubMed] [Google Scholar]
- 7.Larsen RA, Cusumano C, Fujioka A, Lim-Fong G, Patterson P, Pogliano J. Treadmilling of a prokaryotic tubulin-like protein, TubZ, required for plasmid stability in Bacillus thuringiensis. Genes Dev. 2007;21:1340–1352. doi: 10.1101/gad.1546107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simpson AE, Skurray RA, Firth N. A single gene on the staphylococcal multiresistance plasmid pSK1 encodes a novel partitioning system. J. Bacteriol. 2003;185:2143–2152. doi: 10.1128/JB.185.7.2143-2152.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tinsley E, Khan SA. A novel FtsZ-like protein is involved in replication of the anthrax toxin-encoding pXO1 plasmid in Bacillus anthracis. J. Bacteriol. 2006;188:2829–2835. doi: 10.1128/JB.188.8.2829-2835.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Golovanov AP, Barillà D, Golovanova M, Hayes F, Lian LY. ParG, a protein required for active partition of bacterial plasmids, has a dimeric ribbon-helix-helix structure. Mol. Microbiol. 2003;50:1141–1153. doi: 10.1046/j.1365-2958.2003.03750.x. [DOI] [PubMed] [Google Scholar]
- 11.Murayama K, Orth P, de la Hoz AB, Alonso JC, Saenger W. Crystal structure of omega transcriptional repressor encoded by Streptococcus pyogenes plasmid pSM19035 at 1.5 Å resolution. J. Mol. Biol. 2001;314:789–796. doi: 10.1006/jmbi.2001.5157. [DOI] [PubMed] [Google Scholar]
- 12.Møller-Jensen J, Ringgaard S, Mercogliano CP, Gerdes K, Löwe J. Structural analysis of the ParR/parC plasmid partition complex. EMBO J. 2007;26:4413–4422. doi: 10.1038/sj.emboj.7601864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schumacher MA, Glover TC, Brzoska AJ, Jensen SO, Dunham TD, Skurray RA, Firth N. Segrosome structure revealed by a complex of ParR with centromere DNA. Nature. 2007;450:1268–1271. doi: 10.1038/nature06392. [DOI] [PubMed] [Google Scholar]
- 14.Schumacher MA, Funnell BE. Structures of ParB bound to DNA reveal mechanism of partition complex formation. Nature. 2005;438:516–519. doi: 10.1038/nature04149. [DOI] [PubMed] [Google Scholar]
- 15.Delbrück H, Ziegelin G, Lanka E, Heinemann U. Src homology 3-like domain is responsible for dimerization of the repressor protein KorB encoded by the promiscuous IncP plasmid RP4. J. Biol. Chem. 2002;277:4191–4198. doi: 10.1074/jbc.M110103200. [DOI] [PubMed] [Google Scholar]
- 16.Khare D, Ziegelin G, Lanka E, Heinemann U. Sequence-specific DNA binding determined by contacts outside the helix-turn-helix motif of the ParB homolog KorB. Nat. Struct. Mol. Biol. 2004;11:656–663. doi: 10.1038/nsmb773. [DOI] [PubMed] [Google Scholar]
- 17.Schumacher MA, Mansoor A, Funnell B. Structure of a four-way bridged ParB-DNA complex; implications for segrosome assembly. J. Biol. Chem. 2007;282:10456–10464. doi: 10.1074/jbc.M610603200. [DOI] [PubMed] [Google Scholar]
- 18.Barillà D, Rosenberg MF, Nobbmann U, Hayes F. Bacterial DNA segregation dynamics mediated by the polymerizing protein ParF. EMBO J. 2005;24:1453–1464. doi: 10.1038/sj.emboj.7600619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bouet JY, Ah-Seng Y, Benmeradi N, Lane D. Polymerization of SopA partition ATPase: regulation by DNA binding and SopB. Mol. Microbiol. 2007;63:468–481. doi: 10.1111/j.1365-2958.2006.05537.x. [DOI] [PubMed] [Google Scholar]
- 20.Lim GE, Derman AI, Pogliano J. Bacterial DNA segregation by dynamic SopA polymers. Proc. Natl Acad. Sci. USA. 2005;102:17658–17663. doi: 10.1073/pnas.0507222102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunham TD, Xu W, Funnell BE, Schumacher MA. Structural basis for ADP-mediated transcriptional regulation by P1 and P7 ParA. EMBO J. 2009;28:1792–1802. doi: 10.1038/emboj.2009.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatano T, Yamaichi Y, Niki H. Oscillating focus of SopA associated with filamentous structure guides partitioning of F plasmid. Mol. Microbiol. 2007;64:1198–1213. doi: 10.1111/j.1365-2958.2007.05728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van den Ent F, Møller-Jensen J, Amos LA, Gerdes K, Löwe J. F-actin like filaments formed by plasmid segregation protein ParM. EMBO J. 2002;21:6935–6943. doi: 10.1093/emboj/cdf672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mori H, Mori Y, Ichinose C, Niki H, Ogura T, Kato A, Hiraga S. Purification and characterization of SopA and SopB proteins essential for F plasmid partitioning. J. Biol. Chem. 1989;264:15535–15541. [PubMed] [Google Scholar]
- 25.Yates P, Lane D, Biek DP. The F plasmid centromere, sopC, is required for full repression of the sopAB operon. J. Mol. Biol. 1999;290:627–638. doi: 10.1006/jmbi.1999.2909. [DOI] [PubMed] [Google Scholar]
- 26.Bouet JY, Bouvier M, Lane D. Concerted action of plasmid maintenance functions: partition complexes create a requirement for dimer resolution. Mol. Microbiol. 2006;62:1447–1459. doi: 10.1111/j.1365-2958.2006.05454.x. [DOI] [PubMed] [Google Scholar]
- 27.Bouet JY, Lane D. Molecular basis of the supercoil deficit induced by the mini-F plasmid partition complex. J. Biol. Chem. 2009;284:165–173. doi: 10.1074/jbc.M802752200. [DOI] [PubMed] [Google Scholar]
- 28.Castaing J-P, Bouet JY, Lane D. F plasmid partition depends on interaction of SopA with non-specific DNA. Mol. Microbiol. 2008;70:1000–1011. doi: 10.1111/j.1365-2958.2008.06465.x. [DOI] [PubMed] [Google Scholar]
- 29.Adachi S, Hori K, Hiraga S. Subcellular positioning of F plasmid mediated by dynamic localization of SopA and SopB. J. Mol. Biol. 2006;356:850–863. doi: 10.1016/j.jmb.2005.11.088. [DOI] [PubMed] [Google Scholar]
- 30.Hirano M, Mori H, Onogi T, Yamazoe M, Niki H, Ogura T, Hiraga S. Autoregulation of the partition genes of the mini-F plasmid and the intracellular localization of their products. Mol. Gen. Genet. 1998;257:392–403. doi: 10.1007/s004380050663. [DOI] [PubMed] [Google Scholar]
- 31.Terwilliger TC, Berendzen J. automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu S, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D: Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 33.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. Sect. A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 34.Anderson BJ, Larkin C, Guja K, Schildbach JF. Using fluorophore-labeled oligonucleotides to measure affinities of protein-DNA interactions. Methods in Enzymol. 2008;450:253–272. doi: 10.1016/S0076-6879(08)03412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perez-Iratxeta C, Andrade-Navarro MA. K2D2: estimate of protein secondary structure from circular dichroism spectra. BMC Struct. Biol. 2008;8:1–5. doi: 10.1186/1472-6807-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ravin NV, Rech J, Lane D. Mapping of functional domains in F plasmid partition proteins reveals a bipartite SopB-recognition domain in SopA. J. Mol. Biol. 2003;329:875–889. doi: 10.1016/s0022-2836(03)00525-4. [DOI] [PubMed] [Google Scholar]
- 37.Hanai R, Liu R, Benedetti P, Caron PR, Lynch AS, Wang JC. Molecular dissection of a protein SopB essential for Escherichia coli F plasmid partition. J. Biol. Chem. 1996;271:17469–17475. doi: 10.1074/jbc.271.29.17469. [DOI] [PubMed] [Google Scholar]
- 38.Xing Y, Takemaru KI, Liu J, Berndt JD, Zheng JJ, Moon RT, Xu W. Crystal structure of a full-length β-catenin. Structure. 2008;16:478–487. doi: 10.1016/j.str.2007.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brownlie P, Ceska T, Lamers M, Romier C, Stier G, Teo H, Stuck D. The crystal structure of an intact human Max-DNA complex: new insights into mechanisms of transcriptional control. Structure. 1997;5:509–520. doi: 10.1016/s0969-2126(97)00207-4. [DOI] [PubMed] [Google Scholar]
- 40.Kim SK, Shim J. Interaction between F plasmid partition proteins SopA and SopB. Biochem. Biophys. Res. Commun. 1999;263:113–117. doi: 10.1006/bbrc.1999.1317. [DOI] [PubMed] [Google Scholar]
- 41.Dasgupta S, Iyer GH, Bryant SH, Lawrence CE, Bell JA. Extent and nature of contacts between protein molecules in crystal lattices and between subunits of protein oligomers. Proteins. 1997;28:494–514. doi: 10.1002/(sici)1097-0134(199708)28:4<494::aid-prot4>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 42.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J. Mol. Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 43.Marsden BJ, Shaw GS, Sykes BD. Calcium binding proteins. Elucidating the contributions to calcium affinity from an analysis of species variants and peptide fragments. Biochem. Cell Biol. 1990;68:587–601. doi: 10.1139/o90-084. [DOI] [PubMed] [Google Scholar]
- 44.Edgar R, Chattoraj DK, Yarmolinsky M. Pairing of P1 plasmid partition sites by ParB. Mol. Microbiol. 2001;42:1363–1370. doi: 10.1046/j.1365-2958.2001.02717.x. [DOI] [PubMed] [Google Scholar]
- 45.Rodionov O, Lobocka M, Yarmolinsky M. Silencing of genes flanking the P1 plasmid centromere. Science. 1999;283:546–549. doi: 10.1126/science.283.5401.546. [DOI] [PubMed] [Google Scholar]
- 46.Ravishanker G, Swaminathan S, Beveridge DL, Lavery R, Sklenar H. Conformational and helicoidal analysis of 30 PS of molecular dynamics on the d(CGCGAATTCGCG) double helix: ‘curves’, dials and windows. J. Biomol. Struct. Dyn. 1998;6:669–699. doi: 10.1080/07391102.1989.10507729. [DOI] [PubMed] [Google Scholar]
- 47.Biek DP, Shi J. A single 43-bp sopC repeat of plasmid mini-F is sufficient to allow assembly of a functional nucleoprotein partition complex. Proc. Natl Acad. Sci. USA. 1994;91:8027–8031. doi: 10.1073/pnas.91.17.8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Biek DP, Strings J. Partition functions of mini-F affect plasmid DNA topology in Escherichia coli. J. Mol. Biol. 1995;246:388–400. doi: 10.1006/jmbi.1994.0094. [DOI] [PubMed] [Google Scholar]
- 49.Lynch AS, Wang JC. SopB-protein mediated silencing of genes linked to the sopC locus of Escherichia coli F plasmid. Proc. Natl Acad. Sci. USA. 1995;92:1896–1900. doi: 10.1073/pnas.92.6.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garner EC, Campbell CS, Weibel DB, Mullins RD. Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science. 2007;315:1270–1274. doi: 10.1126/science.1138527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Møller-Jensen J, Borch J, Dam M, Jensen RB, Roepstorff P, Gerdes K. Bacterial mitosis: ParM of plasmid R1 moves plasmid DNA by an actin-like insertional polymerization mechanism. Mol. Cell. 2003;12:1477–1487. doi: 10.1016/s1097-2765(03)00451-9. [DOI] [PubMed] [Google Scholar]
- 52.Popp D, Narita A, Oda T, Fujisawa T, Matsuo H, Nitanai Y, Iwasa M, Maeda K, Onishi H, Maeda Y. Molecular structure of the ParM polymer and the mechanism leading to its nucleotide-driven dynamic instability. EMBO J. 2008;27:570–579. doi: 10.1038/sj.emboj.7601978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salje J, Löwe J. Bacterial actin: architecture of the ParMRC DNA partitioning complex. EMBO J. 2008;27:2230–2238. doi: 10.1038/emboj.2008.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Delano WL. The PyMOL Molecular Graphics System. California: DeLano Scientific, San Carlos; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.