Abstract
Domains in Arabidopsis proteins NRPE1 and SPT5-like, composed almost exclusively of repeated motifs in which only WG or GW sequences and an overall amino-acid preference are conserved, have been experimentally shown to bind multiple molecules of Argonaute (AGO) protein(s). Domain swapping between the WG/GW domains of NRPE1 and the human protein GW182 showed a conserved function. As classical sequence alignment methods are poorly-adapted to detect such weakly-conserved motifs, we have developed a tool to carry out a systematic analysis to identify genes potentially encoding AGO-binding GW/WG proteins. Here, we describe exhaustive analysis of the Arabidopsis genome for all regions potentially encoding proteins bearing WG/GW motifs and consider the possible role of some of them in AGO-dependent mechanisms. We identified 20 different candidate WG/GW genes, encoding proteins in which the predicted domains range from 92aa to 654aa. These mostly correspond to a limited number of families: RNA-binding proteins, transcription factors, glycine-rich proteins, translation initiation factors and known silencing-associated proteins such as SDE3. Recent studies have argued that the interaction between WG/GW-rich domains and AGO proteins is evolutionarily conserved. Here, we demonstrate by an in silico domain-swapping simulation between plant and mammalian WG/GW proteins that the biased amino-acid composition of the AGO-binding sites is conserved.
INTRODUCTION
The sequencing of an increasing number of complete genomes during the past 20 years from a variety of organisms has led, within the limits of genome annotation efficiency, to the availability of catalogs of amino-acid sequences for all protein-coding genes from species representing all kingdoms of life, from bacteria to man. Sequence comparison with established, expertized proteins or ab initio analysis of amino-acid sequences has allowed the definition of conserved functional and/or structural motifs, which are available in specialized databases (1,2). It is thus possible to examine newly-acquired sequences for the presence of such motifs and obtain an idea as to the potential functions of a protein. Furthermore, ‘blind’ classifications have been established, which define only ‘Domains of Unknown Function’, or DUFs, which are conserved in several proteins, in an attempt to carry out exhaustive identification of potential functional motifs. However, these classifications are based either on sequence comparisons or analysis of multiple amino-acid sequence alignments and are therefore subject to the limits of these approaches, notably the exploitation of linear, primary sequences. This makes poorly-conserved domains difficult to define.
In plants, analysis of the Arabidopsis genome sequence led to the discovery, in addition to the known RNA polymerases I, II and III, of two distinct plant-specific RNA polymerases, polIV and polV that are implicated in RNA-directed DNA methylation (RdDM), an endogenous RNAi-mediated chromatin silencing pathway (3–6). PolIV and PolV have distinct largest subunits, NRPD1/NRPD1a and NRPE1/NRPD1b, respectively, but share with PolII and/or with each other numerous additional subunits (7–10). The PolV large subunit, NRPE1, is distinguished from that of PolIV, NRPD1, by the presence of a specific C-terminal domain (CTD) composed almost exclusively of divergent repeated motifs containing conserved WG or GW sequences (henceforth called WG/GW motifs) (11).
In agreement with the proposed role of PolV in small RNA (sRNA)-mediated gene silencing, it has been shown that this WG/GW region is able to bind multiple molecules of ARGONAUTE4 (AGO4) protein, an sRNA-binding effector of RdDM in plants (12,13), in a tryptophan-dependent manner (11). Argonaute (AGO) proteins are involved in small RNA-directed regulatory pathways in most eucaryotes. The Arabidopsis genome contains 10 genes potentially encoding AGO proteins, that have been implicated in both transcriptional and post-transcriptional silencing pathways (TGS and PTGS respectively) (14) and are thus essential actors in control of gene expression. Identification of their cellular partners will shed light on their roles in the different silencing pathways.
The WG/GW domains in NRPE1 have a biased amino-acid composition, being rich in glycine, serine and tryptophan and, to a lesser extent, glutamic acid, aspartic acid and asparagine, with low levels of cysteine, phenylalanine, histidine, methionine and tyrosine (11). Comparison of the Arabidopsis NRPE1 sequence with those of other plants shows little sequence conservation in the repeats other than the WG/GW pairs, even between relatively closely related species. Interestingly, sequence alignments of the specific domain of NRPE1 using the PSI-BLAST algorithm (15) to take into account the biased composition revealed sequence similarity with WG/GW repeat regions in a number of proteins from organisms from yeast to man, most of which have been implicated in targeted genome modification (11).
Despite this widespread conservation, the motifs in WG/GW proteins are not defined in any of the protein motif databases and in fact warranted little mention in the original description of the proteins which contain them. The canonical WG/GW protein is human GW182 (16), which is found in cytoplasmic structures involved in the post-transcriptional regulation of eukaryotic gene expression known as P-/GW182 bodies and multivesicular bodies (17,18). The GW182 family members have been shown to interact with all four human AGO proteins (HsAGO1-4) and have been linked to the RNA interference process (19–21). In these proteins, the repetitive WG/GW motifs are even less structured than in NRPE1 and are rather found in a GW-rich N-terminal region of the protein, which was first shown by Behm-Ansmant et al. (22) to interact with AGO1. There is little conservation of primary sequence in this region, although it shows a biased amino-acid composition similar to that of the WG/GW motifs of NRPE1 (11). Three GW182 paralogs have been identified in vertebrates (TNRC6A, TNRC6B and TNRC6C), as well as a single ortholog in insects (GAWKY) (23). They have all been linked to RNA interference (24) and shown to be important for short-interfering-RNA- and microRNA-mediated mRNA decay and translational repression (25). A conserved AGO-binding activity of the WG/GW domains of NRPE1 and GW182 was demonstrated through domain-swapping experiments (11).
AGO-binding properties have been demonstrated for several other WG/GW motif-containing proteins. The Caenorhabditis elegans proteins AIN-1 and AIN-2 have similar amino-acid composition to human GW182-related proteins and also associate with AGO proteins through WG/GW motifs (26,27). However, a lack of common domain architecture suggests that AIN-1 and AIN-2 are not members of the GW182 protein family, but rather represent functional analogs (24). Targeted mutations in the WG/GW repeats of a putative SPT5-type transcription elongation factor in Arabidopsis, known as KTF1/RDM3/SPT5-like have clearly shown that the presence of these motifs is essential for AGO4-dependent RNA-directed DNA methylation (28,29). The Schizosaccharomyces pombe RNA-induced transcriptional silencing complex (RITS) component, Tas3, contains a short WG/GW-repeat-rich region, in which mutation of one WG is sufficient to abolish the interaction with AGO1, which is necessary to establish heterochromatin at centromeric loci (30,31). Moreover, in the ciliated protozoan Tetrahymena thermophila, two WG/GW repeat proteins, Wag1p and CnjBp, interact with the AGO family protein member (Twi1p) and overlap functionally in RNA interference-mediated genome rearrangement (32). Taken together, these results clearly suggest that the WG/GW repeats constitute an evolutionarily-conserved but sequence-divergent AGO-binding platform.
These observations demonstrate that weakly-conserved, functional domains in proteins remain to be identified, even when such motifs have been conserved in widely divergent organisms and over several hundred million years. However, as classical sequence comparison methods are poorly-adapted to the detection of sequence conservation in such loosely-structured motifs, we decided to develop a tool to carry out a systematic search in Arabidopsis thaliana for genes potentially encoding proteins containing WG/GW motifs and which have a similar biased amino-acid composition. The genome of Arabidopsis thaliana is an ideal model for such a search, as the complete genome sequence has now been available for several years (33) and both the sequence and the annotation are of very high quality. EST (34) and full-length cDNA (35) sequences are freely available (36), as are several collections of mutant lines, making possible functional studies on genes encoding identified proteins. Here we describe the exhaustive analysis of the Arabidopsis proteome and genome for all regions potentially encoding proteins bearing WG/GW motifs and consider the possible role of some of them in AGO-dependent mechanisms. Furthermore, we have carried out virtual domain-swapping simulations to identify biologically confirmed mammalian and plant AGO-binding proteins.
MATERIALS AND METHODS
All calculations were carried out under a Linux operating system. The method was implemented in Perl and Python scripting languages and statistical analysis was carried out with the R analysis environment (37).
Calculation of WG/GW domain-specific scoring matrix
The initial sequence dataset contained a manually selected collection of 26 proteins with WG/GW motifs from various plants (NRPE1 sequences from Arabidopsis (GenBank accession NP_181532), grape (XP_002265533), nightshades (AAY89359.1), spinach (AAX12374), tomato (AAY89359), rice (EEE56320, misannotated as a PolII subunit), Physcomitrella patens (XP_001766256), poplar (XP_002303926) and corn NRPE1 sequence (identified by TBLASTN (15) on the genomic sequence), Arabidopsis SPT5-like (NP_196049), GTB1 (NP_176723) and their orthologs in other plant species. The sequences were identified in public databases using a PSI-BLAST (15) based approach and pairwise reciprocal best-hit analyses. The scoring matrix was calculated by compositional analysis of this sequence dataset and subsequently used for the detection of domain boundaries in novel proteins. The scoring table contains values for each amino acid and reflects compositional differences between the domain and the whole protein (Table 1). The following formula was used for the calculation of values for each residue present in manually identified domains: Di = 2 × log2[(Nid/Nd)/(Nip/Np)], where i—each of the amino acids present in the domain sequence; Nip—number of occurrences of amino acid i in the whole protein; Np—number of amino-acid residues in the protein; Nid—number of occurrences of amino acid i in the domain; Nd—number of amino-acid residues in the domain. The values expressed in half-bits were rounded to three decimal places. If the amino acid was not present in the domain, the corresponding value in the table was set to zero, which ensured there was no effect on domain extension and dos scores.
Table 1.
Scoring matrix used to define domain boundaries and calculate dos score, representing likelihood for a given amino acid to be found in GW domain
| Amino acid | Score [half-bits] | Score [bits] | Ratio | Frequency | Count |
|---|---|---|---|---|---|
| W | 2.666 | 1.333 | 2.520 | 0.063:0.025 | 743:1062 |
| G | 2.068 | 1.034 | 2.048 | 0.213:0.104 | 2490:4447 |
| N | 1.510 | 0.755 | 1.688 | 0.081:0.048 | 949:2051 |
| S | 1.236 | 0.618 | 1.535 | 0.152:0.099 | 1774:4213 |
| A | 0.280 | 0.140 | 1.102 | 0.065:0.059 | 762:2537 |
| D | 0.184 | 0.092 | 1.066 | 0.081:0.076 | 950:3254 |
| T | 0.000 | 0.000 | 1.000 | 0.040:0.040 | 467:1718 |
| Q | −0.076 | −0.038 | 0.974 | 0.038:0.039 | 440:1686 |
| K | −0.120 | −0.060 | 0.959 | 0.070:0.073 | 821:3136 |
| R | −0.590 | −0.295 | 0.815 | 0.044:0.054 | 518:2319 |
| P | −0.644 | −0.322 | 0.800 | 0.032:0.040 | 373:1726 |
| E | −1.288 | −0.644 | 0.640 | 0.048:0.075 | 560:3219 |
| V | −2.408 | −1.204 | 0.434 | 0.023:0.053 | 274:2260 |
| F | −2.558 | −1.279 | 0.412 | 0.014:0.034 | 169:1443 |
| H | −3.324 | −1.662 | 0.316 | 0.006:0.019 | 76:796 |
| C | −3.398 | −1.699 | 0.308 | 0.004:0.013 | 43:568 |
| Y | −4.792 | −2.396 | 0.190 | 0.004:0.021 | 50:890 |
| M | −5.012 | −2.506 | 0.176 | 0.003:0.017 | 39:743 |
| I | −5.030 | −2.515 | 0.175 | 0.007:0.040 | 79:1705 |
| L | −5.252 | −2.626 | 0.162 | 0.011:0.068 | 132:2925 |
The amino acids are sorted by the score value, from highest to lowest. The second and third columns were used in domain identification calculations. The last two colums contain counts and frequencies of a given amino acid found in the whole protein sequence versus the domain (format–domain: entire protein).
Domain boundary identification algorithm
The algorithm used for the identification of domain boundaries uses as a starting point (seed position) each WG/GW motif location. By progressing in both directions, it calculates the cumulative score for each position using values from the previously prepared scoring matrix, which represents the likelihood of a given amino acid to be part of the domain. The domain extension is terminated when the calculated linear progression score for the current position drops from its last maximum below the value given by the dec threshold (see later for rationale of threshold value calculation). Because the method uses the position of each occurrence of a WG/GW motif for domain calculation, it sometimes detects overlapping domains, which are part of a domain containing more than one WG/GW motif. In this case, the overlapping domains are joined and the final dos score value for the assembled domain is calculated.
Calculation of internal composition score
The dos score represents the preference that a given amino acid will be found in the analyzed domain rather than in other parts of the sequence or in unrelated proteins. During the analysis, domains representing sequences with biased composition for tryptophan or glycine (the two highest scoring amino acids in the scoring table) often showed relatively high scores without having any obvious relation to the WG/GW motif. Such false positives were mostly observed during analysis of six-frame translation of raw genomic and transcript DNA sequences. To overcome this limitation a new type of score—‘internal composition score’ (ics)—was introduced. This new score allowed more accurate representation of amino-acid composition for domains present in the initial dataset (Supplementary Data—ics scoring matrix). The calculation of ics scores involves two steps: (i) creation of an ics table representing ratios of all amino acids present in a given domain to each other, and (ii) calculation of a difference for each amino acid between ics tables from the reference sequence set and the currently analyzed domain. The final value for each ics score was converted into an absolute number and normalized for the domain sequence length. In contrast to the dos score, where higher values represent better candidates, in this approach values tending towards zero represent a closer compositional relationship with the reference dataset.
The values in the ics table for each amino acid were calculated according to the following formula: Iij = abs[log2(Ni/Nj)], where i and j represent two amino acids present in the same domain; Ni—number of amino acids i in the domain; Nj—number of amino acids j in the domain.
Selection of statistical threshold values
The dec score was estimated by comparison of manually identified domain sequences with the spectrum of domains defined with various dec values by an automatic algorithm search in the initial dataset. The dec value directly influences the domain size: lowering the value identified smaller and more fragmented domains in the proteins. Highest sensitivity (SN = 0.947) and selectivity (SP = 0.946) scores for automatic domain detection were obtained when the dec value was set to 8.6 half-bits (Supplementary Data).
The threshold value for the dos score was selected on the basis of distribution fitting analysis. The Kolmogorov–Smirnov non-parametric distribution-independent test (KS test) (38) was applied to quantify a distance between the dos score distribution function of the Arabidopsis genome and the cumulative distribution function of the reference hypothetical distributions. In addition, the information criteria (SIC, AIC, HQIC) were employed to assess the fit of a model based on its optimum log-likelihood value, after applying a penalty for the parameters that were estimated in fitting the model. Among more than 40 continuous probability distributions fitted, the three-parameter log-logistic distribution, LLD3, (α = 5.77; β = 10.859; γ = –17.061) is the most correct model describing dos scores in Arabidopsis (SIC = 99545.9, AIC = 99514.6, HQIC = 99524.9) and the discrepancy between the observed and LLD3 cumulative frequencies is not significantly different (Kolmogorov-Smirnov P-value ≤ 0.15). In addition, the P–P plot was used as a graphical adjunct to assess the fit of probability distributions (Supplementary Data). A significant dos score of 6.99 half-bits (P-value = 0.01) was selected.
Another approach was used to estimate the cut-off value for the ics score. Here we used the initial set of 26 domains and divided it into two groups: test sequences—used to calculate the ics score, and reference sequences—used to calculate the reference ics table needed for calculation of the ics score of test sequences. Seven series of calculations were performed and in each of them a different number of reference and test sequences were used. Each run involved all combinations of sequences, and at the end scores for a given set were averaged. The estimated threshold was calculated by linear extrapolation of maximal ics values obtained in all series of the analysis. The regression analysis of five data points resulted in setting the value for ics score threshold to 2.14.
Virtual domain swapping
Two separate scoring matrix sets (dos and ics) were calculated using the same method as employed for identification of Arabidopsis WG/GW proteins. The ‘plant-specific’ and ‘mammalian-specific’ scoring tables were calculated using plant and mammalian proteins with experimentally verified AGO-binding activities, respectively. In subsequent steps, the ‘plant-specific’ matrix was used to scan for WG/GW proteins in representative mammalian genomes (human, chimp, mouse, rat, cow, horse, dog, opossum, platypus and rhesus). In a symmetric but opposite approach, the ‘mammalian-specific’ scoring table was used to screen the Arabidopsis sequences.
RNA isolation and RT–PCR analysis
Total RNA was isolated from Arabidopsis seedlings (ecotype Columbia) using the Trizol reagent (Invitrogen). After DNAse treatment, cDNA was obtained with an Affinity Multitemperature cDNA synthesis kit (Agilent Technologies) using an oligodT primer with 500 ng of RNA according to manufacturer’s instructions. RT–PCR amplifications were carried out with primers WGRP1-5′ ATGGGAAAGTGGAATCATCGA and WGRP1-3′ TTACTTAGTTGAGAAATTGAC, respectively.
GST-WGRP1 pull-down experiments
The WG/GW-rich domain (aa 258–395) of WGRP1 was RT–PCR amplified using primers WGRP1int-5′ GGATCCAATCCTTGGGAAGCCCAGCC and WGRP1int-3′ CTCGAGTTGCCAATCACCTGCATTTGC, cloned into a BamHI-XhoI digested pET41a vector and expressed in E. coli BL21 cells. The GST-fusion and GST control proteins were purified on glutathione Sepharose 4B beads (GE Healthcare) and used in pull-down assays as described (11) using myc-AGO4 or Flag-AGO1 lysates from Arabidopsis flowers.
RESULTS
Genome-wide computational identification of WG/GW-motif proteins in Arabidopsis
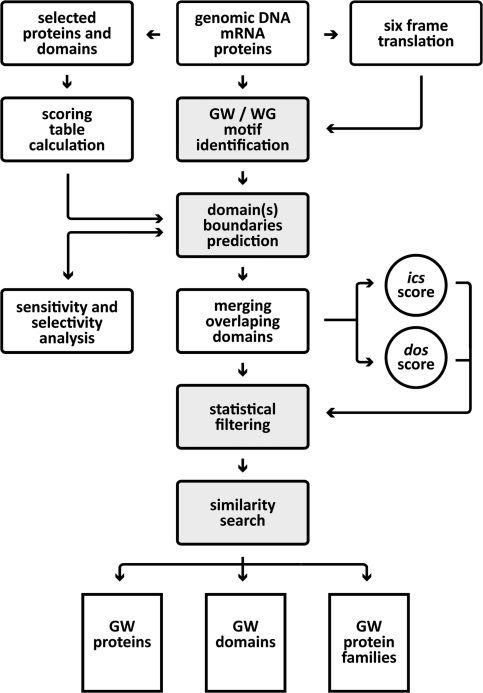
The low level of sequence conservation detected by classical methods of comparison based on dynamic programming algorithms inspired us to look for alternative ways to identify WG/GW-motif proteins encoded by the Arabidopsis genome. Analysis of the domain in previously-discovered WG/GW proteins indicated that, in addition to these residues, the other characteristic is the presence or absence of certain other amino acids (11). Therefore, the most direct approach in designing a strategy for WG/GW protein identification was to use this property and base the method on the amino-acid composition specificity of the domain. The identification procedure developed during this work can be divided into four discrete steps (indicated in Figure 1 by filled boxes). The first phase includes localization of all occurrences of WG/GW motif(s) in the genomic/protein sequence. Secondly, putative domain boundaries are identified using the WG/GW positions as starting points. Thirdly, the final domain score, representing the composition of identified motifs, is calculated and used for filtering out sequences based on statistical criteria. The final step includes a BLAST-based search for putative paralogs, where more conserved parts of the protein(s) can be used to infer sequence relationships.
Figure 1.
Schematic representation of the WG/GW protein identification pipeline. Grey-filled boxes represent the four major steps in the identification procedure.
Arabidopsis is currently the best annotated plant genome available. However, even now some gene models are not correct and, most probably, some genes still await discovery. Therefore our analysis was simultaneously carried out directly on the genomic sequence as well as on the latest Arabidopsis annotation (TAIR 9.0: http://arabidopsis.org). We took this precaution as the biased amino-acid composition of the WG/GW motifs induces an atypical codon usage. This can lead to mis- or non-identification of WG/GW-coding regions, with the loss in certain cases of complete exons (data not shown) and incorrect annotation of the corresponding genes. At the final stage of the analysis pipeline, both genomic and protein based pools were mapped onto each other and, where possible, annotated genes were identified.
A de novo search for new, as yet uncharacterized protein domains, which do not show clear similarity to other sequences presents many challenges, among which one of the most difficult is the assessment of biological significance of the new findings. One solution to this problem is the development of a scoring system which allows clear discrimination between various motifs. Our system is based on the limited number of protein sequences with clear WG/GW domains in proteins identified by PSI-BLAST (15). As only a few such proteins had been identified in Arabidopsis, we included proteins from other plant species—rice, cottonwood, poplar, tomato, grape, corn and spinach in this data set (Supplementary Data) to make the analysis more universal. The resulting scoring table provides information about the preference for a given amino acid to be present in the domain compared with the whole protein (Table 1). As could be expected, the highest scoring amino acids are tryptophan and glycine, followed by asparagine, serine, alanine and aspartic acid. The least expected amino acids in the WG/GW motifs are leucine, isoleucine, methionine, tyrosine and cystein. According to these calculations threonine is ‘neutral’, occurring at the same frequency in the domain as in any other part of the proteins. The properties of the side groups indicate a slight preference for small, hydrophilic and charged amino acids with, of course, the exception of the highest scoring hydrophobic tryptophan. The large number of amino acids having negative values (13 negative versus 6 with positive scores) and higher value range for negative scores (5.252 for leucine versus 2.666 for tryptophan) suggests rather a stronger negative selection against the presence of a number of amino acids rather than positive selection for others.
The precise annotation of domain boundaries is another challenge in the analysis of domains without clear sequence similarity. Our analysis was carried out using a score table (Table 1) and the location of all WG/GW (and combinations thereof—see ‘Materials and Methods’ section) motifs. During the process of domain identification the created algorithm moves from one residue to the next in both directions from the initial WG/GW motif, summing the values from the scoring table for each additional amino acid. The domain is extended as long as the calculated local score maximum (highest calculated value) does not drop below a given value. Precisely defined boundaries of the domain allow the calculation of cumulative score (dos), by summing the values from the scoring matrix for each amino acid. By careful selection of the «decay» value it was possible to achieve high precision in domain boundary identification, with 0.947 sensitivity and 0.946 selectivity scores (Supplementary Data).
The qualification of the new WG/GW motifs based only on the presented scoring schema has one disadvantage—it depends solely on the length of the domain. Because no information was available at the moment of the analysis about the preferred size of the domain it was necessary to develop a second measure (internal domain composition score—ics), which is independent of the length of the sequence but, as for the first score, reflects the composition of the domain (see ‘Material and Methods’ section). The ics scoring table (Supplementary Data) shows the highest relative difference of amino acid presence in the motif between glycine and phenylalanine (4.032 half-bits). The most homogeneous distribution in composition shows tryptophan and asparagine—indicated by the lowest value in the scoring table (0.336 half-bits). The components of the WG/GW motif—tryptophan and glycine—show a middle range value of 1.868 half-bits—indicating that one of them (glycine) is present at a higher level in the domain than the other (tryptophan).
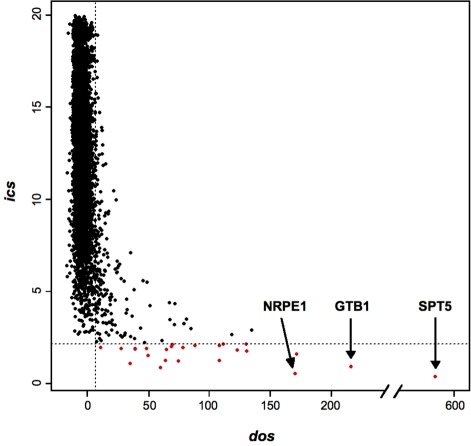
Finally, the introduction of both scoring systems, which provides a measurement of the degree of compositional compatibility of the new domains with the source domains used to calculate the scoring tables, allowed us to address the question of biological significance of the new findings. Based on simple statistical analysis it was possible to select new WG/GW motif-containing proteins which constitute good candidates for further experimental verification. Figure 2 presents the distribution of both scores (dos and ics) for all Arabidopsis proteins which contain at least one WG/GW motif. The sequences that fulfil the calculated cut-off criteria are indicated in red. The calculation of statistical significance was based on the selection of the mathematical model which best represents score values. Distribution fitting involved modeling of the probability distribution of a dos score variable in the Arabidopsis genome (see ‘Materials and Methods’ section). The very stringent score value of 6.99 half-bits was selected, which corresponds to a P-value of 0.01 in the Arabidopsis dos score dataset. The ics score of 2.14 half-bits was chosen based on stringent formal criteria and linear parameter estimation (see ‘Materials and Methods’ section). This guaranteed that the domains showing score values above the selected thresholds (indicated in Figure 2. in red) will have the same properties as source sequences used for the analysis, and therefore most likely can be classified in the same functional group of proteins (Supplementary Data—unfiltered list of all identified proteins).
Figure 2.
Distribution of ics and dos scores of all identified proteins in Arabidopsis. Each point represents a WG/GW-containing protein. Grey dashed lines indicate dos and ics score threshold values revealing WG/GW protein candidates marked in red.
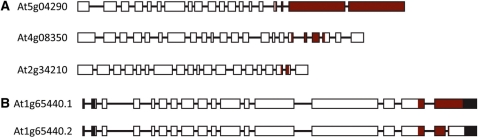
The AGI identifiers of genes encoding proteins containing WG/GW motifs are presented in Table 2. In this table, we have only shown results for regions which were identified in both genomic and protein data screens after applying a cut-off score. Direct analysis of the genomic sequence identified only two potential WG/GW-coding regions which did not map to protein data. Both have an unusual compositional bias and most probably do not code for biologically functional proteins. Overall, we identified 20 different genes with two products of alternatively-spliced messengers for two of them. The alternative spliced forms of At1g65440, which are both supported by several full-length cDNA or EST sequences, are interesting in that they encode proteins which are identical in the N-terminal region, differing only in the length of the WG/GW platform (Figure 3B).
Table 2.
GW motif proteins identified in Arabidopsis genome after applying threshold filters on dos and ics scores
| AGI locus code | dos score | P-value | ics score | TAIR annotation (partial) |
|---|---|---|---|---|
| AT1G04800.1 | 78.26 | 3.55E–06 | 1.96 | Glycine-rich protein; FUNCTIONS IN: molecular_function unknown; INVOLVED IN: N-terminal protein myristoylation; LOCATED IN: endomembrane system; EXPRESSED IN: 17 plant structures; |
| AT1G05460.1 | 74.53 | 4.47E–06 | 1.14 | SDE3—SILENCING DEFECTIVE: a protein with similarity to RNA helicases; mutantsare defective in post-transcriptional gene silencing. |
| AT1G10270.1 | 108.26 | 7.30E–07 | 1.25 | GRP23—GLUTAMINE-RICH PROTEIN 23: InterPro IPR011990—tetratricopeptide-like helical domain; InterPro IPR002885—pentatricopeptiderepeat; InterPro IPR013026—tetratricopeptideregion. |
| AT1G13020.1 | 63.96 | 9.07E–06 | 1.27 | EIF4B2—eukaryotic initiation factor 4B2; Plant specific eukaryotic initiation factor 4B:IPR010433 |
| AT1G15840.1 | 88.09 | 2.01E–06 | 2.06 | Unknown protein; FUNCTIONS IN: molecular_function unknown; INVOLVED IN: biological_process unknown; LOCATED IN: cellular_component unknown; EXPRESSED IN: 11 plant structures |
| AT1G65440.1 | 215.95 | 2.03E–08 | 0.96 | GTB1—GLOBAL TRANSCRIPTION FACTOR GROUP B1: related to yeast Spt6 protein, whichfunctions as part of a protein complex in transcription initiation and also plays a role in chromatinstructure/assembly. |
| AT1G65440.2 | 69.07 | 6.37E–06 | 2.01 | Same as above |
| AT2G16470.1 | 59.91 | 1.22E–05 | 0.89 | DNA binding/nucleic-acid binding/protein binding/zinc ion binding; Zinc finger (CCCH-type) family protein/GYF domain-containing protein: InterPro:IPR000571—CCCH-type zinc-finger domain; InterPro IPR003169—GYF domain. |
| AT2G33410.1 | 27.71 | 2.79E–04 | 1.9 | Heterogeneous nuclear ribonucleoprotein/hnRNP: contains InterPro domain RNA recognition motif, RNP-1; (InterPro:IPR000504); contains InterPro domain Nucleotide-binding, alpha-beta plait; (InterPro:IPR012677) |
| AT2G15780.1 | 107.99 | 7.39E–07 | 2.07 | Glycine-rich protein; FUNCTIONS IN: electron carrier activity, copper ion binding; LOCATED IN: endomembrane system; CONTAINS InterPro DOMAIN/s: Plastocyanin-like (InterPro:IPR003245), Cupredoxin (InterPro:IPR008972). |
| AT2G40030.1 | 170.3 | 7.15E–08 | 0.54 | NRPE1—the largest subunit of nuclear DNA-dependent RNA polymerase V; Required for normal RNA-directed DNA methylation at non-CG methylation sites and transgene silencing. |
| AT3G26400.1 | 49.64 | 2.79E–05 | 1.53 | EIF4B—eukaryotic initiation factor 4B; Plant specific eukaryotic initiation factor 4B:InterPro:IPR010433 |
| AT3G51940.1 | 10.83 | 4.28E–03 | 1.95 | Oxidoreductase/transition metal ion binding: InterPro domain Ferritin/ribonucleotide reductase-like; (InterPro:IPR009078) |
| AT4G16830.1 | 38.91 | 7.69E–05 | 1.87 | Nuclear RNA-binding protein (RGGA): InterPro domain Hyaluronan/mRNA binding protein (InterPro:IPR006861) |
| AT4G16830.3 | 38.95 | 7.66E–05 | 1.9 | Same as above |
| AT4G33930.1 | 130.58 | 2.83E–07 | 1.78 | Glycine-rich protein; LOCATED IN: endomembrane system; CONTAINS InterPro DOMAIN/s: Cupredoxin (InterPro:IPR008972) |
| AT4G36230.1 | 171.65 | 6.86E–08 | 1.62 | Unknown protein; hypothetical protein |
| AT4G38710.1 | 11.05 | 4.09E–03 | 1.97 | Glycine-rich protein: InterPro domain Plant specific eukaryotic initiation factor 4B (InterPro:IPR010433) |
| AT5G03990.1 | 35.08 | 1.16E–04 | 1.09 | Similar to oxidoreductase/transition metal ion binding |
| AT5G04290.1 | 585.79 | 8.37E–11 | 0.4 | KTF1—KOW DOMAIN-CONTAINING TRANSCRIPTION FACTOR 1; SPT5-Like, a member of the nuclear SPT5 (Suppressor of Ty insertion 5) RNA polymerase (RNAP) elongation factor family that is characterized by the presence of a carboxy-terminal extension with more than 40 WG/GW motifs. Interacts with AGO4. Required for RNA-directed DNA methylation. |
| AT5G07540.1 | 122.96 | 3.85E–07 | 1.82 | GLYCINE-RICH PROTEIN 16 (GRP16); Oleosin (InterPro:IPR000136); FUNCTIONS IN: lipid binding, nutrient reservoir activity; INVOLVED IN: sexual reproduction, lipid storage; |
| AT5G61660.1 | 64.68 | 8.62E–06 | 1.84 | Glycine-rich protein; FUNCTIONS IN: molecular_function unknown; INVOLVED IN: biological_process unknown; LOCATED IN: endomembrane system; |
Genes are sorted by AGI identifiers (localization on the genome).
Figure 3.
Domain architectures of selected WG/GW proteins. Gene structures of two small gene families are shown. Exons are represented as boxes and introns by lines. The WG/GW motif-containing region is colored in brown. (A) Three members of the SPT5-like transcription elongation factor family, showing the extensive platform in the At5g04290 gene product. (B) Variable motif domain length illustrated by alternative splicing in the SPT6 global transcription elongation factor family.
The size of the WG/GW domains ranges from 92 to 654aa (for At3g51940 and At5g04290 respectively). As could be expected, genes coding for proteins with already known WG/GW-dependent AGO-binding function (NRPE1 and KFT1/RDM3/SPT5-like) are among the highest scoring proteins. GTB1 is a member of the SPT6 family, implicated in transcription elongation in yeast and animals (39,40), while SDE3 is a protein already known to be involved in silencing mechanisms in Arabidopsis (41), although there is as yet no direct evidence for an interaction of the last two candidates with AGO proteins. There are also two plant-specific translation initiation factors (genes At1g13020 and At3g26400). Overall, the genes identified as encoding WG/GW proteins correspond in the majority of cases to a limited number of families: RNA-binding proteins, transcription factors, glycine-rich proteins, translation initiation factors and known silencing-associated proteins such as SDE3.
AGO binding to the WG motifs of a candidate protein
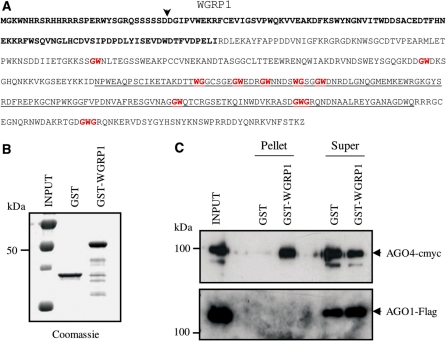
We previously demonstrated in vitro binding of AGO4 proteins to the WG/GW motifs of proteins NRPE1 and SPT5-LIKE (11,28). To validate our bioinformatics analysis, we used the same approach to test the AGO-binding capacity of one of our candidate proteins, encoded by gene At3g51940 and annotated by TAIR as an oxidoreductase/transition metal ion binding protein (Table 2). We focused on this candidate as At3g51940 is an evolutionarily-conserved novel spliced gene which is expressed in Arabidopsis and whose product harbors a large WG/GW-rich platform containing 10 WG/GW motifs (Figure 4A and data not shown). Comparison of At3g51940 with sequences in the protein databases did not support the annotation proposed by TAIR, suggesting that the very limited similarity to a ferritin/ribonucleotide reductase-like domain is probably artefactual. This led us to rename this gene WGRP1, for WG/GW-Rich Protein 1. WGRP1 is ubiquitously expressed in Arabidopsis and contains 2 exons that encode a predicted WGRP1 protein of 454 amino acids (Figure 4A). Putative poplar and castor bean orthologs (XM_002332380 and XM_002513070 accession numbers for poplar and castor bean sequences, respectively) showing strong sequence conservation in the N-terminal ∼120 amino acids and significant enrichment of WG/GW motifs at their C-terminus were identified by BLAST (15) searching (data not shown). Conservation of WG/GW motifs in the WGRP1 orthologs from other plants suggested that they could be functionally significant and prompted us to test the Arabidopsis WGRP1 WG/GW-rich domain for AGO-binding capacity in vitro. We produced a fusion protein containing the WG/GW-rich domain (aa258–396) fused to GST (Figure 4B; GST-WGRP1) and monitored the ability of this construct to interact with AGO1 and AGO4 compared with the GST control protein. Pull-down assays indicated that GST-WGRP1, but not the GST control protein, specifically interacts with AGO4 but not AGO1, indicating a binding specificity of the WG/GW-rich domain of WGRP1 toward AGO4 in vitro (Figure 4C). Interestingly, this specificity in AGO4-binding capacity is similar to that recently demonstrated for SPT5-like in Arabidopsis (28). Although a possible role for WGRP1 in a plant silencing pathway remains to be clarified, this validation strongly supports the output of our bioinformatics screen.
Figure 4.
WGRP1 protein has Argonaute-binding capacity. (A) Primary sequence of the Arabidopsis WGRP1 sequence. The evolutionarily conserved N-terminal sequence is bolded and the location of the intron relative to the open reading frame is indicated by a vertical arrowhead. The WG/GW motifs in the WGRP1 CTD are in red and the WGRP1 sequence fused to GST is underlined. (B) Coomassie staining of the purified GST and GST-WGRP1 recombinant proteins used in the Argonaute-binding assay. (C) Preferential binding of AGO4 to the WG/GW-rich domain of WGRP1 protein. Myc-AGO4 or Flag-AGO1 extracts were applied to equimolar amounts of GST and GST-based fusion protein beads and the bound protein (Pellet) and supernatant (Super) fractions detected by immunoblotting with anti-Myc or anti-M2 antibodies. The GST protein was used as control.
Identification of gene family members
The use of very stringent threshold values could lead to the loss of some of the ‘weak’ signals in the analysis. To avoid this, two strategies were employed: (i) the raw results representing all the WG/GW-motifs were presented in graphical form for manual verification and (ii) a BLAST-based re-screen of the Arabidopsis proteome was carried out to identify related sequences and compare families by multiple sequence alignment using MAFFT (42). The second approach resulted in identification of sequences related to candidate WG/GW-motif proteins based on the similarity of the sequence outside the variable domain. This data ‘explosion’ step enriched the pool of WG proteins by including gene products which apparently did not contain any signatures of the domain of interest. This is the case for NRPD1 and NRPE1, in which the WG platform is internal, and for the putative transcription elongation factors SPT5-like (Figure 3A) and GTB1 (Figure 3B), in which the motifs are found in a C-terminal extension. The presence of WG/GW motifs as a C-terminal extension is found in several other candidate proteins.
Virtual domain swapping
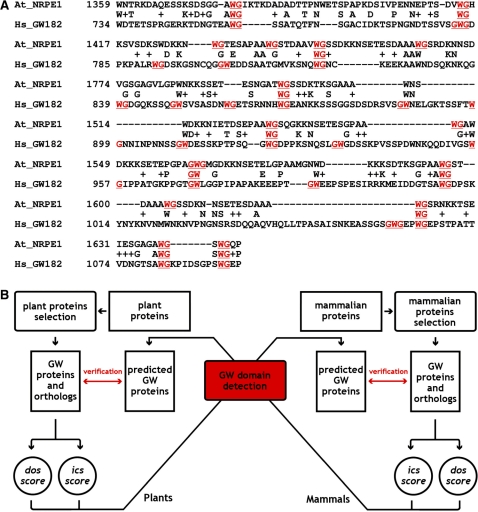
In the original experiment carried out by El-Shami et al. (11), the WG/GW-repeat region in the NRPE1 protein was exchanged with the related region of human GW182 protein (Figure 5A) to show functional conservation of the domain. They showed that the chimeric construct binds to AGO4 in vitro and is able to restore most of its DNA-methylation activity, while site-directed mutations of a tryptophan residue in the WG/GW motifs to phenylalanine or alanine disrupts AGO-binding activity in vitro. To verify whether the AGO-binding activity depends on the amino-acid composition of WG/GW domains, we designed a simulation of domain swapping experiments between plant and mammalian WG/GW proteins (Figure 5B), using plant or animal matrices. In this way, application of the heterologous matrices to screen in silico for WG/GW proteins simulates the original domain swapping experiment.
Figure 5.
Domain-swapping experiment simulation. (A) Pairwise alignment of WG/GW-rich domains from Arabidopsis largest polV subunit, NRPE1 and Human GW182. (B) Outline of the virtual domain swapping experiment between plant and mammalian WG/GW proteins. Dos/ics score tables were calculated based on experimentally-verified plant/mammalian WG/GW proteins and subsequently used to search for WG/GW domains in mammalian/plant proteomes. Detected putative WG/GW motif proteins were compared with experimentally verified AGO-binding proteins. Reciprocal best protein hits of such a bidirectional procedure share a conserved amino-acid composition of WG/GW-rich AGO-binding sites.
Based on the score table calculated on plant proteins with experimentally proven AGO-binding activities, an exhaustive search for WG/GW domains was carried out in representative mammalian genomes (human, chimp, mouse, rat, cow, horse, dog, opossum, platypus and rhesus). The highest scoring proteins identified in this screen (Table 3—list of highest scoring mammalian proteins sorted by ics score and selected with plant specific cut-off threshold values) include all AGO-binding GW182-related proteins, including human TNRC6A (P-value: 1.94E–06), TNRC6B (P-value: 7.11E–07) and TNRC6C (P-value: 1.24E–06). The full, unfiltered list of all identified mammalian proteins is presented as a Supplementary Data. Most of the identified top scoring proteins in the list have not been tested for interaction with AGO proteins (Table 3). This group contains large number of keratinocyte-associated proteins (e.g. highest scoring hornerin and dermatokine) which are know to form large molecular complexes but are unlikely involved in AGO-mediated gene silencing. Other proteins identified in the screen include splicing factors, zinc-finger, homeobox proteins and kinases, which can be considered as potential AGO-binding candidates. There are similarities between the functional categories of the mammalian proteins and the list of Arabidopsis putative WG/GW proteins, although in both cases further experimental work is essential to validate the bioinformatic screen.
Table 3.
List of mammalian proteins identified with plant-specific scoring matrices and selected using thresholds calculated for plant proteins
| Description (partial) | Ago-binding activitya | dos score | P-value | ics score | Organism | NCBI GI |
|---|---|---|---|---|---|---|
| hypothetical protein | nt | 237.7 | 1.21E–8 | 0.41 | Human | 239758013 |
| TNRC6A: trinucleotide repeat containing 6A | + | 92.04 | 1.63E–6 | 0.54 | Human, Cattleb, Horse, Rhesus, Dog, Platypus, Rat, Mouse | 119916998 |
| TNRC6C: trinucleotide repeat containing 6C | + | 89.98 | 1.82E–6 | 0.67 | Human, Cattleb, Horse, Rhesus, Dog, Mouse | 194676322 |
| HRNR: hornerin—intermediate filament-associated protein | nt | 161.44 | 9.46E–8 | 0.69 | Human | 57864582 |
| Hypothetical protein | nt | 265.69 | 6.64E–9 | 0.70 | Human | 169173184 |
| TNRC6B: trinucleotide repeat containing 6B | + | 106.81 | 7.81E–7 | 0.77 | Human, Cattle, Horse, Rhesus, Dogb, Platypus, Rat, Mouse | 73969036 |
| DMKN—dermokine | nt | 182.7 | 4.94E–8 | 0.78 | Rhesusb, Cattle, Dog, Human | 109124494 |
| Microsomal dipeptidase | nt | 187.96 | 4.25E–8 | 0.80 | Cattle | 194687044 |
| Similar to Flag | nt | 47.25 | 3.45E–5 | 1.08 | Platypus | 149631903 |
| ADP-ribosylation factor GTPase activating protein 1 | nt | 31.75 | 1.69E–4 | 1.09 | Cattleb, Dog, Mouse, Platypus, Rat | 115497314 |
| Similar to Repetin | nt | 30.22 | 2.04E–4 | 1.10 | Rat | 27692337 |
| Similar to splicing coactivator subunit SRm300 | nt | 60.94 | 1.13E–5 | 1.13 | Human, Ratb, Platypus | 109497194 |
| Hypothetical protein | nt | 37.81 | 8.62E–5 | 1.14 | Mouse | 149258285 |
| FLG-2: flaggrin-2; similar to ifapsoriasin | nt | 110.16 | 6.69E–7 | 1.18 | Platypus | 149515391 |
| Fibrinogen alpha-chain | nt | 74.37 | 4.51E–6 | 1.19 | Horseb, Cattle, Dog, Rhesus, Human | 194208383 |
| Hypothetical protein | nt | 21.52 | 6.59E–4 | 1.31 | Dog | 74001559 |
| Serine/arginine repetitive matrix 3 | nt | 62.74 | 9.90E–6 | 1.32 | Humanb, Mouse, Rat | 158854042 |
| Collagen, type VI, alpha 6 precursor | nt | 13.74 | 2.42E–3 | 1.33 | Humanb, Mouse | 156616290 |
| Similar to splicing factor, arginine/serine-rich 2 | nt | 7.33 | 9.24E–3 | 1.37 | Rat | 109481239 |
| SCY1-like 1 isoform A; N terminal kinase like protein | nt | 5.53 | 1.43E–2 | 1.38 | Human | 115430241 |
| Zinc finger protein 106 homolog; FOG: WD40 repeat | nt | 50.03 | 2.70E–5 | 1.39 | Cattleb, Mouse, Rhesus | 194670681 |
| Procollagen, type VII, alpha 1 | nt | 28.36 | 2.57E–4 | 1.46 | Rat | 157819015 |
| CDSN—Corneodesmosin | nt | 152 | 1.30E–7 | 1.47 | Platypus | 156602049 |
| Similar to Nucleoporin like 2 | nt | 8.95 | 6.39E–3 | 1.53 | Horseb, Dog, | 149705615 |
| Paired mesoderm homeobox protein 2B | nt | 32.54 | 1.54E–4 | 1.62 | Ratb, Human, Mouse, Rhesus, Dog | 109499673 |
| Hypothetical protein | nt | 26.28 | 3.37E–4 | 1.63 | Human | 239758008 |
| Similar to ribosomal protein S2 | nt | 15.06 | 1.90E–3 | 1.69 | Rhesus | 109073249 |
| Hypothetical protein | nt | 90.01 | 1.81E–6 | 1.72 | Rat | 109510645 |
| Hypothetical protein | nt | 71.91 | 5.28E–6 | 1.80 | Rat | 109511723 |
| Insulin receptor substrate 4 | nt | 127.03 | 3.26E–7 | 1.83 | Dogb, Horse | 74008591 |
| Similar to Mucin-19 | nt | 29.98 | 2.10E–4 | 1.93 | Human | 239755776 |
| Hypothetical protein | nt | 31.99 | 1.65E–4 | 1.93 | Platypus | 149610906 |
| Leukocyte receptor tyrosine kinase isoform 1 precursor | nt | 41.14 | 6.13E–5 | 1.99 | Human | 42544153 |
| Keratin 24 | nt | 66.13 | 7.79E–6 | 2.01 | Mouse | 122425580 |
| Myeloblastin precursor (Proteinase 3) (PR-3) | nt | 22.37 | 5.81E–4 | 2.01 | Horse | 194238637 |
| Epsin 1 isoform b | nt | 33.32 | 1.41E–004 | 2.11 | Human | 194248095 |
The items are sorted accorind to ics score. Only the values for the highest scoring orthologous sequence are presented (marked byb).
ant: not tested AGO-binding activity.
bValues presented in table correspond to orthologous gene from marked organism.
It should also be noted that more than 40% of the highest scoring proteins presented in Table 3 have no experimentally defined function, and are mostly annotated as putative/predicted or ‘similar to’ other known proteins. An analogous, symmetric experiment involving calculation of scoring tables basing solely on characterized mammalian WG/GW proteins and screening the Arabidopsis genome, resulted in identification of known AGO interacting proteins. In this case at the top of the list were: the largest subunit of polV, NRPE1 (P-value = 3.29E–07), SPT5-like (P-value = 1.10E–08), GTB1 (P-value: 2.21E–06) and SDE3 (P-value: 1.31E–05), which precisely mirrors the list of known WG/GW motif containing proteins from Arabidopsis (Supplementary Data—unfiltered list of all identified Arabidopsis proteins).
DISCUSSION
Functionally and evolutionarily conserved WG/GW motifs are found in different protein families from a wide variety of organisms: from protozoa to man. The highly-divergent sequence of WG/GW domains as well as their variable amino-acid lengths and exact number of repeats make WG/GW platform detection very difficult. Here we have developed a tool to carry out an exhaustive search for WG/GW motif genes or proteins in raw genomic sequence or annotated protein libraries respectively, looking for potential AGO-binding platforms in the well-annotated genome of Arabidopsis thaliana. Our computational screening identified a small, well defined group of proteins. The identified proteins are good candidates for implication in AGO-related mechanisms, although direct experimental evidence is required to demonstrate their biological function. While direct analysis of the Arabidopsis genomic sequence identified no additional candidates, this probably reflects the high quality of annotation of this genome. Analysis based on nucleotide sequence will probably be more fruitful on recently-annotated or unannotated sequences.
Global analysis
The raw results from the search for proteins containing WG/GW motifs identified a considerable number of proteins. The motif by itself is very short and is also found very often in randomly-generated protein sequences. The filtering of candidate domains based on pure statistical criteria allowed us to remove the majority of false positives from the final list, however several (later manually excluded) passed the stringent (P ≤ 0.01) statistical criteria. Most of the ‘false’ domains that acquired significant dos scores represented compositionally biased protein fragments, rich in amino acids from the top of the scoring table (most frequently glycine). Such compositionally biased sequences were less frequent in the randomly shuffled data, and highly stringent statistical criteria were necessary to take into account these random compositionally biased fragments. False positives are frequently identified in protein motif searches, particularly for loosely-conserved functional sequences such as the WG/GW motif.
The ics score—which represents the measure of difference in composition between domains—was used as an additional criterion during the data filtering step. The cut-off value for the ics score represents the properties of the group of domains selected as initial data set for this analysis. It is important to note that both scores are sensitive to the length of the domain. The dos score has linear dependency—longer domains have more chance of acquiring higher scores. The ics score shows size dependency in situations in which not all of the amino acids are present in the domain (each domain shorter than 20aa). This is due to the penalty value for a lack of a given amino acid in the domain. In general, none of the manually or automatically identified domains which passed the first filtering criterion was shorter than 20aa (the average size of WG domains identified in this analysis is 184 aa), which is consistent with the proposed function of the domain as a molecular platform interfacing the assembly of functional biological complexes (11).
Domain boundaries are usually predicted based on conservation of sequence and structure between proteins having the same function. In this example it was especially difficult to define the borders of the domain precisely, because of the lack of sequence conservation. The initial, manual identification was based on sequence alignment and therefore was most probably error prone. Our automatic algorithm achieved on average very high sensitivity and selectivity scores when compared with manual detection. In some examples, however, it seems that computational detection performed better and identified boundaries which covered fragments of proteins with more amino acids located at the top of the scoring list. Additional experimental analyses will be needed to verify the correctness of the new predictions.
Using the dos score as a measure of compatibility of the new identified WG/GW motifs and source sequences selected at the beginning of the analysis, we can roughly group the domains into three clusters: strong (dos score > 150 half-bits), medium (dos score in range from 25 to 150 half-bits) and weak (dos score below 25 half-bits). As mentioned already, this classification represents not only the compositional structure of the domain but also reflects the size of the identified fragment. The ‘strong’ group contains only three members: NRPE1, a subunit of polymerase V, SPT5-like and GTB1 transcriptional factors—two of which have confirmed functions in RdDM in an AGO-dependent manner. Fourteen genes classified as ‘medium’ constitute a more diversified group, which includes proteins annotated as translation initiation factors, RNA-binding proteins, and the majority of proteins annotated as glycine-rich domain-containing. Even more divergent genes are classified in the ‘weak’ group. Here we find protein kinases, nucleotide/protein-binding proteins, ATP-dependent helicases, TPR-containing proteins as well as other examples of hnRNP and transcription factors. Beside the first group and highest scoring genes from the second group it is difficult to judge at this moment, if the other genes represent variants of the WG/GW domain (with different affinity for the interacting components) or should be classified as independent domains with a limited number of similar features to the WG/GW motif. However, we previously showed that, at least in vitro, only one WG/GW repeat is sufficient to bind Arabidopsis AGO4. Furthermore, the Arabidopsis genome encodes 10 different AGO proteins, the roles of all of which have not yet been clearly defined, and different AGOs may interact with different WG/GW regions (43). Obviously, experimental work will be needed to answer this question.
Overall, our exhaustive search identified 22 candidate AGO-interacting proteins (two of them representing splicing variants), most of which can be classed as either DNA or RNA-interacting proteins (Table 2). It is important to note that the procedure presented here successfully identified the entire source Arabidopsis proteins used as the starting point for the analysis. However, not all of them were highest scoring sequences—some of the newly-identified proteins showed higher or equal score values. This indicates that our composition-based approach to the identification of WG/GW-platform proteins was a successful strategy and identified proteins representing good candidates to be involved in AGO-mediated silencing of endogenous DNA. Among the newly-identified proteins, the implication of SDE3 in silencing mechanisms in Arabidopsis has already been demonstrated (41). It is clear that these proteins can only be classified for the moment as potential AGO-binding proteins and that further work is necessary to demonstrate a possible implication in AGO-related pathways. However, we have tested one candidate protein, encoded by gene At3g51940, and clearly demonstrated the AGO-binding capacity of its specific WG/GW-rich region.
Characteristics of WG/GW proteins
There are several striking features in the protein families presented here. First, we find members with and without the WG motifs in most families. The most likely explanation is that the ancestral genes lacked WG/GW motifs and that these were acquired by a process such as exon shuffling or exon capture. This hypothesis is supported by the structure of certain genes, such as NRPE1, SPT5-like and GTB1 proteins, in which the complete WG platform is encoded by separate exons in the 3′ region of the gene. SPT5-like proteins are conserved throughout eukaryotes and have been proposed to play closely related roles associated with active transcription (44,45). Reiterated WG/GW repeats of SPT5-like are already known to be sufficient and necessary for interaction with AGO4 proteins implicating their role in RNA-directed DNA methylation (28,29). There are in fact three genes in the Arabidopsis genome encoding SPT5-type proteins. Two have similar structure, with no detectable WG/GW motifs, only one (At5g04290) being identified by our approach (with application of stringent filtering criteria) and which encodes a protein having a long C-terminal extension (Figure 3A).
Alternative splicing is also observed in the WG/GW protein families and in two of them—GTB1 (At1g65440) (Figure 3B) and RNA-binding protein RGGA (AT4G16830)—it has an impact on the structure of the WG/GW domain itself. In other cases, the alternative transcripts encode unmodified WG/GW domains, but usually such families are composed of many proteins, which show a broad spectrum of compositional similarity scores.
Most of the WG/GW regions are composed of repeats which are poorly conserved apart from the WG/GW sequences themselves, both within individual families and with similar proteins in other plants ((11) and our unpublished observations). A notable exception to this is the SPT5-like protein. In contrast to the considerable divergence of WG/GW motifs in NRPE1, the protein encoded by At5g04290 contains 45 highly-conserved repeated motifs. However, in the majority of WG/GW proteins only the WG/GW residues are essential and conserved, and only the nature of the surrounding residues is important. These are mostly small hydrophilic, charged residues. As tryptophan is a hydrophobic amino acid it is possible that the hydrophilic regions in which the WG/GW motifs are located maintain the tryptophan residues accessible at the surface of the proteins rather than being embedded in hydrophobic regions.
Domain swapping
The results of the simulation of domain swapping clearly show the evolutionary conservation of the amino-acid composition of the WG/GW AGO-binding domain in organisms separated by long evolutionary distances. Despite very low sequence similarity between the Arabidopsis and human sequences (Figure 5A), the functional activity of the WG/GW domain is preserved. The algorithm developed in this work, which is based on the compositional scoring system, is capable of identifying such proteins and the best characterized proteins found during this bidirectional analysis are top reciprocal hits. Interestingly, plant WG/GW domain-containing proteins (NRPE1, SPT5, GTB1) identified by the use of the mammalian scoring table, showed higher score values than source mammalian domains. As this experiment shows, the composition of WG/GW platforms is a sufficient signal for identification of AGO-binding proteins and is a conserved feature of these proteins from Arabidopsis to human.
Several proteins identified by screening animal genomes with the plant specific scoring matrix represent predicted/hypothetical proteins which are good candidates as new, as yet undescribed, WG/GW-domain containing proteins. For example, the predicted hypothetical protein XP_001715475, which is located high on the list, contains several WG/GW octapeptide repeats with properties characteristic of the AGO-binding sites of TNRC6A and TNRC6B proteins in its N-terminal region. Apart from keratinocyte-related proteins, remaining mammalian genes identified as encoding WG/GW proteins correspond in the majority of cases to a limited number of families: ADP-ribosylation factor GTPase activating proteins, SCY1-like family of kinase-like proteins and serine/arginine rich proteins. Proteomic analysis of AGO-associated proteins by immunoprecipitation recently identified putative AGO-interacting proteins in man (46). These include RRM proteins, hnRNPs, helicases, among which is MOV10 [the human homolog of SDE3 (47)], TNRC6B (a GW182 paralog), zinc finger proteins and translation initiation factors. However, it must be noted that MOV10 sequence does not contain WG/GW motif (and therefore could not be identified in this screen) and other proteins may not directly interact with AGO protein.
The hornerin and dermatokine keratinocyte-associated proteins are probably false positives, identified due to the biased composition of the repetitive fragments, which represent low-complexity sequences rich in amino acid located at the top of the scoring matrix. For example, nearly 95% of the whole HRNR protein consists of tandem quasi-repetitive, glycine/serine-rich peptide sequences which maintain eight GW motif occurrences. The presence of proteins which probably have no AGO-binding capacity in these results with plant scoring matrices may be the result of three phenomena: (i) The plant matrix is not specific enough to distinguish between the genuine WG/GW proteins and other molecules involved in the formation of large molecular complexes and the proper screen should be conducted with the use of specific mammalian scoring tables. (ii) The signal coming only from the amino-acid composition of the domain, despite using two scores and various statistical measures, is too weak to be used to achieve a high level of specificity of the analysis. When higher threshold values are used it strongly affects the sensitivity of the algorithm. The basis of the method was to use very simple measures which very closely represent compositional properties of the WG/GW domain, but maybe more sophisticated approaches, like the use of artificial intelligence techniques, would produce more selective results by reducing the noise. It should also be noted that in the Arabidopsis screening with plant specific matrices some of the proteins may also not be genuine WG/GW domain containing molecules but rather a product of random bias of composition in the amino-acid sequence. (iii) The third option would be due to very low similarity which is hard to identify between the domains. Manual inspection failed to find any correlation between the keratinocyte-asscociated and WG/GW domains, but both classes of molecules are clearly involved in the formation of large molecular complexes—so they could represent a superclass of general protein–protein interaction domain.
As all commonly-available bioinformatics tools fail to identify the WG/GW domain in systematic analyses (e.g. BLAST, HMMER, Gibbs sampler—data not shown), the approach presented here fills the gap in the annotation tools for prediction of AGO-interacting proteins. By working with very weak signals of positionally-independent amino-acid sequence composition of the functional domain we should be able to identify the WG/GW proteins across all the major groups of living organisms. Some background noise present in the results may indicate common properties of the proteins or may result from method imperfections. However it should be noted, that even the well established bioinformatics methods sometime fail to identify related sequences (orthologs and paralogs) (48) or produce false positives.
Finally, although the original plant WG/GW platform was first identified in the polV NRPE1 gene, we cannot conclude that the WG/GW proteins we have identified are necessarily involved in polV-related mechanisms exclusively. Family members for a number of the proteins we have identified (SPT5-like, GTB1, the glutamine rich protein and the two translation elongation factors) are rather associated with other silencing mechanisms. Experimental validation of two of these proteins, SPT5-like and the product of gene At3g51940, indicates, however, that our approach is efficient in identification of potential AGO-interacting proteins in a wide variety of organisms. Careful examination of the identified domains in fact suggested the possibility of similar but distinct motifs, in which only the tryptophan residue is conserved and the glycine is replaced by various other amino acids. Recent studies have shown that WD-repeat-containing proteins present in all eukaryotes play important role in signal transduction, transcription regulation, cell cycle control and apoptosis. Repeated WD motifs are known to serve as platform for the assembly of protein complexes or mediators of transient interplay among other proteins (49–51). However, to extend this analysis and include such ‘degenerate’ tryptophan-containing motifs, we first need to obtain experimental evidence showing their biological function.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
WMK acknowledges financial support from Marie Curie Host Fellowships for the Transfer of Knowledge (MTKD-CT-2004). JC, DP, TL and RC acknowledge financial support from the Centre National de la Recherche Scientifique and the Agence Nationale de la Recherche, project ANR-08-BLAN-0206-01. Funding for open accesscharge: ANR-08-BLAN-0206-01.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Mulder NJ, Apweiler R. The InterPro database and tools for protein domain analysis. Current Protocols in Bioinformatics. 2008 doi: 10.1002/0471250953.bi0207s21. Chapter 2, Unit 2.7. [DOI] [PubMed] [Google Scholar]
- 2.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz H.-R, Ceric G, Forslund K, Eddy SR, Sonnhammer ELL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pontier D, Yahubyan G, Vega D, Bulski A, Saez-Vasquez J, Hakimi M.-A, Lerbs-Mache S, Colot V, Lagrange T. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 2005;19:2030–2040. doi: 10.1101/gad.348405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- 5.Onodera Y, Haag JR, Ream T, Nunes PC, Pontes O, Pikaard CS. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 6.Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kreil DP, Matzke M, Matzke AJ. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat. Genet. 2005;37:761–765. doi: 10.1038/ng1580. [DOI] [PubMed] [Google Scholar]
- 7.Ream TS, Haag JR, Wierzbicki AT, Nicora CD, Norbeck AD, Zhu J.-K, Hagen G, Guilfoyle TJ, Pasa-Tolić L, Pikaard CS. Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol. Cell. 2009;33:192–203. doi: 10.1016/j.molcel.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang L, Jones AME, Searle I, Patel K, Vogler H, Hubner NC, Baulcombe DC. An atypical RNA polymerase involved in RNA silencing shares small subunits with RNA polymerase II. Nat. Struct. Mol. Biol. 2009;16:91–93. doi: 10.1038/nsmb.1539. [DOI] [PubMed] [Google Scholar]
- 9.He X-J, Hsu Y-F, Pontes O, Zhu J, Lu J, Bressan RA, Pikaard C, Wang C-S, Zhu J-K. NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation. Genes Dev. 2009;23:318–330. doi: 10.1101/gad.1765209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lahmy S, Pontier D, Cavel E, Vega D, El-Shami M, Kanno T, Lagrange T. PolV(PolIVb) function in RNA-directed DNA methylation requires the conserved active site and an additional plant-specific subunit. Proc. Natl Acad. Sci. USA. 2009;106:941–946. doi: 10.1073/pnas.0810310106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El-Shami M, Pontier D, Lahmy S, Braun L, Picart C, Vega D, Hakimi M.-A, Jacobsen SE, Cooke R, Lagrange T. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 2007;21:2539–2544. doi: 10.1101/gad.451207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zilberman D, Cao X, Johansen LK, Xie Z, Carrington JC, Jacobsen SE. Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr. Biol. 2004;14:1214–1220. doi: 10.1016/j.cub.2004.06.055. [DOI] [PubMed] [Google Scholar]
- 13.Qi Y, He X, Wang XJ, Kohany O, Jurka J, Hannon GJ. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature. 2006;443:1008–1012. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- 14.Vaucheret H. Plant ARGONAUTES. Trends Plant Sci. 2008;13:350–358. doi: 10.1016/j.tplants.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 15.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eystathioy T, Chan EKL, Tenenbaum SA, Keene JD, Griffith K, Fritzler MJ. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol. Biol. Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell. Biol. 2009;11:1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 19.Eulalio A, Helms S, Fritzsch C, Fauser M, Izaurralde E. A C-terminal silencing domain in GW182 is essential for miRNA function. RNA. 2009;15:1067–1077. doi: 10.1261/rna.1605509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lazzaretti D, Tournier I, Izaurralde E. The C-terminal domains of human TNRC6A, TNRC6B, and TNRC6C silence bound transcripts independently of Argonaute proteins. RNA. 2009;15:1059–1066. doi: 10.1261/rna.1606309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takimoto K, Wakiyama M, Yokoyama S. Mammalian GW182 contains multiple Argonaute-binding sites and functions in microRNA-mediated translational repression. RNA. 2009;15:1078–1089. doi: 10.1261/rna.1363109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneider MD, Najand N, Chaker S, Pare JM, Haskins J, Hughes SC, Hobman TC, Locke J, Simmonds AJ. Gawky is a component of cytoplasmic mRNA processing bodies required for early Drosophila development. J. Cell. Biol. 2006;174:349–358. doi: 10.1083/jcb.200512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eulalio A, Tritschler F, Izaurralde E. The GW182 protein family in animal cells: new insights into domains required for miRNA-mediated gene silencing. RNA. 2009;15:1433–1442. doi: 10.1261/rna.1703809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jakymiw A, Pauley KM, Li S, Ikeda K, Lian S, Eystathioy T, Satoh M, Fritzler MJ, Chan EKL. The role of GW/P-bodies in RNA processing and silencing. J. Cell Sci. 2007;120:1317–1323. doi: 10.1242/jcs.03429. [DOI] [PubMed] [Google Scholar]
- 26.Ding L, Spencer A, Morita K, Han M. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol. Cell. 2005;19:437–447. doi: 10.1016/j.molcel.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 27.Ding XC, Grosshans H. Repression of C. elegans microRNA targets at the initiation level of translation requires GW182 proteins. EMBO J. 2009;28:213–222. doi: 10.1038/emboj.2008.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bies-Etheve N, Pontier D, Lahmy S, Picart C, Vega D, Cooke R, Lagrange T. RNA-directed DNA methylation requires an AGO4-interacting member of the SPT5 elongation factor family. EMBO Rep. 2009;10:649–654. doi: 10.1038/embor.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He X-J, Hsu Y-F, Zhu S, Wierzbicki AT, Pontes O, Pikaard CS, Liu H-L, Wang CS, Jin H, Zhu J-K. An effector of RNA-directed DNA methylation in arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell. 2009;137:498–508. doi: 10.1016/j.cell.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Partridge JF, DeBeauchamp JL, Kosinski AM, Ulrich DL, Hadler MJ, Noffsinger VJP. Functional separation of the requirements for establishment and maintenance of centromeric heterochromatin. Mol. Cell. 2007;26:593–602. doi: 10.1016/j.molcel.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 31.Till S, Lejeune E, Thermann R, Bortfeld M, Hothorn M, Enderle D, Heinrich C, Hentze MW, Ladurner AG. A conserved motif in argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat. Struct. Mol. Biol. 2007;14:897–903. doi: 10.1038/nsmb1302. [DOI] [PubMed] [Google Scholar]
- 32.Bednenko J, Noto T, Desouza L, Siu K, Pearlman R, Mochizuki K, Gorovsky M. Two GW repeat proteins interact with the tetrahymena argonaute and promote genome rearrangement. Mol. Cell. Biol. 2009;29:5020–5030. doi: 10.1128/MCB.00076-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 34.Cooke R, Raynal M, Laudié M, Grellet F, Delseny M, Morris PC, Guerrier D, Giraudat J, Quigley F, Clabault G, et al. Further progress towards a catalogue of all Arabidopsis genes: analysis of a set of 5000 non-redundant ESTs. Plant J. 1996;9:101–124. doi: 10.1046/j.1365-313x.1996.09010101.x. [DOI] [PubMed] [Google Scholar]
- 35.Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, Sakurai T, Nakajima M, Enju A, Akiyama K, Oono Y, et al. Functional annotation of a full-length Arabidopsis cDNA collection. Science. 2002;296:141–145. doi: 10.1126/science.1071006. [DOI] [PubMed] [Google Scholar]
- 36.Schoof H, Karlowski WM. Comparison of rice and Arabidopsis annotation. Curr. Opin. Plant Biol. 2003;6:106–112. doi: 10.1016/s1369-5266(03)00003-7. [DOI] [PubMed] [Google Scholar]
- 37.R Development Core Team. R: A Language and Environment for Statistical Computing. 2009. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 38.Hollander M, Wolfe D. Nonparametric Statistical Methods 2nd edn. Hoboken, NJ: Wiley-Interscience, John Wiley & Sons, Inc.; 1999. p. 787. [Google Scholar]
- 39.Eitoku M, Sato L, Senda T, Horikoshi M. Histone chaperones: 30 years from isolation to elucidation of the mechanisms of nucleosome assembly and disassembly. Cell. Mol. Life Sci. 2008;65:414–444. doi: 10.1007/s00018-007-7305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 41.Dalmay T, Horsefield R, Braunstein TH, Baulcombe DC. SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J. 2001;20:2069–2078. doi: 10.1093/emboj/20.8.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinformatics. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 43.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell. Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 44.Kaplan CD, Morris JR, Wu C, Winston F. Spt5 and spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 2000;14:2623–2634. doi: 10.1101/gad.831900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ardehali MB, Yao J, Adelman K, Fuda NJ, Petesch SJ, Webb WW, Lis JT. Spt6 enhances the elongation rate of RNA polymerase II in vivo. EMBO J. 2009;28:1067–1077. doi: 10.1038/emboj.2009.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Höck J, Weinmann L, Ender C, Rüdel S, Kremmer E, Raabe M, Urlaub H, Meister G. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haussecker D, Cao D, Huang Y, Parameswaran P, Fire AZ, Kay MA. Capped small RNAs and MOV10 in human hepatitis delta virus replication. Nat. Struct. Mol. Biol. 2008;15:714–721. doi: 10.1038/nsmb.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuzniar A, van Ham RC, Pongor S, Leunissen JA. The quest for orthologs: finding the corresponding gene across genomes. Trends Genet. 2008;24:539–551. doi: 10.1016/j.tig.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 49.Zeng CJT, Lee Y-RJ, Liu B. The WD40 repeat protein NEDD1 functions in microtubule organization during cell division in Arabidopsis thaliana. Plant Cell. 2009;21:1129–1140. doi: 10.1105/tpc.109.065953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lau C-k, Bachorik JL, Dreyfuss G. Gemin5-snRNA interaction reveals an RNA binding function for WD repeat domains. Nat. Struct. Mol. Biol. 2009;16:486–491. doi: 10.1038/nsmb.1584. [DOI] [PubMed] [Google Scholar]
- 51.Smith TF. Diversity of WD-repeat proteins. Subcell. Biochem. 2008;48:20–30. doi: 10.1007/978-0-387-09595-0_3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.