Abstract
While in the last decade mRNA expression profiling was among the most popular research areas, over the past years the study of non-coding RNAs, especially microRNAs (miRNAs), has gained increasing interest. For almost 900 known human miRNAs hundreds of pretended targets are known. However, there is only limited knowledge about putative systemic effects of changes in the expression of miRNAs and their regulatory influence. We determined for each known miRNA the biochemical pathways in the KEGG and TRANSPATH database and the Gene Ontology categories that are enriched with respect to its target genes. We refer to these pathways and categories as target pathways of the corresponding miRNA. Investigating target pathways of miRNAs we found a strong relation to disease-related regulatory pathways, including mitogen-activated protein kinase (MAPK) signaling cascade, Transforming growth factor (TGF)-beta signaling pathway or the p53 network. Performing a sophisticated analysis of differentially expressed genes of 13 cancer data sets extracted from gene expression omnibus (GEO) showed that targets of specific miRNAs were significantly deregulated in these sets. The respective miRNA target analysis is also a novel part of our gene set analysis pipeline GeneTrail. Our study represents a comprehensive theoretical analysis of the relationship between miRNAs and their predicted target pathways. Our target pathways analysis provides a ‘miRNA-target pathway’ dictionary, which enables researchers to identify target pathways of differentially regulated miRNAs.
INTRODUCTION
Despite recent advances in sequencing methodology, microarray expression profiling is still a major technique for studying natural and pathogenic biochemical processes. While in the past decade the analysis of coding RNA molecules, mostly messenger RNAs (mRNAs) were in the focus of research, the relevance of non-coding RNAs has not been realized as of recent years. Especially microRNAs (miRNAs) are of increased interest. Currently, 922 human miRNAs are annotated in the Sanger miRBase (1–3) version 14 and the amount of miRNAs is steadily increasing.
These endogenous non-coding small RNAs usually of length 19–23 nt are known to regulate the translation of the coding mRNAs in a sequence-specific manner. miRNAs seem to be involved in almost all biological processes, including cellular development, differentiation, proliferation or apoptosis (4,5). Evidently, these molecules also play an important role in cancer, as recently reviewed by Drakaki et al. (6). A variety of studies describe that miRNAs can function either on tumor suppressor genes or on oncogenes and thus acting as major regulators of gene expression. While they were so far considered to be negative regulators, recent studies impressively demonstrate that miRNAs can also have positive effects on gene expression (7).
Similar to transcription factors, miRNAs can bind perfectly or imperfectly in the 3′ untranslated region (UTR) of target genes and thereby regulate their expression. For gene regulation via miRNAs, mainly three different mechanisms are known, including (i) translation repression, (ii) direct mRNA degradation and (iii) miRNA-mediated mRNA decay (8). Mostly, miRNAs bind with imperfect complementarity to their targeted mRNAs and thereby guide mRNA translation repression. They interact with targeted mRNAs primarily through the so-called seed, a 6–8 nt long region at their 5′-end. This seed is known to be highly conserved in miRNA families across different species (9).
There is a steady progress in detecting the biological functions of miRNAs, with the target identification mediating the respective functions as the most challenging task. One commonly applied approach is to measure the reduction in target mRNA levels caused by an exogenously added miRNA, as described by Lim et al. (10). Potential targets are typically validated by using luciferase sensors containing the target 3′-UTR.
In addition to experimental approaches, a variety of computer-aided prediction algorithms have been developed (11–14). These algorithms are trained by well-known miRNA–mRNA interaction rules gained from microarray data in order to identify novel miRNA targets. Among the most popular algorithms is miRanda that is freely available at http://www.microrna.org (11). One of the most comprehensive resources for miRNA targets is MicroCosm, a web resource developed by the Wellcome Trust Sanger Institute and now hosted by the European Bioinformatics Institute (EBI) containing computationally predicted targets for microRNAs across many species. The targets of MicroCosm have been predicted with the miRanda algorithm. Beside miRanda, several other tools for target prediction are available. For a review on such tools see Bartel (15). While TargetScan (16), PicTar, (17) or PITA (18) use conservation information, other tools as RNA22 (19) do not rely on such data. The analyses in this work is based on MicroCosm since this algorithm acknowledges complementarity at the 5′-end of the microRNA, where a rather strict complementarity is required, excludes non-stable conformations by using the Vienna RNA folding approach and, in addition, checks whether the site is conserved in orthologous transcripts from other species. However, most of the proposed methods suffer from either high false positive or false negative rates thus showing insufficient specificity or sensitivity. One reason for the low specificity (and sensitivity) may be the lack of negative examples or miRNA-target pairs that are required for a suitable training of the classificators. To improve the false discovery rate, Bandyopadhyay and Mitra identified 300 negative examples using expression profiling of miRNAs and mRNAs and miRNA–mRNA structural interactions together with seed site conservation (20). Based on these data, their ‘TargetMiner’, which relies on a support vector machine, achieved a specificity of 69% and and a sensitivity of 67.8%. Recently, the challenges related to miRNA target prediction have been summarized by Barbato et al. in their work ‘Computational Challenges in miRNA Target Predictions: To Be or Not to Be a True Target?’ (21).
To further improve our understanding of the mode of action of miRNAs and their function, gene set analysis based approaches can be used. Most recently, the group of Hatzigeorgiou proposed two approaches, DIANA-microT (22) and DIANA-mirPath (23). The DIANA-mirPath software performs an enrichment analysis of multiple miRNA target genes comparing each set of miRNA targets to all known KEGG pathways (24,25) and thus is a valuable tool for elucidating targets that are affected by deregulated miRNAs.
The increasing amount of mRNA and miRNA expression data induces a strong demand for computer-aided tools that facilitate the integrative analysis of these data. Among the most popular tools developed for this purpose is ‘microRNA and mRNA Integrated Analysis’ developed by Nam et al. (26) that interprets miRNA and mRNA data in the context of gene ontologies and biochemical pathways.
Our study aims at an improved understanding of miRNA and mRNA relations by addressing three issues. First, as a sequel of the study by Hatzigeorgiou et al., we carry out a comprehensive gene set analysis of the miRNA target sets by considering not only KEGG pathways but also TRANSPATH networks (27), TRANSFAC (28) transcription factors and Gene Ontologies (GO) categories (29). Second, we perform a network analysis of all target genes of all miRNAs. Third, we propose a tool for screening differentially expressed mRNAs for enrichment of specific miRNA targets and apply this tool to expression profiles of 13 data sets of different cancer types containing together over 1.000 microarrays. Our tool is freely accessible as part of our comprehensive gene set analysis pipeline GeneTrail (30,31).
Taken together, our study contributes to an improved understanding of the interactions between miRNAs and putative target genes and also provides a comprehensive ‘miRNA-target pathway’ dictionary. This dictionary enables researchers to readily identify target pathways of differentially regulated miRNAs. The study also provides further evidence that miRNAs are key players in the regulation of oncogenic processes.
MATERIAL AND METHODS
Information resources and databases
As a resource for predicted miRNA targets, we downloaded the MicroCosm Targets (‘miRBase Targets Release Version v5’) (1–3) from the EBI. This information was integrated in our gene set analysis tool GeneTrail to have direct access to its different statistical evaluation methods and predefined biological categories as described in (30,32). For the network analysis, we imported the information for Homo sapiens from KEGG (24,25) into our biochemical network database (BNDB) (33) and implemented a graph data structure based on the boost graph library (34) for efficient usage of the network topology.
Network analysis
The regulatory network is modeled as a directed graph G = (V, E), where the vertices (nodes) V = {v1, … , vn} represent proteins, protein families, protein complexes or other participants and the directed edges e = (vi, vj) ∈ E represent reactions or interactions between these participants. For analyzing the connectivity of miRNA targets in detail, we computed the average distance between all targets of each miRNA. Since our considered regulatory network is directed, the distance of two nodes dist(vi, vj) is not necessarily equal to dist(vj, vi). Therefore, we chose for each pair (vi, vj) the minimum of these distances for the computation of the average distance. If there exists no path between two nodes, the distance was set to the diameter of the complete regulatory network to penalize the absence of a path. The sum of the pair distances is finally divided by the number of pairs considered. To estimate if the average distance of the m targets of one miRNA is significant, we carried out 1000 permutation tests for each target set size m. To this end, we randomly selected m nodes from the complete network and calculated the average distance for the random node set. Finally, the overall distribution of the average distances of the randomly selected nodes and the miRNA targets was compared by performing an unpaired two-tailed t-test.
Gene set enrichment analysis
For computing the statistical significance of an arbitrary biological category C given a sorted list of genes of size n, we apply the so-called unweighted ‘Gene Set Enrichment Analysis’ (GSEA) as proposed by Lamb et al. (35). Using a Kolmogorov–Smirnov-like test that computes whether the genes in C are equally distributed in the sorted list or accumulate on top or on bottom of the list, we determine if the considered category is significantly enriched or depleted. If l genes of the sorted list belong to C, we compute the running sum by processing the input list from top to bottom adding n − l to the running sum if the considered gene belongs to C or subtracting l otherwise. The value of interest is the running sum's maximal deviation from zero, denoted as RSC. The significance value of the score RSC can be calculated by a dynamic programming algorithm that computes the exact number of possible running sum statistics with higher deviation than RSC. For details on the implemented algorithm we refer to (32).
RESULTS AND DISCUSSION
The results presented in this work rely on predicted miRNA targets for Homo sapiens as annotated by MicroCosm that are based on the miRanda algorithm (11). The possible targets of each miRNA are tagged with a significance value, the lower this value is the higher the chance that the respective gene is actually targeted by the respective miRNA. In order to balance sensitivity versus specificity, we considered three significance levels (α-levels): 0.01, 0.001 and 0.0001. Notably, the highest specificity (fewest false positives) are reached at the lowest threshold value.
Furthermore, we provide (i) a comprehensive gene set analysis of the miRNA target sets, (ii) a network analysis of all target genes of all miRNAs and (iii) Gene Set Enrichment analyses of 13 cancer data sets, where we study the enrichment of miRNA targets in sets of genes (mRNA) that are differentially expressed in the corresponding tumor biopsies.
miRNA target enrichment analysis
In order to detect target pathways of miRNAs, we carried out standard over-representation analysis as described in (30). In brief, for each of the human miRNAs in the Sanger miRBase (1,2,4) we extracted all targets with significance value below a given α-level t. The resulting ∼800 gene sets for Homo sapiens were separately evaluated using GeneTrail (30) analyzing about 13 000 biological pathways and categories including KEGG pathways, TRANSPATH pathways, Gene Ontology terms and others using all human genes as background set. The resulting significance values have been adjusted by applying the Benjamini–Hochberg approach (36,37).
As expected, the number of overall target genes drops from 16 200 over 13 200 to 8500 for the target significance levels of 0.01, 0.001 and 0.0001, respectively. The following detailed analysis has been carried out with the most sensitive significance threshold of 0.0001. The respective tables for all thresholds can be found in the Supplementary Data.
Of 13 160 screened biological categories, 1766 are significant for at least a single miRNA. The highest number of hits are achieved by the categories ‘Metabolic Pathways’ (30), ‘Cell Cycle’ (23) and ‘Pathways in cancer’ (20) followed by a long list of disease relevant pathways including TGF-beta and MAPK signaling cascade (see also Table 1, categories which are significant for >10 miRNA target sets).
Table 1.
Categories that are most frequently enriched with miRNA target gene sets
| Category | Number of significant miRNA target gene sets |
|---|---|
| Metabolic pathways | 30 |
| Cell cycle | 23 |
| Pathways in cancer | 22 |
| Focal adhesion | 15 |
| TGF-beta signaling pathway | 13 |
| Fatty acid metabolism | 13 |
| Catalytic activity | 12 |
| Cellular ketone metabolic process | 12 |
| ECM-receptor interaction | 11 |
| Fc epsilon RI signaling pathway | 11 |
| Organic acid metabolic process | 11 |
| Carboxylic acid metabolic process | 11 |
| MAPK signaling pathway | 11 |
| Substrate-specific transporter activity | 11 |
| Substrate-specific transmembrane transporter activity | 11 |
| Oxoacid metabolic process | 11 |
| Transporter activity | 10 |
| E2F network | 10 |
| Valine, leucine and isoleucine degradation | 10 |
| p53 signaling pathway | 10 |
| Colorectal cancer | 10 |
| Toll-like receptor signaling pathway | 10 |
For target sets of 254 miRNAs, at least one significant category has been found. On average each miRNA has five significant categories. The miRNAs with the highest number of significant categories was miR-202 (90) followed by miR-101 (65). A list of miRNAs whose targets are enriched in >40 significant categories is provided in Table 2.
Table 2.
miRNAs with highest number of significant categories
| miRNA | Number of significant categories |
||||
|---|---|---|---|---|---|
| Gene Ontology | KEGG | TRANSFAC | TRANSPATH | Total | |
| hsa-miR-202 | 89 | 1 | 0 | 0 | 90 |
| hsa-miR-101 | 64 | 0 | 0 | 1 | 65 |
| hsa-miR-613 | 55 | 6 | 0 | 0 | 61 |
| hsa-miR-936 | 58 | 0 | 0 | 0 | 58 |
| hsa-miR-196a | 54 | 0 | 2 | 0 | 56 |
| hsa-miR-1 | 53 | 1 | 1 | 0 | 55 |
| hsa-let-7f | 49 | 0 | 1 | 0 | 50 |
| hsa-miR-302b* | 48 | 1 | 0 | 0 | 49 |
| hsa-miR-23b | 47 | 0 | 1 | 0 | 48 |
| hsa-miR-212 | 43 | 4 | 0 | 0 | 47 |
| hsa-miR-23a | 47 | 0 | 0 | 0 | 47 |
| hsa-miR-196b | 44 | 0 | 2 | 0 | 46 |
| hsa-miR-29c | 40 | 5 | 1 | 0 | 46 |
| hsa-miR-191 | 45 | 1 | 0 | 0 | 46 |
| hsa-miR-181c* | 45 | 0 | 0 | 0 | 45 |
| hsa-let-7a | 44 | 0 | 1 | 0 | 45 |
| hsa-miR-801 | 43 | 0 | 0 | 0 | 43 |
| hsa-miR-29a | 37 | 3 | 1 | 0 | 41 |
| hsa-miR-199b-5p | 39 | 1 | 0 | 0 | 40 |
| hsa-miR-29b | 36 | 3 | 1 | 0 | 40 |
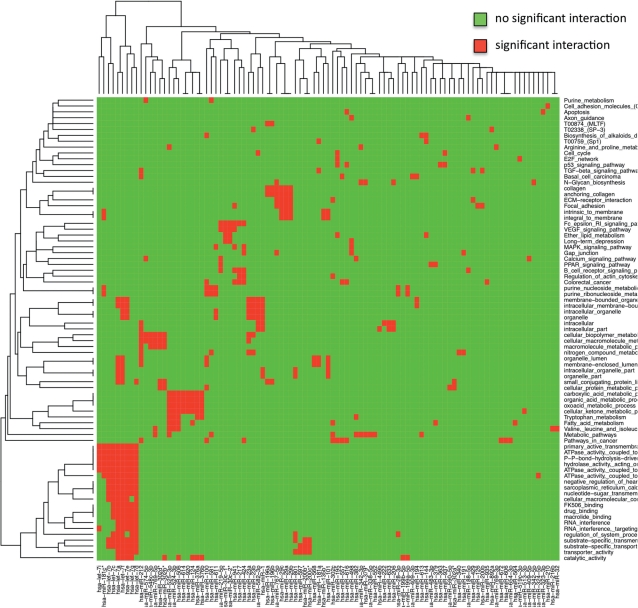
To improve our understanding of the putative pathways or biological categories that miRNAs may regulate or influence, we carried out a clustering approach. First, we removed miRNAs with <5 significant categories and categories that are enriched for <5 miRNA target sets. The clustering is based on a binary matrix that describes which categories (rows) are enriched with respect to the corresponding miRNA target sets (columns), i.e. the matrix contains a 1 at position (i, j) if the targets of miRNA j are enriched in category i and a 0 otherwise. Based on this matrix, we carried out a hierarchical clustering of miRNAs and categories separately. In more detail, we applied bottom-up hierarchical clustering using the Euclidian distance for measuring the distances between pairs of column and row vectors. The result of this clustering is shown in Figure 1. In the lower left corner of the heatmap, a cluster containing the let-7 family can be detected. These miRNAs seem to control, among others, categories as ‘transporter activity’, ‘RNA interference’, ‘macrolide binding’ or ‘drug binding’. The second cluster in the lower left corner contains miRNAs hsa-miR-525-3p, hsa-miR-524-3p, hsa-miR-506, hsa-miR-614, hsa-miR-920, hsa-miR-124, hsa-miR-376a and hsa-miR-376b that control metabolic pathways.
Figure 1.
This heatmap presents significant miRNA to putative pathway or category correspondences. The heatmap has a red spot at position (i, j) if the targets of an miRNA j are significantly enriched in category i. In the bottom left corner, a cluster containing the let-7 family can be detected.
We also addressed the question how specific the detected pathways or categories are and whether there are pathways or categories that are triggered by miRNAs in general. To this end, we set up three lists, containing genes that are targets of at least one miRNA at a target threshold level of 0.01, 0.001 and 0.0001. These lists containing 16.217, 13.168 and 8.508 genes have been processed using GeneTrail.
For the most unspecific miRNA target threshold of 0.01 no significant KEGG pathways have been detected, indicating that this threshold may lead to too many false positive miRNA–mRNA target relations. In contrast, the target threshold values of 0.001 and 0.0001 showed increased numbers of pathways and additionally entailed a significant overlap between both sets. For 0.001, we detected 12 putative target pathways while for 0.0001, 10 such pathways have been detected. Of these, five pathways were significant for both sets including ‘Basal cell carcinoma’, ‘ECM-receptor interaction’, ‘MAPK signaling pathway’, ‘Metabolic pathways’, and ‘Pathways in cancer’. A summary of all pathways and all threshold values is presented in Table 3. The four columns of this table describe the pathway name followed by the three different significance values used for the target prediction. The significance value for the gene set enrichment analysis has been hold constantly at 0.05 for all three analyses. Remarkably, this table shows that for the most unspecific miRNA target sets (P < 0.01), no significant pathways were detected while for the more specific miRNA target sets (P < 0.001 and P < 0.0001) several significant pathways were found.
Table 3.
KEGG pathways targeted by all miRNAs for different thresholds
| Pathway | 0.01 | 0.001 | 0.0001 |
|---|---|---|---|
| ABC transporters | – | 0.0362 | – |
| Aminoacyl-tRNA biosynthesis | – | – | 0.0050 |
| Basal cell carcinoma | – | 0.0154 | 0.0250 |
| Complement and coagulation cascades | – | 0.0447 | – |
| ECM-receptor interaction | – | 0.0447 | 0.0056 |
| Epithelial cell signaling in Helicobacter pylori infection | – | 0.0495 | – |
| Focal adhesion | – | – | 0.0090 |
| Glycine, serine and threonine metabolism | – | 0.0154 | – |
| Lysosome | – | 0.0362 | – |
| MAPK signaling pathway | – | 0.0018 | 0.0103 |
| Metabolic pathways | – | 0.0119 | 0.0173 |
| p53 signaling pathway | – | – | 0.0420 |
| Pathways in cancer | – | 0.0236 | 0.0269 |
| Purine metabolism | – | – | 0.0003 |
| Steroid biosynthesis | – | 0.0447 | – |
| Toll-like receptor signaling pathway | – | 0.0109 | – |
| TGF-beta signaling pathway | – | – | 0.0239 |
The values in the cells of the table correspond to the False Discovery Rate (FDR) adjusted P-values computed for the pathway. –= not significant.
miRNA target network analysis
For the network analysis of miRNAs, we retrieved the KEGG regulatory network for Homo sapiens from our BNDB (33). The resulting graph contains 1679 nodes and 2509 edges in total. Since not all predicted targets of the available 851 human miRNAs could be mapped onto the regulatory network, we removed those miRNAs where <10% of the targets could be mapped or the overall number of mapped targets was <3, resulting in 695 remaining miRNAs. In the following analyses, we used the threshold value of 0.001 for the miRNA targets.
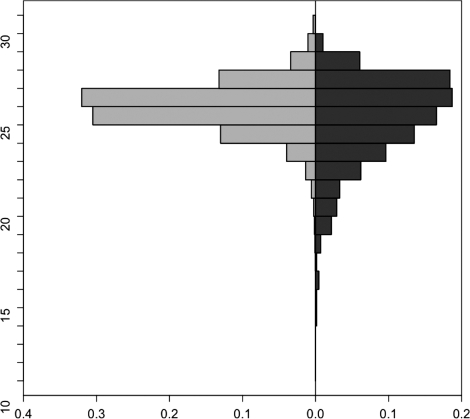
For the considered miRNAs, we wanted to investigate if the average distance between pairs of targets for the different miRNAs is significantly lower in comparison to randomly selected nodes from the complete network. To this end, we computed for each pair of targets or randomly selected nodes their distance. If no path between the considered nodes existed, we added the diameter of the complete graph as penalty term. The distribution of the average distances of randomly selected nodes against the average distances of the miRNA targets is shown in Figure 2. For testing the significance, we performed an unpaired two-tailed t-test, which yielded a P-value <10−9 confirming that miRNA target pairs have a lower average distance than randomly selected nodes.
Figure 2.
Comparison of the distributions of the average distances between randomly selected nodes on the left hand side and the miRNA targets on the right hand side. The y-axis of this back-to-back histogram presents the distance between nodes and the x-axis shows how many percent of random node pairs and of miRNA targets have this distance. The distribution of the miRNA targets is slightly shifted toward smaller distances.
Furthermore, we analyzed the coverage of all miRNA targets and the complete regulatory network considering only such nodes that are proteins (not protein families or complexes). When regarding the union of the targets of each of the 695 miRNAs that can be mapped to proteins in the network, we reach a coverage of the regulatory network of 640/825 (78%). If we take the number of all human genes having an amino acid sequence as reference set (25 673), we would expect to find about 414 proteins mapped on the network instead of 640, if we choose 12 885 miRNA targets from the reference set coding for proteins. The hypergeometric distribution test yields a P-value of <10−60 for obtaining such a coverage per chance. This finding significantly points out the crucial role these miRNAs play in the regulation of biochemical processes and indicates that the regulation takes place on basis of balance and interplay of concentrations of miRNAs rather than by regulating some few important targets or hubs in the network.
Deregulated cancer mRNAs as potential miRNA targets
In this section, we analyze whether the deregulation of genes in cancer could be caused by miRNAs. More exactly, we investigated if genes that are deregulated in cancer are statistically significant enriched with targets of certain miRNAs for a sophisticated set of different cancer entities and a total of over 1.000 microarray experiments. These arrays, extracted from the gene expression omnibus (GEO) (38), are measured using the same platform, GPL96. Here, a large control cohort is also available, the data set GDS596 (39) containing 158 samples from 79 physiologically normal tissues obtained from various sources.
We considered three slightly different scenarios. For most cancer data sets, we compared diseased to samples of the control data set GDS596. This has been done in 10 data sets. For two data sets where matched controls have been provided, we compared diseased samples with the matched controls. Finally, we also investigated one data set, where paired controls are available. In this case, healthy and diseased lung tissue of lung cancer patients were available. The three blocks in Table 4 show the results for the three different scenarios.
Table 4.
Overview of cancer miRNAs
| Entity | # samples cancer | # samples controls | # significant | # over represented | # under represented |
|---|---|---|---|---|---|
| Pheochromocytoma | 75 | 158 | 29 | 24 | 5 |
| Glioma (I) | 85 | 158 | 115 | 74 | 41 |
| Glioma (II) | 100 | 158 | 168 | 108 | 60 |
| Breast | 49 | 158 | 1 | 0 | 1 |
| Myeloma (I) | 50 | 158 | 68 | 57 | 11 |
| Sarcoma | 40 | 158 | 78 | 74 | 4 |
| Acute myeloid leukemia (AML) | 43 | 158 | 17 | 17 | 0 |
| Color. adenoc. | 37 | 158 | 90 | 64 | 26 |
| Prostate cancer | 22 | 158 | 92 | 88 | 4 |
| Lung cancer (I) | 129 | 158 | 130 | 96 | 34 |
| Malignant pleural mesothelioma (MPM) | 44 | 10 | 32 | 28 | 4 |
| Lung cancer (II) | 97 | 90 | 292 | 157 | 135 |
| Lung cancer (III) | 5 | 5 | 0 | 0 | 0 |
For all 13 data sets, we carried out the following analysis procedure by using GeneTrailExpress (31). First, we computed for each gene on the microarray the fold quotient of medians in the control and diseased group. The resulting list of genes sorted by the fold quotient serves as input for GeneTrail. On the basis of this list, we carried out analyses for detecting miRNAs whose targets are significantly up- or downregulated using standard GSEA. For the analysis of all 13 data sets, we set the target threshold to 0.001. In addition, we also investigated the influence of varying this threshold between 0.01 and 0.0001 for two data sets (glioma and lung cancer) exemplarily. The reported findings are finally compared in order to identify miRNAs that are either specific for certain cancer types or common to certain groups of cancer types.
All results are summarized in Table 4. Because of space restrictions we only discuss the most interesting results. The complete result set is available as supplementary Data of this article and will be integrated in a comprehensive database.
Pheochromocytoma
We extracted the data set GDS2113 containing 75 tumors and compared with to the control set GDS596. Here, we found 29 miRNAs, 24 over- and 5 underrepresented. The most significant miRNAs were hsa-miR-615-5p, hsa-miR-615-3p and hsa-miR-127-3, which are related to a manifold of cancer entities [breast neoplasms, colonic neoplasms, prostatic neoplasms, acute myeloid leukemia, ovarian cancer and others; (40)].
High-grade glioma
For high-grade gliomas (WHO grade III and IV astrocytomas), we considered two data sets of the GEO, GDS1975 and GDS1815, that have been analyzed separately.
We extracted the data set GDS1975 containing 85 tumors for comparison against the control set GDS596. Here, we found 115 miRNAs, 74 over- and 41 underrepresented. The most significant miRNAs were hsa-miR-101, hsa-miR-200b and hsa-miR-200c.
For the data set GDS1815 that contains 100 samples, we carried out the same analysis. Here, we detected by far more significant miRNAs, 168 of which 108 are over and 60 under represented. In addition, we compared the two sets of significant miRNAs. The first set contained 115 miRNAs, the second set 168 miRNAs. The overlap between both sets was 103, i.e. of the 115 miRNAs detected for the smaller set, 90% were also significant for the independent second set.
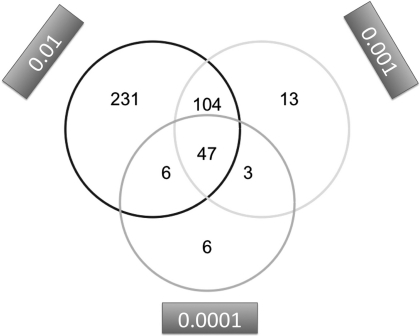
For the larger data set containing 100 samples, we also investigated the influence of the miRNA–mRNA target threshold. For the target gene thresholds of 0.01, 0.001 and 0.0001, 388 (205 up, 183 down), 168 (108 up, 60 down) and 62 (53 up, 9 down) genes have been identified. To reveal the similarity between the three target gene threshold sets, we produced a three-way Venn diagram, which is shown in Figure 3. This diagram outlines that, e.g. the 62 significant miRNAs for threshold 0.0001 split in the following four groups: (i) six are significant only for this threshold, (ii) three are also contained in the set for threshold 0.001, (iii) six are also contained in the set for threshold 0.01 and (iv) the majority of 47 miRNAs is significant for all three thresholds.
Figure 3.
Three-way Venn diagram for the three glioma data sets computed for the miRNA target thresholds 0.01, 0.001 and 0.0001.
The three miRNAs with highest significance values included hsa-miR-1, miR-200b and miR-144. These miRNAs are known to be deregulated in various human neoplasm's (40). Looking specifically at miRNAs known to be related to glioma tumors, we find several occurrences among the significant miRNAs in the analyzed data sets, including hsa-miR-181a and hsa-miR-181b. However, some other popular miRNAs connected to glioma are not detected to be significant in our study, including hsa-miR-221 and hsa-miR-222.
Non-autologous lung cancer samples
For this analysis, we considered the data sets GDS2771 containing lung cancer samples and controls and the data set GDS2373 containing lung cancer samples that have been compared to our standard control set GDS596. As Table 4 [lung cancer (I) and lung cancer (II)] shows, we detected 130 and 292 miRNAs, respectively. Computing the overlap between both sets, we found 101 miRNAs (77% of set lung cancer II) to be consistent between both sets. Many of these 101T miRNAs could be related to lung cancer according to secondary literature (40), including hsa-miR-181c, hsa-miR-18a, hsa-miR-19a, hsa-miR-203, hsa-miR-210, hsa-miR-30b, hsa-miR-30d and hsa-miR-30e. Similar to the results for glioma presented in the previous section, we do not detect some miRNAs to be significant for lung cancer that are described to be lung cancer related, including several members of the let-7 family (e.g. hsa-let-7b, hsa-let-7c, hsa-let-7d and hsa-let-7e) or hsa-miR-17.
Autologous lung cancer samples
As described above, we extracted expression profiles of squamous lung cancer biopsy specimens and paired normal specimens from 5 different patients [GDS1312, (41)] from the GEO (38). For this data set, a standard GSEA has already revealed a manifold of deregulated pathways, including core regulatory pathways as the cell cycle (31).
The GDS1312 data set contains 10 samples, 5 normal lung tissue expression profiles and 5 profiles of cancer patients. Here, we considered targets with thresholds of 0.01, 0.001 and 0.0001 separately.
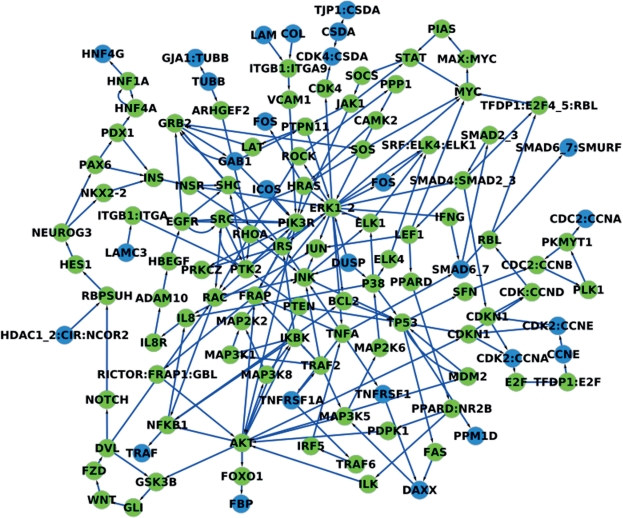
For the threshold value of 0.01 we detected 44 miRNAs to be significant. For 42 of these miRNAs, the targets were significantly upregulated in tumor tissue and for two downregulated. Most of these miRNAs have been linked to cancer in the literature, e.g. the most significant miRNA of these, hsa-miR-146b, is known to be downregulated in lung cancer (42). For the miRNA target threshold of 0.001, we detected no significant miRNAs (see Table 4), while for the threshold of 0.0001 we detected the three miRNAs miR-29a, miR-29b and miR-29c as significant. Notably, these miRNAs are also known to be downregulated in lung cancer [miRNAs miR-29a (43,44), miR-29b (42–44), miR-29c (43,44)). In addition, we carried out a blood screening of healthy individuals and lung cancer patients as described by Keller et al. (45) using the Geniom RT Analyzer (febit biomed gmbh, Heidelberg, Germany) and found these miRNAs at least four times downregulated compared with the control. For the most down-regulated miRNA, miR-29c, the target network is presented in Figure 4 and the significant categories for its target genes are listed in Table 5.
Figure 4.
This figure presents the target network of the miRNA hsa-miR-29c. The subgraph consists of the nodes of the shortest paths between the miRNA targets. The targets of the miRNA are colored in blue.
Table 5.
Overview of the significant categories for the target genes of miR-29c for a threshold value of 0.0001
| Gene Ontology | KEGG | TRANSFAC |
|---|---|---|
| Collagen | ECM-receptor interaction | T09836 (hsa-miR-29c) |
| Extracellular matrix part | Focal adhesion | |
| Proteinaceous extracellular matrix | Primary immunodeficiency | |
| Extracellular matrix | Small cell lung cancer | |
| Extracellular matrix structural constituent | Lysine degradation | |
| Structural molecule activity | ||
| Anchoring collagen | ||
| Extracellular region part | ||
| Basement membrane | ||
| Collagen type IV | ||
| Sheet-forming collagen | ||
| Fibrillar collagen | ||
| Extracellular region | ||
| Extracellular matrix organization | ||
| Membrane part | ||
| Intrinsic to membrane | ||
| Membrane | ||
| Integral to membrane | ||
| Chromatin | ||
| Microfibril | ||
| Protein binding, bridging | ||
| Localization | ||
| FACIT collagen | ||
| Collagen fibril organization | ||
| Androgen receptor binding | ||
| Cell adhesion | ||
| Biological adhesion | ||
| Fibril | ||
| Lysine N-methyltransferase activity | ||
| Protein-lysine N-methyltransferase activity | ||
| Histone-lysine N-methyltransferase activity | ||
| Extracellular structure organization | ||
| S-adenosylmethionine-dependent methyltransferase activity | ||
| Nuclear chromatin | ||
| Nuclear hormone receptor binding | ||
| Androgen receptor signaling pathway | ||
| Steroid hormone receptor binding | ||
| Histone methyltransferase activity | ||
| Hormone receptor binding | ||
| Protein methyltransferase activity |
If we now go back to our primary analysis of target pathways presented in ‘miRNA target enrichment analysis’ section 3.1, we detected for miRNAs miR-29b and miR-29c the KEGG pathway ‘Small cell lung cancer’ to be enriched with targets of these miRNAs. This means that we can find the predicted target pathway directly in the expression data providing evidence for the performance of the target pathway prediction.
Clustering of data sets
Finally, we compared the different miRNA data sets identified with considered cancer entities. To minimize a potential bias due to the usage of different control sets, we only used the 10 data sets with the same control set, GDS596. Analyzing these 10 data sets we found that the majority of miRNAs were rather specific. In more detail, 30.5% were significant for only one data set, 17.1% were significant for two data sets and 9.6% were significant for three data sets. However, some miRNAs have been detected in almost all data sets, including hsa-miR-548a-3p (8 of 10 sets), hsa-miR-200b, hsa-miR-200c, hsa-miR-1, hsa-miR-548c-3p, hsa-let-7f-2*, hsa-miR-548a-5p, hsa-miR-590-3p, hsa-miR-548d-3p and hsa-miR-548b-5p (all in 7 of 10 data sets). Looking into the literature, we find that, especially for miRNA 200b and 200c, a manifold of deregulations in different cancer entities is known [Cholangiocarcinoma, Colonic Neoplasms, Thyroid Neoplasms, Breast Neoplasms, Melanoma, Pancreatic Neoplasms, Adrenocortical Carcinoma, Ovarian Neoplasms, Meningioma (40)]. Looking back to the target pathways of these miRNAs, we find among other categories different metabolic and regulatory pathways, for miR-548a-3p e.g. the Cell cycle, Nucleotide excision repair, Aurora-B cell cycle regulation and some transcription factors [T00874 (MLTF) and T02338 (SP-3)]. The cell cycle is also significant for hsa-let-7f-2* and hsa-miR-548a-5p. Especially miR-200b that is known to be deregulated in cancers has also relevant target pathways, including basal transcription factors, focal adhesion, TGF-beta signaling pathway, c-Jun N-terminal kinase pathway, stress-associated pathways, the FBJ osteosarcoma oncogene, anatomical structure formation and cellular component assembly.
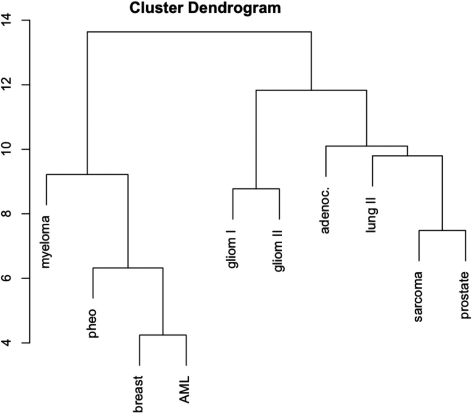
As for the target pathway analysis, we again carried out a hierarchical clustering using the binary matrix where each row represents a miRNA and each column a data set. A matrix entry equals ‘1’ if the respective miRNA has been detected for the corresponding data set, otherwise the entry equals ‘0’. As the cluster dendrogram in Figure 5 shows, the two independently measured glioma data sets cluster well together in the middle of the dendrogram. On the left of this dendrogram, the entities with smaller sets of significant miRNAs cluster together, where the overlap between the respective sets is comparably small and the miRNAs are rather specific. On the right side of the dendrogram, prostate cancer (showing 92 miRNAs) and soft tissue sarcoma samples (showing 78 miRNAs) cluster together. Remarkably, the sarcoma data set GDS1209 contains a mixture of gastrointestinal stromal sarcoma, leiomyosarcoma, dedifferentiated liposarcoma, pleomorphic liposarcoma, malignant fibrous histiocytoma and synovial sarcoma samples. Both data sets, the prostate and sarcoma set shown an overlap of 57 miRNAs.
Figure 5.
Dendrogram of significant miRNAs. The dendrogram shows the similarity of different miRNA cancer sets. The two independently measured glioma data sets cluster well together in the middle of the dendrogram.
CONCLUSION
Our computational analysis deepens the understanding of miRNAs and their putative targets in biochemical networks. We provide a comprehensive ‘dictionary’ of miRNAs to possible target pathways that may be regulated by these miRNAs. This dictionary enables researchers to look up the target pathways of differentially regulated miRNAs that can be used, e.g. for functional studies. As an additional key result, our study also provides further evidence that miRNAs are key players in the regulation of oncogenic processes by interpreting the results of 13 cancer microarray data sets. Thus, our results also provide evidence that an integrative screening of miRNAs and mRNAs can contribute to an improved understanding of human diseases, finally furthering disease diagnosis, prognosis and monitoring.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported in parts by funding of the German Ministry of Research Education (BMBF) under contract 01EX0806. Funding for open access charge: German Ministry of Research Education (BMBF) under contract 01EX0806.
Conflict of interest statement. A.K. is affiliate of febit biomed.
Supplementary Material
REFERENCES
- 1.Griffths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griffths-Jones S. miRBase: the microRNA sequence database. Methods Mol. Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- 3.Griffths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Medina PP, Slack FJ. microRNAs and cancer: an overview. Cell Cycle. 2008;7:2485–2492. doi: 10.4161/cc.7.16.6453. [DOI] [PubMed] [Google Scholar]
- 5.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 6.Drakaki A, Iliopoulos D. MicroRNA gene networks in Oncogenesis. Curr. Genomics. 2009;10:35–41. doi: 10.2174/138920209787581299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 8.Guarnieri DJ, DiLeone RJ. MicroRNAs: a new class of gene regulators. Ann. Med. 2008;40:197–208. doi: 10.1080/07853890701771823. [DOI] [PubMed] [Google Scholar]
- 9.Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, Mandel G, Hannon GJ. A biochemical approach to identifying microRNA targets. Proc. Natl Acad. Sci. USA. 2007;104:19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 11.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 13.Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z, Hatzigeorgiou A. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often anked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 15.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lall S, Grün D, Krek A, Chen K, Wang YL, Dewey CN, Sood P, Colombo T, Bray N, Macmenamin P, et al. A genome-wide map of conserved microRNA targets in C. elegans. Curr. Biol. 2006;16:460–471. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 18.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 19.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 20.Bandyopadhyay S, Mitra R. TargetMiner: MicroRNA target prediction with systematic identification of tissue specific negative examples. Bioinformatics. 2009;25:2625–2631. doi: 10.1093/bioinformatics/btp503. [DOI] [PubMed] [Google Scholar]
- 21.Barbato C, Arisi I, Frizzo ME, Brandi R, Da Sacco L, Masotti A. Computational challenges in miRNA target predictions: to be or not to be a true target? J. Biomed. Biotechnol. 2009;2009:803069. doi: 10.1155/2009/803069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, et al. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res. 2009;37:W273–W276. doi: 10.1093/nar/gkp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papadopoulos GL, Alexiou P, Maragkakis M, Reczko M, Hatzigeorgiou AG. DIANA-mirPath: Integrating human and mouse microRNAs in pathways. Bioinformatics. 2009;25:1991–1993. doi: 10.1093/bioinformatics/btp299. [DOI] [PubMed] [Google Scholar]
- 24.Kanehisa M. The KEGG database. Novartis Found. Symp. 2002;247:91–101. [PubMed] [Google Scholar]
- 25.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nam S, Li M, Choi K, Balch C, Kim S, Nephew KP. MicroRNA and mRNA integrated analysis (MMIA): a web tool for examining biological functions of microRNA expression. Nucleic Acids Res. 2009;37:W356–W362. doi: 10.1093/nar/gkp294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krull M, Pistor S, Voss N, Kel A, Reuter I, Kronenberg D, Michael H, Schwarzer K, Potapov A, Choi C, et al. TRANSPATH: an information resource for storing and visualizing signaling pathways and their pathological aberrations. Nucleic Acids Res. 2006;34:D546–D551. doi: 10.1093/nar/gkj107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Backes C, Keller A, Kuentzer J, Kneissl B, Comtesse N, Elnakady YA, Müller R, Meese E, Lenhof HP. GeneTrail-advanced gene set enrichment analysis. Nucleic Acids Res. 2007;35:W186–W192. doi: 10.1093/nar/gkm323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keller A, Backes C, Al-Awadhi M, Gerasch A, Küntzer J, Kohlbacher O, Kaufmann M, Lenhof HP. GeneTrailExpress: a web-based pipeline for the statistical evaluation of microarray experiments. BMC Bioinformatics. 2008;9:552. doi: 10.1186/1471-2105-9-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keller A, Backes C, Lenhof H.-P. Computation of significance scores of unweighted gene set enrichment analyses. BMC Bioinformatics. 2007;8:290. doi: 10.1186/1471-2105-8-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuentzer J, Backes C, Blum T, Gerasch A, Kaufmann M, Kohlbacher O, Lenhof H.-P. BNDB - The Biochemical Network Database. BMC Bioinformatics. 2007;8:367. doi: 10.1186/1471-2105-8-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee LQ, Lumsdaine A, Siek JG. Boost Graph Library, The: User Guide and Reference Manual. 1st edn. Amsterdam: Addison-Wesley Longman; 2001. [Google Scholar]
- 35.Lamb J, Ramaswamy S, Ford HL, Contreras B, Martinez V, Kittrell S, Zahnow CA, Patterson N, Golub TR, Ewen ME. A mechanism of cyclin d1 action encoded in the patterns of gene expression in human cancer. Cell. 2003;114:323–334. doi: 10.1016/s0092-8674(03)00570-1. [DOI] [PubMed] [Google Scholar]
- 36.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B. 1995;57:289–300. [Google Scholar]
- 37.Hochberg Y. A sharper bonferroni procedure for multiple tests of significance. Biometrika. 1988;75:800–802. [Google Scholar]
- 38.Barrett T, Edgar R. Gene expression omnibus: microarray data storage, submission, retrieval, and analysis. Meth. Enzymol. 2006;411:352–369. doi: 10.1016/S0076-6879(06)11019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl Acad. Sci. USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu M, Zhang Q, Deng M, Miao J, Guo Y, Gao W, Cui Q. An analysis of human microRNA and disease associations. PLoS ONE. 2008;3:e3420. doi: 10.1371/journal.pone.0003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wachi S, Yoneda K, Wu R. Interactome-transcriptome analysis reveals the high centrality of genes differentially expressed in lung cancer tissues. Bioinformatics. 2005;21:4205–4208. doi: 10.1093/bioinformatics/bti688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 43.Dalmay T, Edwards DR. MicroRNAs and the hallmarks of cancer. Oncogene. 2006;25:6170–6175. doi: 10.1038/sj.onc.1209911. [DOI] [PubMed] [Google Scholar]
- 44.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl Acad. Sci. USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keller A, Leidinger P, Borries A, Wendschlag A, Wucherpfennig F, Scheffler H, Huwer H, Lenhof HP, Meese E. miRNAs in lung cancer - Studying complex fingerprints in patient's blood cells by microarray experiments. BMC Cancer. 2009;9:353. doi: 10.1186/1471-2407-9-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.