Abstract
Three Rat1/Xrn2 homologues exist in Arabidopsis thaliana: nuclear AtXRN2 and AtXRN3, and cytoplasmic AtXRN4. The latter has a role in degrading 3′ products of miRNA-mediated mRNA cleavage, whereas all three proteins act as endogenous post-transcriptional gene silencing suppressors. Here we show that, similar to yeast nuclear Rat1, AtXRN2 has a role in ribosomal RNA processing. The lack of AtXRN2, however, does not result in defective formation of rRNA 5′-ends but inhibits endonucleolytic cleavage at the primary site P in the pre-rRNA resulting in the accumulation of the 35S* precursor. This does not lead to a decrease in mature rRNAs, as additional cleavages occur downstream of site P. Supplementing a P-site cleavage-deficient xrn2 plant extract with the recombinant protein restores processing activity, indicating direct participation of AtXRN2 in this process. Our data suggest that the 5′ external transcribed spacer is shortened by AtXRN2 prior to cleavage at site P and that this initial exonucleolytic trimming is required to expose site P for subsequent endonucleolytic processing by the U3 snoRNP complex. We also show that some rRNA precursors and excised spacer fragments that accumulate in the absence of AtXRN2 and AtXRN3 are polyadenylated, indicating that these nucleases contribute to polyadenylation-dependent nuclear RNA surveillance.
INTRODUCTION
The degradation and processing of several classes of eukaryotic RNAs require the 5′→3′ exoribonucleases, cytoplasmic Xrn1 and nuclear Rat1/Xrn2. For example, normal and specialized mRNA turnover, such as non-sense mediated, ‘No-Go’ and ARE-dependent pathways, involve the action of Xrn1 in different organisms from yeast Saccharomyces cerevisiae to Drosophila melanogaster to humans (1,2). In addition, the connection between RNA interference (RNAi) and 5′-end degradation was recently established: after endonucleolytic cleavage by RISC the resulting 3′ mRNA fragment is degraded by AtXRN4 in Arabidopsis thaliana and dXrn1 in Drosophila (3,4). Interestingly, yeast Xrn1 was reported to degrade some unspliced pre-mRNAs and CUT RNAs (cryptic unstable transcripts) in the cytoplasm (5–7). Although Xrn1 mainly localizes to the cytoplasm, it has the ability, at least in yeast, to contribute to some of the nuclear activities carried out by Rat1 (8).
The function of Rat1 was initially described in S. cerevisiae, involving the maturation of 5′-ends of 5.8S and 25S rRNAs and precursors of small nucleolar RNAs (snoRNAs) (9–11). More recently, degradation of defective mRNAs and some non-coding RNAs (ncRNAs), including rRNAs, hypomodified tRNAs and telomeric repeat-containing TERRA RNA, by Rat1 in the yeast nucleus has been reported (12–15). Interestingly, nuclear RNAs are targeted for destruction by the exosome and Rat1p as a result of becoming polyadenylated by a TRAMP complex containing the specific poly(A) polymerases Trf4/Trf5 (2,16,17). Xrn2/Rat1 was also shown in yeast and humans to be required for transcription termination of RNA polymerase II, not only due to degradation of the cleaved-off nascent transcript but also by promoting the recruitment of 3′-end processing factors (18–20). Yeast Rat1 is also important for efficient termination of RNA polymerase I at rDNA genes (21,22). These examples show the widespread involvement of 5′→3′ exonucleases in different aspects of RNA metabolism.
Plants only contain Rat1/Xrn2-like members of the XRN family, of which there are three in Arabidopsis thaliana: AtXRN2, AtXRN3 and AtXRN4 (3,23). AtXRN2 and AtXRN3 possess nuclear localization signals and are targeted to the nucleus, whereas AtXRN4 localizes to the cytoplasm. In contrast to yeast Xrn1, AtXRN4 is not a general mRNA decay enzyme, but degrades only specific transcripts, and the main role in mRNA decay in Arabidopsis is mediated by the exosome, a large complex with 3′→5′ exonuclease activity (24). Another important function of AtXRN4 is in miRNA-mediated RNA decay, where it degrades 3′ products of mRNA cleavage and affects the accumulation of small RNA-generating parent transcripts (3,25). In addition, AtXRN4 removes aberrant, uncapped mRNAs that may otherwise trigger RNA-dependent RNA polymerase-mediated post-transcriptional gene silencing (26). Two nuclear exonucleases, AtXRN2 and AtXRN3, may also be involved in the RNAi pathway, as mutations in these genes were reported to suppress the transgene silencing in hypomorphic mutants of AGO1, one of the major players in this pathway (27). The set of key AtXRN2 and AtXRN3 substrates, however, are still unknown, even though AtXRN3 is essential for proper plant development [(27), this study]. As only a limited number of transcripts are affected in the xrn4 mutant and the general 5′→3′ decay pathway is present in all eukaryotes studied to date, it seems possible that the remaining nuclear exonucleases AtXRN2 and AtXRN3 may substitute for, or assist AtXRN4.
The 18S, 5.8S and 25–28S rRNAs are transcribed by RNA polymerase I (Pol I) as a large precursor (35S in yeast, 40S in Xenopus laevis and 45S in mammals) that undergoes complex post-transcriptional processing to remove external transcribed spacers (5′ETS and 3′ETS) and internal transcribed spacers (ITS1 and ITS2) to release mature rRNAs. This process involves multiple endonucleolytic and exonucleolytic steps carried out by ribonucleoprotein complexes (RNP) mainly in the nucleolus. Processing events and participating factors are most precisely defined in yeast and to some degree in Xenopus, human and mouse cells (28–30). General principles of the processing pathway are conserved throughout eukaryotes. However, several events may vary substantially between different organisms, including the order of processing steps and the position of cleavages. For example, depending on the organism or even cell type, the initial steps of the pathway proceed either via cleavages in the 5′ ETS (yeast, Drosophila, Xenopus somatic cells), ITS1 (HeLa cells) or sometimes by both (Xenopus oocytes). The primary cleavage in the 5′ ETS is located either at site A′ near the precursor 5′-end in mammals and plants or at site A0 in yeast and Xenopus, closer to the 5′-end of the 18S, whereas two cleavages, A′ and A0, have recently been described in mouse (30–34). In turn, co-transcriptional processing in the 3′ETS by RNase III-type enzymes, Rnt1 and AtRTL2, has been shown in S. cerevisiae and Arabidopsis, respectively (35,36).
Early 5′ETS cleavages that generate mature 18S strictly require the interaction of pre-rRNA with U3 snoRNA and the presence of other snoRNAs, including U14, snR10 and snR30 in yeast and U14, U22, U17, E2 and E3 in vertebrates (28,30). In yeast and human cells, a large 80S small ribosomal subunit (SSU) processome complex or, alternatively, the 90S pre-ribosome, containing U3 snoRNP and several additional proteins, were reported to be involved in the synthesis of 18S (37,38). In crucifer plants, Arabidopsis, cauliflower, cabbage and radish, primary endonucleolytic cleavage at a conserved site P in the 5′ETS, corresponding to A′, was also shown to be snoRNP-dependent. A 600-kDa U3 snoRNP (NF D) complex, that was demonstrated to carry out this cleavage in vitro, contains nucleolar factors that are involved in pre-rRNA processing, including U3 and U14 snoRNAs, fibrillarin, NOP5/Nop58, Diskerin/Cbf5p and the the nucleolin-like protein (33,39,40). In addition to snoRNP complexes, maturation of rRNA subunits requires several endonucleases (including RNases III and MRP and Nob1) and exonucleases: 5′→3′ Rat1/Xrn2 and Xrn1 and the 3′→5′ multicomponent exosome (28,29,41).
Here, we report that one of the Arabidopsis nuclear 5′→3′ exonucleases, AtXRN2, contributes to the primary processing event in the 5′ETS. Endonucleolytic cleavage at site P is inhibited in the absence of AtXRN2 leading to strong accumulation of the entire pre-rRNA molecule and various intermediates, including a precursor to 18S that is not processed in the 5′ETS. This defect, however, does not affect the level of mature rRNAs as additional cleavages take place downstream of P. We propose a model in which exonucleolytic shortening by AtXRN2 of the exceptionally long Arabidopsis 5′ETS enables endonucleolytic processing at site P by the U3 snoRNP complex. In addition, AtXRN2 and AtXRN3 are involved in the 5′-processing of 5.8S rRNA and degrade some fragments excised from pre-rRNA during processing as well as precursors and intermediates that are targeted for destruction by polyadenylation.
Our work adds to the description of the pre-rRNA processing and nuclear surveillance pathways in plants, which, in comparison to other organisms, are relatively uncharted.
MATERIALS AND METHODS
Plant material and growth conditions
The wild-type ecotype Columbia (Col-0) of Arabidopsis thaliana was used in this study. Three homozygous xrn2 mutant lines, SALK_041148 (xrn2-1), SALK_114258 (xrn2-3) and SALK_073255 (xrn2-4), containing T-DNA insertions in the gene At5g42540 were identified. Seeds were surface-sterilized with 30% bleach/0.02% Triton-X100 solution and grown on Murashige and Skoog (MS) medium supplemented with 3% (w/v) sucrose and 0.6% agar (42), under a 16 h light/8 h dark photoperiod. Transgenic xrn3 RNAi lines were generated with the XRN3-RNAi stem-loop construct formed by sense and antisense sequences of the AtXRN3 cDNA in opposing orientations and separated by an 800-bp PDK intron (43). The AtXRN3 cDNA fragment (611-bp between 2135 and 2745 nt relative to the ATG) was amplified using a p1941 plasmid (23) and primers from X3-1 to X3-8 (oligonucleotides are listed in Supplementary Table S1) and the resulting PCR products were cloned into the pKANNIBAL vector (43). The cassette containing the CaMV 35S promoter, XRN3-RNAi construct and the OCS terminator was then moved using NotI sites into the binary vector pART27 (44). Agrobacterium tumefaciens strain GV3101 carrying the resulting plasmid with the XRN3-RNAi construct was used to transform Arabidopsis plants as described (45). Seeds from A. tumefaciens-treated plants were selected on MS plates containing 70 mg/l kanamycin and kanamycin-resistant plantlets were transferred to soil. The level of AtXRN3 mRNA was tested by northern blot on total RNA extracted from homozygous 14-day-old xrn3 seedlings. Mutant lines xrn3-3 and xrn2-1 xrn3-3 were a kind gift of Allison Mallory (INRA, Versailles).
Yeast strains, plasmids and media
Yeast strains yRP841 (MATα, trp1-Δ1, ura3-52, leu2-3,112, lys2-201, cup1::LEU2/PGK1pG/MFA2pG) and yRP884 (MATα, trp1-Δ1, ura3-52, leu2-3,112, lys2-201, cup1::LEU2/PGK1pG/MFA2pG, XRN1::URA3) were kindly provided by Roy Parker (University of Arizona). For expression of AtXRN2-TAP protein, AtXRN2 cDNA (restriction sites KpnI-NotI) with the N-terminal TAP-tag (restriction sites HindIII-KpnI) was cloned into a pYES2 vector under the control of the galactose-inducible GAL1/10 promoter and transformed into the xrn1Δ strain (yRP884) as described (46). Expression of AtXRN2-TAP was induced by shifting cells grown to early exponential phase at 30°C in synthetic medium (SC, 0.67% yeast nitrogen base, supplemented with required amount of amino acids and nucleotide bases) without uracil supplemented with 2% raffinose and 0.8% glucose to SC medium containing 2% galactose for 18 h. The expression of AtXRN2-TAP fusion protein was confirmed by western blot using peroxidase–anti-peroxidase antibody. For complementation tests in yeast rat1-1 and rat1-1 xrn1Δ strains, full-length AtXRN2, AtXRN3 and AtXRN4 cDNAs were cloned into a p415TEF vector (47) between BamHI-PstI, PstI and SpeI-Sal sites, respectively.
AtXRN2 activity assay in plant cell extracts
AtXRN2-TAP was purified as described (48) with rabbit IgG Fast Flow Sepharose (GE Healthcare) from whole cell extracts of xrn1Δ yeast expressing the fusion protein or carrying an empty vector. The purified activity was tested in plant extracts prepared from 14-day-old A. thaliana seedlings by homogenization using a mortar and pestle in liquid nitrogen and extraction buffer (150 mM NaCl; 50 mM Tris–HCl pH = 8; 2.5 mM MgCl2; 0.1% Triton X-100; 1 mM DTT; 1 mM PMSF; 10 mM Vanadyl ribonucleoside complex, Sigma; Complete Protease Inhibitor Cocktail, Roche). AtXRN2 nuclease activity assay contained 150 µl of cell extract and 50 µl of purified IgG-bound AtXRN2-TAP protein. The reaction mixture was incubated at 30°C for 30 min and stopped with phenol–chloroform. Total RNA was extracted and analysed by primer extension.
RNA methods
Total RNA was isolated from 14-day-old seedlings using Trizol reagent (Sigma) according to the manufacturer’s instructions. Low-molecular weight RNAs were separated on 6% acrylamide/7 M urea gels and transferred to a Hybond N+ membrane (GE Healthcare) by electro-transfer. High-molecular-weight RNAs were analysed on 1.1% agarose/6% formaldehyde gels and transferred to a Hybond N+ membrane by capillary elution. γ-32P 5′-end-labelled p1–p12 and p34–p34 oligonucleotides (Supplementary Table S1) were used as probes against precursor and mature RNAs. 7SL probe (p13) was used as a loading control. Random primed probes were amplified on a cDNA template using respective primers and DECAprimeTM II labeling kit (Ambion). Quantification of northern blots was performed using Storm 860 PhosphorImager and ImageQuant software (Molecular Dynamics). Primer extension was performed as described (49) using 8 µg of total RNA and γ-32P 5′-end-labelled primers, p23 and p24 for detection of cleavage sites P and P1, respectively, and p37–p41 for detection of sn/snoRNAs. For the 5′ RACE, cDNA was synthesised using total RNA, p1 primer and SuperScript III reverse transcriptase (Invitrogen). Second-strand synthesis and adapter ligation was performed using Marathon cDNA Amplification Kit (BD Biosciences). PCR was amplified with primer p25 and a primer complementary to the adaptor ligated to cDNA. Resulting PCR product was cloned into pCR-BluntII-TOPO (Invitrogen) and sequenced. Circular RT-PCR (CRT-PCR) was carried out on total or poly(A)+ RNA extracted from xrn2-3 seedlings that had been circularized using T4 RNA ligase (NEB). RT-PCR (reverse-transcription PCR) reactions were carried out with primers p25 and p26, products were purified, cloned into pCR-BluntII-TOPO vector (Invitrogen) and sequenced using M13 Reverse Primer.
Bioinformatics
Protein sequence alignment was generated using the CLUSTAL W algorithm (http://www.ebi.ac.uk/clustalw/) with default parameters (50). Phylogenetic analysis was performed using PHYML (http://mobyle.pasteur.fr/cgi-bin/portal.py?form=phyml) and the unrooted tree was drawn with TreeDyn198.3. Protein motifs in PROSITE and Pfam databases and biochemical parameters were analysed using Motif Scan (http://myhits.isb-sib.ch/cgi-bin/motif_scan/) and ProtParam (http://www.expasy.ch/tools/protparam.html), respectively.
Additional methods are described in Supplementary Data.
RESULTS
Isolation of mutant lines in AtXRN2 and AtXRN3 genes
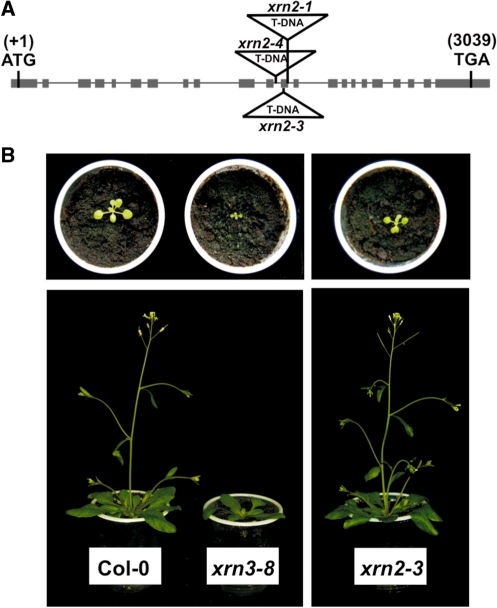
Of the three members of the Arabidopsis XRN family of Xrn2/Rat1 orthologues, cytoplasmic AtXRN4 functions in degradation of a relatively small number of transcripts (3,23,51,52). To find out whether the remaining nuclear exonucleases, AtXRN2 and AtXRN3, also contribute to mRNA decay, three homozygous xrn2 mutants with T-DNA inserts in their genes from the SALK collection (Figure 1A) were identified by PCR screening and tested by Southern hybridization (Supplementary Results). Neither of these mutants had any discernible morphological phenotype (Figure 1B). Based on Southern blot results, the analysis of the expression of AtXRN2 mRNA and the in vivo complementation tests for the possible residual enzymatic AtXRN2 activity in the mutants (Supplementary Results), the xrn2-3 line was selected for detailed analysis and the remaining two xrn2 lines were used at times to confirm observed phenotypes.
Figure 1.
Characterization of xrn2 and xrn3 mutant lines. (A) Structure of the AtXRN2 (At5g42540) gene. Exons are represented by grey bars, the location of T-DNA insertions are indicated. (B) 14-day-old (top) and 30-day-old (bottom) wild-type, xrn2 and xrn3 plants.
In contrast to xrn2 T-DNA insertions, we were unable to obtain homozygous plants from four heterozygous xrn3 lines, which is consistent with the reported embryo-lethality of xrn3 loss-of-function mutants (27). To circumvent this problem, expression of AtXRN3 was silenced using a double-stranded RNAi approach that is known to result in varying degrees of silencing in independent transformants (53). The obtained xrn3-4 and xrn3-8 RNAi plants had significantly delayed growth, but did not show developmental defects, such as serrated leaves, similar to those described for the xrn3-3 hypomorphic mutant (27) (Figure 1B). One homozygous line, xrn3-8, that had a 70% reduction of AtXRN3 in the T3 generation was chosen for further analysis.
AtXRN2 degrades polyadenylated rRNAs and pre-rRNAs but appears non-essential for mRNA decay
Expression and mRNA stability profiles in xrn2-3 mutant were analysed using Affymetrix Arabidopsis ATH1 Gene-Chip microarrays. Total RNA was isolated from xrn2-3 and corresponding wild-type (Col-0 ecotype) seedlings at times 0 and 120 min following transcriptional inhibition in the presence of cordycepin. After statistical analysis of two independent biological replicates, no unstable mRNA was found to have significantly altered level or stability in the mutant compared to the wild-type (data not shown). Since the probes on the ATH1 array are predominantly at the 3′ end of the genes these results represent the sum of full-length transcripts and 3′ fragments derived from mi-RNA dependent cleavage. These data indicate that AtXRN2 on its own is not necessary for general mRNA decay. Also the data did not provide evidence of AtXRN2 involvement in the specialized degradation of miRNA targets. Analysis of mRNA decay in the xrn3-8 mutant will be published elsewhere.
Surprisingly, the signal originating from the 3′-end of 25S rRNA, used as one of the Arabidopsis control genes in the Affymetrix Gene-Chip, showed a marked 3.7-fold increase in the xrn2-3 mutant for the two biological replicates, but not in xrn3-8 (data not shown). In contrast, the signal from another rRNA, 5S, which is transcribed by RNA polymerase III, was not elevated (data not shown). To verify this effect, the level of mature 25S rRNA in xrn2 mutants was evaluated using northern hybridization with oligonucleotide probes p8 and p9 complementary to the 5′- and 3′-ends of the molecule (regions starting at positions +56 and +3057, respectively; Figure 2A and B and data not shown). eIF-4A mRNA, which was not affected in the mutant, was used as a loading control. In contrast to the microarray data, there was no change in 25S rRNA in plants lacking AtXRN2. Similar results were obtained for 18S and 5.8S rRNAs using probes p10, p11 and p7 that are complementary to these species. The level of mature rRNAs was also not affected in xrn3-8, xrn3-3 and double xrn2-1 xrn3-3 mutants (Figure 2A and data not shown).
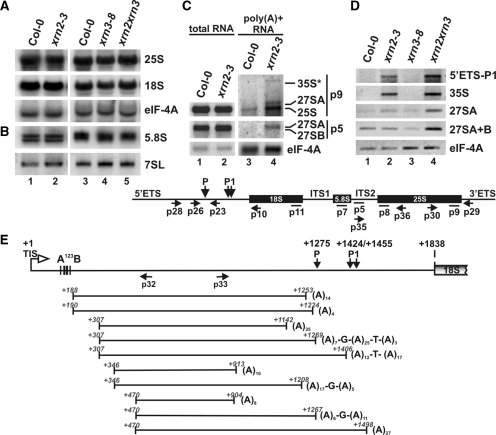
Figure 2.
Polyadenylated mature, precursor and intermediate rRNAs accumulate in the absence of AtXRN2. (A and B) Northern analysis of mature rRNAs in xrn2 and xrn3 mutants. Total RNA was extracted from wild-type, xrn2-3, xrn3-8 and xrn2-1 xrn3-3 seedlings and separated on 1.1% agarose (A) or 6% polyacrylamide (B) gels and hybridized with probes specific for 5′ regions of 25S (p8) and 18S rRNAs (p10) as well as 5.8S (p7). Hybridizations for eIF-4A mRNA and 7SL RNA (p13) were used as loading controls. (C) Northern analysis of total and poly(A)+ RNA extracted from wild-type and xrn2-3 inflorescences, using probes p9 (25S rRNA) and p5 (27S pre-rRNA). rRNA, pre-RNA and intermediates detected in (A–C) are indicated on the right. (D) RT-PCR on total RNA from wild-type, xrn2-3, xrn3-8 and xrn2-1 xrn3-3 lines. Reverse transcription was performed using oligo(dT)20 and PCR using primers p28 and p23 for detection of the 5′ETS fragment, p10 and p26 for 35S, p29 and p30 for 27SA, and p35 and p36 for both 27SA/27SB pre-rRNAs. RT-PCR for eIF-4A was used as a control. The structure of the pre-rRNA with location of probes (solid lines) and PCR primers (arrows) used is shown below. (E) CRT-PCR on poly(A)+ RNA extracted from xrn2-3 inflorescences. Top diagram represents the structure of 5′ETS with primers used for PCR. Indicated are: transcription initiation site TIS (+1); conserved cluster A123B; cleavage sites P (+1275) and P1 (+1423/+1454); 5′-end of 18S rRNA (+1836). RNA fragments identified as sequenced clones are represented below as horizontal lines, their 5′ -and 3′-ends are shown in italics and the number of adenine residues is indicated at the 3′-end of each molecule.
Affymetrix arrays are performed on poly(A)+ cRNA samples synthesized from cDNA that is reverse transcribed from total RNA using oligo(dT). The observed result may therefore correspond to the increase in the abundance of polyadenylated 25S rRNA species. The existence of polyadenylated stable RNAs, including rRNAs and pre-rRNAs, which are targeted for degradation by the exosome, and above all by its nuclear component Rrp6, was recently reported in yeast, humans and plants (16,17,24,54–56). A minor role of yeast Rat1 in these processes has also been shown, particularly in the absence of Rrp6 (13). To test whether this is also the case for AtXRN2 in Arabidopsis, the level of polyadenylated 25S rRNA and different pre-rRNAs in xrn2-3 versus Col-0 was assessed by northern analysis of the poly(A)+ fraction selected on an oligo(dT) column (Figure 2C). Hybridization with probe p9 revealed that polyadenylated 25S rRNA was 2.7-fold more abundant in xrn2-3 than in Col-0. At the same time some poly(A)+ pre-rRNAs, such as a full-length 35S or 27SA, an extended precursor to mature 25S and 5.8S, also increased, as shown by hybridization using probes p9 and p5 (Figure 2C and data not shown).
These results were verified by RT-PCR using oligo(dT) for the synthesis of cDNA and primers specific for the 5′ETS, 35S/33S, 27SA and 27SA/27SB RNAs (see Figure 3A for the description of pre-rRNAs). Polyadenylated forms of the 5′ETS, 35S/33S and 27SA were increased in xrn2-3 plants, but not in either of the xrn3 lines (Figure 2D and data not shown). Interestingly, much stronger accumulation of all tested poly(A)+ species was observed in the double xrn2-1 xrn3-3 mutant, indicating some level of redundancy between these two nucleases.
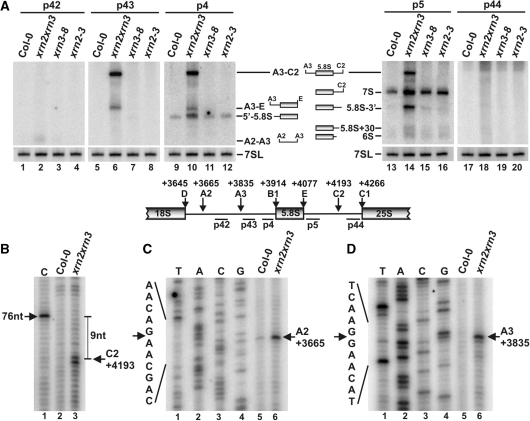
Figure 3.
5′ processing of 5.8S and 25S rRNAs involves AtXRN2/3. (A) Northern analysis of 5.8S processing in wild-type (Col-0) and xrn2-3, xrn3-8 and xrn2-1 xrn3-3 mutants. Total RNA isolated from seedlings was separated on 6% polyacrylamide gels and hybridized with probes located along ITS1 (probes p42, p43 and p4; lanes 1–12) and ITS2 (probes p5 and p44; lanes 13–20). Names and schematic representations of rRNA precursors and intermediates are shown between the two sections. 7SL RNA (p13) was used as a loading control. (B–D) Mapping cleavages at sites C2 in ITS2 (B) and A2 and A3 in ITS1 (C and D) by primer extension in wild-type (Col-0) and xrn2-1 xrn3-3 plants. Primers p44, p4 and p42 were used to detect cleavage at C2, A3 and A2, respectively. Primer extension reactions were separated on 6% sequencing polyacrylamide gels alongside DNA sequencing (shown on the left) using the same primers on a PCR product encompassing ITS1 (lanes 1–4, C and D). Cleavage at site C2 was mapped relative to RNA ladder of a product 76 nt in length (B). Positions of cleavages relative to the TIS are shown on the right. The structure of the relevant region of the pre-rRNA with location of cleavage sites, probes and primers used is shown between (A) and (B).
In a complementary approach, Circular RT-PCR (CRT-PCR) on poly(A)+ RNA isolated from Col-0 and xrn2-3 seedlings, followed by sequencing, was carried out for 5′ processing products enclosing the 5′ETS. In this technique total RNA circularized with T4 RNA ligase is used as a template for RT-PCR using primers against 5′ and 3′ regions of RNA of interest, designed in such a way that only circularized molecules are amplified. This analysis did not yield any products for Col-0 samples but 10 out of 14 sequenced clones obtained for the xrn2-3 mutant contained poly(A) tails ranging from 4 to 35 nt with the occasional occurrence of non-adenosine residues (Figure 2E). Intermediates identified in this manner started at a few different but recurring sites (e.g. +188–190, +307, +346, +470 relative to the start of transcription) downstream of the A123B cluster (33) and terminated at diverse positions in the region of site P. Such semi-defined 5′ ends suggests that they might be generated by endonucleolytic processing, whereas the heterogeneous nature and location of 3′ ends points to exonucleolytic trimming probably initiated from an entry site downstream of P. The appearance of polyadenylated species strongly implicates participation of the plant counterpart of the yeast TRAMP complex, which was reported to target excised rRNA fragments and aberrant processing intermediates for degradation through their polyadenylation (16,17).
These data clearly show that AtXRN2 and, to some extent, AtXRN3 contribute to the nuclear surveillance pathway in plants, in addition to the probably predominant 3′→5′ activity of the exosome and its nuclear cofactor AtRRP6L2 (24,55).
AtXRN nucleases play a role in the 5′-end processing of 5.8S and 25S rRNAs
It has long been established that the 5′-end maturation of several stable yeast RNAs, including 25S and 5.8S rRNAs and some snoRNAs, is carried out by 5′→3′ exonucleases, Rat1p and Xrn1p, and that inactivation of these enzymes leads to a reduction in mature RNAs and the accumulation of their 5′-extended forms (9–11). In plants, rRNAs are also processed from a large precursor and most snoRNAs are synthesized as polycistronic or intronic transcripts and are presumably released by endonucleolytic cleavages and exonucleolytic trimming (57,58).
Northern hybridization and primer extension analyses of rRNA (25S, 5.8S and 18S) and selected snoRNA (U3, U4, U14, snR10 and snR30) 5′ ends did not reveal 5′-unprocessed precursors in xrn2 or xrn3 plants (Figure 3A, Supplementary Figure S2A–C and data not shown). There was also no difference in the quantity of mature RNAs. In the double xrn2-1 xrn3-3 mutant 5′ processing of snoRNAs was unaffected (Supplementary Figure S2C), but 5′-extended precursors and intermediates of 5.8S accumulated (Figure 3A).
In most eukaryotes the 5′ ends of the two mature 5.8S species are generated via alternative pathways. The shorter 5.8SS is produced by exonucleolytic trimming by Rat1/Xrn1 from cleavage at site A3 in ITS1, while direct endonucleolytic cleavage gives rise to the longer 5.8SL. In turn, the 3′ end of both variants is generated by the exosome, primarily by its nuclear cofactor Rrp6, from cleavage at site C2 in ITS2 (28,29,41). Little is known about these events in plants, except that inactivation of exosome components leads to a defect in the 3′-processing of 5.8S (24,55).
Northern analysis of the 5.8S processing pathway using probes located in different regions of ITS1 and ITS2 revealed clear accumulation of various precursors and intermediates in the xrn2-1 xrn3-3 mutant. Their sizes and hybridization pattern permitted the identification of a 5′-extended 7S pre-rRNA (A3–C2, Figure 3A, probes p43, p4 and p5) and a much less abundant 5′-unprocessed 5.8S (A3–E, probes, p43 and p4), as well as their processing intermediate 5′-5.8S. In addition, regular 3′-extended 5.8S intermediates known from yeast, namely a major 7S and less common 5.8S + 30 and 6S pre-rRNAs, as well as an unusual 5.8S-3′ species (Figure 3A, probe p5), were increased in plants lacking both nucleases. None of the detected RNAs stretched to site A2 in ITS1 or C1 (5′ end of 25S) (Figure 3A, probes p42 and p44). These phenotypes indicate that, as in yeast, processing steps in ITS1 and ITS2 are to some extent also linked in plants (21,59). In contrast to yeast, however, a defect in the 5.8S 5′-processing did not lead to a visible decrease in the 5.8SS form, the mature product of exonucleolytic trimming (Figure 2B, lane 5 and Supplementary Figure S3A). This observation, together with accumulation of the 5′-5.8S intermediate in xrn2-1 xrn3-3 plants, suggests a lesser role of AtXRN2/3 in this process and the contribution of an additional 5′→3′ exonuclease.
A similar conclusion can be reached for the 5′ processing of 25S rRNA, as northern hybridization using probe p44, which is located directly upstream of the mature 25S, did not reveal accumulation of a 5′-extended precursor in the xrn2-1 xrn3-3 mutant (Supplementary Figure S3B), although this is readily observed in yeast cells lacking exonucleases Rat1 and Xrn1 (11,59). However, primer extension with the same probe detected a faint stop, visible only in xrn2-1 xrn3-3, located 72 nt from the mature 5′-end of 25S, which probably corresponds to cleavage at site C2 (Figure 3B). In turn, primer extension over ITS1 using probe p4 confirmed accumulation of 5′-unprocessed 5.8S species in the xrn2-1 xrn3-3 mutant and enabled identification of cleavages at positions +3665 and +3835 relative to the transcription initiation site (TIS), corresponding to sites A2 and A3 (Figures 3C and D). Detection of an A2–A3 fragment in the xrn2-1 xrn3-3 line by northern hybridization using probe p42, which is located between the two sites, corroborated the hypothesis that these sites are generated by endonucleolytic cleavage (Figure 3A).
Thus, inactivation of both AtXRN2 and AtXRN3 confirms their joint involvement in the 5′-end processing of 5.8S and 25S, however, as the level of mature rRNAs is not affected, the major activity in this pathway, as well as in the formation of mature snoRNAs, is probably due to another 5′→3′ exonuclease.
Normal pre-rRNA processing pathway is affected in the absence of AtXRN2
In addition to the analysis of rRNA 5′-processing, northern hybridizations using probes located in different regions of pre-rRNA revealed specific alterations in the pattern of rRNA precursors in the xrn2 mutant, which is indicative of pre-rRNA processing defects (Figure 4A). The sequence of events and the identity of precursors and intermediates of the rRNA synthesis pathway in plants, unlike those in yeast, are poorly resolved. The pattern of RNA species detected in Arabidopsis shows that some, but not all, are similar to those observed in yeast. For this reason throughout this paper we will refer to those RNAs as established for yeast counterparts, or name them according to the region they span.
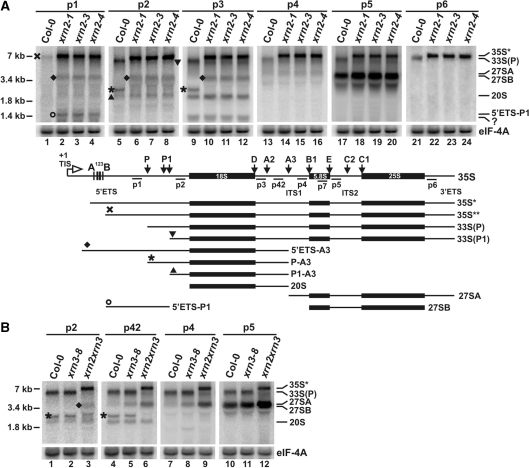
Figure 4.
AtXRN2 participates in pre-rRNA processing. Northern analysis of rRNA precursors and intermediates in xrn2 (A) and xrn3 (B) mutants. Total RNA isolated from wild-type (Col-0), xrn2-1, xrn2-2, xrn2-3, xrn3-8 and xrn2-1 xrn3-3 seedlings were separated on 1.1% agarose gels and hybridized with probes indicated above each panel. rRNA precursors and intermediates are described on the right; molecular weight of 35S, 25S and 18S (7, 3.4 and 1.8 kb) are on the left. Intermediates designated with symbols together with the structure of all detected rRNA species and location of probes along the 35S precursor are shown between (A) and (B). A band detected with probe p3 designated with a question mark probably does not correspond to a nuclear pre-RNA but results from cross-hybridization to organellar rRNA. eIF-4A mRNA was used as a loading control.
The highest molecular weight RNAs present in the wild-type (Col-0) are a 35S** precursor that was detected at a relatively low level by probe p1 located in the 5′ETS upstream of primary cleavage at site P, and a more abundant 33S(P) pre-rRNA that migrated faster and was detected by the remaining probes (p2–p6). These species were essentially absent in three independent xrn2 lines. Instead, one major pre-rRNA was identified in xrn2 plants with all probes directed against the external and internal spacers (ETS and ITS1). This species, designated 35S*, is longer than 35S** and 33S(P) in Col-0 and extends from the beginning of the 5′ETS. In addition, a smaller minor species, 33S(P1), was detected by probes p2–p5, but not with p6, indicating that it may be 3′-processed to the mature 25S 3′ termini. None of these RNAs was detected with probes against the 3′ETS past the AtRTL2 cleavage site (36), indicating that all precursors had been efficiently processed (data not shown).
Other clear differences between wild-type and mutant lines, in addition to the strong accumulation of the 35S* precursor in xrn2 plants, were the absence of the P-A3 intermediate, which migrates faster than 3.4 kb (the size of mature 25S) and slower than 1.8 kb (the size of mature 18S), and the presence of two RNAs, one of ∼1.4 kb (5′ETS-P1) and another (5′ETS-A3) located above 3.4 kb. The P-A3 intermediate that was present in Col-0 but missing from xrn2 mutants hybridized to probes p2 and p3, which are located directly up- and down-stream of 18S, respectively, and thus probably stretches from site P to cleavage at A3 in the ITS1. This indicates that, unlike in yeast in which the P-A3 species has not been described, in plants events in the ITS1 can take place prior to maturation of the 5′ETS. The P1-A3 fragment, which was detected using probes p2 and p3 and is present at a low level in both Col-0 and xrn2 plants, migrates faster than the P-A3 intermediate, and therefore it probably represents 5′- and 3′-extended 18S resulting from processing in the ITS1 and a potential, but previously unidentified cleavage in the 5′ETS, denoted P1, downstream of P. Unexpectedly, the P-A3 and P1-A3 species, as well as 5′ETS-A3 and 20S (see below), extend to site A3, since they were also detected by a probe located between A2 and A3 (data not shown). This result suggests that, in contrast to yeast, usual order of processing in the ITS1 in Arabidopsis may entail cleavage at A3 prior to that at A2.
An aberrant 5′ETS-P1 species was detected in xrn2 lines by probe p1, which is specific for the 5′ETS upstream of site P, and by probe p23, which is situated 53 nts downstream of site P, but not by probe p2, which is located closer to the 5′ end of 18S, suggesting that this fragment covers the 5′ETS past site P and extends to a novel cleavage at site P1. This pattern strongly suggests that 5′ETS-P1 RNA is a processing intermediate that had not been cleaved at site P. In addition, the 5′ETS-A3 fragment that was present only in xrn2 plants and was observed using probes p1, p2 and p3, indicates a defect in processing at site P. No 5′ products of cleavages in the 5′ETS were seen in the wild-type, most likely because they are normally processed or degraded by AtXRN2. We conclude that the pattern, sizes and relative abundance of pre-rRNAs and processing intermediates observed in the absence of AtXRN2 are most consistent with the inhibition of cleavage at site P.
Direct precursors to 18S and 25S, covering regions 5′18S-A3 and A3-3′ETS, which correspond to 20S, 27SA and 27SB in S. cerevisiae respectively, were also detected in A. thaliana using probes p3–p5 (Figure 4A). 27SA, the longer, 5′-extended form of the two 27S precursors, is present at a relatively low level but can be distinguished from 27SB with the mature 5′ 5.8S end, by hybridizing to probe p4, which is located directly upstream of the 5.8S rRNA. Both 27SA and 27SB were detected in Col-0 and xrn2 plants, showing that subsequent processing steps in ITS1 and ITS2 are not inhibited in the mutant. However, the level of 27SB and 7S precursors, but not of 20S, was increased by 1.5–1.7-fold in xrn2 plants (Figure 4A and see Figure 3A). In contrast, most of the effects seen in xrn2 plants, including the elevated level of 35S* and the appearance of aberrant intermediates that had not been cleaved at site P, were not observed in the xrn3-8 mutant, except for some increase in the 33S(P), 7S and 27S species (Figure 4B, probes p4 and p5 and see Figure 3A, probe p5), this being more pronounced for 27SA than for 27SB (1.9- versus 1.2-fold). Similar but considerably weaker phenotypes were observed for another available xrn3 line, xrn3-3 (27) (data not shown).
The accumulation of 7S and 27S pre-rRNAs was much greater in the double xrn2-1 xrn3-3 line (2.4-fold for 7S, 6.4-fold for 27SA, 3-fold for 27SB; Figure 4B and see Figure 3A), which is consistent with at least partial redundancy between the two enzymes. Such effects in plants lacking AtXRN2, AtXRN3 or both may result from a defect in the 5′ trimming of 27SA to 27SB and a subsequent delay in the processing in ITS2. Alternatively, both nucleases may participate in RNA quality control by eliminating the surplus of precursors and intermediates that failed to be converted to ensuing products in a timely fashion. Interestingly, different 27S species are affected in the absence of AtXRN2 (27SB) and AtXRN3 (27SA), indicating that their substrate specificities may vary to some extent.
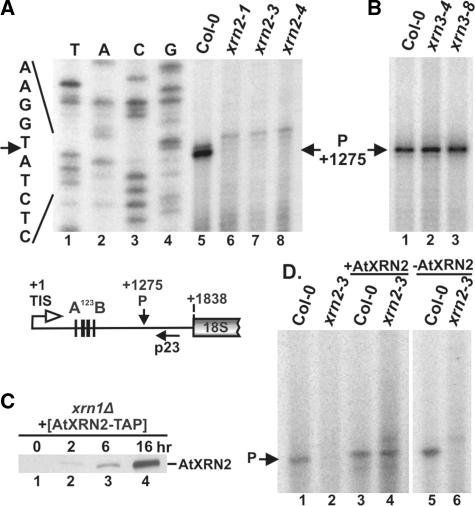
AtXRN2 is required for processing at site P in vivo and restores the cleavage in xrn2 plant extracts
To confirm that the primary cleavage at site P in the 5′ETS is inhibited in plants lacking AtXRN2, primer extension was carried out on total RNA from Col-0 and xrn2 plants using primer p23 located 53 nt downstream from site P. Analysis clearly showed that this cleavage was missing in all xrn2 lines (Figure 5A). A faint primer extension stop a few nucleotides upstream of this position was most likely derived from the RNA secondary structure, especially since regular processing intermediates resulting from P cleavage, e.g. the P-A3 fragment, were not detected in the mutant. This finding confirms that two intermediates in xrn2 plants, 5′ETS-A3 and 5′ETS-P1 (Figure 4A), accumulate because processing at site P does not occur. In agreement with northern data, primer extension showed that P cleavage is not affected in xrn3-4 and xrn3-8 mutants (Figure 5B). In addition, primer extension analysis did not reveal cleavage sites upstream of P either in wild-type or xrn2 plants (Supplementary Figure S3C).
Figure 5.
Cleavage at site P is inhibited in xrn2 mutants and is restored in plant extracts by the purified AtXRN2. (A and B) Mapping the cleavage at site P by primer extension in wild-type, xrn2 and xrn3 plants. The location of p23 primer used to detect the cleavage is shown on the schematic below. Primer extension reactions were separated on 6% sequencing polyacrylamide gels alongside DNA sequencing using the same primers on a PCR product encompassing 5′ETS (A, lanes 1–4). The sequence with primer extension stops is shown on the left and the position of cleavage relative to the TIS on the right. (C) Western blot of AtXRN2-TAP expressed under the control of the GAL1/10 promoter in the xrn1Δ yeast strain carrying a pYES2-GAL1/10::AtXRN2-TAP plasmid. Cells were pre-grown in medium containing 2% raffinose and 0.8% glucose and shifted to medium containing 2% galactose for times indicated when samples were harvested and analysed by western blot using peroxidase-anti-peroxidase antibody. (D) Analysis of cleavage at site P by primer extension using primer p23 on RNA isolated from wild-type and xrn2-3 plant cell extracts incubated alone (lanes 1 and 2) or supplemented with IgG-purified fraction from yeast expressing (lanes 3 and 4) or not expressing (lanes 5 and 6, mock controls) AtXRN2-TAP.
To assess the role of AtXRN2 in site P processing, in vitro reactions using the AtXRN2 enzyme purified from yeast were performed on extracts from xrn2-3 plants. To this end, AtXRN2 fused to the N-terminal tandem affinity purification (TAP) tag (48) was expressed under the control of the inducible GAL1/10 promoter in the yeast xrn1Δ strain (Figure 5C). The fusion protein was purified by affinity purification with IgG Sepharose and, as it was not susceptible to cleavage by TEV protease, the protein was immobilized on beads in the assays. As a control an equivalent amount of the IgG-bound fraction from xrn1Δ cells not expressing AtXRN2-TAP was used. Purified AtXRN2, or the mock control, was incubated with cell extracts from wild-type or xrn2-3 plants, and RNA was extracted from each reaction and analysed for cleavage at site P using primer extension with primer p23 (Figure 5D). To control for the quality of RNA samples, primer extensions were performed on RNA purified from untreated extracts (Figure 5D, lanes 1 and 2). Cleavage at site P was detected in Col-0 extract regardless of the reaction conditions. Remarkably, the addition of purified AtXRN2, but not of the mock sample, efficiently restored the processing at site P in the otherwise cleavage deficient xrn2-3 mutant extract (Figure 5D, lanes 4 and 6). In contrast, in vitro processing with purified AtXRN2 alone, or the mock control, performed on total RNA from the xrn2-3 mutant, using an amount of RNA equivalent to that present in reactions with plant extracts, did not re-establish the P cleavage (data not shown).
These results show that AtXRN2 is required for processing at site P to occur, but that the activity leading to this cleavage, probably within the U3 snoRNP complex, is inherent to the plant extract and not to AtXRN2. It appears that pre-rRNP assembled in xrn2 cells is competent for P cleavage except for the missing AtXRN2, and thus addition of this enzyme leads to the completion of this processing step.
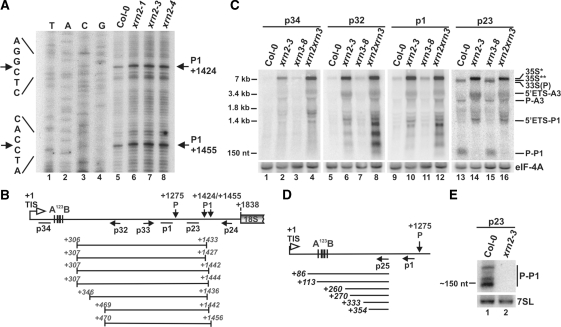
Cleavage at a novel site P1 in the 5′ ETS does not involve AtXRN2
The accumulation of the 5′ETS-P1 fragment in xrn2 plants indicates the occurrence of an additional cleavage in the 5′ETS downstream of site P. In turn, the presence of the P1-A3 species in both wild-type and mutant plants indicates that this cleavage, termed P1, is a regular pre-rRNA processing event. To map the position of cleavage(s) located between site P and 18S rRNA, primer extension was performed using primer p24 located 280 nt upstream of the 18S 5′ end. This analysis revealed two major cleavages, at the same positions in Col-0 and xrn2, +1424 and +1455 relative to the TIS (Figure 6A). These cleavages were not inhibited in the absence of AtXRN2 or in plants with a reduced level of AtXRN3 (data not shown). Pre-rRNA appears to be cut more frequently at site P1 in xrn2 compared to Col-0, since primer extension signals corresponding to these cleavages and the level of the P1-A3 intermediate were more pronounced in the mutants. This reaction, however, is not sufficiently effective, leading to an apparent delay in the processing and accumulation of the 35S* precursor.
Figure 6.
AtXRN2 is not required for processing at site P1 but is involved in the 5′ETS trimming that precedes cleavage at site P. (A) Mapping cleavages at sites P1 by primer extension in wild-type and xrn2 plants using primer p24. Description is as for Figure 5A. (B) Mapping the 5′ and 3′ ends of 5′ETS-P1 fragment by CRT-PCR on total RNA from the xrn2-3 mutant. Description is as for Figure 2E. Identified termini are shown in italics. (C) Northern analysis of the 5′ETS-containing pre-rRNAs in wild-type, xrn2-3, xrn3-8 and xrn2-1 xrn3-3 plants. Total RNA was separated on a 1.1% agarose gel and hybridized with probes distributed along 5’ETS, shown in the panel below. Pre-rRNA and intermediate species are designated as in Figure 4. eIF-4A mRNA was used as a loading control. (D) Mapping 5′ ends of 35S* pre-rRNA in the xrn2-3 mutant by 5′ RACE. cDNA synthesis was performed with primer p1 and PCR with p25 and a primer complementary to an adaptor ligated to cDNA. Identified 5′ ends are shown in italics. (E) Excised P-P1 fragment in wild-type and xrn2-3 plants detected by northern analysis. Total RNA was separated on a 6% polyacrylamide gel and hybridized with probe p23. 7SL RNA was used as a control. Location of primers used for primer extension, northern blots and CRT-PCR are shown in the schematics below panel A.
The occurrence of cleavages at site P1 was confirmed by CRT-PCR designed to map the 5′ and 3′ extremities of the 5′ETS-P1 fragment. Since this species was not detectable in Col-0 plants, the procedure was carried out on total RNA from the xrn2-3 mutant. Most 5′ ends mapped to positions downstream of the A123B cluster, and 3′ ends to the region between the two main cleavages at site P1 (Figure 6B). Surprisingly, this analysis failed to detect pre-rRNAs reaching position +1 (TIS), indicating that a fast processing event(s) may take place in the 5′ETS prior to or concomitantly with cleavages at sites P and P1.
To look at this possibility in more detail, northern analysis was carried out for Col-0, xrn2-3, xrn3-8 and xrn2-1 xrn3-3 using probes p34, p32, p1 and p23 covering different regions of the 5′ETS. In wild-type plants, the largest pre-rRNAs corresponded to a set of heterogeneous species detected with probes p32 and p1, but no RNA was visible using p34 located upstream of the A123B motif (Figure 6C) or a probe directly downstream from the TIS (data not shown). These precursors, jointly designated 35S**, became progressively shorter and slightly more abundant (compare hybridizations with p32 and p1) but their level was still very low. The most prominent of the 5′ETS-containing precursors corresponded to 33S(P) pre-RNA processed at site P, indicating that this reaction is fairly rapid. In contrast, the major 35S* pre-rRNA in the xrn2-3 mutant migrated more slowly than any species in the wild-type and was observed using all probes in Figure 6C. However, only a small fraction reached the TIS as the level of the full-length transcript detected with probes located upstream of the A123B motif was significantly lower than that of the 5′-truncated precursors recognized by the remaining probes (Figure 6C, compare p34 and p32). Since only 35S* in xrn2, and not 35S** in Col-0, hybridized to probe p34, these two species represent distinct precursors. In agreement with CRT-PCR the cut off 5′ETS-P1 fragment was not observed using probe p34, though a faint diffused set of larger RNAs was visible (Figure 6C, lane 2). This strongly indicates that the majority of the 5′ETS-P1 terminates around the A123B region with a small number of heterogeneous species extending beyond this motif. Together, these observations are consistent with shortening of the primary precursor 5′ end prior to cleavage in the 5′ETS. Since cleavage at site P and trimming of the 5′ETS are inhibited in the absence of AtXRN2, both processes may be linked via AtXRN2. We postulate that without the removal of the defined portion of 5′ETS the next processing step, at site P, cannot take place.
The initial shortening of the primary 35S to 35S*/35S** pre-rRNA was verified by assessing its 5′ ends by 5′ RACE on total RNA from Col-0 and xrn2-3 plants. This analysis did not yield any sequences when performed on the wild-type, probably due to a very low level of this RNA. In contrast, sequences obtained for the xrn2-3 mutant revealed heterogeneous 5′ ends of the 35S* transcript, which did not reach the TIS but in most cases terminated downstream of the A123B cluster (Figure 6D). The extension of the precursor 5′ end to the TIS was observed only by primer extension using primers located upstream, but not downstream, of the A123B box [(39) and data not shown], indicating that these species are exceptionally rare. Based on these data we conclude that even in the absence of AtXRN2, when fast early processing steps in the 5′ETS leading to the 33S(P) pre-rRNA cleaved at site P are blocked, there is some initial trimming of the primary pre-rRNA.
Another interesting observation was detection of a small RNA of ∼150 nt in Col-0 and xrn3-8 lines using probe p23 (downstream of P and upstream of P1; Figures 6C, lanes 13 and 15, and Figure 6E, lane 1). This RNA probably corresponds to the excised P-P1 fragment, which corroborates the authenticity of cleavage at site P1. The occurrence of this species also supports that cleavages P and P1 may occur on the same molecule, whereas detection of the P-A3 intermediate (Figure 6C, lanes 13 and 15) indicates that processing at site P possibly precedes that at site P1. In addition, accumulation of the P-P1 species in wild-type plants shows that it is not degraded by AtXRN2/3, the exosome or any other nuclease, probably as a result of its compact, resistant secondary structure. Alternatively, it is protected from digestion by a bound U3 snoRNP complex.
Inspection of 5′ETS processing in the xrn3-8 mutant confirmed that defects due to the inhibition of cleavage at site P were not detectable in the absence of AtXRN3, though the level of various 35S pre-rRNAs was slightly higher, indicating that this nuclease may be involved in their processing or decay. This observation is supported by the greater accumulation of these precursors in the double xrn2-1 xrn3-3 mutant relative to plants lacking only AtXRN2 for probes located upstream, but not downstream, of the A123B cluster (Figure 6C, compare probes p34 and p32, and data not shown). Such a feature can be accounted for when initial shortening of the 5′ETS is carried out by both nucleases while digestion of the 35S* beyond the A123B box is executed exclusively by AtXRN2.
In addition, accumulation of degradation products below the 5′ETS-P1 species is apparent in xrn2-3 and is even more robust in the double xrn2-1 xrn3-3 line (Figure 6C, probes p32 and p1). This result is consistent with the participation of both enzymes in the decay of processing intermediates and cleaved-off products.
AtXRN2 and AtXRN3 are distinct nucleases
XRN 5′→3′ exonucleases are highly homologous and they show 65% identity and 90% similarity within conserved XRN family domains. In addition, they exhibit a comparable level of exonucleolytic activities when tested for complementation of yeast Xrn1 [(23) and data not shown]. Although AtXRN2 and AtXRN3 may show some functional redundancy (27), their major cellular roles are distinct. Their unique features are underscored by the essential nature of AtXRN3 and participation of AtXRN2 in pre-rRNA processing. This raises a question which physical protein properties are accountable for these differences.
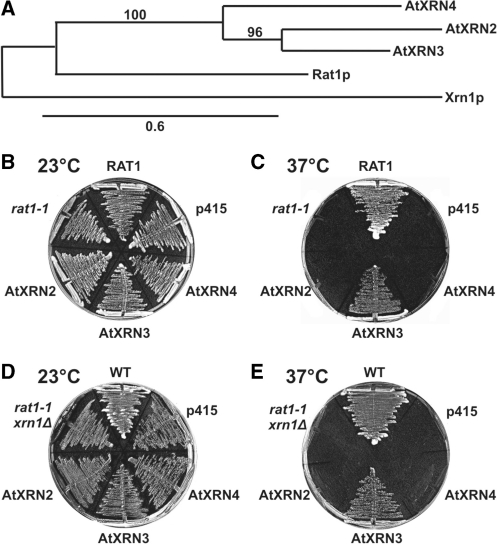
The phylogenetic tree of yeast Rat1 and AtXRN2-4 shows that the closest homologue of yeast Rat1 is Arabidopsis XRN3 (Figure 7A). In turn, protein sequence alignment points to at least two significant differences between AtXRN2 and AtXRN3 (Supplementary Figure S4). The first covers residues 430–480 immediately following the nuclear localization signal (NLS), where there are only a small number of identical or similar amino acids and where a short 8 amino acid deletion in AtXRN2 is situated. A more substantial variation that spans a larger area is located in the C-termini of both proteins, encompassing residues 870–1020. In general, the C-terminal portion of the XRN family is not well conserved and does not contain any recognized domains, though it has four highly conserved regions, of which two acidic N-terminal domains confer catalytic activity (23,60,61). Analyses of characteristic motifs within the C-terminal sequences of the two proteins and biochemical properties based on their amino acid content revealed that AtXRN3 contains a Glu-rich region between positions 943 and 993, while in AtXRN2 the 907–1002 area is rich in basic Lys residues. This difference is reflected in the pI values of AtXRN2 and AtXRN3 (9.04 versus 6.53, respectively). Note that the AtXRN3 pI value is closest to that of yeast Rat1 (6.37). Such disparities in biochemical properties of their C-terminal sequences may result in a diverse set of interacting partners for these two nuclear exonucleases, and, as a consequence, mediate their distinct cellular functions.
Figure 7.
(A) Bootstrapped neighbour-joining phylogram representing evolutionary relationships between yeast Rat1 and Xrn1, and Arabidopsis AtXRN2, AtXRN3 and AtXRN4 nucleases. Bootstraps values are indicated along the branches. The scale bar shows the evolutionary distance (amino acid substitutions per site). (B–E) Complementation tests of the temperature sensitive phenotypes of rat1-1 (B and C) and rat1-1, xrn1Δ (D and E) yeast strains by AtXRN2, AtXRN3 or AtXRN4 expressed from a low copy p415TEF plasmid. RAT1, control strain containing the wild-type RAT1 allele; p415, strain containing an empty p415TEF vector; WT, wild-type strain. Plates were incubated at 23°C (B and D) or 37°C (C and E).
Surprisingly, AtXRN2 and AtXRN3 show different complementation results when expressed in rat1-1 and rat1-1 xrn1Δ mutants on a single-copy plasmid under the control of the TEF promoter (47). Although mRNAs of both proteins are expressed at a comparable level (data not shown), only the presence of AtXRN3 rescues the temperature-sensitive phenotype of this strain at the non-permissive temperature (Figure 7B), in agreement with higher similarities in sequence and properties between AtXRN3 and Rat1. This effect was not observed when the AtXRN2 and AtXRN3 genes were present on a multicopy plasmid [(23) and data not shown], showing that in this case phenotyptic differences can be suppressed by high levels of either of the proteins. The opposing impact of AtXRN2 and AtXRN3 on restoring the essential function of Rat1 is consistent with their non-redundant functions.
We conclude that the two nuclear XRN nucleases have distinct key cellular activities, which are possibly conferred through different interacting partners binding with their non-conserved C-terminal domains.
DISCUSSION
Arabidopsis XRN proteins belong to the 5PX family of 5′→3′ exoribonucleases and are orthologues of yeast and human Rat1/Xrn2 enzymes (23,62). Members of this family are present only in eukaryotes and, what is more, until recently bacteria were believed to be totally devoid of exonucleases capable of degrading RNAs from their 5′ end. Even though two novel bacterial 5′→3′ exoribonucleases, essential RNase J1 and its non-essential paralogue RNase J2, were identified in Bacillus subtilis, they belong to the metallo β-lactamase family (63). While functions of 5′→3′ exoribonucleases have been thoroughly characterized in yeast, relatively little is known about their plant counterparts, particularly the nuclear AtXRN2/3. Their roles as endogenous silencing suppressors and in degradation of miRNA loops excised from miRNA precursors are not by any means key activities, particularly given that AtXRN3 is critical for plant development (27). Since xrn2 xrn3 plants show a stronger morphological and molecular phenotype than single mutants, some functions of these two nucleases are likely to overlap.
This work describes an unexpected role of exonuclease AtXRN2 in the early endonucleolytic cleavage of the pre-rRNA at site P that is specific for this enzyme and cannot be replaced by the other nuclear nuclease, AtXRN3. This processing step is inhibited in plants lacking AtXRN2 but not AtXRN3, which is consistent with the notion that both enzymes, besides sharing potential common substrates, have distinct cellular functions. We have also established that processing in the 5′ETS involves its initial shortening by AtXRN2 and an additional cleavage downstream of P that does not involve AtXRN2. These data make an important contribution to defining the pathways of pre-rRNA processing in plants. In addition, we have demonstrated the involvement of both nuclear exonucleases AtXRN2/3 in polyadenylation-mediated RNA surveillance in Arabidopsis.
An important question is whether AtXRN2 involvement in early pre-rRNA processing is direct or secondary. The first event in the 35S precursor in plants was reported to occur by an endonucleolytic cleavage at site P that can be accurately reproduced in vitro using the purified U3 snoRNP complex (33,40). However, the co-purifying endonucleolytic activity has not been identified and, predictably, AtXRN2 is not associated with this complex (M. Echeverria, personal communication), indicating that their actions are separate. On the other hand, the levels of mature snoRNAs required for cleavage at site P, including components of the U3 snoRNP, U3 and U14, are not affected in xrn2 mutants (this work), indicating that the role of AtXRN2 is not mediated through their defective 5′ processing. Moreover, a defect in P-site cleavage is present in three independent xrn2 mutants and can be restored by the addition of the purified AtXRN2.
These results indicate a direct contribution of AtXRN2 to endonucleolytic processing at site P, but its exact mechanism is not immediately apparent. It is highly unlikely that AtXRN2 acts as an endonuclease since all known members of this family were characterized as 5′→3′ exonucleases (8,60,61) and a recent crystallographic structure of Rat1 implies that its active site is incompatible with an endonucleolytic activity (64). An alternative possibility leading to the defects observed in xrn2 mutants would be that the processing event at site P is not endonucleolytic, but results from exonucleolytic digestion by AtXRN2 from an unidentified cleavage upstream. Several lines of evidence, however, conflict with this hypothesis. Site P is highly conserved in crucifer plants and occurs just upstream of a conserved hairpin structure that, except for Arabidopsis, is located close to the 5′ end of the 35S precursors. Moreover, the products of the in vitro cleavage using a short precursor fragment encompassing site P and the purified U3 snoRNP complex corroborate the endonucleolytic nature of this event (33). Finally, we failed to detect potential cleavages upstream of site P. We propose that the function of AtXRN2 is to shorten the 5′ETS to expose site P for endonucleolytic processing by the U3 snoRNP complex.
It is difficult to predict whether this role of AtXRN2 is conserved in higher plants. Among crucifers, only the Arabidopsis 5′ETS contains the 1083 nt insertion immediately upstream of site P. This region forms multiple putative secondary structures [predicted with Mfold, data not shown; (65)] that may mask or disturb the hairpin following site P, which is conserved in crucifers and probably contributes to the cleavage (33). It is possible that a long distance from the TIS to the major primary processing site necessitates participation of the 5′→3′ exonuclease in Arabidopsis.
An important observation in this study is the 5′ETS-shortening mechanism prior to cleavage at site P. In wild-type plants, the largest pre-rRNAs detectable at a low level (35S**) consist of species with heterogeneous 5′ ends that are indicative of their rapid but gradual shortening until site P is made available. In addition, the major 35S* pre-rRNA in the xrn2 mutant, though larger than species in the wild-type, is still not a full-length precursor. Instead of extending to the TIS it reaches the region surrounding the A123B motif, indicating that some initial trimming also takes place in the absence of AtXRN2. This observation is supported by the analysis of the double xrn2 xrn3 mutant showing that AtXRN3 is also involved in this shortening activity. However, in contrast to AtXRN2, the other nuclease does not participate in the subsequent truncation of the 5′ETS beyond the A123B cluster, as defects related to site P processing are not observed in xrn3 mutants and no exacerbation of this phenotype is seen in xrn2 plants also lacking AtXRN3.
Our thorough analysis of events taking place in the 5′ETS revealed the existence of an additional cleavage at site P1 located downstream of P. Processing at this position occurs in wild-type plants and is independent of AtXRN2, such that it permits the synthesis of mature rRNAs in xrn2 mutants when cleavage at site P is inhibited. Even so, in this case processing at site P1 is not sufficient to release further intermediates, since high levels of the truncated 35S* precursor accumulate. Otherwise, only a small fraction of pre-rRNA undergoes this event as an alternative pathway, and cleavage at site P to some extent can be followed directly by processing at the 5′ end of 18S, without cleavage at P1. Other cleavages in the 5′ETS, dubbed P′, have been mapped closer to the mature 18S, however, these may be optional, as they were observed in the absence of one of the nuclear Arabidopsis Rrp6 homologues, AtRRP6L2 (55), and we failed to detect them by primer extension in wild-type plants (M.Z-P. and J.K., unpublished results).
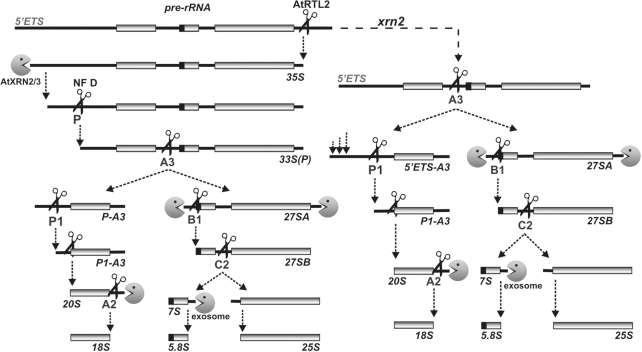
Although the picture of the pre-rRNA processing pathway in Arabidopsis is still far from complete and factors that contribute to different steps are mostly unknown, our analyses using wild-type, xrn2 and xrn2 xrn3 plants provided important information concerning processing events in the 5′ETS, ITS1 and ITS2. Based on these data and previous reports we propose a model of this process in A. thaliana (Figure 8). Following co-transcriptional cleavage of the nascent pre-rRNA in the 3′ETS by RNase III-like enzyme AtRTL2 (36), the 5′ETS undergoes major exonucleolytic shortening (this work), which is most likely biphasic with initial trimming carried out jointly by AtXRN2/3 and subsequent truncation solely by AtXRN2. As a result, site P becomes accessible for the U3 snoRNP complex, which carries out the endonucleolytic cleavage (33,39), thus releasing the 33S(P) precursor. The order of the next two steps is most likely not obligatory: some fraction of the 33S(P) precursor is cleaved at P1 and then at a site located in ITS1, generating a P-P1 fragment and the P1-A3 intermediate, but not P-A3, while the remaining molecules are processed in the reverse order, with cleavage at A3 in ITS1 preceding that at P1, resulting in the P-P1, P1-A3 and P-A3 species all together. It is also possible that P1 cleavage is optional and only occurs on some substrates. In xrn2 plants, 35S* pre-rRNA, which is not processed at site P but has a slightly shortened 5′ETS, is cleaved either at A3 or at P1, probably in a similar manner as in the wild-type. The ambiguities related to the sequence of steps could be resolved by metabolic labelling of precursor and mature rRNAs or by visualization of pre-rRNPs in Miller chromatin spreads (66). However, these methods have not been successfully adapted for plants.
Figure 8.
Model of early pre-rRNA processing steps in Arabidopsis thaliana. Alternative pathway in the xrn2 mutant is shown on the right.
Processing of Arabidopsis ITS1 and ITS2 is only partially analogous to the pathway characterized in yeast. The major cleavage at site A3 in ITS1 followed by exonucleolytic digestion to site B1, together with the alternative pathway via direct cleavage at site B1, generate two mature 5′ ends of 5.8S rRNA, whereas cleavage at site C2 accompanied by exonucleolytic trimming produces the mature 5′ end of 25S rRNA. The involvement of nuclear exonucleases AtXRN2/3 in the 5′ processing of 5.8S and 25S rRNAs is less prominent than is the case for the yeast homologue Rat1. Although AtXRN2/3 have redundant roles in this process, this phenotype is of little consequence and does not affect the level of mature species. These observations suggest the existence of an additional enzyme, which, in fact, may be a major player. Recent analysis of yeast Rrp17 provided evidence that this novel, atypical but highly conserved 5′→3′ exonuclease, is required for the formation of mature 5′ ends of 5.8S and 25S, along with previously characterized Rat and Xrn1 (59). It is therefore probable that the Arabidopsis homologue of Rrp17 makes the most important contribution to the maturation of 5.8S and 25S rRNAs. Other major differences in Arabidopsis pre-rRNA processing compared to yeast include 5′ETS shortening prior to the primary cleavage, the independence of events in the 5′ETS and ITS1, a less well-defined order of cleavages in ITS1 and the existence of the alternative 5′ETS pathway via site P1 that enables the synthesis of mature rRNAs in cases in which the major cleavage is inhibited.
Our data also show that another redundant function for AtXRN2 and AtXRN3 concerns nuclear RNA surveillance, in which they contribute to degradation of polyadenylated pre-rRNA processing intermediates. These data confirm the existence of polyadenylation-dependent RNA quality control in plants and demonstrate that 5′→3′ decay plays a part in this mechanism, as shown previously for yeast Rat1 (13). The major contribution is probably made by the exosome and its nuclear-specific component Rrp6 (AtRRP6L2 in Arabidopsis) via the 3′→5′ pathway (16,17,24,54–56). It remains possible, however, that different substrates will be more efficiently degraded by one of these pathways.
Issues that are still open for investigation concern the essential function(s) of AtXRN3, which to date has not been demonstrated to participate in any important process that has been examined [(27) and our work]. Although yeast Rat1 has been studied extensively for many years, its major role is also unknown. It has been reported that the essential function of Rat1 can be complemented by expressing cytoplasmic Xrn1 carrying a NLS (8). Since NLS-Xrn1 is not able to fulfill the role of Rat1 in transcription termination (20), this is not its central activity. In addition, both AtXRN2 and AtXRN3 rescue the snoRNA and rRNA processing defects resulting from Rat1 deficiency in yeast (G.J.S. and J.K., unpublished), while only AtXRN3 complements the temperature-sensitive phenotype of rat1 mutants when expressed on a single-copy plasmid. Moreover, due to a functional interchangeability of Rat1 and Xrn1 (8), several molecular defects, such as in snoRNA and rRNA 5′-processing and in degradation of excised RNA fragments, are relatively weak in the strain lacking Rat1 and are fully manifested only when combined with deletion of XRN1 (9–11,21). Together, these observations and our work suggest that 5′-processing of rRNA and snoRNA substrates are also not the crucial function of Rat1 or AtXRN3. Identification of this activity is necessary for full perception of essential RNA processes that take place in the cell nucleus.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Wellcome Trust to J.K. (067504/Z/02/Z); Ministry of Science and Higher Education to M.Z. (3781/B/P01/2007/33); National Science Foundation Grant to P.J.G. (MCB#0445638). Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Allison Mallory (INRA,Versaille) for xrn3-3 and xrn2-1 xrn3-3 seeds, Roy Parker (University of Arizona) for yeast strains, Andrzej Dziembowski (IBB PAN, Warsaw) for pYES2-TAP vector and Dr Linda Rymarquis for comments on the manuscript and helpful discussions.
REFERENCES
- 1.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell. Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 2.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 3.Souret FF, Kastenmayer JP, Green PJ. AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol. Cell. 2004;15:173–183. doi: 10.1016/j.molcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 4.Orban TI, Izaurralde E. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA. 2005;11:459–469. doi: 10.1261/rna.7231505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheth U, Parker R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 2006;125:1095–1109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sayani S, Janis M, Lee CY, Toesca I, Chanfreau GF. Widespread impact of nonsense-mediated mRNA decay on the yeast intronome. Mol. Cell. 2008;31:360–370. doi: 10.1016/j.molcel.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson DM, Parker R. Cytoplasmic decay of intergenic transcripts in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007;27:92–101. doi: 10.1128/MCB.01023-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson AW. Rat1p and Xrn1p are functionally interchangeable exoribonucleases that are restricted to and required in the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 1997;17:6122–6130. doi: 10.1128/mcb.17.10.6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henry Y, Wood H, Morrissey JP, Petfalski E, Kearsey S, Tollervey D. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 1994;13:2452–2463. doi: 10.1002/j.1460-2075.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petfalski E, Dandekar T, Henry Y, Tollervey D. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol. Cell. Biol. 1998;18:1181–1189. doi: 10.1128/mcb.18.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geerlings TH, Vos JC, Raue HA. The final step in the formation of 25S rRNA in Saccharomyces cerevisiae is performed by 5′→3′ exonucleases. RNA. 2000;6:1698–1703. doi: 10.1017/s1355838200001540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bousquet-Antonelli C, Presutti C, Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000;102:765–775. doi: 10.1016/s0092-8674(00)00065-9. [DOI] [PubMed] [Google Scholar]
- 13.Fang F, Phillips S, Butler JS. Rat1p and Rai1p function with the nuclear exosome in the processing and degradation of rRNA precursors. RNA. 2005;11:1571–1578. doi: 10.1261/rna.2900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ, Phizicky EM. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5′-3′ exonucleases Rat1 and Xrn1. Genes Dev. 2008;22:1369–1380. doi: 10.1101/gad.1654308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol. Cell. 2008;32:465–477. doi: 10.1016/j.molcel.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 16.LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 17.Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 18.Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, Buratowski S. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- 19.West S, Gromak N, Proudfoot NJ. Human 5′→3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- 20.Luo W, Johnson AW, Bentley DL. The role of Rat1 in coupling mRNA 3′-end processing to transcription termination: implications for a unified allosteric-torpedo model. Genes Dev. 2006;20:954–965. doi: 10.1101/gad.1409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El Hage A, Koper M, Kufel J, Tollervey D. Efficient termination of transcription by RNA polymerase I requires the 5′ exonuclease Rat1 in yeast. Genes Dev. 2008;22:1069–1081. doi: 10.1101/gad.463708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawauchi J, Mischo H, Braglia P, Rondon A, Proudfoot NJ. Budding yeast RNA polymerases I and II employ parallel mechanisms of transcriptional termination. Genes Dev. 2008;22:1082–1092. doi: 10.1101/gad.463408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kastenmayer JP, Green PJ. Novel features of the XRN-family in Arabidopsis: Evidence that AtXRN4, one of several orthologs of nuclear Xrn2p/Rat1p, functions in the cytoplasm. Proc. Natl Acad. Sci. USA. 2000;97:13985–13990. doi: 10.1073/pnas.97.25.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chekanova JA, Gregory BD, Reverdatto SV, Chen H, Kumar R, Hooker T, Yazaki J, Li P, Skiba N, Peng Q, et al. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell. 2007;131:1340–1353. doi: 10.1016/j.cell.2007.10.056. [DOI] [PubMed] [Google Scholar]
- 25.Gregory BD, O'Malley RC, Lister R, Urich MA, Tonti-Filippini J, Chen H, Millar AH, Ecker JR. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev. Cell. 2008;14:854–866. doi: 10.1016/j.devcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Gazzani S, Lawrenson T, Woodward C, Headon D, Sablowski R. A link between mRNA turnover and RNA interference in Arabidopsis. Science. 2004;306:1046–1048. doi: 10.1126/science.1101092. [DOI] [PubMed] [Google Scholar]
- 27.Gy I, Gasciolli V, Lauressergues D, Morel JB, Gombert J, Proux F, Proux C, Vaucheret H, Mallory AC. Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell. 2007;19:3451–3461. doi: 10.1105/tpc.107.055319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Ann. Rev. Gen. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 29.Fatica A, Tollervey D. Making ribosomes. Curr. Opin. Cell Biol. 2002;14:313–318. doi: 10.1016/s0955-0674(02)00336-8. [DOI] [PubMed] [Google Scholar]
- 30.Gerbi SA, Borovjagin AV, Lange TS. The nucleolus: a site of ribonucleoprotein maturation. Curr. Opin. Cell Biol. 2003;15:318–325. doi: 10.1016/s0955-0674(03)00049-8. [DOI] [PubMed] [Google Scholar]
- 31.Kass S, Craig N, Sollner-Webb B. Primary processing of mammalian rRNA involves two adjacent cleavages and is not species specific. Mol. Cell. Biol. 1987;7:2891–2898. doi: 10.1128/mcb.7.8.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Savino R, Gerbi SA. In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J. 1990;9:2299–2308. doi: 10.1002/j.1460-2075.1990.tb07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saez-Vasquez J, Caparros-Ruiz D, Barneche F, Echeverria M. A plant snoRNP complex containing snoRNAs, fibrillarin, and nucleolin-like proteins is competent for both rRNA gene binding and pre-rRNA processing in vitro. Mol. Cell. Biol. 2004;24:7284–7297. doi: 10.1128/MCB.24.16.7284-7297.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kent T, Lapik YR, Pestov DG. The 5′ external transcribed spacer in mouse ribosomal RNA contains two cleavage sites. RNA. 2009;15:14–20. doi: 10.1261/rna.1384709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abou Elela S, Igel H, Ares MJ. RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell. 1996;85:115–124. doi: 10.1016/s0092-8674(00)81087-9. [DOI] [PubMed] [Google Scholar]
- 36.Comella P, Pontvianne F, Lahmy S, Vignols F, Barbezier N, Debures A, Jobet E, Brugidou E, Echeverria M, Saez-Vasquez J. Characterization of a ribonuclease III-like protein required for cleavage of the pre-rRNA in the 3′ETS in Arabidopsis. Nucleic Acids Res. 2008;36:1163–1175. doi: 10.1093/nar/gkm1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dragon F, Gallagher JE, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prieto JL, McStay B. Recruitment of factors linking transcription and processing of pre-rRNA to NOR chromatin is UBF-dependent and occurs independent of transcription in human cells. Genes Dev. 2007:2041–2054. doi: 10.1101/gad.436707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pontvianne F, Matia I, Douet J, Tourmente S, Medina FJ, Echeverria M, Sáez-Vásquez J. Characterization of AtNUC-L1 reveals a central role of nucleolin in nucleolus organization and silencing of AtNUC-L2 gene in Arabidopsis. Mol. Biol. Cell. 2007;18:369–379. doi: 10.1091/mbc.E06-08-0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samaha H, Delorme V, Pontvianne F, Cooke R, Delalande F, Van Dorsselaer A, Echeverria M, Sáez-Vásquez J. Identification of protein factors and U3 snoRNAs from a Brassica oleracea RNP complex involved in the processing of pre-rRNA. Plant J. 2010;61:383–398. doi: 10.1111/j.1365-313X.2009.04061.x. [DOI] [PubMed] [Google Scholar]
- 41.Strunk BS, Karbstein K. Powering through ribosome assembly. RNA. 2009;15:2083–2104. doi: 10.1261/rna.1792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962;15:473–497. [Google Scholar]
- 43.Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 2001;27:581–590. doi: 10.1046/j.1365-313x.2001.01105.x. [DOI] [PubMed] [Google Scholar]
- 44.Gleave AP. A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 1992;20:1203–1207. doi: 10.1007/BF00028910. [DOI] [PubMed] [Google Scholar]
- 45.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 46.Gietz D, St Jean A, Woods RA, Schiestl RH. Improved method for high efficient transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 48.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Séraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 49.Beltrame M, Tollervey D. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 1992;11:1531–1542. doi: 10.1002/j.1460-2075.1992.tb05198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 51.Olmedo G, Guo H, Gregory BD, Nourizadeh SD, Aguilar-Henonin L, Li H, An F, Guzman P, Ecker JR. ETHYLENE-INSENSITIVE5 encodes a 5′→3′ exoribonuclease required for regulation of the EIN3-targeting F-box proteins EBF1/2. Proc. Natl Acad. Sci. USA. 2006;103:13286–13293. doi: 10.1073/pnas.0605528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Potuschak T, Vansiri A, Binder BM, Lechner E, Vierstra RD, Genschik P. The exoribonuclease XRN4 is a component of the ethylene response pathway in Arabidopsis. Plant Cell. 2006;18:3047–3057. doi: 10.1105/tpc.106.046508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kerschen A, Napoli CA, Jorgensen RA, Muller AE. Effectiveness of RNA interference in transgenic plants. FEBS Lett. 2004;566:223–228. doi: 10.1016/j.febslet.2004.04.043. [DOI] [PubMed] [Google Scholar]
- 54.Kuai L, Fang F, Butler JS, Sherman F. Polyadenylation of rRNA in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2004;101:8581–8586. doi: 10.1073/pnas.0402888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lange H, Holec S, Cognat V, Pieuchot L, Le Ret M, Canaday J, Gagliardi D. Degradation of a polyadenylated rRNA maturation by-product involves one of the three RRP6-like proteins in Arabidopsis thaliana. Mol. Cell. Biol. 2008;28:3038–3044. doi: 10.1128/MCB.02064-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Slomovic S, Laufer D, Geiger D, Schuster G. Polyadenylation of ribosomal RNA in human cells. Nucleic Acids Res. 2006;34:2966–2975. doi: 10.1093/nar/gkl357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leader DJ, Clark GP, Watters J, Beven AF, Shaw PJ, Brown JW. Clusters of multiple different small nucleolar RNA genes in plants are expressed as and processed from polycistronic pre-snoRNAs. EMBO J. 1997;16:5742–5751. doi: 10.1093/emboj/16.18.5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leader DJ, Clark GP, Watters J, Beven AF, Shaw PJ, Brown JW. Splicing-independent processing of plant box C/D and box H/ACA small nucleolar RNAs. Plant. Mol. Biol. 1999;39:1091–1100. doi: 10.1023/a:1006157022319. [DOI] [PubMed] [Google Scholar]
- 59.Oeffinger M, Zenklusen D, Ferguson A, Wei KE, El Hage A, Tollervey D, Chait BT, Singer RH, Rout MP. Rrp17p is a eukaryotic exonuclease required for 5′ end processing of Pre-60S ribosomal RNA. Mol. Cell. 2009;36:768–781. doi: 10.1016/j.molcel.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Page AM, Davis K, Molineux C, Kolodner RD, Johnson AW. Mutational analysis of exoribonuclease I from Saccharomyces cerevisiae. Nucleic Acids Res. 1998;26:3707–3716. doi: 10.1093/nar/26.16.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Solinger JA, Pascolini D, Heyer WD. Active-site mutations in the Xrn1p exoribonuclease of Saccharomyces cerevisiae reveal a specific role in meiosis. Mol. Cell. Biol. 1999;19:5930–5942. doi: 10.1128/mcb.19.9.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zuo Y, Deutscher MP. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mathy N, Bénard L, Pellegrini O, Daou R, Wen T, Condon C. 5′-to-3′ exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5′ stability of mRNA. Cell. 2007;129:681–692. doi: 10.1016/j.cell.2007.02.051. [DOI] [PubMed] [Google Scholar]
- 64.Xiang S, Cooper-Morgan A, Jiao X, Kiledjian M, Manley JL, Tong L. Structure and function of the 5′-3′ exoribonuclease Rat1 and its activating partner Rai1. Nature. 2009;458:784–788. doi: 10.1038/nature07731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zucker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miller OLJ, Beatty BR. Visualization of nucleolar genes. Science. 1969;164:955–957. doi: 10.1126/science.164.3882.955. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.