Abstract
We report that combining a DNA analog (2′F-ANA) with rigid RNA analogs [2′F-RNA and/or locked nucleic acid (LNA)] in siRNA duplexes can produce gene silencing agents with enhanced potency. The favored conformations of these two analogs are different, and combining them in a 1–1 pattern led to reduced affinity, whereas alternating short continuous regions of individual modifications increased affinity relative to an RNA:RNA duplex. Thus, the binding affinity at key regions of the siRNA duplex could be tuned by changing the pattern of incorporation of DNA-like and RNA-like nucleotides. These heavily or fully modified duplexes are active against a range of mRNA targets. Effective patterns of modification were chosen based on screens using two sequences targeting firefly luciferase. We then applied the most effective duplex designs to the knockdown of the eIF4E binding proteins 4E-BP1 and 4E-BP2. We identified modified duplexes with potency comparable to native siRNA. Modified duplexes showed dramatically enhanced stability to serum nucleases, and were characterized by circular dichroism and thermal denaturation studies. Chemical modification significantly reduced the immunostimulatory properties of these siRNAs in human peripheral blood mononuclear cells.
INTRODUCTION
RNA interference (RNAi), an endogenous gene silencing process, can be triggered by dsRNA to elicit specific gene silencing (1). The discovery that 21-nt siRNAs act as exogenous synthetic triggers of RNAi in mammalian cells (2) incited rapid development of siRNA-based therapeutic candidates. Yet the fact remains that siRNAs are not ideal drug candidates. Cellular uptake of siRNA is poor, rapid nuclease-mediated degradation in vivo reduces duration of activity, and off-target effects (OTEs) arising from partial complementarity to unintended genes and nonspecific immune responses to siRNAs are important considerations for the clinical development of siRNA therapeutics. A variety of chemical modifications have been developed to address these issues (3). In some cases, modification of siRNA leads to increased activity. However, patterns of modification are typically heavily sequence dependent, forcing optimization of the modification strategy for each different siRNA sequence and target. A universally active heavily modified siRNA design has thus far proven elusive but remains an attractive goal. We have argued that siRNA designs comprising combinations of chemical modifications are more likely to achieve this goal (3,4).
When siRNA is introduced into mammalian cells, it is recognized by the RNA-induced silencing complex (RISC), a multiprotein complex containing the endonuclease Argonaute2 (Ago2). One strand of the duplex (the so-called passenger strand) is removed, while the other remains associated with RISC and guides it to complementary mRNA. For an active duplex, the guide strand is antisense to the target mRNA, thus the terms guide strand or antisense strand are often used interchangeably.
OTEs commonly arise when the guide RNA directs RISC to partially complementary sequences within unintended mRNAs, in a fashion similar to the control of gene expression exerted by endogenous microRNAs (miRNAs) (5). Still other OTEs can arise when cellular receptors such as RIG-I, MDA5 and toll-like receptors (TLRs) bind to siRNAs and trigger innate immune responses, resulting in cytokine production (6).
A wide variety of nucleotide modifications can enhance potency, improve serum stability and reduce OTEs in certain siRNA sequence contexts (3,4).
We have a long-standing interest in 2′-deoxy-2′-fluoroarabinonucleic acid (2′F-ANA) (7–11). 2′F-ANA is a modification that closely mimics DNA with respect to duplex structure (12), and favors an eastern sugar conformation (11,13–15). 2′F-ANA can also mimic DNA in its ability to trigger RNase H-mediated RNA degradation and has applications in antisense technology (16–18). Interestingly, although a DNA-mimic, 2′F-ANA is also compatible and often stabilizing within a dsRNA context. 2′F-ANA has been successfully incorporated into siRNAs, and is especially well tolerated in the sense strand which can be fully 2′F-ANA modified with little effect on potency while enhancing siRNA serum stability (19,20). Given the DNA-like conformation of 2′F-ANA, we hypothesized that combining 2′F-ANA with RNA-like analogs would further enhance gene silencing potency by forcing A-form modified siRNA structures similar to the native siRNA substrates recognized by RISC. To this end, we chose to investigate chimeric siRNAs composed of 2′F-ANA with the RNA analogs 2′F-RNA and/or locked nucleic acid (Figure 1).
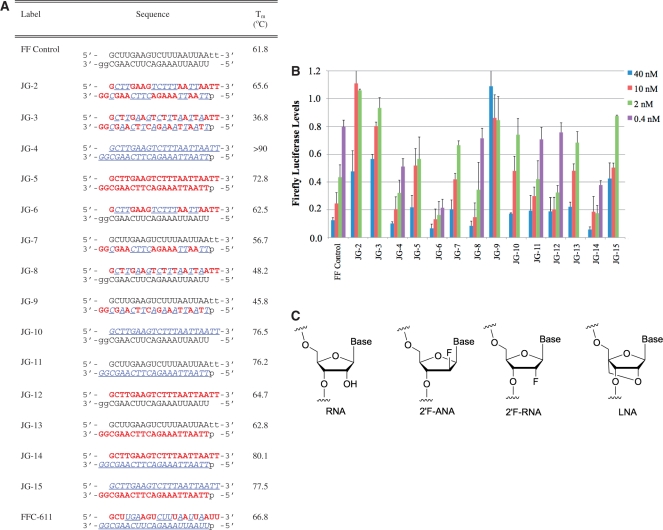
Figure 1.
(A) Sequences and thermal denaturation studies of siRNAs targeting firefly luciferase mRNA (the JG series) from position +1818 to + 1836. (B) Activity of siRNAs modified with 2′F-ANA and 2′F-RNA (Sequences shown in (A)). mRNA target is firefly luciferase. n = 2 for select siRNAs at the 0.4 nM concentration, n = 4 for FF Control, JG-4, 6, 8, 11, 12, 14 at the 0.4 nM, 2 nM, 10 nM and 40 nM concentrations, n = 2 for JG-2, 3, 5, 7, 9, 10, 13, 15 at the 2 nM, 10 nM and 40 nM concentrations. Firefly luciferase levels were normalized to total cellular protein and luciferase counts of mock treated cells. Bars indicate standard deviation. (C) Chemical structures of ribonucleic acid (RNA), 2′-fluoroarabinonucleic acid (2′F-ANA), 2′-fluororibonucleic acid (2′F-RNA) and locked nucleic acid (LNA). Legend: RNA, dna,  , p = 5′ phosphorylation.
, p = 5′ phosphorylation.
We designed a large number of modified duplexes targeting two regions of the firefly luciferase gene. Active modification designs that emerged from this screen were used to target the eIF4E-binding proteins 1 and 2 (4E-BP1 and 4E-BP2) using previously reported siRNA sequences (21).The eukaryotic initiation factor 4F (eIF4F) complex recognizes mRNA cap structures and facilitates translation by recruiting the 43S initiation complex to the mRNA (22). 4EBPs inhibit eIF4F complex assembly by binding eIF4E, a component of the eIF4F complex, preventing translation. It has been reported that 4EBPs are negative regulators of type-I interferon production because they repress translation of interferon regulatory factor 7 (IRF-7) mRNA (23), an essential transcription factor for activation of interferon responses to viral infections (24). Thus, efficient knockdown of these factors could have beneficial consequences on human health.
MATERIALS AND METHODS
Oligonucleotide synthesis and siRNA preparation
Standard phosphoramidite solid-phase synthesis conditions were used for the synthesis of all modified and unmodified oligonucleotides (25). Syntheses were performed on an Applied Biosystems 3400 DNA Synthesizer at a 1-µmol scale using Unylink CPG support (ChemGenes). All phosphoramidites were prepared as 0.15 M solutions in acetonitrile (ACN), except DNA, which was prepared as 0.1 M. 5-ethylthiotetrazole (0.25 M in ACN) was used to activate phosphoramidites for coupling. Detritylations were accomplished with 3% trichloroacetic acid in CH2Cl2 for 110 s. Capping of failure sequences was achieved with acetic anhydride in tetrahydrofuran (THF) and 16% N-methylimidazole in THF. Oxidation was done using 0.1 M I2 in 1:2:10 pyridine:water:THF. Coupling times were 600 s for RNA, 2′F-ANA, 2′F-RNA, and LNA (5-methyl C and U) phosphoramidites, with the exception of their guanosine phosphoramidites which were allowed to couple for 900 s. 5′-phosphorylation of chemically modified antisense strands was achieved using bis-cyanoethyl-N,N-diisopropyl-2-cyanoethyl phosphoramidite at 0.15 M (600 s coupling time). Deprotection and cleavage from the solid support was accomplished with either 3:1 NH4OH:EtOH for 48 h at room temperature (RT), or with 40% methylamine for 10 min at 65°C (26). Oligonucleotides containing RNA were synthesized with standard 2′-TBDMS phosphoramidites, and desilylation was achieved with either neat triethylamine trihydrofluoride for 48 h at RT (27), or with triethylamine trihydrofluoride/N-methyl pyrrolidone/triethylamine (1.5:0.75:1 by volume) for 2.5 h at 65°C (26,28). Purification of crude oligonucleotides was done by preparative denaturing polyacrylamide gel electrophoresis (PAGE) using 24% acrylamide gels. Gel bands were extracted overnight in DEPC-treated autoclaved Millipore water, and lyophilized to dryness. Purified oligonucleotides were desalted with Nap-25 Sephadex columns from GE Healthcare. Sequences were verified by analytical denaturing PAGE and ESI-LCMS. siRNAs were prepared by annealing equimolar quantities of complementary oligonucleotides in siRNA buffer [100 mM KOAc, 30 mM HEPES-KOH, 2 mM Mg(OAc)2, pH 7.4] by slowly cooling from 96°C to RT.
Thermal denaturation and circular dichroism studies
Complementary sequences (1 nmol) were combined, dried and rediluted in pH 7.2 buffer (1 ml) containing 140 mM KCl, 1 mM MgCl2 and 5 mM NaHPO4. After heating to 94°C, samples were slowly cooled to RT and refrigerated overnight. Samples were transferred into cold cuvettes in a Varian Cary 300 UV spectrophotometer. The change in absorbance at 260 nm was monitored upon heating from 10°C to 94°C (ramped at 1°C/min). Melting temperatures were determined using the baseline method, as implemented in the Cary software.
CD spectra were obtained on a Jasco J-810 spectropolarimeter at 20°C using samples annealed in the same buffer and under the same conditions as for the thermal denaturation studies. Spectra were baseline-corrected with respect to a blank containing the buffer but no duplex. Smoothing and adjustment for duplex concentration were effected using the Spectra-Manager program (Jasco).
Fetal bovine serum stability assays
Fetal bovine serum (FBS) stability assays were conducted on unmodified and modified siRNAs. siRNAs were prepared by annealing equimolar amounts of complementary oligonucleotides as described above with the exception that water was used instead of siRNA buffer. Samples were then dried, redissolved in 10% FBS (Wisent) in Dulbecco's Modified Eagle Medium (DMEM; Invitrogen), vortexed and incubated at 37°C. Aliquots of 0.15 OD units were withdrawn at various times, flash-frozen on dry ice and stored in the freezer. Extent of degradation was determined by analytical native 20% PAGE. Bands were visualized by Stains-All (Sigma-Aldrich).
Luciferase siRNA assays
Firefly luciferase siRNA assays were carried out in HeLa X1/5 cells stably expressing firefly luciferase, which were grown as previously described (19). The JG-series duplexes targeted firefly luciferase mRNA from position +1818 to +1836 and the ff6 series from +515 to +533. The day prior to transfection, 0.5 × 105 cells were plated in each well of a 24-well plate. The next day, the cells were incubated with increasing amounts of siRNAs premixed with Lipofectamine-Plus (Invitrogen) using 1 µl of lipofectamine and 4 µl of the Plus reagent per 20 pmol of siRNA (for the highest concentration tested). For the siRNA titrations, each siRNA was diluted into buffer [30 mM HEPES-KOH, 100 mM KOAc, 2 mM Mg(OAc)2, pH 7.4] and the ratio of Lipofectamine-Plus to siRNA remained constant. Twenty-four hours after transfection, the cells were lysed in hypotonic lysis buffer (15 mM K3PO4, 1 mM EDTA, 1% TritonX-100, 2 mM NaF, 1 mg/ml BSA, 1 mM DTT, 100 mM NaCl, 4 mg/ml aprotinin, 2 mg/ml leupeptin and 2 mg/ml pepstatin) and the firefly light units were determined using a Fluostar Optima 96-well plate bioluminescence reader (BMG Labtech) using firefly substrate as described (29). The luciferase counts were normalized to the protein concentration of the cell lysate as determined by the DC protein assay (BioRad). Cotransfecting the siRNAs and the plasmid pCI-hRL-con expressing the Renilla luciferase mRNA (30) in the same cell line showed no difference in expression of this reporter, suggesting specificity of the RNAi effects (data not shown).
4EBP siRNA assays
4EBP siRNA transfections were performed in HEK293T cells using Lipofectamine Plus reagent on cells plated at 60–70% confluence in 24-well plates. siRNAs were serially diluted in 100 µl Opti-MEM and 1 µl Plus reagent and incubated for 5 min at RT. A mixture of 5 µl Lipofectamine and 100 µl Opti-MEM was then added to the precomplexed RNA mix and incubated for 20 min at RT before adding to cells. Five hours later, 500 μl of complete medium (DMEM with 10% FBS) was added to the transfection mixture. Cells were harvested 48 h after transfection and proteins were extracted and used for western blots using antibodies against 4E-BP1 (Cell Signalling) or 4E-BP2 (Cell Signalling), or Actin (Sigma).
Immunostimulation assays in human peripheral blood mononuclear cells
Peripheral blood mononuclear cells (PBMCs) were transfected using either DOTAP or Lipofectamine 2000 at a final siRNA concentration of 80 nM. siRNA (2 µl of 20 µM stock) was diluted with 50 µl OptiMEM and combined with 1 µl Lipofectamine 2000 in 50 ul OptiMEM then incubated for 20 min. For transfection with DOTAP, siRNA (2 μl of 20 uM stock) was diluted with 13 μl HEPES-buffered saline (HBS) and mixed with 5 μl DOTAP in 25 μl of HBS, then incubated for 15 min. The mixture was then added to 400 µl (Lipofectamine 2000) or 460 μl (DOTAP) of cells in culture medium (RPMI supplemented with 3% FBS) and incubated for 24 h. Control (Mock) cells were incubated with lipid alone. Supernatants (20 μl) from untreated or treated cells were mixed with HEK-Blue cells (Invivogen) in Hek-Blue detection medium and incubated overnight. Hek-Blue cells carry a reporter gene expressing a secreted alkaline phosphatase under the control of the ISRE9 (interferon stimulated response element 9) promoter. In response to IFN production, the HEK blue cells release soluble alkaline phosphatase. The colored product of the alkaline phosphatase and Hek-Blue detection medium reaction is quantitated at 650 nm. Results are presented as relative IFN units when background was subtracted. As a positive control for the assay Hek-Blue cells were incubated with various doses of purified interferon α 2b (GeneScript, NJ, USA).
RESULTS AND DISCUSSION
Tuning duplex binding affinity with combinations of modifications
We and others have previously shown that 2′F-ANA and 2′F-RNA adopt different preferential conformations (11,12,31). While both fluorinated analogs have increased binding affinity to RNA (relative to either DNA or RNA), combinations of the two can be either stabilizing or destabilizing depending on how they are combined. Thus, replacing either strand of an siRNA duplex with 2′F-ANA or 2′F-RNA increased the duplex Tm value with respect to the native duplex (Figure 1, duplexes JG-10–13). In contrast, a 1–1 altimer design significantly decreased duplex stability (Figure 1, duplexes JG-8,9), possibly arising from the introduction of A/B junctions within the duplex (12). Chimeric strands with groups of each modification such as JG-2 and JG-6, which have 2′F-ANA purines and 2′F-RNA pyrimidines, contain fewer A/B junctions and exhibit slightly better stability than the unmodified siRNA control. A second set of siRNAs of a different sequence, the ff6 series (Figure 2A), supports these observations.
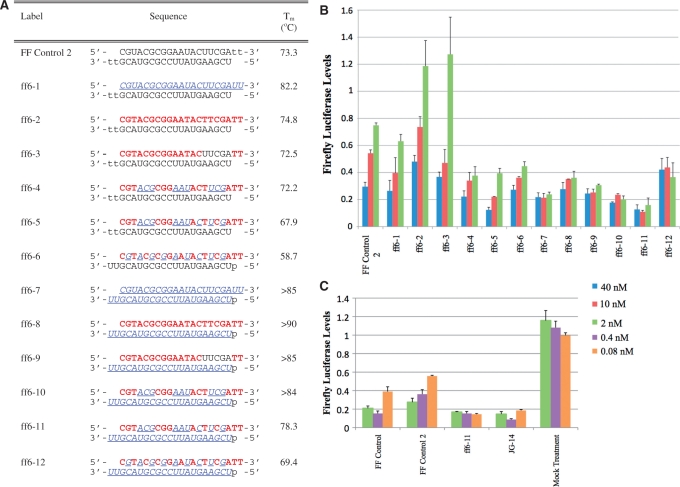
Figure 2.
(A) Sequences and thermal denaturation studies of siRNAs targeting firefly luciferase mRNA (the ff6 series) from position +515 to +533. (B) Assays demonstrating activity of modified siRNAs. n = 2. Firefly luciferase levels were normalized to total cellular protein and luciferase counts of cells treated with scrambled (non-targeting) siRNAs. (C) Gene silencing assays to directly compare siRNAs targeting the two different regions of luciferase mRNA. The two unmodified siRNAs were tested, along with ff6-11 and JG-14 modification designs. n = 2. Firefly luciferase levels were normalized to total cellular protein and luciferase counts of cells treated with scrambled (nontargeting) siRNA. Bars indicate standard deviation. Legend: RNA, dna,  , p = 5′ phosphorylation.
, p = 5′ phosphorylation.
To further investigate the effects of contrasting conformations on duplex stability, we replaced 2′F-RNA inserts with LNA as shown in Figure 3. for these sequences, LNAs are flanked by RNA (no A/B junctions) and/or 2′F-ANA (potential A/B junction). However, LNA is so strongly preorganized that it may influence flanking 2′F-ANA units into an RNA-like conformation (32–34) and so A/B junctions may be reduced. When LNA is flanked by 2′F-ANA at the 3′-end (L-FL 2), effects on stability are minimal; however, LNA does not increase duplex stability by the characteristic >3°C typically observed (35). Alternating 2′F-ANA with RNA at the 3′-end did reduce stability (L-FL 4).
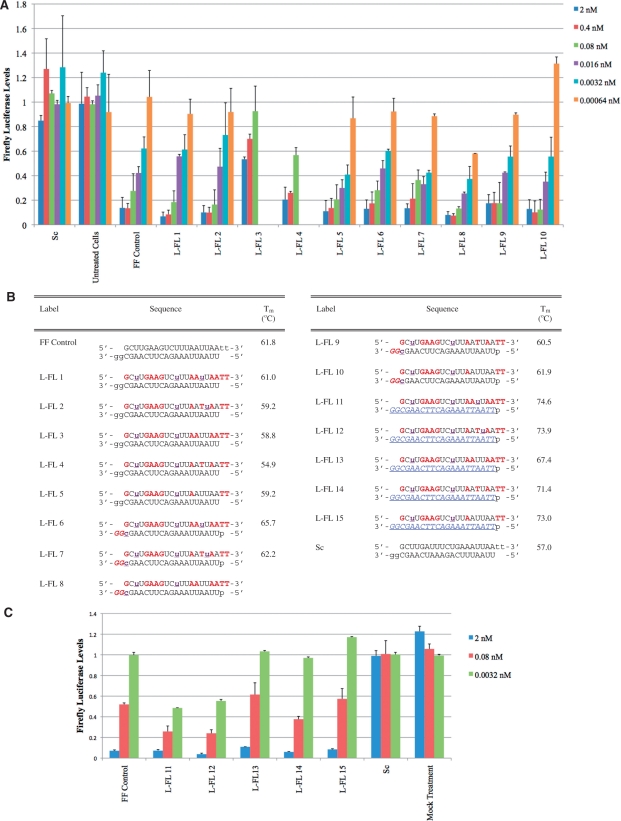
Figure 3.
(A) Assays demonstrating activity of siRNAs modified with 2′F-ANA, and LNA [sequences shown in (B)]. mRNA target is the firefly luciferase gene. Firefly luciferase levels were normalized to total cellular protein and average luciferase counts of untreated cells. For 2, 0.4 and 0.08 nM concentrations, n = 2 (for Sc, L-FL 3, and L-FL4) or n = 4 (for FF Control, L-FL 1, 2, 5–10 and untreated cells). n = 2 for 0.016, 0.0032, 0.00064 nM concentrations. L-FL 3 and 4 were not tested at the bottom three concentrations. (C) Assays demonstrating activity of siRNAs modified with 2′F-ANA and LNA, with a 2′F-RNA antisense stand [sequences shown in (B)]. Firefly luciferase levels were normalized to total cellular protein and luciferase counts of cells treated with scrambled (nontargeting) siRNA. n = 2. Bars indicate standard deviation. Legend: RNA, dna, 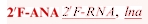 , p = 5′ phosphorylation.
, p = 5′ phosphorylation.
RNAi activity of modified siRNAs targeting firefly luciferase mRNA
We chose a firefly luciferase model system to assess the activity of our first set of chemically modified siRNAs (Figure 1A) because it allowed rapid determination of gene silencing activity. This first set of siRNAs combined 2′F-ANA with a well-known RNA analog used in siRNA, 2′F-RNA (36–38). We chemically 5′-phosphorylated all 5′-modified antisense strands in this study, since a 5′ phosphate is required for RISC loading (39) and cellular kinases may not be effective at phosphorylating modified nucleosides (20,40). Modified sense strands were not chemically phosphorylated, potentially inhibiting sense strand loading into RISC.
Firefly luciferase levels following treatment with siRNAs at a range of concentrations are shown in Figure 1B. In Figure 1B, the gene silencing activity of the full set of siRNAs from Figure 1A are provided. Many of the chemically modified siRNAs are of equal or greater potency than the unmodified siRNA (FF control). The most notable architectures in terms of potency enhancement are JG-4, 6, 8, 11, 12 and 14, with JG-6 and 14 being the most potent of the set in this experiment.
Several trends emerged from this first set of siRNA designs: (i) comparing JG-6 with JG-7 and JG-8 with JG-9, we inferred that chimeric 2′F-ANA/2′F-RNA strands were best used on the sense strand of the siRNA. (ii) By comparing JG-10 with JG-11, we observed that a fully 2′F-RNA strand works better as an antisense strand than as a sense strand. In addition, a fully 2′F-RNA-modified siRNA (JG-4) was potent, which has precedence in at least one other siRNA study (41). (iii) Comparing JG-12 with JG-13, we observed that 2′F-ANA strands work better as the sense strand than as the antisense strand. Surprisingly, a fully 2′F-ANA modified duplex (JG-5) had gene silencing activity, albeit less than the unmodified siRNA. Based on our observations of the preferences of 2′F-ANA for the sense strand and 2′F-RNA for the antisense strand, JG-14 is expected to have good gene silencing activity and JG-15 should be ineffective, which was the case.
A major challenge in the development of chemically modified siRNA designs is the heavy sequence dependence exhibited by many chemical modifications. When a potent modification architecture is identified for one siRNA sequence, there is no assurance that the architecture will be amenable to other siRNA sequences without loss of potency.
To examine the applicability of our modification designs to a range of targets, we first chose to target a second region of the firefly luciferase mRNA (from +515 to +533) (2) which allowed us to examine the sequence dependence of our designs. The sequences and architectures tested against this second mRNA region, the ff6 series, are shown in Figure 2A, and their RNAi activity is illustrated in Figure 2B.
The majority of 2′F-ANA/2′F-RNA modifications tested in this experiment were more potent than the unmodified control. The lower potency of siRNAs ff6-2 and 3 suggests that fully 2′F-ANA sense stands are less tolerated for this siRNA sequence than the previous one. Chimeric 2′F-ANA/2′F-RNA designs, however, retained their high potency across both sequences. The chimeric sense strands in ff6-4 and 10 are of a 3–3 altimeric design, roughly mimicking the sense strands of JG-2 and 6 (which were designed to use 2′F-RNA pyrimidines which are less expensive than purines), but in a manner that can be used regardless of siRNA sequence. Duplex ff6-8, which is analogous to JG-14, was more potent than the unmodified siRNA in this system as well. The sense strand of ff6-5 and ff6-11 was designed to take advantage of our observations that the 1–1 altimer design decreased duplex stability, but the 2′F-ANA purine/2′F-RNA pyrimidine design did not. By placing a 1–1 altimer stretch at the 3′-end of the sense strand, we hoped to destabilize the duplex at the 5′-end of the antisense strand and thus favor loading of the antisense strand into RISC (42,43). Notably, ff6-11 was the most potent siRNA of this data set, suggesting our rational design strategy improved siRNA activity. To further verify our rational design, we made siRNA FFC-611 (Figure 1A), which targets position +1818 to + 1836 of the firefly luciferase mRNA. As shown in Supplementary Figure S3, FFC-611 was as potent as the unmodified sequence.
These results and previous studies (20) hinted at synergy between 2′F-ANA and northern analogs in siRNA. To explore whether the synergy between 2′F-ANA and northern nucleotide analogs was general, we replaced the 2′F-RNA inserts in our sense strands with LNA, another northern nucleoside analog with siRNA activity (36,44–48). Because of the very high binding affinity increase induced by a single LNA, we used mixtures of LNA and RNA to replace the 2′F-RNA regions of our duplexes. Figure 3 shows the five LNA-modified sense strands we designed to mimic the sense strand of JG-2.The five sense strands were paired with either a normal RNA antisense strand with DNA overhangs, an antisense strand with a double 2′F-ANA overhang and a single LNA insert opposite the 5′-end of the sense strand (to favor loading of the correct antisense strand), or a fully 2′F-RNA antisense strand. As shown in Figure 3A, these LNA inserts were also able to produce heavily modified siRNAs with potencies equivalent or better than the unmodified controls despite heavy chemical modification.
We are particularly interested in siRNAs L-FL 6-15, which have both modified sense and antisense strands. L-FL 6-10 have modification of the antisense 3′-end, protecting from 3′ exonucleases, and L-FL 11-15 have fully modified antisense strands for even greater stability.
Combining 2′F-ANA with strongly preorganized northern nucleotides in siRNA duplexes appears to give a significant advantage over RNA itself or combinations of 2′F-ANA with RNA. These improvements may arise for a number of reasons: (i) heavily modified siRNAs are degraded more slowly than unmodified (or less-modified) siRNA; (ii) using the 1–1 altimer design (ff6-5 and ff6-11), we have lowered the binding affinity at the 3′-end of the sense strand to favor antisense strand loading; (iii) correct strand loading may be further encouraged by the selective phosphorylation of the antisense strand combined with sense strands that are modified at the 5′-end; and (iv) we may speculate that a subtle structural or dynamic effect is in operation whereby RISC prefers duplexes that are A-like but not as rigidly preorganized in the north as duplexes containing only 2′F-RNA or LNA.
Gene silencing activity of modified siRNAs targeting 4EBPs
To further determine the scope of application of our chimeric siRNAs, we decided to target endogenous therapeutically relevant genes. The 4E-binding proteins 4E-BP1 and 4E-BP2 repress translation by inhibiting formation of the eIF4F complex. 4EBPs downregulate interferon production by repressing translation of interferon regulatory factor 7 (23), a transcription factor essential for interferon responses to viral infections (24). A mouse knockout of 4E-BP1 and 4E-BP2 showed enhanced immune responses (23).
We synthesized 2′F-ANA/2′F-RNA chimeric siRNAs to target 4E-BP1 and 4E-BP2 in human cells. Effective siRNA sequences targeting the 4EBPs have been previously reported in a study examining hepatitis C virus mRNA translation and replication (21). Two of the most promising modified siRNA designs, corresponding to JG-14 and ff6-11, were selected from our previous luciferase gene silencing results and used for targeting 4E-BP1 and 4E-BP2 in human (HEK293T) cells (see the BP series in Table 1 and Supplementary Figure S4).
Table 1.
Sequences and thermal denaturation studies of siRNAs targeting 4E-BP2 mRNA (the BP2 series), and siRNAs used for studying the effects of modification on immunostimulation (the 728 and 728UU series). Legend: RNA, dna,  ,
,  , p = 5′ phosphorylation
, p = 5′ phosphorylation
 |
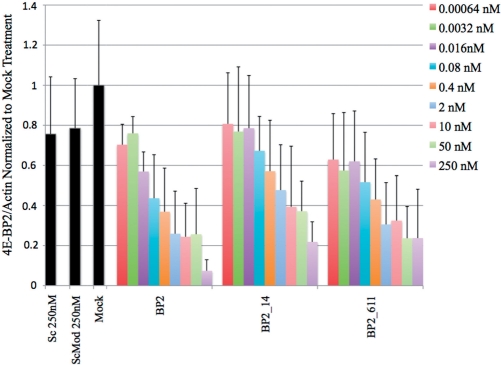
Levels of 4E-BP2 protein after treating cells with siRNA are shown in Figure 4 (representative western blots can be found in Supplementary Figure S6). Both modifications designs were very effective at silencing human 4E-BP2. Both BP2_14 and BP2_611, siRNA designs derived from JG-14 and ff6-11, were potent silencing agents for 4E-BP2, with potencies comparable to the unmodified siRNA (BP2). 4E-BP2 knockdown was assayed over a wide range of siRNA concentrations, from 0.64 pM to 250 nM. Gene silencing activity became detectable in the 3–80-pM range, and both native and modified duplexes had IC50 values in the low nanomolar. The unmodified BP2 siRNA was the most potent of the three, with the BP2_611 and BP2_14 modified siRNAs demonstrating only slightly less potency. Gene silencing experiments against 4E-BP2 included controls in which cells were treated with a nontargeting, scrambled siRNA (Sc) and chemically modified scrambled siRNA (FFC-611), which were simply selected from our studies of siRNAs targeting firefly luciferase. The scrambled controls behaved as expected, exerting little effect on 4E-BP2 gene expression.
Figure 4.
Densitometry analysis of western blot results following cell treatment with siRNAs modified with 2′F-ANA and 2′F-RNA (sequences shown in Table 1). mRNA target is 4E-BP2 in human HEK293T cells. siRNA concentrations ranged from 0.00064 to 250 nM. n = 10 for Sc 250 nM and ScMod 250 nM treatments, n = 13 for Mock treatment, n = 4 for BP2 0.00064–0.016 nM, n = 5 for BP2 0.08–250 nM, and n = 3 for all BP2_14 and BP2_611 treatments. Representative western blots are shown in Supplementary Figure S6. Bars represent standard deviation.
Similar results were obtained when we targeted 4E-BP1 with the modified siRNAs shown in Supplementary Figure S4. The unmodified BP1 siRNA was the most potent of the set, with BP1_611 (an siRNA based on ff6-11) a close second, followed by BP1_14 (based on JG-14) which was the least potent but still demonstrated good gene silencing activity. Results suggest that these modification designs will be widely applicable for silencing a range mRNA targets.
Immunostimulation
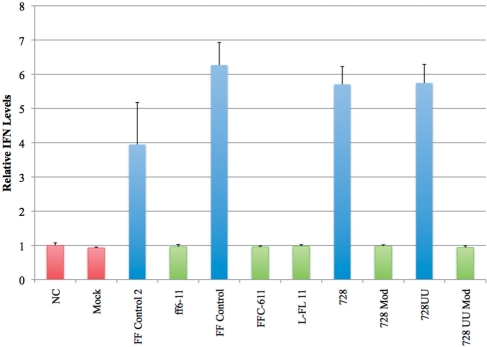
siRNAs can elicit strong interferon-α (IFN-α) responses in human PBMCs (47,49). To determine if our modified siRNAs can reduce non-specific immunostimulatory activity in PBMC cells, we treated PBMCs with unmodified or modified siRNAs (80 nM) and measured IFN production 24 h later. siRNAs were delivered using DOTAP (Figure 5). We tested the two sequences used in our luciferase screen as well as an siRNA sequence previously reported to have strong immunostimulatory activity in PBMCs (728UU, and the blunt-ended version, 728) (49). PBMCs treated with the four unmodified siRNAs (FF Control 2, FF Control, 728 and 728UU) showed significantly increased IFN production compared to the mock treatment (3–7-fold increases were observed). Modification of the four sequences based on the ff6-11 design pattern resulted in immunostimulation levels equivalent to the negative control measurements. The immunostimulatory profiles of the siRNAs were equivalent when transfected with Lipofectamine 2000 instead of DOTAP (Supplementary Figure S5A).
Figure 5.
IFN levels in PBMC cells 24 h after treatment with siRNAs transfected using DOTAP, as measured by the HEK-Blue IFN assay. IFN levels in response to unmodified siRNAs are shown in blue, modified siRNAs in green, and control treatments in red. NC is the negative control (cells alone). Mock treatment was transfection without siRNA. Data were collected in duplicate for each of two donors. Results were normalized to the negative control.
Therefore, the immunostimulatory effects of all siRNAs tested in this experiment can be eradicated by modification with 2′F-ANA/2′F-RNA. Our LNA constructs have been less extensively studied, but the results suggest that these modification designs also reduce immunostimulation in human PBMCs to a similar extent (L-FL 11 and data not shown).
Circular dichroism studies of 2′F-ANA/2′F-RNA-modified siRNAs
To further characterize the 2′F-ANA/2′F-RNA-modified siRNAs, we utilized circular dichroism (CD) to investigate the helical structure of modified siRNAs in the JG series (Supplementary Figure S2). In general, RNA or 2′F-RNA-modified duplexes were strongly A-form, the fully 2′F-ANA duplex JG-5 was more B-form, and chimeric duplexes maintained a more A-form conformation. In some cases, we observed significant differences in CD curves depending on whether equivalent modifications appear on the sense strand or the antisense strand. This is in spite of the fact that the purines and pyrimidines are evenly distributed among the two strands. Thus, duplex structure is related to the interplay of chemical modifications with duplex sequence.
FBS stability assays
Enhanced serum stability is a major advantage of chemically modified siRNAs. We expected the heavily modified nature of many of the 2′F-ANA/2′F–RNA/LNA architectures in this report to have significant serum stability, which might play a role in enhancing the potency and duration of activity of these siRNAs. We compared the serum stability of the native FF Control duplex with its fully modified analog, FFC-611. As shown in Supplementary Figure S1, the modified duplex remained intact for over 1 h, and slightly degraded duplex (perhaps missing only the single-stranded 3′ overhangs) was still observed after 4 days. In contrast, most of the unmodified siRNA was degraded within 30 min. FBS stability studies on heavily modified 2′F-ANA–RNA chimeric siRNAs, under similar conditions to the ones used here, have been previously described (19). In the previous study, 2′F-ANA–RNA chimeric siRNAs had a serum half-life of about 5 h. Results shown in Supplementary Figure S1 are in agreement with stability experiments of 2′F-ANA–RNA chimeric siRNAs, and suggest that siRNAs made of 2′F-ANA and 2′F-RNA are even more stable than those made of 2′F-ANA and RNA.
CONCLUSIONS
siRNAs that contain combinations of 2′F-ANA with the rigid RNA analogs 2′F-RNA or LNA show superior properties to native duplexes or duplexes with these modifications alone. Each of these nucleoside analogs has previously been used for siRNA-mediated gene silencing, but to the best of our knowledge, this is the first report of chimeric siRNAs composed of combinations of these modifications. Our results show knockdown of several genes with high potency, surpassing even the native duplexes in some cases. Furthermore, dramatically reduced immunostimulatory activity and excellent serum stability was observed.
Previous work by Ui-Tei et al. (50) showed that combinations of DNA and RNA gave siRNAs with potent silencing ability and reduced OTEs. The reduction in OTEs observed by these chimeric siRNAs was attributed in part to the lower binding affinity at the DNA-rich 5′-end of the duplex (50). Our results are similar in at least two respects: (i) we show that combining analogs of DNA and RNA imparts superior properties to siRNAs than are obtained by either analog taken alone; and (ii) we also attribute part of this success to the lower binding affinity at the 5′-end of the duplex. However, many differences exist between the two designs. For example, where Ui-Tei et al. used stretches of dsDNA to reduce OTEs, we propose a region of alternating nucleotides with different conformations in one strand. Furthermore, their duplexes are entirely constructed from native DNA and RNA, while ours are mostly or entirely made of modified nucleotides. Thus, the two designs may have different strengths and different spheres of application.
Several heavily modified siRNA designs have been reported recently (19,20,41,51–55), a few of which are entirely devoid of RNA. The sequence dependence of many of these architectures has yet to be fully determined. Our results indicate that siRNAs fully modified with rational combinations of 2′F-ANA, 2′F-RNA and/or LNA are advantageous gene silencing agents with a broad range of applications.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Sciences and Engineering Research Council of Canada (NSERC) (Vanier CGS to G.F.D. and CGS to J.K.W.); Canadian Institutes for Health Research (CIHR) (grants to M.J.D. and N.S.); National Cancer Institute of Canada (NCIC) (grant 17099 to J.P.); National Institutes of Health (NIH) (PO1-CA72765 to A.M.G.); and a grant from the PA Dept. of Health, Tobacco Settlement Award (to A.M.G.). Funding for open access charge: Canadian Institutes for Health Research.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 3.Watts JK, Deleavey GF, Damha MJ. Chemically modified siRNA: tools and applications. Drug Discov. Today. 2008;13:842–855. doi: 10.1016/j.drudis.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 4.Deleavey GF, Watts JK, Damha MJ. Chemical modification of siRNA. Curr. Protoc. Nucleic Acid Chem. 2009:16.3.1–16.3.21. doi: 10.1002/0471142700.nc1603s39. [DOI] [PubMed] [Google Scholar]
- 5.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotech. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 6.Judge A, MacLachlan I. Overcoming the innate immune response to small interfering RNA. Hum. Gene Ther. 2008;19:111–124. doi: 10.1089/hum.2007.179. [DOI] [PubMed] [Google Scholar]
- 7.Wilds CJ, Damha MJ. 2′-Deoxy-2′-fluoro-β-D-arabinonucleosides and oligonucleotides (2′F-ANA): synthesis and physicochemical studies. Nucleic Acids Res. 2000;28:3625–3635. doi: 10.1093/nar/28.18.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Damha MJ, Wilds CJ, Noronha A, Brukner I, Borkow G, Arion D, Parniak MA. Hybrids of RNA and arabinonucleic acids (ANA and 2′F-ANA) are substrates of ribonuclease H. J. Am. Chem. Soci. 1998;120:12976–12977. [Google Scholar]
- 9.Rosenberg I, Soler JF, Tocik Z, Ren WY, Ciszewski LA, Kois P, Pankiewicz KW, Spassova M, Watanabe KA. Synthesis of oligodeoxynucleotides containing the C-nucleoside and 2′-deoxy-2′-fluoro-ara-nucleoside moieties by the H-phosphonate method. Nucleosides Nucleotides. 1993;12:381–401. [Google Scholar]
- 10.Kois P, Tocik Z, Spassova M, Ren WY, Rosenberg I, Soler JF, Watanabe KA. Synthesis and some properties of modified oligonucleotides. 2. Oligonucleotides containing 2′-deoxy-2′-fluoro-β-D-arabinofuranosyl pyrimidine nucleosides. Nucleosides Nucleotides. 1993;12:1093–1109. [Google Scholar]
- 11.Watts JK, Damha MJ. 2′F-arabinonucleic acids (2′F-ANA) - history, properties, and new frontiers. Can. J. Chem. 2008;86:641–656. [Google Scholar]
- 12.Ikeda H, Fernandez R, Wilk A, Barchi J.J., Jr, Huang X, Marquez VE. The effect of two antipodal fluorine-induced sugar puckers on the conformation and stability of the Dickerson–Drew dodecamer duplex [d(CGCGAATTCGCG)]2. Nucleic Acids Res. 1998;26:2237–2244. doi: 10.1093/nar/26.9.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger I, Tereshko V, Ikeda H, Marquez VE, Egli M. Crystal structures of B-DNA with incorporated 2′-deoxy-2′-fluoro-arabino-furanosyl thymines: implications of conformational preorganization for duplex stability. Nucleic Acids Res. 1998;26:2473–2480. doi: 10.1093/nar/26.10.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denisov AY, Noronha AM, Wilds CJ, Trempe J.-F, Pon RT, Gehring K, Damha MJ. Solution structure of an arabinonucleic acid (ANA)/RNA duplex in a chimeric hairpin: comparison with 2′-fluoro-ANA/RNA and DNA/RNA hybrids. Nucleic Acids Res. 2001;29:4284–4293. doi: 10.1093/nar/29.21.4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trempe J.-F, Wilds CJ, Denisov AY, Pon RT, Damha MJ, Gehring K. NMR solution structure of an oligonucleotide hairpin with a 2′F-ANA/RNA stem: implications for RNase H specificity toward DNA/RNA hybrid duplexes. J. Am. Chem. Soc. 2001;123:4896–4903. doi: 10.1021/ja003859p. [DOI] [PubMed] [Google Scholar]
- 16.Mangos MM, Min KL, Viazovkina E, Galarneau A, Elzagheid MI, Parniak MA, Damha MJ. Efficient RNase H-directed cleavage of RNA promoted by antisense DNA or 2′F-ANA constructs containing acyclic nucleotide inserts. J. Am. Chem. Soc. 2003;125:654–661. doi: 10.1021/ja025557o. [DOI] [PubMed] [Google Scholar]
- 17.Kalota A, Karabon L, Swider CS, Viazovkina E, Elzagheid M, Damha MJ, Gewirtz AM. A novel 2′-deoxy-2′-fluoro (2′F-ANA) ribose modification significantly enhances the duration, and efficiency, of nucleic acid mediated gene silencing. ASH Annu. Meeting Abstr. 2005;106:3062. [Google Scholar]
- 18.Kalota A, Karabon L, Swider CR, Viazovkina E, Elzagheid M, Damha MJ, Gewirtz AM. 2′-deoxy-2′-fluoro-β-D-arabinonucleic acid (2′F-ANA) modified oligonucleotides (ON) effect highly efficient, and persistent, gene silencing. Nucleic Acids Res. 2006;34:451–461. doi: 10.1093/nar/gkj455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dowler T, Bergeron D, Tedeschi AL, Paquet L, Ferrari N, Damha MJ. Improvements in siRNA properties mediated by 2′-deoxy-2′-fluoro-β-D-arabinonucleic acid (FANA) Nucleic Acids Res. 2006;34:1669–1675. doi: 10.1093/nar/gkl033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watts JK, Choubdar N, Sadalapure K, Robert F, Wahba AS, Pelletier J, Mario Pinto B, Damha MJ. 2′-Fluoro-4′-thioarabino-modified oligonucleotides: conformational switches linked to siRNA activity. Nucleic Acids Res. 2007;35:1441–1451. doi: 10.1093/nar/gkl1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murata T, Hijikata M, Shimotohno K. Enhancement of internal ribosome entry site-mediated translation and replication of hepatitis C virus by PD98059. Virology. 2005;340:105–115. doi: 10.1016/j.virol.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 22.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colina R, Costa-Mattioli M, Dowling R.JO, Jaramillo M, Tai L.-H, Breitbach CJ, Martineau Y, Larsson O, Rong L, Svitkin YV, et al. Translational control of the innate immune response through IRF-7. Nature. 2008;452:323–328. doi: 10.1038/nature06730. [DOI] [PubMed] [Google Scholar]
- 24.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 25.Damha MJ, Ogilvie KK. Oligoribonucleotide synthesis: the silyl-phosphoramidite method. In: Agrawal S, editor. Protocols for Oligonucleotides and Analogs: Synthesis and Properties, Methods in Molecular Biology. Vol. 20. Totowa, NJ: The Humana Press Inc.; 1993. pp. 81–114. [DOI] [PubMed] [Google Scholar]
- 26.Bellon L. Oligoribonucleotides with 2′-O-(tert-butyldimethylsilyl) groups. Curr. Protoc. Nucleic Acid Chem. 2000:3.6.1–3.6.13. doi: 10.1002/0471142700.nc0306s01. [DOI] [PubMed] [Google Scholar]
- 27.Westmanu E, Stromberg R. Removal of t-butyldimethylsilyl protection in RNA-synthesis. Triethylamine trihydrofluoride (TEA, 3HF) is a more reliable alternative to tetrabutylammonium fluoride (TBAF) Nucleic Acids Res. 1994;22:2430–2431. doi: 10.1093/nar/22.12.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wincott F, DiRenzo A, Shaffer C, Grimm S, Tracz D, Workman C, Sweedler D, Gonzalez C, Scaringe S, Usman N. Synthesis, deprotection, analysis and purification of RNA and ribosomes. Nucleic Acids Res. 1995;23:2677–2684. doi: 10.1093/nar/23.14.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Novac O, Guenier A.-S, Pelletier J. Inhibitors of protein synthesis identified by a high throughput multiplexed translation screen. Nucleic Acids Res. 2004;32:902–915. doi: 10.1093/nar/gkh235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 microRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 31.Kawasaki AM, Casper MD, Freier SM, Lesnik EA, Zounes MC, Cummins LL, Gonzalez C, Cook PD. Uniformly modified 2′-deoxy-2′-fluoro-phosphorothioate oligonucleotides as nuclease-resistant antisense compounds with high affinity and specificity for RNA targets. J. Med. Chem. 1993;36:831–841. doi: 10.1021/jm00059a007. [DOI] [PubMed] [Google Scholar]
- 32.Petersen M, Bondensgaard K, Wengel J, Jacobsen JP. Locked nucleic acid (LNA) recognition of RNA: NMR solution structures of LNA:RNA hybrids. J. Am. Chem. Soc. 2002;124:5974–5982. doi: 10.1021/ja012288d. [DOI] [PubMed] [Google Scholar]
- 33.Kent B, Michael P, Sanjay KS, Vivek KR, Ravindra K, Jesper W, Jens Peter J. Structural studies of LNA:RNA duplexes by NMR: conformations and implications for RNase H activity. Chem. Eur. J. 2000;6:2687–2695. doi: 10.1002/1521-3765(20000804)6:15<2687::aid-chem2687>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 34.Kaur H, Arora A, Wengel J, Maiti S. Thermodynamic, counterion, and hydration effects for the incorporation of locked nucleic acid nucleotides into DNA duplexes. Biochemistry. 2006;45:7347–7355. doi: 10.1021/bi060307w. [DOI] [PubMed] [Google Scholar]
- 35.Koshkin AA, Nielsen P, Meldgaard M, Rajwanshi VK, Singh SK, Wengel J. LNA (locked nucleic acid): an RNA mimic forming exceedingly stable LNA:LNA duplexes. J. Am. Chem. Soc. 1998;120:13252–13253. [Google Scholar]
- 36.Braasch DA, Jensen S, Liu Y, Kaur K, Arar K, White MA, Corey DR. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry. 2003;42:7967–7975. doi: 10.1021/bi0343774. [DOI] [PubMed] [Google Scholar]
- 37.Chiu YL, Rana TM. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harborth J, Elbashir SM, Vandenburgh K, Manninga H, Scaringe SA, Weber K, Tuschl T. Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense Nucleic Acid Drug Dev. 2003;13:83–105. doi: 10.1089/108729003321629638. [DOI] [PubMed] [Google Scholar]
- 39.Weitzer S, Martinez J. The human RNA kinase hClp1 is active on 3′ transfer RNA exons and short interfering RNAs. Nature. 2007;447:222–226. doi: 10.1038/nature05777. [DOI] [PubMed] [Google Scholar]
- 40.Fisher M, Abramov M, Van Aerschot A, Xu D, Juliano RL, Herdewijn P. Inhibition of MDR1 expression with altritol-modified siRNAs. Nucleic Acids Res. 2007;35:1064–1074. doi: 10.1093/nar/gkl1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blidner RA, Hammer RP, Lopez MJ, Robinson SO, Monroe WT. Fully 2′-Deoxy-2′-Fluoro substituted nucleic acids induce RNA interference in mammalian cell culture. Chem. Biol. Drug Des. 2007;70:113–122. doi: 10.1111/j.1747-0285.2007.00542.x. [DOI] [PubMed] [Google Scholar]
- 42.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 43.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 44.Petersen M, Wengel J. LNA: a versatile tool for therapeutics and genomics. Trends Biotechnol. 2003;21:74–81. doi: 10.1016/S0167-7799(02)00038-0. [DOI] [PubMed] [Google Scholar]
- 45.Bramsen JB, Laursen MB, Damgaard CK, Lena SW, Babu BR, Wengel J, Kjems J. Improved silencing properties using small internally segmented interfering RNAs. Nucleic Acids Res. 2007;35:5886–5897. doi: 10.1093/nar/gkm548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elmen J, Thonberg H, Ljungberg K, Frieden M, Westergaard M, Xu Y, Wahren B, Liang Z, Urum H, Koch T, et al. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33:439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, Noronha A, Manoharan M, Akira S, de Fougerolles A, et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 48.Mook OR, Baas F, de Wissel MB, Fluiter K. Evaluation of locked nucleic acid-modified small interfering RNA in vitro and in vivo. Mol. Cancer Ther. 2007;6:833–843. doi: 10.1158/1535-7163.MCT-06-0195. [DOI] [PubMed] [Google Scholar]
- 49.Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat. Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 50.Ui-Tei K, Naito Y, Zenno S, Nishi K, Yamato K, Takahashi F, Juni A, Saigo K. Functional dissection of siRNA sequence by systematic DNA substitution: modified siRNA with a DNA seed arm is a powerful tool for mammalian gene silencing with significantly reduced off-target effect. Nucleic Acids Res. 2008;36:2136–2151. doi: 10.1093/nar/gkn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choung S, Kim YJ, Kim S, Park H.-O, Choi Y.-C. Chemical modification of siRNAs to improve serum stability without loss of efficacy. Biochem. Biophys. Res. Commun. 2006;342:919–927. doi: 10.1016/j.bbrc.2006.02.049. [DOI] [PubMed] [Google Scholar]
- 52.Kraynack BA, Baker BF. Small interfering RNAs containing full 2′-O-methylribonucleotide-modified sense strands display Argonaute2/eIF2C2-dependent activity. RNA. 2006;12:163–176. doi: 10.1261/rna.2150806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.David VM, Karin B, Lucinda S, Kristi J, Jennifer AL, Brent D, James AM, Chandra V, Keith B, Chris SS, et al. Activity of stabilized short interfering RNA in a mouse model of hepatitis B virus replication. Hepatology. 2005;41:1349–1356. doi: 10.1002/hep.20702. [DOI] [PubMed] [Google Scholar]
- 54.Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, Hartsough K, Machemer L, Radka S, Jadhav V, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat. Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 55.Allerson CR, Sioufi N, Jarres R, Prakash TP, Naik N, Berdeja A, Wanders L, Griffey RH, Swayze EE, Bhat B. Fully 2′-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J. Med. Chem. 2005;48:901–904. doi: 10.1021/jm049167j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.