Abstract
TRAF1 is a member of the TRAF family, which plays important roles in signal transduction that mediate cell life and death in the immune response, inflammatory and malignant diseases. It is known that TRAF1 transcription is inducible by various cytokines, but little is known about the regulation of its mRNA translation. In the present study, we demonstrated that the human TRAF1 mRNA has an unusually long 5′-UTR that contains internal ribosome entry segment (IRES) regulating its translation. By performing gene transfection and reporter assays, we revealed that this IRES sequence is located within the 572 nt upstream from the AUG start codon. An element between nt −392 and −322 was essential for the IRES activity. Interestingly, we found that the TRAF1 expression is induced in cancer cells by chemotherapeutic drug vincristine that regulates cytoplasmic localization of polypyrimidine tract binding protein, which may contribute to the IRES-dependent translation of TRAF1 during vincristine treatment. These results indicate that TRAF1 translation is initiated via the IRES and regulated by vincristine, and suggest that regulation of the IRES-dependent translation of TRAF1 may be involved in effecting the cancer cell response to vincristine treatment.
INTRODUCTION
TRAF1 is a member of the TRAF family that was originally identified based on its ability to interact with the cytosolic domain of tumor necrosis factor (TNF) receptor type 2 (TNFR2) (1). To date, six members of the TRAF family have been identified. All of these proteins share a C-terminal TRAF domain, which is required for the binding of these signal-transducing adaptors to TNFRs. A more variable N-terminal domain, containing a ring finger and several zinc finger motifs, is also found in all TRAFs except TRAF1 (2). TRAF1 differs from the other TRAFs not only at the structural level, but also by its tissue-specific expression (2,3). TRAF1 expression is restricted to the normal cells of the spleen, lung and testis; however, expression and even overexpression of this protein occurs in many cancer cells, in particular the lymphoid malignancies (4–8). TRAF1 expression can be induced by stimulation with various cytokines, such as TNF-α, IL-1 and CD40L (4,9). It has been reported that cytokine-induced expression of TRAF1 occurs at the transcriptional level (10,11). So far, no reports have studied the regulation of TRAF1 expression at the translational level.
It is postulated that the regulation of gene expression at the level of translation plays an important role in controlling gene expression that provides the cell with the plasticity needed to respond to rapid changes in the environment, such as cellular stress or apoptosis (12,13). Initiation of translation can occur by two distinct mechanisms, cap-dependent scanning and internal ribosome entry. The latter requires an internal ribosome entry segment (IRES), located in the 5′-UTR of the mRNA (14). IRES elements are mainly found within the mRNAs of proteins involved in regulating gene expression during development, differentiation, cell growth and apoptosis (15,16). The cap-independent (i.e. IRES-dependent) translation represents a fail-safe mechanism for protein expression to progress when cap-dependent translation is prevented or when the leader sequence contains structural elements that inhibit scanning by ribosomes (17,18). In particular, IRES becomes activated under conditions in which cap-dependent protein synthesis is greatly reduced, such as upon cellular stress and DNA damage, whereupon the activated IRES initiates translation of only those specific proteins that are able to protect cells from stress (19). It is basically the energy-saving, cell-saving option for stressed cells.
The IRES-dependent translation requires the presence of an additional complex set of trans-acting factor/ribonucleoprotein (RNP) participants for translational activity to occur (14,20). Previous studies have shown that several trans-acting factors and RNPs can regulate IRES activity, including the polypyrimidine tract binding (PTB) protein, which induces the translation of many mRNA with IRES during cellular stress (21–24). The trans-acting factor/RNP complexes bind to the IRES region of mRNA, inducing the conformational changes that facilitate recruitment of the ribosome to the IRES.
Because TRAF1 is an inducible protein that mediates signals during cellular stress and apoptosis, we studied TRAF1 to see whether its translation was regulated by an IRES-dependent mechanism. In searching the TRAF1 gene from the Human Genome database, we found that TRAF1 mRNA contains an unusually long 5′-UTR. Thus, we evaluated the potential for IRES elements within the 5′-UTR of TRAF1 and looked for possible regulators of IRES-mediated TRAF1 translation in cancer cells of lymphoid origin, following treatment with several chemotherapeutic drugs that are cell stress inducers.
MATERIALS AND METHODS
Cell lines and primary leukemia cells
Two lymphoid neoplastic cell lines, Raji and HDLM, originating from human Burkitt lymphoma and Hodgkin’s lymphoma, respectively, were used in this study. Primary leukemia cells were derived at diagnosis, from patients who were treated for acute lymphoblastic leukemia (ALL) at Emory University’s clinical facilities. Following informed consent, pre-treatment bone marrow specimens containing primary leukemia cells were obtained from eight patients with B-cell precursor (BCP) ALL, as diagnosed by standard immunologic, morphologic and cytochemical criteria. The mononuclear cells were separated by centrifugation on Ficoll-Hypaque (1.077 g ml−1), washed twice in phosphate-buffered saline (PBS) and resuspended at a density of 106 cells ml−1 in RPMI 1640 medium containing 10% fetal bovine serum (FBS). The cells were incubated on plastic petri dishes for one hour at 37°C to remove the monocytes, and then the desired non-adherent cells were recovered by gently washing the dishes. Following purification, all specimens collected contained more than 90% blasts.
Cell culture, reagents and treatment
Both the cell lines and primary leukemia cells were grown in standard culture medium (RPMI 1640 containing 10% FBS, 2 mmol l−1 of l-glutamine, 50 units penicillin and 50 µg ml−1 streptomycin) while incubated at 37°C in air containing 5% CO2. All chemotherapeutic drugs tested, including dexamethasone (DEXA), vincristine (VCR), doxorubicin (Dox), methotrexate (MTX), cytarabine (Ara-c) and l-asparaginase (l-Asp), were purchased from Sigma. Cells were treated with a clinically relevant range of concentration of each drug for the time period indicated. Controlled experiments were performed using one set of cells, and then repeated at least three times.
Plasmid construction
The full length as well as various deletions of the TRAF1 5′-UTR cDNA were obtained by Reverse transcriptase PCR (RT-PCR) from RNA extracted from the Raji cell line. These TRAF1 5′-UTR cDNAs were each inserted immediately upstream from the translation start codon of the firely luciferase (FL) gene in pGL3-Control (pGL3C) plasmid at Hind III and Nco I sites, to generate the monocistronic TRAF1 5′-UTR-luciferase plasmids. The dicistronic vector pRL-FL was kindly provided by Dr. Steve Haines (University of Nottingham, UK). Also, the full length and various deletions of TRAF1 5′-UTR cDNA were each inserted between RL and FL of the pRL-FL vector at EcoR I and Nco I sites, to generate dicistronic reporter plasmids. Many control plasmids were made simultaneously, including 5′-UTR sequences with mutations in the pGL3C and pRL-FL vectors and insertion of an inverted repeat sequence, which forms a very stable RNA hairpin structure, into the pRL-FL vector. All the generated plasmids were DNA sequenced in order to confirm the presence and correctness of the TRAF1 5′-UTR cDNAs in pGL3C and pRL-FL vectors. The pSUPER/PTB siRNA plasmid was constructed by inserting a specific 19 nt PTB sequence 5′-CAAAGGAGCCCAGGGCTCT-3′ into an expression plasmid, pSUPER-neo, purchased from OligoEngine (Seattle, WA).
Gene transfection and reporter assay
Transient transfections were performed to identify the presence of IRES activity in the TRAF1 5′-UTR. Cells were transfected with either monocistronic or dicistronic TRAF1 5′-UTR reporter plasmids by electroporation at 320 V, 950 µF, using a Gene Pulser II System (Bio-Rad, Hercules, CA). As an internal control, the pRL-CMV vector was co-transfected with monocistronic plasmids. Transfected cells were resuspended in 10 ml of RPMI containing 10% FBS, and then incubated 24–36 h. Cell extracts were prepared with 1× lysis buffer, then 20 µl aliquots of the supernatant were mixed first with 100 µl of Luciferase Assay Reagent II (Promega) to measure the FL activity and next, the RL activity was determined by adding Stop & Glo® Reagent to the same sample. These luciferase activities were analyzed via Microplate Instrumentation (BioTek).
Protein extraction and western blot analyses
Whole cell protein samples were prepared by lysing cells for 30 min at 4°C in a lysis buffer composed of 150 mM NaCl, 50 mM Tris (pH 8.0), 5mM EDTA, 1% (v/v) Nonidet p-40, 1 mM phenylmethylsulfonyl fluoride, 20 µg ml−1 aprotinin and 25 µg ml−1 leupeptin. To detect the cellular localization of protein, both nuclear and cytoplasmic fractions were isolated using the NE-PER kit (Pierce), according to the instructions of the manufacturer. Equal amounts of protein extracts were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and transferred to a nitrocellulose filter. After blocking with buffer containing 5% non-fat milk in 20 mM Tris–HCl (pH 7.5) with 500 mM NaCl for 1 h at room temperature, the filter was incubated with specific antibodies for 1 h at room temperature, followed by HRP-labelled secondary antibody. The blots were developed using a chemiluminescent detection system (ECL, Amersham Life Science, Buckinghamshire, England).
RNA isolation and RT–PCR
The total RNA was extracted with the RNeasy Mini Kit (Qiagen, Hilden, Germany) and was digested with DNAse. RT–PCR was performed with an Access RT–PCR Kit (Promega Corporation) according to the manufacturer’s protocols. Reactions without addition of reverse transcriptase (RT-) served as controls in experiments to amplify the TRAF1 5′-UTR. To specifically amplify the 5′-UTR of TRAF1 mRNAs, different primers were designed according to the corresponding sequences from the TRAF1 5′-UTR mRNA. After electrophoresis on a 1.5% agarose gel, the PCR products were visualized with ethidium bromide staining under UV light. For testing TRAF1 mRNA expression, First-strand cDNA synthesis was performed with a mixture of random monamers and oligo-dT as primers (Qiagen). Amplification of TRAF1 was performed with a 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA), using a QuantiFast SYBR Green RT–PCR kit (Qiagen), according to the manufacturer’s instructions. The TRAF1 primers and the housekeeper gene GAPDH were purchased from Qiagen (primer sequences not provided). For testing the expression of FL and RL in cells transfected with reporter plasmids, the primers used were: For FL, forward 5′-GGTGGACATCACTTACGC-3′ and reverse 5-GAAGGCTCCTCAGAAACA-3; for RL, forward 5′-AAAGGTGAAGTTCGTCGTCCAAC-3′ and reverse 5′- TTTGAGAACTCGCTCAACGAACG-3′.
Polysome preparation and analysis
Polysomes profiling was carried out essentially as described previously (25) with slight modifications. Briefly, Raji cells, with or without exposure to VCR, were incubated with 100 μg ml−1 cycloheximide for 15 min to arrest polyribosome migration. Cells were then lysed to isolate cytoplasmic extracts in a buffer containing 20 mM Tris–HCl at pH 8.0, 100 mM NaCl, 5 mM MgCl2, 0.5% Triton X-100, 500 U ml−1 RNAsin and a cocktail of protease inhibitors, followed by fractionation on 15–45% (w/v) sucrose gradient. The gradient was centrifuged in a SW41Ti rotor at 39 000 rpm for 1 h. Fractions were collected from each gradient tube by upward replacement with monitored absorption at OD254 by using a fractionator (Isco, Lincoln, NE). The RNA in each fraction was extracted and subjected to real-time PCR as described above.
Expression of GST-tagged PTB protein
The expression and purification of GST-fused PTB protein were performed as described previously (25). Briefly, after transfection of the GST-fused PTB plasmid into BL21 Escherichia coli, the bacteria were incubated in LB medium, and then harvested after incubation with 0.1 mM IPTG for 2 h. The purification of GST-fused PTB protein involved lysing of induced cells with sonication, followed by isolation of the complex with glutathione-agarose beads (Pharmacia). The purity and correct expression of the GST-fused PTB protein were analyzed by gel electrophoresis with Coomassie G250 staining, plus a western blot assay using antibodies against GST or PTB.
UV cross-linking and RNA binding assays
UV cross-linking and RNA binding assays were performed as described previously (25). Briefly, the DNA templates for TRAF1 IRES from −392 to −322 (71 bases) with a T7 promoter for synthesis of the TRAF1 IRES RNA probe (probe 1) was chemically synthesized. The same 71 bases DNA fragment with mutations at the CU-rich sequences (−379 to −370 and −332 to 324) served as control (probe 2). Internally labelled RNA probes were synthesized by in vitro transcription with T7 polymerase (MAXIScript T7 RNA polymerase kit, Ambion) in the presence of [α-32P] UTP (Amersham). Either GST-fused PTB or GST only proteins were mixed with each of the 32P-labelled RNA probes. UV cross-linking of the RNA–protein complexes was performed using a 254-nm UV light source set at 400 000 µJ cm−2. The UV-irradiated RNA–protein complexes were then treated with RNase T1, resolved by 10% SDS–PAGE gel and visualized by autoradiography.
For in vivo association between PTB protein and TRAF1 IRES, TRAF1 mRNA was co-immunoprecipitated from whole-cell extracts by using the modified as described previously (25). Briefly, cultured Raji cells were harvested by low-speed centrifugation at 4°C. The cell pellets were resuspended in 100 μl of RNA binding buffer [20 mM Tris (pH 7.5), 150 mM NaCl, 5 mM MgCl2, 0.1% NP-40, 50 μM ZnCl2, 2% glycerol, 1 mM DTT] that was supplemented with 10U of RNase inhibitor (5′–3′), then cell extracts were prepared by the freeze-thaw method. To whole-cell extracts, 5 μl of monoclonal anti-PTB antibody or the anti-actin antibody, 20 μl of protein A plus G agarose beads, and 5 U of RNase inhibitor were added, after which the samples were incubated for 60 min at room temperature. Next, the beads were washed extensively with RNA-binding buffer supplemented with RNase inhibitor. The RNA associated with the antibody–antigen complexes was isolated by repeat phenol–chloroform extractions and precipitation with 2M ammonium acetate and 3 volumes of cold ethanol. The RNA was then analyzed by RT–PCR, using TRAF1 IRES-specific primers.
RESULTS
Identification of the long 5′-UTR within TRAF1 mRNA
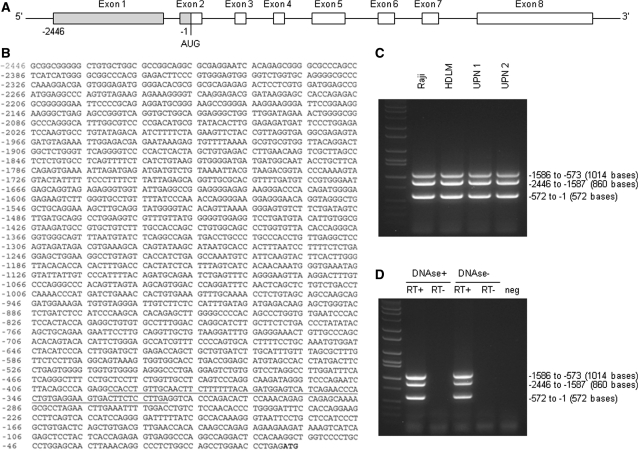
We searched the Human Genome database (http://www.ensembl.org/index.html) and found that only TRAF1, but not other TRAF members, has an unusually long 5′-UTR. The 5′-UTR lengths for the TRAF family are 2446 bases for TRAF1 (Figure 1A and B), 55 bases for TRAF2, 353 bases for TRAF3, 108 bases for TRAF4, 54 bases for TRAF5 and 340 bases for TRAF6. We performed RT–PCR on cancer cell lines and primary leukemia cells, demonstrating and confirming the presence of the 5′-UTR of TRAF1 mRNA in all cancer cells studied (Figure 1C and D).
Figure 1.
(A) Schematic illustration of the TRAF1 mRNA structure containing a long 5′-UTR (from −2446 to −1). The translation start codon AUG located 226 bases from the 5′ border of exon 2. (B) DNA sequence of the TRAF1 5′-UTR. The start codon ATG is highlighted and a potential IRES sequence is underlined. (C) RT–PCR products indicate that TRAF1 5′-UTR was expressed by all cancer cells studied, including the two lymphoma cell lines (Raji and HDLM) and primary ALL cells from two patients (UPN 1 and UPN 2) shown. The primers used for PCR amplification were designed from the corresponding areas on TRAF1 5′-UTR, to synthesize the three fragments indicated. (D) RT–PCR was performed using the same primers as in (C) and total Raji RNA that was digested with or without DNAse, in the presence or absence of RT.
Analysis of IRES activity within the 5′-UTR of TRAF1 mRNA
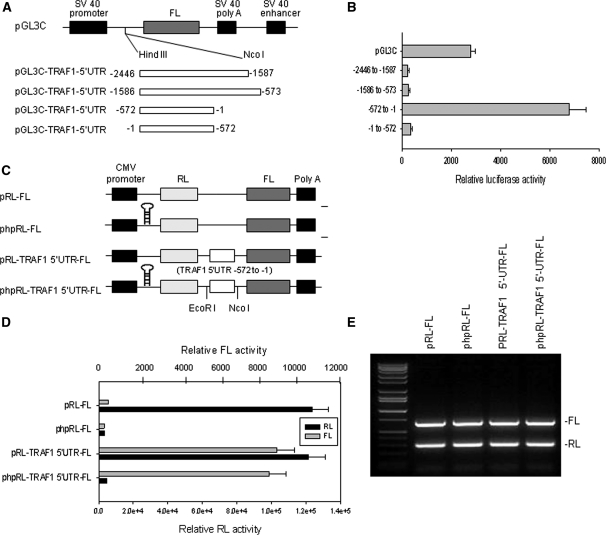
To investigate the possibility that the TRAF1 5′-UTR may have IRES activity, we divided the entire 2446 bases 5′-UTR of TRAF1 into three parts (−2446 to −1587, −1586 to −573 and −572 to −1) and cloned each of them into the pGL3C plasmid immediately upstream of the FL translation start codon, to generate monocistronic plasmids (Figure 2A). The SV40 promoter-driven FL activity in this plasmid could be affected by any insertion between the promoter and FL gene. For example, if the insertions have IRES activity, the FL activity in cells transfected with this plasmid would not be inhibited, or may even be enhanced. As shown in Figure 2(B), when the plasmids were transfected into Raji cells, the insertions with the −2446 to −1587 and −1586 to −573 sequences inhibited FL activities as compared to pGL3C, suggesting that these sequences have no IRES activity. In contrast, the −572 to −1 fragment increased FL activity, suggesting that this fragment has IRES activity. Insertion into pGL3C of the same −572 to −1 fragment in a reverse orientation blocked FL activity, further confirming that the forward −572 to −1 sequence of the TRAF1 5′-UTR has IRES activity.
Figure 2.
Analysis of IRES activity in the 5′-UTR of TRAF1. (A) Schematic representation of the monocistronic plasmids used for the reporter assays. The entire 2446 bases TRAF1 5′-UTR was divided into three sections (−2446 to −1587, −1586 to −573 and −572 to −1) and each was cloned into pGL3C plasmid immediately upstream from the translation start codon of the firefly luciferase (FL) gene. (B) Transfection and reporter assay for IRES activity of TRAF1 5′-UTR. Raji cells were each co-transfected with 5 μg of pGL3C-TRAF1-5′UTR plasmid and 1 μg of pRL-SV40 vector as an internal control, by electroporation, and then quantitative RL and FL activities were detected using the Dual-Luciferase Reporter System. Data represent the mean ± SD of three independent experiments normalized to RL activity. (C) Schematic representation of the dicistronic constructs used for the reporter assays. The phpRL-FL is similar to pRL-FL, but with the addition of a stable hairpin at the 5′-end of the RL transcript to suppress cap-dependent translation of RL. (D) RL and FL activity analyses following transfection of Raji cells with 5 μg of each plasmid indicated, including pRL-FL and phpRL-FL vectors and constructions of these vectors with insertion of the fragment −572 to −1 of the TRAF1 5′-UTR. (E) RL and FL mRNA levels analyses following transfection of cells with various plasmids as described in (D). Total RNA was extracted, and RT–PCR was performed with specific primers targeted to FL and RL.
To further assess the IRES activity of the −572 to −1 fragment, we cloned it into the pRL-FL vector to generate various dicistronic plasmids (Figure 2C). Transfection of the pRL-FL vector alone into Raji cells showed RL activity, but not FL activity (Figure 2D). However, FL activity was stimulated in Raji cells that were transfected with pRL-FL containing the −572 to −1 fragment, while the RL activity level was similar to pRL-FL alone. To ensure that it was an effect of internal ribosome entry rather than enhanced ribosomal read through or re-initiation, an inverted repeat sequence, which forms a very stable RNA hairpin structure, was inserted into the upstream RL cistron (Figure 2C). As shown in Figure 2(D), this inserted hairpin absolutely inhibited the cap-dependent translation of RL, while the presence of the −572 to −1 fragment of TRAF1 5′-UTR in the constructor allows initiation of FL translation. The FL activity stimulated by the −572 to −1 fragment with the hairpin added (phpRL-FL) was comparable to that of the transfected with pRL-FL containing the fragment without the hairpin. These results confirmed that the FL activity induced in the dicistronic plasmid was indeed stimulated by that −572 to −1 fragment that can act as IRES does, and was not a product of enhanced ribosomal read through or re-initiation.
Furthermore, we performed RT–PCR analyses of mRNA expression of FL in cells transfected with dicistronic reporter plasmids. We found that FL mRNA expression was not altered by insertion of the −572 to −1 fragment (Figure 2E), which suggested that the fragment-stimulated FL activity occurs solely at the translational level.
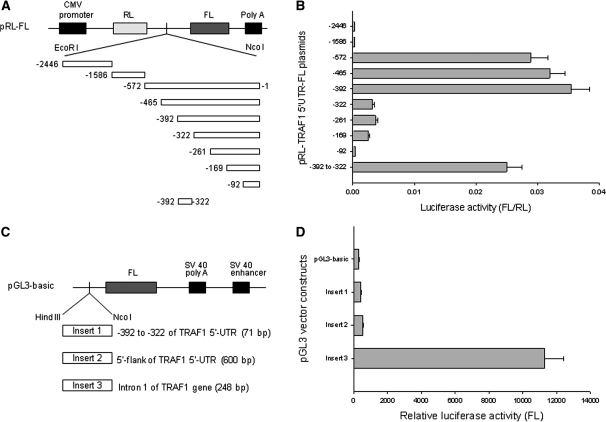
Mapping the TRAF1 IRES
To further determine the core IRES region within the TRAF1 5′-UTR, we generated dicistronic plasmids containing a series of deleted fragments of the −572 to −1 sequence (Figure 3A). Each of these plasmids was transfected into Raji cells. Upon assessment of their luciferase activities, we discovered that the core IRES region contained 71 bases and resided between −392 and −322. As shown in Figure 3B, the construct −392 to −1 expressed maximal FL luciferase activity similar to that of the full-length fragment −572 to −1, while −322 to −1 showed a significant reduction (approximately a 7-fold decrease) of FL activity. To further confirm that the sequence between −392 and −322 is critical for TRAF1 IRES activity, we cloned that 71 bases sequence into the pRL-FL vector, where it stimulated FL activity comparable to that of the larger −572 to −1 fragment.
Figure 3.
Deletion mapping for identification of TRAF1 IRES. (A) Schematic representation of dicistronic pRL-FL constructs containing a series of 5′–3′ or 3′–5′ deleted TRAF1 5′-UTR fragments. (B) Raji cells were transfected with 5 μg of each indicated plasmid. After 24 h, luciferase activities were tested. Data represent the mean ± SD of luciferase activity of FL versus RL in at least three independent experiments. (C) Schematic representation of pGL3-basic vector with different inserts, including the −392 to −322 fragment of the TRAF1 5′-UTR, the putative TRAF1 promoter regions of the 5′-flank of the TRAF1 5′-UTR and the intron 1 of the TRAF1 gene. (D) Transfection and luciferase activity assays were performed as described in Figure 2B. Transfection of pGL3-basic vector without insert served as control. Data represent the mean ± SD of three independent experiments normalized to RL activity.
Previous reports have described the molecular cloning of the promoter of the human TRAF1 gene, and the putative TRAF1 promoter region proposed includes intron 1 and part of the 5′-UTR of TRAF1 mRNA (9,26). To clarify whether the 71 bases sequence between −392 and −322 of the TRAF1 5′-UTR acts as a promoter to stimulate FL activity when it is inserted into the pRL-FL vector, we cloned this 71 bases fragment into the pGL3-basic vector (Figure 3C). We also cloned the intron 1 and the 5′-flank of the 5′-UTR of TRAF1 into the pGL3-basic vector, to serve as controls. By similar transfections and reporter assays in Raji cells, we found that the 71 bases sequence between −392 and −322 did not stimulate FL activity in the pGL3-basic vector. In contrast, the intron 1 DNA sequence of TRAF1 gene stimulated FL activity (Figure 3D), but not the FL activity in the pRL-FL vector when inserted between the RL and FL cistrons (data not shown). Our results also did not show any promoter activity of the 5′-flank of the 5′-UTR mRNA. Taken together, these results clearly demonstrated that only the 71 bases sequence between −392 and −322 is IRES that can regulate TRAF1 translation, and it is not a promoter mediating TRAF1 transcription.
Induction of TRAF1 IRES activity and TRAF1 protein expression by vincristine
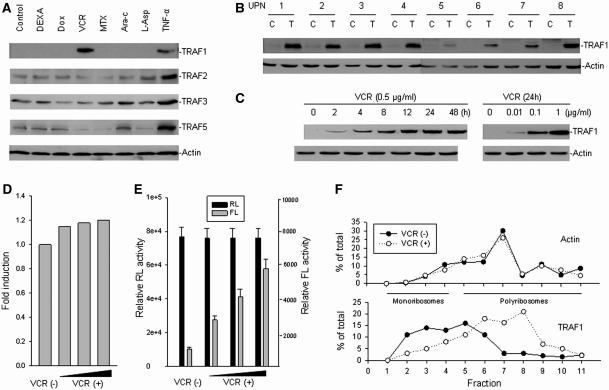
It is known that for certain mRNAs, IRES-mediated initiation of protein translation is activated during conditions such as cellular stress and apoptosis. To investigate whether the IRES-mediated translation of TRAF1 is affected during cellular stress, we examined both TRAF1 IRES activity and TRAF1 protein expression in leukemia cells treated with various stress inducers. Western blot assays were performed to examine TRAF1 protein expression in primary ALL cells treated with the stress cytokine TNF-α, as well as a series of chemotherapeutic drugs (DEXA, VCR, Dox, MTX, Ara-c and L-Asp) that are frequently used in the treatment of ALL. While primary ALL cells derived from patient bone marrow generally express no or very low levels of TRAF1 (Figure 4A and B), TNF-α treatment induced expression of TRAF1 as well as several other TRAFs. Surprisingly, we found that TRAF1 expression in ALL cells was only induced by VCR, but not other chemotherapeutic drugs (Figure 4A). Furthermore, among the TRAF family members tested, only TRAF1 was induced by VCR. After studying cells from eight ALL patients, we confirmed that VCR induced expression of TRAF1 in all of them, although the levels of TRAF1 differed from case to case (Figure 4B). In addition, we found the VCR-induced TRAF1 expression was dose and time dependent (Figure 4C).
Figure 4.
Induction of TRAF1 translation by vincristine in cancer cells. (A) Fresh isolated leukemic cells from an ALL patient was treated with 1 µg ml−1 DEXA, 0.5 µg ml−1 Dox, 0.5 µg ml−1 VCR, 0.5 µg ml−1 MTX, 0.5 µg ml−1 Ara-c, 1 IU/ml L-Asp and 0.05 µg ml−1 TNF-α, respectively, for 24 h. The protein expression for different TRAFs was examined by western blot analysis. (B) Fresh leukemic cells from eight ALL patients (UPN, unique patient number) were incubated with 0.5 µg ml−1 VCR for 24 h (T). The expression of TRAF1 in each was detected by western blot assay as compared with untreated cells (C). (C) Time- and dose-dependent induction of TRAF1 protein expression in ALL cells treated with VCR. Leukemia cells from UPN 1 were treated with 0.5 µg ml−1 VCR for different times or with VCR for 24 h at different doses, as indicated. Protein expression was examined by western blot. (D) mRNA expression levels of TRAF1 after treatment with or without increasing concentrations (0.01, 0.1 and 1 μg ml−1) of VCR in Raji cells, as determined by quantitative RT–PCR. (E) Transfection and reporter assay for the effect of VCR on TRAF1 IRES activity. Raji cells were transfected with 5 µg pRL-392-FL plasmid containing TRAF1 IRES from −392 to −1 fragment and treated with or without increasing concentration (0.01, 0.1 and 1 μg ml−1) of VCR. Data represented the mean ± SD of RL and FL activities in at least three independent experiments. (F) Raji cells were treated with or without 0.5 µg ml−1 VCR for 24 h, and cytoplasmic lysates were fractionated on sucrose gradient. RNA was extracted from each of the fractions and subjected to quantitative RT–PCR for quantitative analysis of the distribution of TRAF1 and Actin mRNAs. Data represent percentage of the total amount of corresponding mRNA on each fraction.
To evaluate whether VCR, like TNF-α, induces TRAF1 mRNA expression, we performed RT–PCR to examine TRAF1 mRNA expression in ALL cells that were treated with VCR. Results, as shown in Figure 4D, demonstrated that VCR did not induce TRAF1 transcription whereas TNF-α induced TRAF mRNA expression (data not shown). These results imply that VCR-induced TRAF1 expression might occur at the translational level. We performed gene transfections and reporter assays to see if VCR induced TRAF1 IRES activity. The lymphoma cell line Raji was transfected with the pRL-FL plasmid containing the TRAF1 5′-UTR sequence from −392 to −1 (pRL-392-FL) and then treated the transfected cells with VCR. We found that VCR increased the FL (TRAF1 IRES) activity (Figure 4E). Furthermore, we performed linear sucrose gradient fractionation to assess the polyribosome association of the TRAF1 mRNA in Raji cells subjected to VCR and mock treatment. We found that TRAF1 mRNA was clearly shifted from fractions containing translation dormant complexes including mRNPs, ribosome subunits, and monosomes (Figure 4F, bottom, fractions 1–4) to fractions enriched of translating polyribosomes (Figure 4F, bottom, fractions 5–11). This observation was likely specific, because VCR treatment had no effect on the polyribosome profile of Actin mRNA (Figure 4F, top), thus demonstrating that induction of TRAF1 by VCR occurred at the translational level.
Subcellular redistribution of PTB induced by vincristine confers TRAF1 translation
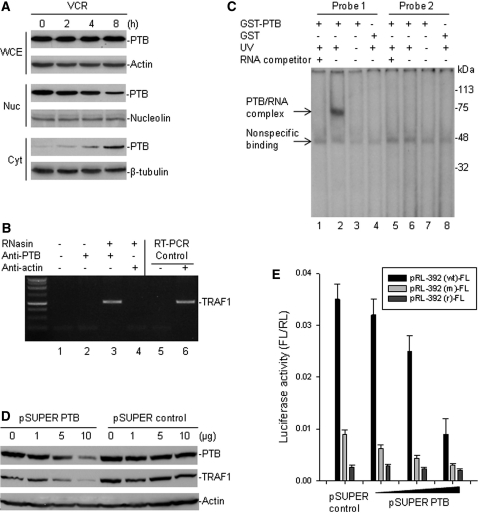
A previous study shows that VCR induces translocation of the PTB protein from the nucleus to the cytoplasm, where it induces BAG-1 IRES in HeLa cells (24). We wondered whether VCR induces TRAF1 IRES-mediated translation through a similar mechanism as for BAG-1 induction. Our study was performed in Raji cells that express high levels of both PTB and TRAF1 proteins. VCR treatment of Raji cells produced similar results as in HeLa cells, as expression of nuclear PTB was decreased and cytoplasmic PTB was significantly increased (Figure 5A). As also shown in Figure 5A, VCR treatment did not affect total cellular PTB expression, suggesting that the increased PTB in the cytoplasm was derived from the translocation of nuclear PTB.
Figure 5.
Vincristine-modulated PTB that binds to the TRAF1 IRES and regulates TRAF1 translation. (A) Raji cells were treated with 0.5 µg/ml VCR for different times, then cell extracts were tested for both nuclear (Nuc) and cytoplasmic (Cyt) expression of PTB, as well as for total PTB in the whole cell extracts (WCE), by western blot assays. (B) cell extracts from Raji were prepared in RNA-binding buffer in the presence of RNase inhibitor (RNasin). Following co-IP with anti-PTB and anti-actin (control), the TRAF1 mRNA was detected by RT–PCR analysis. IP without addition of RNasin and antibody (lane 1) and with anti-PTB but no RNasin (lane 2) were done as additional controls. The negative (no template, lane 5) and positive (Raji RNA as template, lane 6) controls for RT–PCR are also shown. (C) the GST-fused PTB protein and GST only were incubated, respectively, with 32P-labelled RNA probes corresponding to the wild-type (probe 1) and mutant (probe 2) TRAF1 IRES −392 to −322 elements. Protein/RNA complexes were UV cross-linked, run on an SDS–PAGE gel, and imaged by autoradiography. The reactions of GST-PTB protein and probes with prior addition of RNA competitor (lanes 1 and 5) or without UV exposure (lanes 3 and 7) were included as additional controls. (D) Raji cells were transfected with different amounts of pSUPER plasmids containing either PTB siRNA or a control siRNA for 24 h. The expression of endogenous PTB and TRAF1 was detected by western blot assay. (E) Raji cells were co-transfected with 5 µg pRL-392-FL and with increasing amounts (1, 5 and 10 µg) of pSUPER PTB or pSUPER control. Controls also include transfection of a plasmid pRL-392 (m)-FL containing the −392 to −1 fragment with mutations at the CU-rich sequences and a plasmid pRL-392 (r)-FL containing the −392 to −1 fragment in a reverse (r) orientation. Quantitative RL and FL were detected using the Dual-Luciferase Reporter System. Data represents the mean ± SD luciferase activity of FL versus RL, in at least three independent experiments.
Because it is known that PTB physically interacts with specific RNA structures within IRES elements in the cytoplasm to regulate IRES-dependent translation, we tested the ability of PTB to bind to the TRAF1 IRES. We first performed IP and RT–PCR to search for possible association in vivo between the PTB protein and the TRAF1 mRNA, and found that the PTB protein was able to bind TRAF1 mRNA (Figure 5B). Then, a protein–RNA-binding assay with UV cross-linking of the 32P-labelled TRAF1 IRES probes and the recombinant GST-fused PTB protein, we were able to show that the PTB protein specifically bound to the CU-rich sequences of the TRAF1 IRES RNA (Figure 5C).
To further evaluate whether PTB regulates TRAF1 IRES-mediated translation, we performed gene transfections followed by reporter assay as well as western blot assays to examine both TRAF1 IRES activity and the endogenous TRAF1 protein expression under conditions where PTB is silenced by siRNA. For this, we generated a pSUPER/PTB plasmid containing a 19 nt siRNA sequence specific for targeting PTB. Transfection of this plasmid into Raji cells significantly suppressed endogenous PTB expression (Figure 5D). As also seen in Figure 5D, we observed a reduced expression of the endogenous TRAF1 protein in those PTB siRNA-transfected Raji cells. We also found that co-transfection of the PTB siRNA plasmid with the pRL-392-FL reporter plasmid into Raji cells remarkably decreased the activity of FL (Figure 5E).
DISCUSSION
In this report, we describe the identification and analysis of the 5′-UTR region of TRAF1 mRNA as well as its possible role in the regulation of translation, with the goal of gaining insight into the properties of TRAF1 in regulating the life and death of cancer cells. By searching the Human Genome database, we discovered that the TRAF1 mRNA contains an unusually long 5′-UTR. When cloned into the luciferase reporter vectors and transfected into lymphoid cancer cells, the 5′-UTR of the TRAF1 mRNA showed considerable IRES activity. By performing deletion mapping, the core IRES region of the TRAF1 mRNA was found to be located between −392 and −322. It turned out that this IRES region contained two copies of CU-rich sequences that promoted interaction with and regulation by PTB protein. We also found that unlike many other anticancer drugs, the chemotherapeutic drug VCR, which is able to induce subcellular redistribution of PTB, uniquely stimulated TRAF1 translation in cancer cells of a lymphoid origin.
Unlike other TRAF proteins that are more ubiquitously expressed throughout the human body, the in vivo expression of TRAF1 is the most restricted, suggesting that the expression of TRAF1 is highly regulated. The expression of genes is mainly regulated at the transcriptional level; however, evidence is growing in support of the concept that translational regulation also plays an important role in the control of gene expression (27). In particular, the translation of certain mRNAs via a specific cap-independent mechanism is involved in a diverse range of cellular activities, including proliferation, growth and apoptosis (28). Our present study verified that cap-independent (IRES-dependent) mechanism plays a critical role in TRAF1 translation. For this study, we only analyzed the TRAF1 IRES activity in lymphoid cancer cells. Still to be investigated is the significance of IRES-mediated TRAF1 translation in both non-lymphoid cancer cells and in normal cells. For example, evidence must still be collected to demonstrate whether the IRES-mediated translation is responsible for the restricted expression of TRAF1 in various types of normal cells, due to inefficient or lowered levels of cap-dependent translation, and whether IRES-mediated TRAF1 translation could be cell-type specific, helping to explain why there is TRAF1 expression in only certain types of cells.
An important finding of our present study is that IRES-regulated TRAF1 translation is activated during the treatment of lymphoid cancer cells with the chemotherapeutic drug VCR. Treatment with VCR significantly increased the expression of endogenous TRAF1 protein, which was maintained at high levels for up to 48 h following VCR exposure in the BCP ALL cells. Interestingly, VCR did not regulate TRAF1 mRNA expression, suggesting that the observed increase in protein expression occurred at the translational level. Transfection and reporter assay indeed demonstrated that VCR stimulated the TRAF1 IRES activity that mediates a specific cap-independent translation. Even more notable was the fact that only VCR induced TRAF1 expression in ALL cells, but not any other cell-stressing chemotherapeutic drugs including DEXA, Dox, MTX, Ara-c and L-Asp. These observations suggest that not all stress and apoptosis inducers can regulate IRES-dependent TRAF1 translation and that a rather specific or unique cellular mechanism or signaling cascade existed in ALL cells (and potentially other cancer cells) that was able to regulate or promote VCR-induced TRAF1 IRES activity. Translated to an in vivo situation, it could mean that ALL, and perhaps other cancer cells, have a greater ability to evade death during VCR therapy, compared with other drugs.
It has been reported that VCR can induce cytoplasmic translocation of PTB, which can in turn regulate BAG-1 IRES-mediated translation (24). Consistent with that observation, our results showed that VCR induces relocation of PTB from the nucleus to the cytoplasm in Raji cells. Although the mechanism by which PTB regulates IRES-dependent translation has not been completely understood, it is known that cytoplasmic PTB can directly bind to a CU-rich sequence within the cellular IRES and regulate its activity (23,29). By analysing the active 71 bases TRAF1 IRES region, we found that it contains two CU-rich sequences (CUUCUUUUUU and CUUCUCCUU). We then demonstrated that PTB bound to the 71 bases TRAF IRES. In addition, we showed that the silencing of PTB expression in TRAF1-overexpressing Raji cells resulted in the inhibition of TRAF1 IRES activity and TRAF1 protein expression as well. These results indicate that IRES-mediated TRAF1 translation was regulated by PTB, which may be the most likely mechanism by which VCR treatment induced TRAF1 expression, i.e. through its modulation of the PTB protein.
It is reported that TRAF1 is typically overexpressed in B-cell lymphoid malignancies, including Hodgkin and non-Hodgkin lymphomas and chronic lymphoblastic leukemia (4–7). In the present study, we found that the early BCP ALL express no or very low levels of TRAF1 endogenously, although TRAF1 can be induced in these cells by VCR. This appears to imply that constitutive TRAF1 overexpression is associated with the maturation of B cells and the development of B-cell lymphoma. Despite the current observation that it is deregulation of the signaling pathways that regulate NF-kB that causes the overexpression of TRAF1 transcripts in lymphomagenesis (8), it is possible that aberrant regulation of TRAF1 translation through other pathways may also play a role in the development of lymphoma. At least one may have been uncovered here: We, for the first time, have identified that the translation initiation of TRAF1 is regulated by an IRES-dependent mechanism. Further studies evaluating other trans-acting factors, in addition to PTB, that could also regulate TRAF1 IRES activity may allow us to better understand the magnitude of the biological role of TRAF1 in lymphomagenesis and resistance to therapy.
FUNDING
National Institutes of Health (R01 CA123490); the Leukemia and Lymphoma Society (6033-08) and CURE. Funding for open access charge: National Institutes of Health R01 funding.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank Dr Steve Haines for providing us with the dicistronic vector pRL-FL.
REFERENCES
- 1.Rothe M, Wong SC, Henzel WJ, Goeddel DV. A novel family of putative signal transducers associated with the cytoplasmic domain of the 75 kDa tumor necrosis factor receptor. Cell. 1994;78:681–692. doi: 10.1016/0092-8674(94)90532-0. [DOI] [PubMed] [Google Scholar]
- 2.Arch RH, Gedrich RW, Thompson CB. Tumor necrosis factor receptor-associated factor (TRAFs): a family of adapter proteins that regulates life and death. Genes Dev. 1998;12:2821–2830. doi: 10.1101/gad.12.18.2821. [DOI] [PubMed] [Google Scholar]
- 3.Song HY, Rothe M, Goeddel DV. The tumor necrosis factor-inducible zinc finger protein A20 interacts with TRAF1/TRAF2 and inhibits NF-kappa B activation. Pro. Natl Acad. Sci. USA. 1996;93:6721–6725. doi: 10.1073/pnas.93.13.6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zapata JM, Krajewska M, Krajewski S, Kitada S, Welsh K, Monks A, McCloskey N, Gordon J, Kipps TJ, Gascoyne RD, et al. TNFR-associated factor family protein expression in normal tissues and lymphoid malignancies. J. Immunol. 2000;165:5084–5096. doi: 10.4049/jimmunol.165.9.5084. [DOI] [PubMed] [Google Scholar]
- 5.Dürkop H, Foss HD, Demel G, Klotzbach H, Hahn C, Stein H. Tumor necrosis factor receptor-associated factor 1 is overexpressed in Reed-Sternberg cells of Hodgkin's disease and Epstein-Barr virus-transformed lymphoid cells. Blood. 1999;93:617–623. [PubMed] [Google Scholar]
- 6.Dürkop H, Hirsch B, Hahn C, Foss HD, Stein H. Differential expression and function of A20 and TRAF1 in Hodgkin lymphoma and anaplastic large cell lymphoma and their induction by CD30 stimulation. J. Pathol. 2003;200:229–239. doi: 10.1002/path.1351. [DOI] [PubMed] [Google Scholar]
- 7.Munzert G, Kirchner D, Stobbe H, Bergmann L, Schmid RM, Döhner H, Heimpel H. Tumor necrosis factor receptor-associated factor 1 gene overexpression in B-cell chronic lymphocytic leukemia: analysis of NF-kappa B/Rel-regulated inhibitors of apoptosis. Blood. 2002;100:3749–3756. doi: 10.1182/blood.V100.10.3749. [DOI] [PubMed] [Google Scholar]
- 8.Zhang B, Wang Z, Li T, Tsitsikov EN, Ding HF. NF-kappaB2 mutation targets TRAF1 to induce lymphomagenesis. Blood. 2007;110:743–751. doi: 10.1182/blood-2006-11-058446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwenzer R, Siemienski K, Liptay S, Schubert G, Peters N, Scheurich P, Schmid RM, Wajant H. The human tumor necrosis factor (TNF) receptor-associated factor 1 gene (TRAF1) is up-regulated by cytokines of the TNF ligand family and modulates TNF-induced activation of NF-kappaB and c-Jun N-terminal kinase. J. Biol. Chem. 1999;274:19368–19374. doi: 10.1074/jbc.274.27.19368. [DOI] [PubMed] [Google Scholar]
- 10.Wang CY, Mayo MW, Korneluk RG, Goeddel DV, Baldwin A.S., Jr NF-kappaB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science. 1998;281:1680–1683. doi: 10.1126/science.281.5383.1680. [DOI] [PubMed] [Google Scholar]
- 11.Carpentier I, Beyaert R. TRAF1 is a TNF inducible regulator of NF-kappaB activation. FEBS Lett. 1999;460:246–250. doi: 10.1016/s0014-5793(99)01356-3. [DOI] [PubMed] [Google Scholar]
- 12.Lackner DH, Bähler J. Translational control of gene expression from transcripts to transcriptomes. Int Rev Cell. Mol. Biol. 2008;271:199–251. doi: 10.1016/S1937-6448(08)01205-7. [DOI] [PubMed] [Google Scholar]
- 13.Sheikh MS, Fornace A.J., Jr Regulation of translation initiation following stress. Oncogene. 1999;18:6121–6128. doi: 10.1038/sj.onc.1203131. [DOI] [PubMed] [Google Scholar]
- 14.Stoneley M, Willis AE. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- 15.Bonnal S, Boutonnet C, Prado-Lourenco L, Vagner S. IRESdb: the Internal Ribosome Entry Site database. Nucleic Acids Res. 2003;31:427–428. doi: 10.1093/nar/gkg003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray NK, Hentze MW. Regulation of protein synthesis by mRNA structure. Mol. Biol. Rep. 1994;19:195–200. doi: 10.1007/BF00986961. [DOI] [PubMed] [Google Scholar]
- 17.Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 18.Baird SD, Turcotte M, Korneluk RG, Holcik M. Searching for IRES. RNA. 2006;12:1755–1785. doi: 10.1261/rna.157806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komar AA, Hatzoglou M. Internal ribosome entry sites in cellular mRNAs: mystery of their existence. J. Biol. Chem. 2005;280:23425–23428. doi: 10.1074/jbc.R400041200. [DOI] [PubMed] [Google Scholar]
- 20.Pestova TV, Shatsky IN, Hellen CU. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho S, Kim JH, Back SH, Jang SK. Polypyrimidine tract-binding protein enhances the internal ribosomal entry site-dependent translation of p27Kip1 mRNA and modulates transition from G1 to S phase. Mol. Cell. Biol. 2005;25:1283–1297. doi: 10.1128/MCB.25.4.1283-1297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schepens B, Tinton SA, Brynooghe Y, Beyaert R, Cornelis S. The polypyrimidine tract-binding protein stimulates HIF-1alpha IRES-mediated translation during hypoxia. Nucleic Acids Res. 2005;33:6884–6894. doi: 10.1093/nar/gki1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bushell M, Stoneley M, Kong YW, Hamilton TL, Spriggs KA, Dobbyn HC, Qin X, Sarnow P, Willis AE. Polypyrimidine tract binding protein regulates IRES-mediated gene expression during apoptosis. Mol. Cell. 2006;23:401–412. doi: 10.1016/j.molcel.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 24.Dobbyn HC, Hill K, Hamilton TL, Spriggs KA, Pickering BM, Coldwell MJ, de Moor CH, Bushell M, Willis AE. Regulation of BAG-1 IRES-mediated translation following chemotoxic stress. Oncogene. 2008;27:1167–1174. doi: 10.1038/sj.onc.1210723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu L, Zhu N, Zhang H, Durden DL, Feng Y, Zhou M. Regulation of XIAP translation and induction by MDM2 following irradiation. Cancer Cell. 2009;15:363–375. doi: 10.1016/j.ccr.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siemienski K, Peters N, Scheurich P, Wajant H. Organization of the human tumour necrosis factor receptor-associated factor 1 (TRAF1) gene and mapping to chromosome 9q33-34. Gene. 1997;195:35–39. doi: 10.1016/s0378-1119(97)00147-9. [DOI] [PubMed] [Google Scholar]
- 27.Garbarino-Pico E, Green CB. Posttranscriptional regulation of mammalian circadian clock output. Cold Spring Harb. Symp. Quant. Biol. 2007;72:145–156. doi: 10.1101/sqb.2007.72.022. [DOI] [PubMed] [Google Scholar]
- 28.Graber TE, Holcik M. Cap-independent regulation of gene expression in apoptosis. Mol. Biosyst. 2007;3:825–334. doi: 10.1039/b708867a. [DOI] [PubMed] [Google Scholar]
- 29.Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, et al. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]