Abstract
5-Aza-2′-deoxycytidine (5-aza-dC) is a nucleoside analogue with cytotoxic and DNA demethylating effects. Here we show that 5-aza-dC induces the proteasomal degradation of free (non-chromatin bound) DNMT1 through a mechanism which is dependent on DNA synthesis and the targeting of incorporated 5-aza-dC residues by DNMT1 itself. Thus, 5-aza-dC induces Dnmt1 degradation in wild-type mouse ES cells, but not in Dnmt [3a–/–, 3b–/–] mouse ES cells which express Dnmt1 but lack DNA methylation (<0.7% of CpG methylated) and contain few hemi-methylated CpG sites, these being the preferred substrates for Dnmt1. We suggest that adducts formed between DNMT1 and 5-aza-dC molecules in DNA induce a ubiquitin-E3 ligase activity which preferentially targets free DNMT1 molecules for degradation by the proteasome. The proteasome inhibitor MG132 prevents DNMT1 degradation and reduces hypomethylation induced by 5-aza-dC.
INTRODUCTION
5-Aza-2′-deoxycytidine (5-aza-dC, Decitabine) is a nucleoside analogue and DNA demethylating agent. Early studies showed that treatment of cell lines with the ribonucleoside analogue 5-azacytidine lead to a loss of extractable DNA methyltransferase activity (1). It was proposed that this was caused by the entrapment of maintenance DNA methyltransferase DNMT1 by 5-aza-dC residues incorporated into the DNA. The mechanism of action predicts that during synthesis the enzyme would attack and attempt to methylate 5-aza-dC molecules incorporated 5′ to G residues and opposite methylated CpG dinucleotides in the parent strand (azaCpG:mCpG). However, because of the lack of a proton at the N5 position of the aza-substituted pyrimidine ring, the enzyme would not subsequently be released by β-elimination (2). As DNMT1 is a highly processive enzyme, this covalent entrapment of the enzyme would lead to a significant loss of maintenance methyltransferase activity and demethylation of the DNA. A recent study used the FRAP (fluorescence recovery after photobleaching) technique to provide evidence in support of this mechanism (3). The study demonstrated that the half-time of fluorescence recovery (t1/2) after photo bleaching was 15 s for DNMT1 protein, but recovery did not occur in cells treated with 5-aza-dC. Furthermore, photoactivatable DNMT1 (paGFP-DNMT1) did not dissociate from the irradiated site in cells treated with 5-aza-dC indicating that it had indeed been trapped.
Whilst the trapping mechanism has been the accepted paradigm for explaining the demethylation induced by 5-aza-dC, a recent study showed that 5-aza-dC caused the proteasomal degradation of DNMT1 through a process that is not apparently dependent on its incorporation into DNA: the effect could not be abolished by prior treatment of the cells with the DNA synthesis inhibitor aphidicolin (4). The proteasomal degradation of DNMT1 was shown to be dependent on the KEN box, bromo-adjacent domain and nuclear localization domains located in the N-terminus of the enzyme and was mediated by APCCDH1/FZR1-catalysed polyubiquitinylation of DNMT1(4).
Because two potential mechanisms limiting DNMT1 availability in response to 5-aza-dC have been uncovered, it is not clear whether the hypomethylating effects of the drug are due to degradation of the DNMT1 enzyme, entrapment of it by 5-aza-dC residues in the DNA, or both. In this report we have investigated the mechanism of the 5-aza-dC effect on DNMT1 degradation and find that, contrary to the original report uncovering the DNMT1 degradation pathway, DNA synthesis is required. Our data indicate that the DNMT1 degrading effects of 5-aza-dC are very sensitive to and dependent upon the incorporation of 5-aza-dC into DNA. We provide evidence that Dnmt1 itself triggers its own degradation by attacking 5-aza-dC residues incorporated into the DNA, and demonstrate that DNMT1 degradation contributes to the hypomethylation induced by 5-aza-dC.
MATERIAL AND METHODS
Materials
5-Aza-dC (Decitabine), MG132, phosphodiesterase I, alkaline phosphatase, Triton X-100, 5-methyl-2-deoxycytidine (5mdC), 2-deoxycytidine (dC), 2-deoxyguanosine (dG) monohydrate and 2-deoxythymidine (dT) monohydrate were all purchased from Sigma, (Sigma, Gillingham, UK). Benzonase was purchased from Merck (Merck, Nottingham, UK). HPLC grade acetonitrile (MeCN), water and methanol were from Rathburn (Walkerburn, UK). Mass spectrometry grade formic acid was from BDH (BDH, Poole, UK).
Instrumentation
The HPLC system consisted of a Dionex 3000 Ultimate series LC (Sunnyvale, CA, USA) connected to either the photodiode array detector (Dionex) with the Foxy Jr. fraction collector (Dionex) or a 4000 QTrap LC–MS/MS mass spectrometer equipped with an orthogonal electrospray ion source (Applied Biosystems, Foster City, CA, USA). Data were acquired and processed with Chromeleon 6.1 and Analyst 1.4 chromatography manager software, respectively.
HPLC chromatographic and mass spectrometry conditions
For HPLC fractionation of nucleoside standards and digestions of tritium labelled thymidine and 5-aza-dC DNA, a LUNA C18 (150 × 4.6 mm I.D.) and 3 µm particle size (Phenomenex, Torrance, CA, USA) column was used. The HPLC method used isocratic elution; mobile phase solvent A was water with 50 mM sodium phosphate pH 5 (98%) and mobile phase B was methanol (2%) with a total run time of 40 min. The column was set at a flow rate of 0.8 ml min–1, a column temperature of 25°C and an injection volume of 50 μl. Alternatively, digested nucleosides for global DNA methylation analyses were separated on a Gemini C18 (150 × 2.0 mm ID) and 3 µm particle size (Phenomenex, Torrance, CA, USA) protected by a Phenomenex Gemini C18 (4.0 × 2.0 mm ID) and 3 µm particle size guard cartridge. The HPLC method used gradient elution; mobile phase solvent A was water with 0.1% formic acid and mobile phase B was acetonitrile with 0.1% formic acid. The initial mobile phase composition of 99% solvent A and 1% solvent B was maintained for 2 min. Between 2 and 7 min the percentage of mobile phase B was increased to 40% and then back to the initial mobile phase composition within 0.1 min, with a total time of 14 min. The column was set at a flow rate of 0.2 ml min–1 and a column temperature of 35°C. Sample volume of 5 µl was used for all LC–MS experiments. The mass spectrometer was operated in positive electrospray mode. The source temperature was 450°C and a spray voltage 3 kV was used. The collision gas pressure was 1.5 mTorr. All analytes were optimized using the Analyst software auto tune facility for MRM transitions. Quantification was accomplished in multiple reaction monitoring (MRM) mode by monitoring a transition pair of m/z 242.1 (molecular ion)/126.1 (fragment ion) for 5mdC and m/z 268.1/152.1 for dG, which was used as an internal standard for the measurement, with a dwell time of 100 ms for each pair.
Cell culture, DNA isolation and cell lysate preparation
HCT116 and SW620 cell lines were cultured at 37°C in a 5% CO2 incubator using an RPMI medium supplemented with 10% fetal bovine serum (FBS, Invitrogen, Paisley, UK). Mouse J1 (wild-type) and Dnmt[3a–/–, 3b–/–] ES cell lines (5) and were cultured in Glasgow’s MEM/10% FCS supplemented with non-essential amino acids, pyruvate, β-mercaptoethanol and leukaemia inhibitory factor (LIF). Cell lines were treated with 5-aza-dC (Decitabine) ranging from 0.0005 to 10 µM, MG132 (10 μM) and aphidicolin (20 µg/ml). Genomic DNA was isolated from the above cells using QIAamp DNA mini kit (Qiagen, West Sussex, UK), according to the manufacturer’s instructions. For cell lysate preparation fresh cells (106–108 cells) in culture medium were resuspended in 1 ml ice-cold PBS, centrifuged at 1000 × g for 5 min at 4°C, and the supernatant was removed and discarded. The pellet was resuspended in 100–400 µl ice-cold RIPA lysis buffer (150 mM NaCl, 1% NP40, 0.5% (deoxycholic acid), 0.1% SDS, 50 mM Tris pH 7.0 and protease inhibitor cocktail tablet (Roche, West Sussex, UK) and incubated on ice for 10 min. The lysate was centrifuged at 12 000 × g for 10 min at 4°C. The supernatants were stored at –20°C.
DNA hydrolysis
DNA hydrolysis was performed as previously described (6). This method improves the recovery of 5-aza-dC from DNA incorporated with 5-aza-dC because denaturation of the DNA, is avoided which affects 5-aza-dC stability. Briefly, 250 U Benzonase (Merck), 500 mU phosphodiesterase I (Sigma), 200 U alkaline phosphatase (Sigma) was added to 2.5 ml of Tris–HCl buffer (pH 7.9, 20 mM) containing 100 mM NaCl and 20 mM MgCl2. Following the preparation of the DNA 25 μl of the reaction mix was added to 1 μg of genomic DNA. The DNA samples were digested for 6 h. A one-tenth dilution was made of the digest with HPLC grade water and injected into the LC–MS/MS system.
Cell-cycle analysis
HCT116 cells were synchronized by blocking them at the G1/S boundary. The cells were first grown in reduced serum (RPMI/0.5% FBS) for 24 h, followed by growth in aphidicolin (20 µg/ml) and RPMI/10% serum for a further 24 h. This effectively blocked the vast majority of cells at the G1/S boundary. The block was released by washing out the aphidicolin containing medium and replacing it with normal RPMI/10% FBS. Cells were harvested and prepared for flow cytometry by trypsinization to produce a mononuclear cell suspension, followed by quenching of the trypsin with serum and resuspension of the cells in PBS. After counting 106 cells were transferred to 12 × 75 mm polystyrene tubes, centrifuged and resuspended in 300 µl PBS. An amount of 700 µl ice cold ethanol was added (drop wise) and the cells which were allowed to fix on ice for 1 h. The cells were then resuspended in 250 µl PBS containing 5 µl of a 10 mg/ml solution of RNase A and 10 µl of a 1 mg/ml solution of propidium iodide and their fluorescence intensity was measured in a flow cytometer at 488 nm.
Analysis of [5-methyl-3H]-Thymidine and 5-aza-[63H]-dC from DNA
Cells were treated with 5-aza-[63H]-dC purchased from Moravek Biochemicals (1 mCi/ml, Brea, CA) or [5-Methyl-3H]-thymidine (1 mCi/ml, Perkin Elmer, Bucks, UK). Subsequently the cells were incubated for 0.5, 2, 4 and 6 h or replaced in, after 2 h, 5-aza-[63H]-dC or [5-Methyl-3H]-thymidine free medium and incubated for 0.5, 2, 4 and 6 h. DNA was isolated as described above. Aliquots were taken for DNA quantification, using the Quant-it dsDNA kit (Invitrogen, Paisley, UK) according to the manufacturer’s instructions and liquid scintillation (Beckman LS 5000 Liquid Scintillation Counter).
Preparation of detergent-soluble and -insoluble fractions
After the designated drug treatments, Triton X-100 soluble and insoluble fractions were prepared by adding gentle lysis buffer 200 μl (PBS + 0.5% Triton X-100 with protease inhibitor tablets) to cells in six-well plates. The plates were left on a rocking platform for 30 min at 4°C. The cell lysate was transferred and centrifuged at 13 000 rpm for 10 min. The supernatant was collected as the soluble fraction and the remaining pellet resuspended in 50 μl of 1 × SDS loading buffer.
Immunoblotting
Equal amounts of protein for each sample were then separated on 6–12% Bis–Tris acrylamide gels using MOPS buffer and then transferred onto PVDF membranes (Amersham, Buckinghamshire, UK). The blots were blocked in TBS-T (10 mM Tris–HCl, pH 8.0, 150 mM NaCl, 0.1% Tween 20) containing 5% non-fat milk and subsequently incubated with anti-DNMT1 (New England Biolabs, Herts, UK). Anti-CDH1/FZR1 mouse monoclonal was purchased from Genway. The sheep anti-mouse Dnmt3a antibody (cross-reacting with human DNMT3A) was raised in our laboratory. The anti-DNMT3B antibody was purchased from Santa Cruz. Anti Histone H4 antibody was purchased from Millipore. Equal loading was confirmed by probing for β-ACTIN (Cell Signalling), Ku70 (Abcam) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH, Cell Signalling). Horseradish peroxidase conjugated secondary antibodies were detected using a Chemiluminescent detection kit (Amersham).
siRNA experiments
RNA interference assay
CDH1/FZR1 small interfering RNA (siRNA) smart pool and scrambled non-targeting fluorescent control siRNA were obtained from Dharmacon. HCT116 cells were transfected for 48 h with 100 nM siRNA using Lipofectamine 2000 (Invitrogen) following the manufacturers instructions. After 48 h, cells were treated with 5-aza-dC (0.1 and 1 µM) for 12 h, and cell extracts were subjected to western blot analysis.
Quantitative RT–PCR
Quantitive estimation of DNMT1 transcript levels was carried out using the primers and conditions set out in Ghoshal et al. (4).
RESULTS
DNMT1 is degraded by the proteasome upon treatment of 5-aza-dC
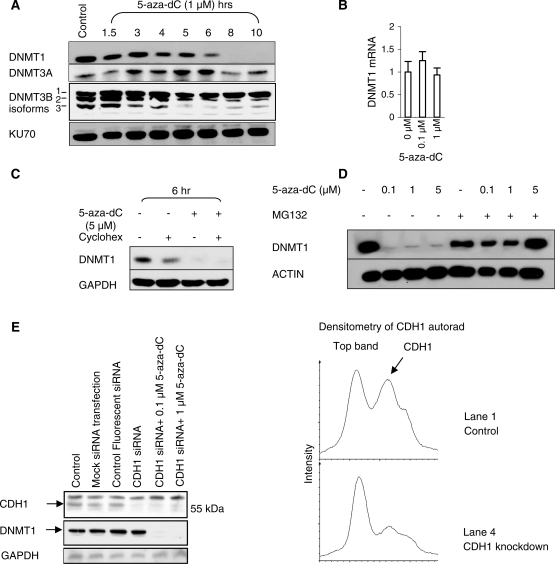
We exposed HCT116 cells to 1 µM 5-aza-dC for 10 h (Figure 1A). A reduction in DNMT1 levels was observed by 6 h and levels were massively reduced by 8 h, as assessed by western blotting for DNMT1 protein, but there was no significant change in DNMT3A or DNMT3B protein levels. Quantitative RT–PCR of DNMT1 mRNA showed that the reduction in DNMT1 protein at 24 h was not caused by reduced transcription (Figure 1B). We demonstrated that 5-aza-dC induced the degradation of preformed DNMT1 protein by pre-treating cells with the protein synthesis inhibitor cycloheximide prior to exposure to 5-aza-dC (Figure 1C). Cycloheximide reduces DNMT1 levels by approximately 50% at 6 h, indicating that DNMT1 protein molecules have a half-life of approximately 6 h (Figure 1C, lane 2). Addition of 5 μM 5-aza-dC resulted in a further loss of DNMT1 protein (Figure 1C, lane 4). This can only be explained by increased DNMT1 protein degradation as DNMT1 protein synthesis had already been inhibited by cycloheximide. Pre-treatment with the proteasome inhibitor MG132 inhibited DNMT1 degradation induced by 5-aza-dC, suggesting that degradation occurred through the proteasome (Figure 1D). As it has previously been reported that the anaphase promoting complex (APC) component CDH1/FZR1 is the ubiquitin ligase responsible for marking DNMT1 for degradation, we knocked down CDH1/FZR1 in HCT116 cells with siRNA and determined the effectiveness of 5-aza-dC at inducing DNMT1 degradation in these cells. Despite knockdown of CDH1/FZR1 protein levels to 40% of the level seen in control cells, we were unable to rescue the DNMT1 degradation induced by 5-aza-dC (Figure 1E). These data do not argue against proteasomal degradation as being the cause of DNMT1 loss in response to 5-aza-dC treatment, but they cast doubt on whether CDH1/FZR1 is the ligase responsible.
Figure 1.
5-Aza-dC induces the proteasomal degradation of DNMT1. (A) Western blot of DNMT1, DNMT3A, DNMT3B and KU70 (loading control) after treatment of HCT116 cells with 1 µM 5-aza-dC. Degradation is induced by 6 h of treatment. (B) Quantitative RT–PCR of DNMT1 relative to 18S RNA in HCT116 cells exposed to 0.1 and 1 µM 5-aza-dC for 24 h. (C) Western blot of DNMT1 levels and GAPDH (loading control) after treatment of HCT116 cells with cycloheximide (protein synthesis inhibitor) and 5-aza-dC. The DNMT1-lowering effect of 5-aza-dC in cycloheximide treated cells indicates that 5-aza-dC acts post-transcriptionally and post-translationally to lower DNMT1 levels. (D) Western blot of DNMT1 and ACTIN (loading control) to show the effect of a 12-h treatment with 5-aza-dC and MG132 (10 μM). The MG132 treatment was commenced 2 h before 5-aza-dC to ensure that proteasome blockade was effective prior to 5-aza-dC exposure. (E) CDH1/FZR1 was knocked down with siRNA in HCT116 cells before being exposed to 0.1 and 1 µM 5-aza-dC. Controls include a mock transfection control (no siRNA), to control for untoward effects of the transfection reagent, and a fluorescent nonsense siRNA, to ensure that transfection had taken place and to exclude non-specific effects. The graphs demonstrate that CDH1/FZR1 was knocked down to 40% of the level seen in the control cells.
Degradation of DNMT1 is highly sensitive to 5-aza-dC
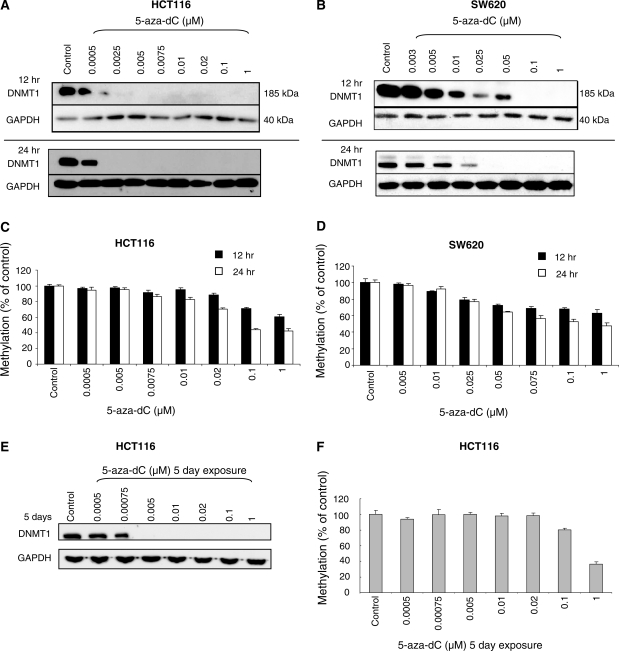
We investigated the sensitivity of cells to DNMT1 degradation induced by 5-aza-dC by exposing HCT116 cells to various concentrations of the drug for 12 and 24 h (Figure 2A). No loss of DNMT1 was observed with 0.0005 μM 5-aza-dC at 24 h, but significant loss was detected in cells exposed to 0.0025 µM. SW620 cells were approximately 10 times less sensitive to the DNMT1-degrading effects of 5-aza-dC. At 24 h a significant reduction was observed at 0.01 µM and DNMT1 protein was undetectable at 0.025 μM (Figure 2B). In both cell lines there appeared to be a 5-aza-dC concentration threshold at which DNMT1 depletion was induced rather than a graduated effect of the drug. The reason for the differences in sensitivity of the two cell lines to 5-aza-dC induced DNMT1 degradation is not known. The cell lines have similar cell-cycle characteristics and grow at the same rates (18-h doubling time, data not shown).
Figure 2.
DNMT1 expression is highly sensitive to 5-aza-dC treatment. (A) Western blots of DNMT1 and GAPDH (loading control) after the indicated doses of 5-aza-dC in HCT116 cells (A) and SW620 cells (B). (C) and (D) Global DNA methylation analysis corresponding to the 5-aza-dC treatments in the HCT116 and SW620 cell lines. (E) DNMT1 protein levels, and (F) global DNA methylation levels, in HCT116 cells exposed to 5-aza-dC for 5 days.
We looked to see whether there was a correlation between DNMT1 loss and hypomethylation. Surprisingly, in HCT116 cells the threshold effect observed for DNMT1 loss was not observed for demethylation (measured by mass spectrometry) which exhibited more of a graduated dose dependent effect (Figure 2C). Interestingly, very low doses of 5-aza-dC (0.0025 and 0.005 µM) caused significant loss of DNMT1 protein but had negligible effects on DNA methylation levels. It appeared therefore that effective maintenance methylation could take place in the presence of significant reductions in DNMT1 protein, either because other DNA methyltransferases (the de novo methyltransferases DNMT3A and DNMT3B) were still present and could support methylation, or simply because maintenance methylation by DNMT1 can occur effectively when DNMT1 levels are low. The capacity for low levels of DNMT1 to maintain methylation was previously reported by Egger et al. (7). The fact that SW620 cells were ∼10 times less sensitive to the DNMT1-depleting effects of 5-aza-dC, but were equally sensitive to its demethylating effects (Figure 2D), indicated that it was the level of incorporation of 5-aza-dC into DNA, rather than the degree of DNMT1 degradation, which was the most important factor determining the amount of demethylation induced.
To determine whether low dose 5-aza-dC might have had a more significant effect on DNA methylation over a longer time course through its effect on DNMT1 levels we performed a separate experiment in which HCT116 cells were exposed to 5-aza-dC for 5 days, replenishing the drug containing medium every 12 h. Here, the effects on DNMT1 protein levels were maintained (Figure 2E), but surprisingly the effects of low dose 5-aza-dC on demethylation were actually less at 5 days than they were at 24 h (Figure 2F). The effects of the highest dose of 5-aza-dC (1 µM) were as expected, methylation levels being slightly lower (36%) at 5 days than they were at 24 h (42%).
5-aza-dC is stably incorporated into DNA and incorporation is reduced but not abolished by aphidicolin
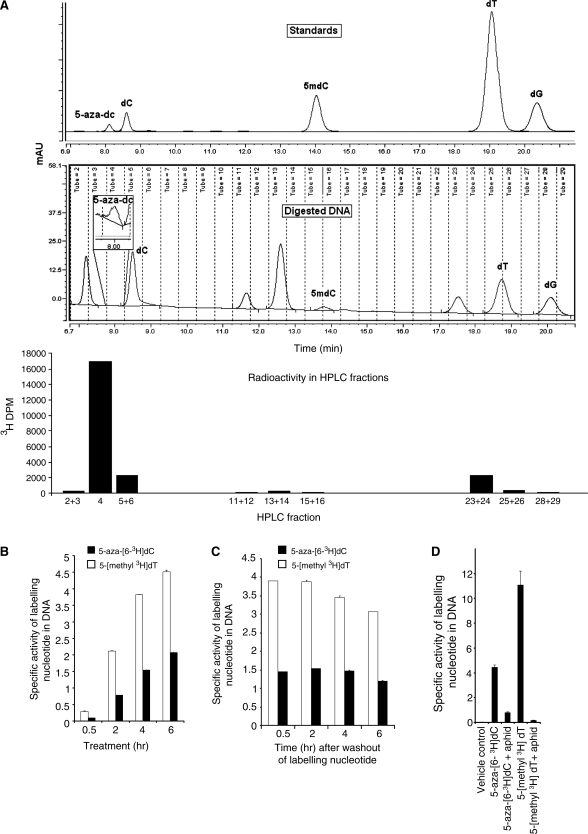
To investigate whether the induction of DNMT1 degradation by 5-aza-dC was dependent on incorporation of 5-aza-dC into the DNA we first needed to demonstrate that 5-aza-dC was incorporated into the DNA as 5-aza-dC, and not some metabolite of it. Previous studies have shown that the tritium label in 5-aza-[6-3H]dC is transferred to the DNA (8), but no studies have demonstrated chromatographically that the label is still present within the incorporated 5-aza-dC molecules. 5-aza-dC is highly labile, being sensitive to acid and base, and this may explain why its direct incorporation into DNA has not yet been shown directly. We used a rapid DNA hydrolysis technique using low temperature (37°C) conditions and reverse-phase HPLC (see methods) to confirm the presence of 5-aza-[6-3H]dC residues in the DNA (Figure 3A). We were also interested to know whether 5-aza-dC might be actively excised from the DNA. A time course experiment demonstrated a gradual increase in 5-aza-[6-3H]dC in the DNA (Figure 3B), and levels were maintained when the 5-aza-[6-3H]dC label was removed (Figure 3C). These data demonstrate that there is no rapid excision of 5-aza-[6-3H]dC from the DNA.
Figure 3.
(A) Chromatogram of nucleoside standards (top panel) and nucleosides from digested DNA of cells treated with 5-aza-[63H]-dC for 4-h post-medium replacement (middle panel). The bottom panel shows 3H-DPM of fractions collected in the panel above. Specific activity of DNA from HCT116 cells treated with aphidicolin (20 μg/ml) for 24 h followed by treatment with 5-aza-[63H]-dC and [5-methyl-3H]-thymidine for 12 h. (B) Specific activity of 5-aza-[63H]-dC and [5-Methyl-3H]-thymidine in HCT116 cells treated for 0.5, 2, 4 and 6 h. (C) Specific activities at 0.5, 2, 4 and 6 h after washout of labeling nucleotides. (D) The incorporation of 5-aza-[63H]-dC and [5-methyl-3H]-thymidine into HCT116 cell DNA, with and without prior treatment with the DNA synthesis inhibitor aphidicolin at 20 μg/ml for 24 h.
We went on to determine the extent to which the DNA polymerase inhibitor aphidicolin could inhibit incorporation of the 5-aza-dC label into the DNA. As expected, prior exposure of the cells to aphidicolin (20 µg/ml for 24 h) reduced 5-aza-[6-3H]dC incorporation dramatically, but incorporation was not completely abolished, falling to 10% of the levels seen without aphidicolin treatment (Figure 3D). Aphidicolin also caused a substantial reduction in [5-methyl-3H]thymidine incorporation, reducing it to 3% of the level seen without aphidicolin treatment (Figure 3D). Thus, DNA synthesis is markedly reduced but is not abolished by aphidicolin.
The degradation of DNMT1 is dependent on DNA synthesis
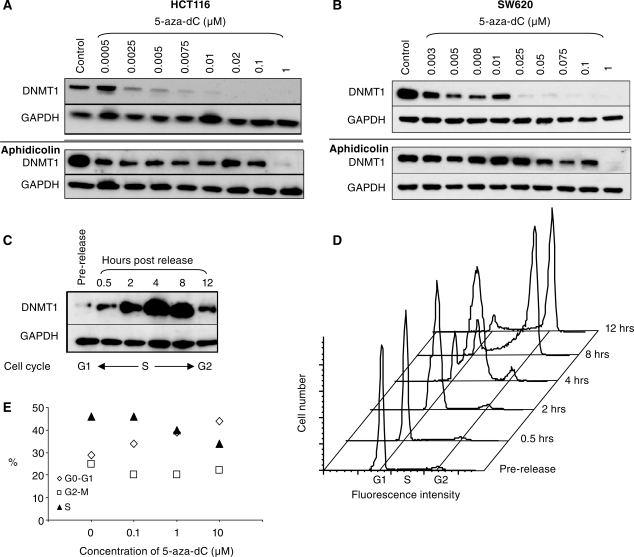
To investigate whether the proteasomal degradation of DNMT1 was dependent upon incorporation of 5-aza-dC into DNA, we determined whether DNMT1 degradation could be inhibited by pre-treatment of HCT116 and SW620 cells with aphidicolin. We examined the effects of aphidicolin over a range of different concentrations of 5-aza-dC. We found that aphidicolin suppressed DNMT1 degradation induced by low doses of 5-aza-dC, but degradation induced by higher doses (1 μM) of 5-aza-dC could not be suppressed (Figure 4A and B). As DNA synthesis is reduced but not completely inhibited by aphidicolin (Figure 3D) we conclude that the effect of 5-aza-dC on DNMT1 degradation is indeed dependent on DNA synthesis. At low concentrations of 5-aza-dC pre-treatment with aphidicolin is sufficient to reduce 5-aza-dC incorporation below the level required to trigger degradation. However at high concentrations of 5-aza-dC (1 µM) pre-treatment with aphidicolin has no effect on the DNMT1 degradation because sufficient 5-aza-dC is still incorporated into the DNA to induce DNMT1 degradation.
Figure 4.
The DNA synthesis inhibitor aphidicolin prevents the 5-aza-dC induced degradation of DNMT1. Western blots of DNMT1 and GAPDH (loading control) in 5-aza-dC treated HCT116 cells (A) and SW620 cells (B) to show the effects of 5-aza-dC on its own (top panels) as compared with cells that had been treated with the DNA synthesis inhibitor aphidicolin 20 μg/ml for 24 h prior to treatment with 5-aza-dC (bottom panels). (C) Western blot and (D) cell-cycle analysis to show DNMT1 levels at various stages after synchronization and release from an aphidicolin block. DNMT1 protein levels are prominent in synthesis, and markedly reduced in G1 and G2 phases of the cell cycle. (E) Cell-cycle analysis of HCT116 cells exposed to the indicated concentrations of 5-aza-dC for 24 h.
We were intrigued to see that the DNMT1 protein appeared to have been substantially lost from the entire culture, whilst our results pointed to a mechanism whereby DNMT1 loss occurred as a result of 5-aza-dC incorporation during DNA synthesis. A synthesis specific effect would have affected approximately 30–40% in the culture but DNMT1 protein should have been detectable in those cells that were not in DNA synthesis. However, if the expression of DNMT1 protein was itself tightly restricted to the DNA synthesis phase of the cell cycle, this might explain why DNMT1 loss was so complete. It has previously been shown that DNMT1 transcription is largely restricted to synthesis (9), but to our knowledge no studies have looked at protein levels during the cell cycle. To look at the expression of DNMT1 protein during different phases of the cell cycle we synchronized HCT116 cells by serum starvation (24 h) followed by aphidicolin treatment in the presence of 10% serum. This synchronized the cells at the G1/S phase boundary. DNMT1 levels were monitored by western blotting following release of the aphidicolin block, and cell-cycle progression was followed by flow cytometry using the DNA stain propidium iodide. Figures 4C and D show that DNMT1 is markedly reduced in G1, highly induced in S-phase (0.5–8 h) and reduced again in G2 (8–12 h). Thus, in protein extracts made from unsynchronized cells, most of the DNMT1 protein comes from the cells that are actively undergoing DNA synthesis.
We investigated whether any of the reduction in DNMT1 expression observed in 5-aza-dC treated cells might be attributable to 5-aza-dC induced changes in the cell cycle. We observed a small reduction in the number of cells undergoing synthesis in unsynchronized HCT116 culture exposed to 5-aza-dC, particularly at doses greater than 0.1 µM (Figure 4E), but even cells treated with 10 µM 5-aza-dC retained 30% of the culture in synthesis (compared with 45% untreated). Thus the modest reduction in the proportion of cells in S phase cannot explain the dramatic loss of DNMT1 protein from these cultures.
5-aza-dC-induced degradation of DNMT1 is DNA methylation dependent
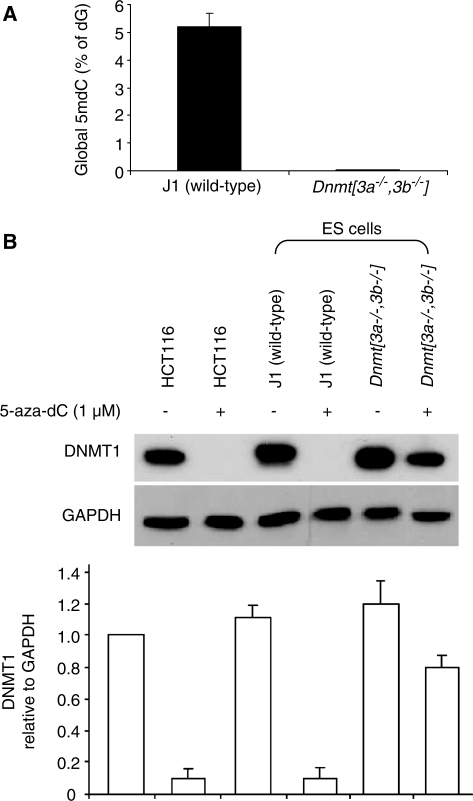
As DNMT1 degradation was dependent on 5-aza-dC incorporation into DNA, we speculated that DNMT1 might be required to attack 5-aza-dC residues in the DNA before DNMT1 degradation was triggered. To test this we needed conditions in which 5-aza-dC would be incorporated into the DNA without being targeted by DNMT1. As DNMT1 targets hemi-methylated DNA in preference to unmethylated DNA, and was recently shown to be preferentially trapped by 5-aza-dC molecules incorporated into a hemi-methylated substrate (10), we reasoned that the effects of 5-aza-dC would be less pronounced on a background of very low genomic DNA methylation as the number of hemi-methylated CpG target sites would also be low. We have previously shown that late passage Dnmt[3a–/–,3b–/–] mouse ES cells retain Dnmt1 but are severely demethylated (11). We confirmed this difference using the mass spectrometry technique to quantify DNA methylation (Figure 5A). We tested the effects of 5-aza-dC on Dnmt1 levels in wild-type (70% of CpG methylated) and severely hypomethylated Dnmt[3a–/–,3b–/–] mouse ES cells (<0.7% of CpG methylated). We found that 5-aza-dC caused marked Dnmt1 degradation in the wild-type cells but not in the mutant cells (Figure 5B). As the Dnmt[3a–/–,3b–/–] mouse ES cells have markedly reduced DNA methylation but retain Dnmt1, it is clear that Dnmt1 does not target the unmethylated CpG sites in these cells (else they would be methylated), but targets and maintains only the small number of hemi-methylated CpG that remain. These data therefore indicate that the targeting of Dnmt1 to 5-aza-dC residues in hemi-methylated DNA is required before Dnmt1 degradation is induced.
Figure 5.
The DNMT1-degrading effects of 5-aza-dC are dependent on pre-existing DNA methylation (A) Global DNA methylation in wild-type (J1) ES cells and Dnmt[3a–/–,3b–/–] ES cells. (B) Dnmt1 protein levels of in wild-type and Dnmt[3a–/–,3b–/–] mouse ES cells treated with 1 μM 5-aza-dC for 12 h. The bar graph shows the averaged quantifications with standard deviations (by densitometry) of western blots from three independent experiments.
A recent study indicated that in HCT116 cells the targeting of DNMT1 to 5-aza-dC residues causes DNA damage as assessed by the number of foci containing histone H2AX (12). We therefore asked whether DNA damage itself might be the trigger for the proteasomal degradation of DNMT1. We looked at the effects of gamma irradiation (Supplementary Figure S1) and UV irradiation (Supplementary Figure S2), but neither of these treatments induced loss of DNMT1, despite the induction of p53 by gamma irradiation. DNMT1 degradation also occurred in p53 deficient HCT116 cells (Supplementary Figure S3) ruling out a function for p53 in the degradation pathway.
Chromatin bound DNMT1 is relatively resistant to DNMT1 degradation
The observation that DNA methylation could be relatively well maintained in the presence of a low dose of 5-aza-dC despite significant degradation of DNMT1 (Figure 2A) prompted us to investigate whether DNMT1, which is a nuclear protein (13,14), might exist in two separate nuclear pools. We postulated that within the nucleus there might be a small chromatin bound pool associated with PCNA (15) that was actively engaged in maintenance methylation and which could become covalently linked to 5-aza-dC during synthesis, and a much larger soluble pool that was not actively engaged in maintaining methylation but which was preferentially degraded in the proteasome following 5-aza-dC treatment. In the absence of 5-aza-dC both forms would be detectable by western blotting, as the chromatin bound form would dissociate in denaturing SDS lysis buffer. After treatment with 5-aza-dC some chromatin-associated DNMT1 would become irreversibly linked (by covalent attachment) to 5-aza-dC molecules in the DNA, to an extent which was dependent on the dosage of the 5-aza-dC. This would lead to a dose dependent reduction in DNA methylation levels. The linked (trapped) DNMT1 would not be detectable by western blotting as it would not migrate out of the gel onto the membrane, but any DNMT1 that was not trapped (because of low 5-aza-dC dosage) would be free to continue maintenance methylation and would be detectable by western blotting.
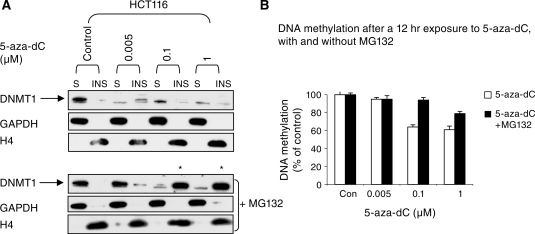
We extracted protein from cells using a gentle lysis buffer so as to preserve any weak association between DNMT1 and the chromatin (see ‘Materials and Methods’ section). After centrifugation the cellular proteins were separated into soluble (the supernatant) and insoluble fractions (the chromatin bound pellet) and re-suspended in 1× SDS loading buffer. We found that most DNMT1 was in the soluble fraction, and after 5-aza-dC treatment, DNMT1 was preferentially lost from this fraction (Figure 6A). At 0.005 µM 5-aza-dC DNMT1 was lost from the soluble fraction but was retained in the insoluble fraction, and at higher doses DNMT1 was progressively lost from the insoluble fraction. To demonstrate that we had fractioned the DNMT1 as envisaged we looked at the levels of GAPDH which is a cytoplasmic protein, explaining why it was hardly detected in the chromatin pellet, but was abundant in the soluble fraction. By contrast Histone H4, a chromatin protein, was only detectable in the pellet. Our findings indicate that whilst DNMT1 is required to target the DNA before the signal for DNMT1 degradation is induced, the mechanism that degrades DNMT1 or that marks it for degradation is induced globally within the nucleus, with the soluble form of DNMT1 being more susceptible to degradation than the chromatin bound form. Interestingly, rescue of the DNMT1 protein by pre-treatment of the cells with MG132 changed the distribution of DNMT1 markedly (Figure 6A, lower panel). At low doses of 5-aza-dC (0.005 µM) treatment with MG132 caused the DNMT1 protein to appear in the soluble fraction (as expected), whereas at higher doses of 5-aza-dC (0.1 and 1 µM) pre-treatment with the same dose of MG132 caused the protein to reappear in the chromatin bound fraction (asterisks, Figure 6A). This result is most probably explained by the rescued DNMT1 homing to increased amounts of hemi-methylated DNA in chromatin. Hemi-methylated DNA is the preferred natural substrate of the enzyme and 5-aza-C treatment results in high levels of hemi-methylated DNA (16). Consistent with this idea, MG132 caused a marked reduction in the degree of hypomethylation induced by 5-aza-dC (Figure 6B), presumably because the rescued DNMT1 was able to restore full methylation to a proportion of the hemi-methylated sites induced by trapping. Because of the relative ineffectiveness of low dose 5-aza-dC at inducing demethylation (even when DNMT1 degradation has been induced) we suggest that dose-dependent trapping is the primary mechanism whereby 5-aza-dC induces hypomethylation. As this also triggers the proteasomal degradation of free non-chromatin associated DNMT1 the cell has no way of compensating for the hypomethylation induced by trapping. The ameliorating effects of MG132 on 5-aza-dC induced hypomethylation indicate that trapping on its own would be relatively ineffective at inducing hypomethylation.
Figure 6.
Soluble DNMT1 is more sensitive to degradation that chromatin bound DNMT1. (A) upper panel, western blot of HCT116 cells treated with the indicated doses of 5-aza-dC for 12 h. Whole cell proteins were fractionated by differential solubility in PBS/0.5% Triton X-100. The soluble fractions (S) fractions were separated from the chromatin bound insoluble (INS) fractions by centrifugation. In the lower panel the effects of pre-treatment of the cells with MG132 are shown. GAPDH and Histone H4 serve as a loading controls for the soluble fraction and insoluble fractions respectively, and serve to verify that chromatin was separated as expected. (B) Pre-treatment with MG132 reduces the DNA hypomethylating effects of 5-aza-dC.
DISCUSSION
In this report we show that incorporation of 5-aza-dC during DNA synthesis causes the proteasomal degradation of DNMT1. The phenomenon is very sensitive to 5-aza-dC incorporation, occurring at much lower doses than are commonly used for inducing demethylation. The high sensitivity of cells to 5-aza-dC-induced DNMT1 degradation explains why, at high doses of 5-aza-dC, the DNA synthesis inhibitor aphidicolin fails to inhibit the effect. Aphidicolin does not completely inhibit DNA synthesis. Therefore, at high doses of 5-aza-dC, sufficient drug is still incorporated into DNA to induce the effect. These findings explain why the authors of the original report which identified the 5-aza-dC-induced DNMT1 degradation pathway concluded that the effect must in some way have been independent of DNA synthesis (4). The lowest dose of 5-aza-dC used in their study was 0.1 µM (range 0.1–10 µM). The ameliorating effect of DNA synthesis inhibition was only seen at doses of 0.1 µM and below in our study.
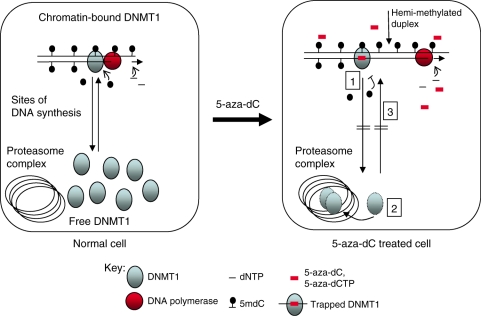
5-aza-dC-induced degradation of Dnmt1 does not occur in severely hypomethylated mouse Dnmt[3a–/–,3b–/–] ES cells. As DNA methylation is essentially absent in these cells, this indicates that targeting of incorporated 5-aza-dC molecules by Dnmt1 (a process known to be dependent on the presence of hemi-methylated target sites) is also essential before Dnmt1 degradation takes place. Our data are therefore consistent with a mechanism whereby, on the incorporation of 5-aza-dC into DNA, the chromatin bound enzyme attacks any 5-aza-dC residues that are incorporated diagonally opposite 5mC residues in the parent strand, and attempts to methylate them. Trapping of the enzyme ensues and the covalent complexes formed activate the proteasomal degradation of the free enzyme (Figure 7). Adduct formation between DNMT1 and DNA could also explain the DNMT1 degradation induced by SGI-1027, a compound which has been shown to compete with S-adenosylmethionine in the methylation reaction (17). Interference with S-adenosylmethionine binding would prevent transfer of the methyl group after covalent adduction of the DNMT1 protein to C6 of the cytosine ring had taken place. The mechanism of action of DNMT1 predicts that failure to transfer the methyl group to C5 would prevent release of the enzyme by β-elimination, mimicking the effect of 5-aza-dC-induced entrapment (2).
Figure 7.
Model to explain the effects of 5-aza-dC on DNA methylation and the proteasomal degradation of DNMT1. In normal cells undergoing replication DNMT1 binds to and remethylates the hemi-methylated DNA duplexes produced as a result of DNA synthesis. There is an exchange of DNMT1 between the chromatin and free compartments. This exchange is indicated by the arrows in the first panel. Maintenance methylation is linked to DNA replication and in normal untreated cells the amount of hemi-methylated DNA is low because of effective maintenance methylation. The addition of 5-aza-dC causes: 1, Incorporation of 5-aza-dC into DNA and trapping of DNMT1 enzymes in a dosage dependent manner. This leads to an increase in the amount of hemi-methylated DNA and fall in proportion of cytosines methylated. 2, 5-aza-dC also induces the proteasomal degradation of free DNMT1, even at very low dosage. The mechanism is unknown but involves the targeting of 5-aza-dC incorporated DNA by DNMT1. 3, Degradation of free DNMT1 prevents the re-methylation of the hemi-methylated DNA that accumulates as a result of trapping. Proteasomal degradation of DNMT1 contributes to the hypomethylation induced by 5-aza-dC.
The mechanism whereby the DNMT1-DNA adducts induce DNMT1 degradation has not been elucidated in this study. The study by Ghoshal et al. (4) provided evidence that the ubiquitin-E3 ligase CDH1/FZR1, a component of the anaphase promoting complex, was responsible for ubiquitinylation of DNMT1. However we were unable to prevent DNMT1 destruction by knocking down CDH1/FZR1 using siRNA (Figure 1E). Our results can not exclude CDH1/FZR1 as being the E3 ligase responsible for targeting DNMT1 for destruction as we can never be certain that our knockdown, though effective, was effective enough. Ghoshal et al. (4) also indicated that the N terminus of DNMT1 was crucial for the degradation. We have not tested this but evidence from transgenic mice indicates that it is likely to be correct. The shorter oocyte specific isoform of Dnmt1 (Dnmt1o) is driven by a separate promoter and is identical to the somatic form (Dnmt1s) apart from the fact that it lacks the extreme 118 N-terminal amino acids (14). When Dnmt1o protein was engineered to be driven by the endogenous Dnmt1s promoter in mice, late embryos heterozygous for both proteins had Dnmt1o protein levels that were five times higher than Dnmt1s levels. Thus under conditions of equal transcription Dnmt1o undergoes less protein degradation presumably because it lacks the N-terminal sequence. These data indicate that the N terminal amino acids are crucial for controlling Dnmt1 protein levels even in the absence of 5-aza-dC (18).
The DNMT1 degrading effects of 5-aza-dC can occur at very low doses of the drug, and at these doses there appears to be little or no effect on demethylation. We can not exclude the possibility that some hypomethylation has occurred at a limited number of specific sites, but at a global level demethylation is not obvious. We can also not rule out the possibility that DNMT3A and DNMT3B compensate for methylation losses due to mild DNMT1 inhibition. However Egger et al. (7) found that treatment of HCT116 cells which are hypomorphic for DNMT1 with siRNA molecules against DNMT1 caused a lowering of DNA methylation levels. As these cells express both DNMT3A and DNMT3B, these enzymes were clearly not able to compensate for methylation losses caused by DNMT1 depletion. We favour the possibility that, as DNMT1 degradation does not affect all DNMT1 molecules (the chromatin bound enzyme which is actively engaged in methylation is relatively spared), there is enough residual DNMT1 activity to support maintenance methylation in the presence of low dose 5-aza-dC.
Our data indicate that most of the cellular DNMT1 is soluble (in the nucleoplasm) and it would appear that cells have considerably more DNMT1 than they need for maintaining DNA methylation. This is also suggested by the results of Egger et al. (7) who found that DNA methylation levels are near normal in HCT116 cells which are hypomorphic for DNMT1. These cells express a truncated form of DNMT1 which is present at 20% of the normal wild-type levels of the protein. DNA methylation levels are also normal in heterozygous Dnmt1+/– ES cells and Dnmt1 mutated heterozygous mice despite the heterozygotes having lower levels of Dnmt1 protein (19). The DNMT1 protein that is not actively engaged in maintenance methylation may fulfil other non-enzymatic functions. Studies have shown that apart from maintenance methylation DNMT1 is also involved in mis-match repair (20) the prevention of apoptosis (21) and the mitotic phase of the cell cycle (22).
As the induction of hypomethylation correlates better with the 5-aza-dC dose than the DNMT1 level (as assessed by western blot) we suggest that it is trapping of chromatin bound DNMT1 molecules by 5-aza-dC which is the primary cause of hypomethylation. Degradation of free DNMT1 exacerbates the effect by preventing the full methylation of hemi-methylated sites created by trapping. As the degradation of DNMT1 occurs at low dosage, our findings raise the possibility that there may be distinct S-phase specific biological effects of 5-aza-dC which are operative at dosages which would not ordinarily be toxic, but which might collaborate with other established therapies to modulate and increase the effectiveness of cancer therapy.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Breakthrough Breast Cancer Charity and Cancer Research UK. Funding for open access charge: Breakthrough Breast Cancer Charity.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Richard Meehan and Nick Gilbert for critical reading of the manuscript.
REFERENCES
- 1.Taylor SM, Jones PA. Mechanism of action of eukaryotic DNA methyltransferase. Use of 5-azacytosine-containing DNA. J. Mol. Biol. 1982;162:679–692. doi: 10.1016/0022-2836(82)90395-3. [DOI] [PubMed] [Google Scholar]
- 2.Wu JC, Santi DV. Kinetic and catalytic mechanism of HhaI methyltransferase. J. Biol. Chem. 1987;262:4778–4786. [PubMed] [Google Scholar]
- 3.Schermelleh L, Spada F, Easwaran HP, Zolghadr K, Margot JB, Cardoso MC, Leonhardt H. Trapped in action: direct visualization of DNA methyltransferase activity in living cells. Nat. Methods. 2005;2:751–756. doi: 10.1038/nmeth794. [DOI] [PubMed] [Google Scholar]
- 4.Ghoshal K, Datta J, Majumder S, Bai S, Kutay H, Motiwala T, Jacob ST. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol. Cell. Biol. 2005;25:4727–4741. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 6.Crain PF. Preparation and enzymatic hydrolysis of DNA and RNA for mass spectrometry. Methods Enzymol. 1990;193:782–790. doi: 10.1016/0076-6879(90)93450-y. [DOI] [PubMed] [Google Scholar]
- 7.Egger G, Jeong S, Escobar SG, Cortez CC, Li TW, Saito Y, Yoo CB, Jones PA, Liang G. Identification of DNMT1 (DNA methyltransferase 1) hypomorphs in somatic knockouts suggests an essential role for DNMT1 in cell survival. Proc. Natl Acad. Sci. USA. 2006;103:14080–14085. doi: 10.1073/pnas.0604602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vesely J, Cihak A, Sorm F. Characteristics of mouse leukemic cells resistant to 5-azacytidine and 5-aza-2′-deoxycytidine. Cancer Res. 1968;28:1995–2000. [PubMed] [Google Scholar]
- 9.Adams RLP, Lindsay H, Reale A, Seivwright S, Kass S, Cummings M, Houlston C. In: DNA Methylaion: Molecular Biology and Biological Significance. 1st edn. Jost JP, Saluz HP, editors. Switzerland: Birkhauser; 1993. pp. 120–144. [Google Scholar]
- 10.Frauer C, Leonhardt H. A versatile non-radioactive assay for DNA methyltransferase activity and DNA binding. Nucleic Acids Res. 2009;37:e22. doi: 10.1093/nar/gkn1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson M, Krassowska A, Gilbert N, Chevassut T, Forrester L, Ansell J, Ramsahoye B. Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells. Mol. Cell. Biol. 2004;24:8862–8871. doi: 10.1128/MCB.24.20.8862-8871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palii SS, Van Emburgh BO, Sankpal UT, Brown KD, Robertson KD. DNA methylation inhibitor 5-Aza-2′-deoxycytidine induces reversible genome-wide DNA damage that is distinctly influenced by DNA methyltransferases 1 and 3B. Mol. Cell. Biol. 2008;28:752–771. doi: 10.1128/MCB.01799-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlson LL, Page AW, Bestor TH. Properties and localisation of DNA methyltransferase in preimplantation mouse embryos: implications for genomic imprinting. Genes Dev. 1992;6:2536–2541. doi: 10.1101/gad.6.12b.2536. [DOI] [PubMed] [Google Scholar]
- 14.Howell CY, Bestor TH, Ding F, Latham KE, Mertineit C, Trasler JM, Chaillet JR. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell. 2001;104:829–838. doi: 10.1016/s0092-8674(01)00280-x. [DOI] [PubMed] [Google Scholar]
- 15.Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 16.Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 17.Datta J, Ghoshal K, Denny WA, Gamage SA, Brooke DG, Phiasivongsa P, Redkar S, Jacob ST. A new class of quinoline-based DNA hypomethylating agents reactivates tumor suppressor genes by blocking DNA methyltransferase 1 activity and inducing its degradation. Cancer Res. 2009;69:4277–4285. doi: 10.1158/0008-5472.CAN-08-3669. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Ding F, Chaillet JR. In vivo stabilization of the Dnmt1 (cytosine-5)- methyltransferase protein. Proc. Natl Acad. Sci. USA. 2002;99:14861–14866. doi: 10.1073/pnas.232565599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lei H, Oh SP, Okano M, Juttermann R, Goss KA, Jaenisch R, Li E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- 20.Guo G, Wang W, Bradley A. Mismatch repair genes identified using genetic screens in Blm-deficient embryonic stem cells. Nature. 2004;429:891–895. doi: 10.1038/nature02653. [DOI] [PubMed] [Google Scholar]
- 21.Ruzov A, Shorning B, Mortusewicz O, Dunican DS, Leonhardt H, Meehan RR. MBD4 and MLH1 are required for apoptotic induction in xDNMT1-depleted embryos. Development. 2009;136:2277–2286. doi: 10.1242/dev.032227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen T, Hevi S, Gay F, Tsujimoto N, He T, Zhang B, Ueda Y, Li E. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat. Genet. 2007;39:391–396. doi: 10.1038/ng1982. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.