Abstract
Twisted intercalating nucleic acid (TINA) is a novel intercalator and stabilizer of Hoogsteen type parallel triplex formations (PT). Specific design rules for position of TINA in triplex forming oligonucleotides (TFOs) have not previously been presented. We describe a complete collection of easy and robust design rules based upon more than 2500 melting points (Tm) determined by FRET. To increase the sensitivity of PT, multiple TINAs should be placed with at least 3 nt in-between or preferable one TINA for each half helixturn and/or whole helixturn. We find that ΔTm of base mismatches on PT is remarkably high (between 7.4 and 15.2°C) compared to antiparallel duplexes (between 3.8 and 9.4°C). The specificity of PT by ΔTm increases when shorter TFOs and higher pH are chosen. To increase ΔTms, base mismatches should be placed in the center of the TFO and when feasible, A, C or T to G base mismatches should be avoided. Base mismatches can be neutralized by intercalation of a TINA on each side of the base mismatch and masked by a TINA intercalating direct 3′ (preferable) or 5′ of it. We predict that TINA stabilized PT will improve the sensitivity and specificity of DNA based clinical diagnostic assays.
INTRODUCTION
Twisted intercalating nucleic acid (TINA) is a novel artificial intercalating nucleic acid, which is capable of stabilizing Hoogsteen-type DNA triplex formations (1). Currently, limited knowledge concerning design of oligonucleotides containing TINA is available.
Preceding data suggest that Hoogsteen-type DNA triplex formations are characterized by a pronounced decrease in melting point (Tm) per base mismatch (1,2), which is in contrast to the relatively low change in Tm (ΔTm) per base mismatch seen in antiparallel duplex (AD) formations (2). Triplex formations are divided into parallel and antiparallel triplexes by the orientation of the Triplex Forming Oligonucleotide (TFO) (3). The TFOs of parallel triplexes (PT) follow the orientation of the homopurine target strand and consist of CT or GT sequences (4), whereas antiparallel triplexes follow the orientation of the homopyrimidine AD strand and consist of GT or GA sequences (5). The formation of triplexes is limited by (i) dependence on DNA homopurine stretches in the target sequence (6–8), (ii) acidic pH for PT formation due to the need of protonated cytosine in the TFO (4,9,10) and (iii) quadruplex formation of guanosine-rich TFOs in antiparallel triplexes (11).
TINA has been shown to increase Tm and to decrease pH dependence of PT seemingly without compromising ΔTm (1). TINA in the TFO should be placed as an intercalator between two bases and not as a substitution for a base, and the stabilizing effect of two TINAs in the TFO increases, when the TINAs are placed with several bases in-between (1,12–14). In vitro gene-expression studies of antiparallel triplex formation have shown that TINA lowers the tendency of potassium induced oligonucleotide aggregation compared with DNA TFO (15). In addition, TINA has been used to flank guanosine stretches within quadruplex oligonucleotides. This lead to an increase Tm of the quadruplex and induced a strong antiproliferative effect in Panc-1 cells by blocking the KRAS promoter (16).
Very little is known about the stabilizing effect of TINA on DNA triplex formations. During conventional oligonucleotide synthesis, TINA is covalently inserted between two bases in the phosphate backbone, and TINA is believed to form a bulge in the phosphate backbone while its pyrene intercalator is inserted into the base stacking of the AD part of the triplex (17). Recently, a number of TINA analogues have been synthesized, but none of them are superior stabilizers of PT compared to TINA (1,14,18–20).
The aim of this study was to verify the previously observed high ΔTm for DNA triplex formations and to determine the optimal design rules and stabilizing effect of TINA in PT. To do so a high-speed melting curve acquisition method has been used and validated towards a slower standard method. If TINA modified Hoogsteen-type PT have increased ΔTm compared to conventional AD formations, this could improve the specificity of future DNA-based clinical assays.
MATERIALS AND METHODS
Fluorescence resonance energy transfer (FRET) system
For determinations of ΔTm, a synthetic polypurine DNA target (5′ AGG GAA AAG AAG GGA GGG ATA AC 3′) was constructed to ensure different base mutations at position 4, 7, 10, 13 and 16 (GGA, AAA, GAA, GGG and AGG) from the 5′-end of the polypurine target strand. To visualize PT formation, target oligonucleotides were labelled with ATTO495 and TFOs were labelled with ATTO590. For visualization of AD formation the ATTO590 fluorophore was placed on the non-target strand of the AD. Target oligonucleotides labelled with ATTO495 were synthesized as oligonucleotides with 5′-amino-modifier-C6 and linked to the ATTO495 NHS-ester. ATTO495 is a modification of Acridine Orange with excitation maximum at 495 nm and emission maximum at 527 nm. Oligonucleotides labelled with ATTO590 were synthesized with a 5′-amino-modifier-C6 or 3′-amino-modifier-C7 and linked to ATTO590 NHS-ester. ATTO590 is a derivative of Rhodamine dyes with excitation maximum at 594 nm and emission maximum at 624 nm. The ATTO495 and ATTO590 FRET pair was excitated at 470 nm on a LightCycler 2.0 and fluorescence emission was detected at 640 nm.
Oligonucleotides
Unlabelled and ATTO495/ATTO590-labelled oligonucleotides were purchased from IBA GmbH (Göttingen, Germany) and DNA Technology A/S (Risskov, Denmark) on a 0.2 µmol synthesis scale with high performance liquid chromatography (HPLC) purification and quality control. A 23-nt target sequence as 5′ ATTO495-AGG GAA AAG AAG GGA GGG ATA AC 3′ (D-200) with a base mismatch at either position 4, 7, 9, 10, 11, 13 or 16 or two base mismatches at position 9 and 11 from the 5′-end were combined with a 23-nt AD sequence (AD) of 5′ GTT ATC CCT CCC TTC TTT TCC CT 3′(D-256) with zero to two TINA modifications and a TFO as 5′ ATTO590-TCC CTT TTC TTC CCT CCC T 3′ (D-284) with zero to four TINA modifications and a TFO length of 7–19 nt counted from 5′ to 3′. The TINA modification is a (R)-1-O-[4-(1-pyrenylethynyl)phenylmethyl]glycerol and is inserted as an internal bulge in the DNA oligonucleotides. An overview of most oligonucleotides used in this study can be found in Supplementary Tables S1, S2 and S3.
Melting curve acquisition
Melting curve experiments were performed on a LightCycler 2.0 (Roche Applied Science, Basel, Switzerland) as previously described and validated (21,22). In short, PT experiments were performed in 20 µl LightCycler capillaries using 1.0 µM of each oligonuclotide in sodium acetate buffer (50 mM NaOAc, 100 mM NaCl and 10 mM MgCl2) at pH 4.5 (experiments with shorter TFOs), pH 5.0 (standard conditions), pH 5.5 or pH 6.0 (pH validation and comparison of ΔTm for AD and PT). AD experiments were performed with 0.5 µM of each oligonucleotide in sodium phosphate buffer (50 mM NaH2PO4/Na2HPO4, 100 mM NaCl and 0.1 mM EDTA) at pH 5.5 or pH 6.0 (comparison of ΔTm for AD and PT) and pH 7.0 (standard conditions). All chemicals were purchased from Sigma-Aldrich Denmark (Brøndby, Denmark). Tm measurements were carried out using a high-speed standard program of (i) a dissociation step from 37 to 95°C with a ramp rate of 0.2°C/s and hold for 15 s at 95°C, (ii) annealing from 95 to 37°C with a ramp rate of 0.2°C/s and hold for 5 min at 37°C and (iii) dissociation step from 37 to 95°C with a ramp rate of 0.05°C/s and continued measurement of fluorescence. Tm was identified using the LightCycler Software 4.1 for melting curve analysis and defined as the peak of the first derivative.
All melting curve determinations were conducted as single capillary measurements. A setup control composed of D-200, D-256 and D-284 was included in all runs. Prior to Tm identification, runs were colour compensated by subtraction of the fluorophore background fluorescence.
Method validation
We have previously validated the use of ATTO fluorophores for PT and AD FRET on the LightCycler and compared it to UV absorbance measurements (22). The present high-speed LightCycler method uses a ramp rate of 3°C/min, which is too fast to allow the PT to be in thermodynamic equilibrium under dissociation. Thus, the reported Tm values are overestimates. Consequently, the results were validated doing annealing curves by (i) heating to 95°C at a ramp rate of 0.2°C/s and hold for 10 min, (ii) cooling from 95 to 37°C leaving the samples to equilibrate for 5 min before each fluorescence reading, leading to one fluorescence reading per 1°C. In addition dissociation curves where done by (i) leaving the capillaries for 30 min at 37°C, (ii) heating from 37 to 95°C leaving the samples to equilibrate for 5 min before each fluorescence reading. This method is routinely used on the LightCycler to avoid significant hysteresis between the dissociation and annealing curves (23). Validation data for (i) dissociation speed (3, 1, 0.5 and 0.2°C/min), (ii) time before dissociation (5 min, 30 min, 1 h and 24 h), (iii) Tm comparisons for TINA oligonucleotides (at pH 5.0, 5.5 and 6.0) and (iv) ΔTm comparisons for AD and PT (at pH 5.0, 5.5 and 6.0 in sodium acetate buffer (50 mM NaOAc, 100 mM NaCl and 10 mM MgCl2) or phosphate buffer (50 mM NaH2PO4/Na2HPO4, 100 mM NaCl and 0.1 mM EDTA) at pH 5.5, 6.0 and 7.0) were done comparing annealing and dissociation curves at a ramp rate of 0.2°C/min and 3°C/min. Data are included in Supplementary Tables S5–S8.
RESULTS
Tm determinations and design rules for parallel triplex with TINA in the TFO
Table 1 highlights the effects of TINAs inserted in the TFO. A complete list of tested TINA positions in the TFO and Tm data can be found in Supplementary Table S1. On average, one TINA in the TFO increased Tm by 3.9°C (range 0.6–5.9°C), whereas two TINAs in the TFO increased Tm by an average of 6.5°C (range 1.9–9.9°C). As illustrated in Table 1 the number of bases in-between two TINAs had a huge impact on Tm. Based on all Tms in Supplementary Table 1 we found that two to four TINAs should be placed with (i) at least three bases in-between, (ii) an optimum of 5–7 or 10–14 bases in-between (equivalent to a half or a full helix-turn) and (iii) a TINA at each end of the oligonucleotide.
Table 1.
TINA in triplex TFO and change in Tm and ΔTm
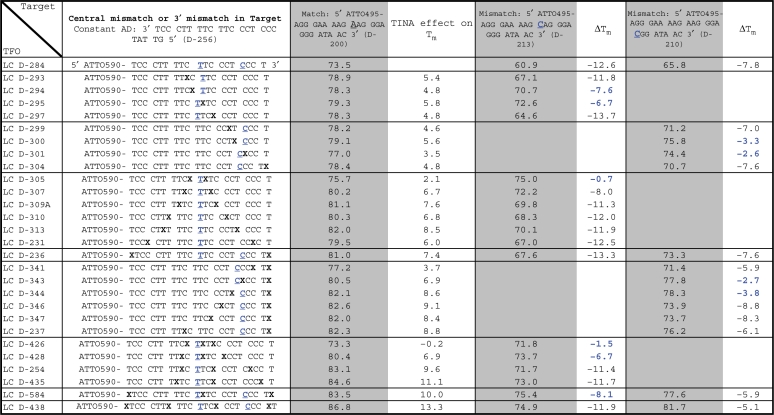 |
Change in triplex Tm by introduction of one to four TINAs in the TFO and change in ΔTm by introduction of a central base mismatch or a base mismatch towards the 3′ end of the target strand. Tm was determined using 1.0 µM of each strand in NaOAc-buffer at pH 5.0. X is TINA. Base mismatches are underlined and marked in bold blue.
The effect of base mismatches in the target sequence on ΔTm of the TFO
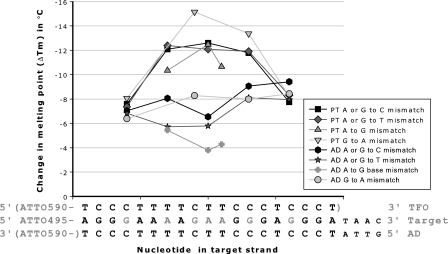
A number of base mismatches at different positions in the target sequence were tested. Figure 1 shows the effect of the base mismatches in the target oligonucleotide on Tm of the TFO. Central base mismatches at base 9–11 in the target oligonucleotide decreased Tm of the TFO with 10.7–15.2°C, whereas base mismatches towards the 5′ (base 4) and 3′-end (base 16) of the target oligonucleotide decreased Tm of the TFO with 7.6–8.3°C. Adenine to guanine mutations were less destabilizing than guanine to adenine base mismatches, whereas purine to either thymine or cytosine base mismatches were equally, but less destabilizing than guanine to adenine mutations. A double mutation of guanine to adenine at position 9 and adenine to guanine at position 11 in the target oligonucleotide had a ΔTm on the TFO of 20.8°C compared with 15.2°C for the guanine to adenine base mismatch at position 9 and 10.7°C for the adenine to guanine base mismatch at position 11 in the target oligonucleotide (Tm data in Supplementary Table S1).
Figure 1.
Introduction of a base mismatch along the target strand and change in melting point (ΔTm) of the AD and the parallel triplex TFO. The ADs consist of the oligonucleotides D-200 to D-222 as target strands with Base mismatches in combination with D-484 as AD strand. The PT consist of the oligonucleotides D-200 to D-222 as target strands with base mismatches in combination with D-256 as AD strand and D-284 as triplex TFO strand. Triplex melting points were determined using 1.0 µM of each strand in sodium acetate buffer at pH 5.0. AD melting points were determined using 0.5 µM of each strand in sodium phosphate buffer at pH 7.0. Base mismatch positions are marked in gray. Fluorophore and TFO strand are placed in brackets as they are not used in all experiments. PT is parallel triplex, AD is antiparallel duplex, A is adenine, G is guanine, C is cytosine and T is thymine.
The effect of base mismatches in the target sequence on ΔTm of the Watson–Crick type AD
Base mismatches in the target sequence decreased Tm of the AD with 3.8–9.4°C (Figure 1). For all positions ΔTm of a base mismatch in the PT was higher compared with a ΔTm of the AD. Loss of A:T base pairs where less destabilizing than loss of G:C base pairs. The double mutation of guanine to adenine at position 9 and adenine to guanine at position 11 in the target oligonucleotide had a ΔTm of 11.1°C compared with 8.3°C for the guanine to adenine base mismatch at position 9 and 4.3°C for the adenine to guanine base mismatch at position 11 in the target oligonucleotide (Supplementary Table S1).
Neutralizing the effect of base mismatches in the target strand by TINAs in the TFO
The ΔTm of a base mismatch in the target strand was reduced by positioning a TINA directly adjacent. The effect was particular pronounced when TINA was placed directly 3′ to the base mismatch and less pronounced at the 5′ position of the base mismatch (Table 1: D-294/D-295, D-300/D-301, D-343/D-344). When TINA was moved away from the base mismatch its effect on ΔTm declined (Table 1: D-299/D-300). Positioning a TINA in the TFO on each side of an adenine to cytosine base mismatch in the target strand almost neutralized the ΔTm of the base mismatch on the TFO (Table 1: D-305 and D-426).
Changes in Tm and ΔTm of the TFO caused by TINAs in the AD strand
To evaluate whether TINAs within the AD strand stabilized the TFO, eight oligonucleotides with one TINA and six oligonucleotides with two TINAs were tested and the results are presented in Table 2 and Supplementary Table S2. On average, one TINA in the AD increased Tm of the TFO with 3.9°C (range 1.5–6.5°C), whereas a TINA placed in the AD outside the triplex forming part only increased Tm with 0.8°C (Table 2: D-452). Two TINAs in the AD stabilized the TFO, particularly when they were placed with at least three bases in-between and preferable with five bases in-between (Table 2: D-499/D-500). A TINA in the AD strand 3′ to a base mismatch in the target strand lowered ΔTm of the base mismatch (Table 2: D-448 and D-450). Two TINAs in the AD flanking an adenine to cytosine base mismatch in the target strand partially concealed the effect of the base mismatch on TFO ΔTm (Table 2: D-498). The best way to mask the base mismatch is to place the TINA in the TFO rather than in the AD strand (Table 2: D-498/Table 1: D-305, Table 2: D-448/Table 1: D-294, Table 2: D-450/Table 1: D-300).
Table 2.
TINA in triplex AD strand and change in Tm and ΔTm
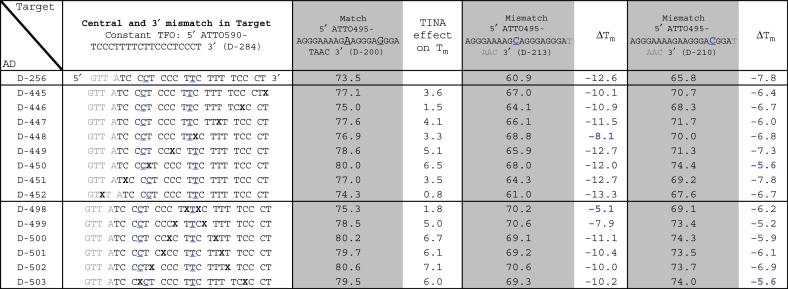 |
Change in triplex Tm by introduction of one or two TINAs in the AD strand and change in ΔTm by introduction of a central base mismatch or a base mismatch towards the 3′ end of the target strand. Tm was determined using 1.0 µM of each strand in NaOAc-buffer at pH 5.0. X is TINA. Base mismatches are underlined and marked in bold blue.
Triplexes with TINA in both the TFO and AD strands
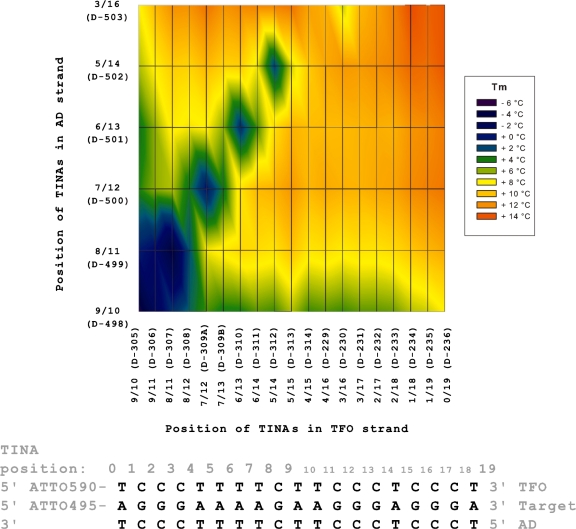
To evaluate triplex stability with TINA in both the AD and TFO strand, we combined oligonucleotides with two TINAs in both the AD and TFO strands. The effect on Tm was compared with Tm of the pure DNA triplex and the changes in Tm are plotted in Figure 2 (Tm data in Supplementary Table S3). As previously seen, Tm increased as the two TINAs in each strand were moved apart and Tm increased up to 12.5°C (Figure 2: red area). As previously observed, positioning of two TINAs with less than three bases in-between only slightly increased Tm (Figure 2: green stretches). All positions of two TINAs directly opposite each other in the AD and TFO strands decreased the stability of the triplex (Figure 2: blue and green spots).
Figure 2.
Position of two TINAs in both the TFO and AD strands of the triplex and change in Tm. Parallel triplex with two TINAs in the AD strand and two TINAs in the TFO strand and change in triplex Tm. On both axes TINAs are moved from the centre to the ends of the oligonucleotide. Triplex Tm was determined using 1.0 µM of each strand in NaOAc-buffer at pH 5.0. TINA positions are indicated above the triplex sequence counted from the 5′ to the 3′ position in the target strand, and the numbers reflect the position of the base 5-prime to the TINA bulge insertion. Colour indicates the change in Tm compared with a pure DNA triplex.
Changes in Tm and ΔTm by the length and number of TINAs in the TFO
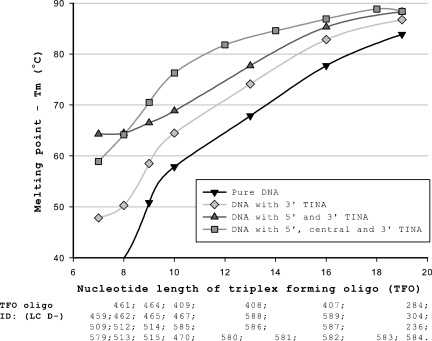
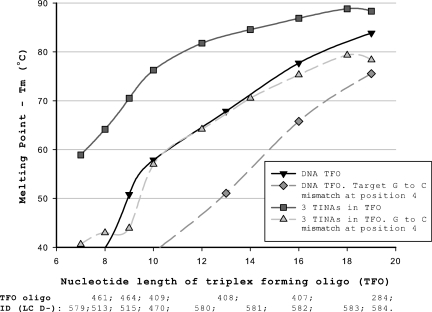
To confirm that the positive effect on Tm of TINA in the TFO was not restricted to a TFO length of 19 nt at pH 5.0, we tried a number of shorter TFOs at pH 4.5. Experiments were conducted at pH 4.5 to ensure the outmost stability of the DNA triplexes and are presented in Figures 3 and 4. Shorter TFOs decreased Tm of the TFOs as seen in Figure 3. The effect of TINA on Tm was enhanced as the TFO were shortened, e.g. three TINAs in the 19 oligonucleotide TFO increased Tm by 4.5°C, whereas three TINAs in the 10-nt TFO increased Tm by 18.4°C compared with the DNA TFO (Figure 3). At a TFO length below 9 nt, the Tm of TFOs with two TINAs was higher than Tm of TFOs with three TINAs, whereas TFOs longer than 9 nt were stabilized by a third TINA in the center of the TFO. Shorter TFOs increased ΔTm of a cytosine to guanine base mismatch at position 4 in the target oligonucleotide (Figure 4). An identical increase in ΔTm was found for DNA TFOs and TFOs with three TINAs. When the TFO length was reduced to 7–8 nt the TINA in the middle of the TFO was placed adjacent at the 3′ position of the base mismatch in the target, which lead to a stabilization and thereby decrease in ΔTm of the TFO as illustrated in Figure 4 and previously demonstrated in Table 1 for TFOs with TINA.
Figure 3.
Effect of TFO length and number of TINAs in the TFO on triplex Tm. The triplexes consisted of D-200 as target strand with D-256 as AD strand and different TFO strands. The length of the TFO is counted from the 5′ to the 3′ position, and oligo IDs for TFOs used in the experiments are given below the oligo length. Oligosequences corresponding to the oligo IDs are found in Supplementary Table S4. Tm was determined using 1.0 µM of each strand in sodium acetate buffer at pH 4.5.
Figure 4.
ΔTm of triplexes of different lengths with a G to C base mismatch at position 4. Length of pure DNA TFO or TFO with three TINAs and effect on Tm by introduction of guanine (G) to cytosine (C) base mismatch at position 4 counted from the 5′-end of the target oligonucleotide. D-200 or D-201 as target strands in combination with D-256 as AD strand and different TFO strands. The length of the TFO is counted from the 5′ to the 3′ position, and oligo IDs for TFOs used in the experiments are given below the oligo length. Oligosequences corresponding to the oligo IDs are found in Supplementary Table S4. Tm was determined using 1.0 µM of each strand in NaOAc-buffer at pH 4.5.
Influence of LightCycler 2.0 program on Tm of PT
Since PT are not in thermodynamic equilibrium at a dissociation ramp rate of 3°C/min, we evaluated the PT shown in Table 1 at slower dissociation and annealing rates (3, 1, 0.5 and 0.2°C/min) to demonstrate that slower ramp rates leads to a parallel shift in Tm. Lowering the ramp rate from 3°C/min to 0.2°C/min for the TINA PT caused a parallel shifted decrease in dissociation Tm of 5.3°C (range 4.6–5.7°C) (Supplementary Table S5). It was also tested, whether an incubation time of 5 min before dissociation experiments was sufficient to form stable PT. To evaluate this we preincubated the PT shown in Table 1 for 5 min, 30 min, 1 h or 24 h before dissociation experiments. Preincubation time did not influence the Tms determinated at a ramp rate of 3°C/min or 0.2°C/min (Supplementary Table S6).
Changes in Tm of PT with TINA at pH above 5.0
To evaluate how increasing pH influenced the Tm determination of PT with TINA we increased pH from 5.0 to 5.5 and 6.0 for the PT shown in Table 1 at a ramp rate of 3°C/min and 0.2°C/min. The stabilizing effect of TINA on Tm increased by increasing pH and was independent of ramp rate. Increasing pH did favour multiple TINAs and internal placement of TINAs with a half or whole helixturn in-between compared to placement of TINA only at the ends of the TFO (Supplementary Table S7).
Comparison of parallel triplex and AD ΔTm in identical buffer at pH 5.0 and 5.5
In Figure 1 we compared ΔTm of PT in sodium acetate buffer at pH 5.0 with 10 mM magnesium chloride and AD in phosphate buffer at pH 7.0 without magnesium chloride. This was done to ensure maximum stability of PT and stringent hybridization conditions for ΔTm of AD. To compare ΔTm of AD and PT at equal pH, we changed the AD buffer to sodium acetate buffer and compared AD and PT at pH 5.0 and 5.5. The introduction of magnesium chloride increased Tm of AD by approximately 4°C, but decreased ΔTm by approximately 0.5–1°C. For PT Tm decreased by approximately 14°C, whereas ΔTm was unaffected with a tendency towards a minor increase (Supplementary Table S8).
DISCUSSION
The ΔTm caused by a base mismatch is affected by buffer composition, the length and the sequence of the oligonucleotide and the nature and position of the base mismatch. We found that ΔTm of base mismatches in the central part of a 23 bp AD at pH 7.0 in phosphate buffer varied between 3.8 and 9.4°C compared with 10.7–15.2°C for a central base mismatch in a 19 nt PT at pH 5.0 in sodium acetate buffer with magnesium chloride. Thus, ΔTm of PT in the current study is remarkably high, and higher than ΔTm of AD for all base mismatch positions tested. Experiments in sodium acetate buffer at identical pH for AD and PT showed similar results. Furthermore, ΔTm of PT increases for shorter TFOs as seen in Figure 3. To our knowledge this is the first study comparing ΔTm of AD and PT for multiple base mismatch positions in the target sequence containing both guanine and adenine nucleotides. The outstanding ΔTm of PT makes PT interesting for design of highly specific assays within microbiology, forensics, cancer diagnostics, clinical genetics and in vivo gene regulation.
Despite the remarkable ΔTm of PT their use is still restricted by the need of DNA homopurine stretches in the target sequence and protonation of cytosine in the TFO at acidic pH.
Since incorporation of TINA decrease the pH dependence of PT, TINAs are an obvious choice for stabilization of PT, especially since ΔTm of PT in general is not changed by introduction of TINAs in the TFO. One exception from this rule is that TINA minimizes the ΔTm of a base mismatch, when TINA in the TFO is placed adjacent to a base mismatch in the target strand. This is especially pronounced when TINA intercalates from the 3′ position of the mismatch base pair. This suggests that the incorporated TINAs in the TFO intercalate mostly with the base pair directly 5′ to it in the base stacking and to a lesser degree with the base pair direct 3′ to it. This masking effect has not been described previously, but is especially interesting for assay development, where masking of possible base mismatches in the target sequence is of importance.
Previous publications describing the synthesis and effect of TINA in PT have indicated that two TINAs should be placed with several bases in-between to improve Tm (1,12–14). In the present study we find that the optimal position of TINAs to be with 5–7 or 10–14 nt in-between, equalling a half to one helix turn of the DNA double helix. Géci et al. (17) have previously demonstrated that the number of pyrene intercalators in the PT decreases the pH dependent drop in Tm of PT. We find that as pH increases the number of TINAs should be increased and TINAs should be included in the central part of the TFO. By optimal placement of TINAs the Tm, and thereby the stability of the PT, is markedly increased and will most likely enable an increase in sensitivity of clinical assays. A complete set of design rules for TINA in the TFO are presented in Table 3.
Table 3.
Design rules for placement of TINA in PT
| For maximum increase in ΔTm for the TFO (increase sensitivity) |
| (1) Place the TINAs in the TFO and internal TINAs as a bulge (ref. no. 1) |
| (2) Place a TINA at each end of the oligonucleotide |
| (3) Always place TINAs with three or more nucleotides in-between |
| (4) Place a TINA for each half helixtum and/or whole helixtum |
| (5) For pH > 6.0 use maximum number of TINAs following rules 3 and 4 |
| (6) For oligonucleotides shorter than 9 nts use rule 2 |
| To increase ΔTm for the TFO in parallel triplex (increase specificity) |
| (1) Position target base mismatches in the centre of the TFO |
| (2) Avoid A, C or T to G target base mismatches when feasible |
| (3) Choose a shorter TFO or higher pH to increase ΔTm of a target base mismatch |
| (4) Place TINAs several nucleotides from the target base mismatch |
| To decrease ΔTm for the TFO in parallel triplex (neutralize effect of base mismatch) |
| (1) Place a TINA direct 3′ of a target base mismatch to reduce ΔTm |
| (2) Place two TINA's direct 5′ and 3′ of a target base mismatch to neutralize ΔTm |
The aim of this study has been to determine the optimal design rules for placement of TINA in TFOs, and a general description regarding the thermodynamic properties of PT is beyond the scope of the present work. Hysteresis is seen for PT at the high-speed ramp rate of 3°C/min, but can as previously observed be eliminated at a ramp rate of 0.2°C/min (23,24). When we compare Tms for PT at both ramp rates, we reproducibly observe a parallel shift in Tm. This parallel shift is equal for matching oligonucleotides and base mismatches in sodium acetate buffers with magnesium chloride at pH 5.0, 5.5 and 6.0 and for oligonucleotides with variable number of TINAs. The comparison of ΔTm between PT and AD and between Tm for oligonucleotides with different number and placement of TINAs is thereby not influenced by the faster ramp rate. We therefore believe that it is appropriate to compare ΔTm and to give design rules for placement of TINA based on the presented data collection.
The different dissociation and annealing Tms for the TINA oligonucleotides in Table 1 are intriguing. TINA has previously been shown not only to stabilize the PT, but also to stabilize the underlying AD (1). It has been demonstrated that dissociation of PT at acidic pH can occur by (i) dissociation of the triplex strand from the PT complex and thereafter dissociation of the AD at higher temperature, (ii) a simultaneously dissociation of all three strands in the PT and (iii) in rare cases a dissociation of the AD strand of the PT followed by dissociation of the parallel duplex at higher temperature (25–30). At pH 5.0 we find that TINA modified TFOs can stabilize the whole PT leading to a dissociation Tm above the AD Tm meaning that the whole PT dissociates simultaneously. At higher pH the Tm of the TFO with TINA drops below the Tm of the underlying AD leading to dissociation of the TFO from the PT complex and thereafter dissociation at a higher temperature of the AD. Annealing of the PT is independent of pH and is based on the formation of an AD and thereafter association of the third strand to form the PT. This means that the dissociation Tms determined at pH 5.0 with TINAs in the TFO reflect a PT to single oligonucleotide dissociation, whereas the annealing Tms reflect the formation of the AD, which is necessary for the PT formation and thereby melting curve determination in the FRET system (Supplementary Table S5).
When working with PT it is necessary to ensure that PT and not parallel duplexes are being measured. In our control measurements, Tm of parallel duplex was 15.5°C lower compared to PT, which validates that the presented Tms are derived from PT (Supplementary Table S1). In our study we observed a lower increase in Tm when TINA was placed within the first three to four bases from the 5′-end of the TFO. This may be an effect due to the TINA, but may also be due to the fluorophores used for FRET. Incorporation of TINA may change the shape of the PT and thereby change the FRET efficiency between the ATTO495 fluorophore on the target and the ATTO590 fluorophore on the TFO. This may lead to an under-estimation of the stabilizing effect of TINA on the PT in the 5′-end of the PT.
Our determinations of the stabilizing effect of TINA in the TFO on PT are based upon a liquid phase system. We are currently developing both liquid and solid phase systems using TINA stabilized PT instead of conventional AD to increase the specificity and sensitivity of hybridization assays. Previous studies have demonstrated that DNA oligonucleotides with TINA extend the operational triplex forming pH range. Furthermore, TINA incorporation in TFOs implies reproducible positive effects on Tm and ΔTm. Based on these qualities we anticipate that TINA modified oligonucleotides for triplex capture and detection, designed accordingly to the design rules for incorporation of TINA in TFOs described in this study, will be able to improve existing clinical diagnostic assay design based upon conventional Watson–Crick AD formations.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: QuantiBact Inc.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank laboratory technician Jette Krogh Severinsen for excellent and well executed laboratory work.
REFERENCES
- 1.Filichev VV, Pedersen EB. Stable and selective formation of hoogsteen-type triplexes and duplexes using twisted intercalating nucleic acids (TINA) prepared via postsynthetic Sonogashira solid-phase coupling reactions. J. Am. Chem. Soc. 2005;127:14849–14858. doi: 10.1021/ja053645d. [DOI] [PubMed] [Google Scholar]
- 2.Fossella JA, Kim YJ, Shih H, Richards EG, Fresco JR. Relative specificities in binding of Watson-Crick base pairs by third strand residues in a DNA pyrimidine triplex motif. Nucleic Acids Res. 1993;21:4511–4515. doi: 10.1093/nar/21.19.4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jain A, Wang G, Vasquez KM. DNA triple helices: biological consequences and therapeutic potential. Biochimie. 2008;90:1117–1130. doi: 10.1016/j.biochi.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Letai AG, Palladino MA, Fromm E, Rizzo V, Fresco JR. Specificity in formation of triple-stranded nucleic acid helical complexes: studies with agarose-linked polyribonucleotide affinity columns. Biochemistry. 1988;27:9108–9112. doi: 10.1021/bi00426a007. [DOI] [PubMed] [Google Scholar]
- 5.Beal PA, Dervan PB. Second structural motif for recognition of DNA by oligonucleotide-directed triple-helix formation. Science. 1991;251:1360–1363. doi: 10.1126/science.2003222. [DOI] [PubMed] [Google Scholar]
- 6.Han H, Dervan PB. Sequence-specific recognition of double helical RNA and RNA:DNA by triple helix formation. Proc. Natl Acad. Sci. USA. 1993;90:3806–3810. doi: 10.1073/pnas.90.9.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Escude C, Francois JC, Sun JS, Ott G, Sprinzl M, Garestier T, Helene C. Stability of triple helices containing RNA and DNA strands: experimental and molecular modeling studies. Nucleic Acids Res. 1993;21:5547–5553. doi: 10.1093/nar/21.24.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts RW, Crothers DM. Stability and properties of double and triple helices – dramatic effects of Rna Or Dna backbone composition. Science. 1992;258:1463–1466. doi: 10.1126/science.1279808. [DOI] [PubMed] [Google Scholar]
- 9.Lee JS, Johnson DA, Morgan AR. Complexes formed by (pyrimidine)n. (purine)n DNAs on lowering the pH are three-stranded. Nucleic Acids Res. 1979;6:3073–3091. doi: 10.1093/nar/6.9.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rajagopal P, Feigon J. Triple-strand formation in the homopurine:homopyrimidine DNA oligonucleotides d(G-A)4 and d(T-C)4. Nature. 1989;339:637–640. doi: 10.1038/339637a0. [DOI] [PubMed] [Google Scholar]
- 11.Sundquist WI, Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989;342:825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- 12.Boutorine AS, Doluca O, Filichev VV. Optimization of the sequence of twisted intercalating nucleic acids (TINA) forming triple helix with the polypurine tract of the proviral HIV DNA. Nucleic Acids Symp. Ser. (Oxf) 2009;53:139–140. doi: 10.1093/nass/nrp070. [DOI] [PubMed] [Google Scholar]
- 13.Filichev VV, Nielsen MC, Bomholt N, Jessen CH, Pedersen EB. High thermal stability of 5′-5′-linked alternate hoogsteen triplexes at physiological pH. Angew. Chem. Int. Ed Engl. 2006;45:5311–5315. doi: 10.1002/anie.200601127. [DOI] [PubMed] [Google Scholar]
- 14.Filichev VV, Gaber H, Olsen TR, Jorgensen PT, Jessen CH, Pedersen EB. Twisted intercalating nucleic acids – intercalator influence on parallel triplex Stabilities. Eur. J. Organic Chem. 2006:3960–3968. [Google Scholar]
- 15.Paramasivam M, Cogoi S, Filichev VV, Bomholt N, Pedersen EB, Xodo LE. Purine twisted-intercalating nucleic acids: a new class of anti-gene molecules resistant to potassium-induced aggregation. Nucleic Acids Res. 2008;36:3494–3507. doi: 10.1093/nar/gkn242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cogoi S, Paramasivam M, Filichev V, Geci I, Pedersen EB, Xodo LE. Identification of a new G-quadruplex motif in the KRAS promoter and design of pyrene-modified G4-decoys with antiproliferative activity in pancreatic cancer cells. J. Med. Chem. 2009;52:564–568. doi: 10.1021/jm800874t. [DOI] [PubMed] [Google Scholar]
- 17.Géci I, Filichev VV, Pedersen EB. Stabilization of parallel triplexes by twisted intercalating nucleic acids (TINAs) incorporating 1,2,3-triazole units and prepared by microwave-accelerated click chemistry. Chem. Eur. J. 2007;13:6379–6386. doi: 10.1002/chem.200700053. [DOI] [PubMed] [Google Scholar]
- 18.Bomholt N, Ostnan AMA, Pedersen EB. High physiological thermal triplex stability optimization of twisted intercalating nucleic acids (TINA) Organic Biomol. Chem. 2008;6:3714–3722. doi: 10.1039/b808564a. [DOI] [PubMed] [Google Scholar]
- 19.Geci I, Filichev VV, Pedersen EB. Synthesis of twisted intercalating nucleic acids possessing acridine derivatives. Thermal stability studies. Bioconjug. Chem. 2006;17:950–957. doi: 10.1021/bc060058o. [DOI] [PubMed] [Google Scholar]
- 20.Osman AMA, Jorgensen PT, Bomholt N, Pedersen EB. Using an aryl phenanthroimidazole moiety as a conjugated flexible intercalator to improve the hybridization efficiency of a triplex-forming oligonucleotide. Bioorganic Med. Chem. 2008;16:9937–9947. doi: 10.1016/j.bmc.2008.10.037. [DOI] [PubMed] [Google Scholar]
- 21.Darby RA, Sollogoub M, McKeen C, Brown L, Risitano A, Brown N, Barton C, Brown T, Fox KR. High throughput measurement of duplex, triplex and quadruplex melting curves using molecular beacons and a LightCycler. Nucleic Acids Res. 2002;30:e39. doi: 10.1093/nar/30.9.e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider UV, Severinsen JK, Geci I, Okkels LM, Johnk N, Mikkelsen ND, Klinge T, Pedersen EB, Westh H, Lisby G. A novel FRET pair for detection of parallel DNA triplexes by the LightCycler. BMC. Biotechnol. 2010;10:4. doi: 10.1186/1472-6750-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rusling DA, Powers VE, Ranasinghe RT, Wang Y, Osborne SD, Brown T, Fox KR. Four base recognition by triplex-forming oligonucleotides at physiological pH. Nucleic Acids Res. 2005;33:3025–3032. doi: 10.1093/nar/gki625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rusling DA, Brown T, Fox KR. DNA triple-helix formation at target sites containing duplex mismatches. Biophys. Chem. 2006;123:134–140. doi: 10.1016/j.bpc.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 25.Bhaumik SR, Chary KV, Govil G, Liu K, Miles HT. NMR characterisation of a triple stranded complex formed by homo-purine and homo-pyrimidine DNA strands at 1:1 molar ratio and acidic pH. Nucleic Acids Res. 1995;23:4116–4121. doi: 10.1093/nar/23.20.4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hashem GM, Wen JD, Do Q, Gray DM. Evidence from CD spectra and melting temperatures for stable Hoogsteen-paired oligomer duplexes derived from DNA and hybrid triplexes. Nucleic Acids Res. 1999;27:3371–3379. doi: 10.1093/nar/27.16.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavelle L, Fresco JR. UV spectroscopic identification and thermodynamic analysis of protonated third strand deoxycytidine residues at neutrality in the triplex d(C(+)-T)6:[d(A-G)6.d(C-T)6]; evidence for a proton switch. Nucleic Acids Res. 1995;23:2692–2705. doi: 10.1093/nar/23.14.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scaria PV, Shafer RH. Calorimetric analysis of triple helices targeted to the d(G3A4G3).d(C3T4C3) duplex. Biochemistry. 1996;35:10985–10994. doi: 10.1021/bi960966g. [DOI] [PubMed] [Google Scholar]
- 29.Umemoto K, Sarma MH, Gupta G, Luo J, Sarma RH. Structure and stability of a Dna triple helix in solution – Nmr-studies on D(T)6.D(A)6.D(T)6 and its complex with a minor groove binding-drug. J. Am. Chem. Soc. 1990;112:4539–4545. [Google Scholar]
- 30.Wan C, Guo X, Liu Z, Liu S. Studies of the intermolecular DNA triplexes of C+.GC and T.AT triplets by electrospray ionization Fourier-transform ion cyclotron resonance mass spectrometry. J. Mass Spectrom. 2008;43:164–172. doi: 10.1002/jms.1277. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.