Abstract
RNA interference (RNAi) has been revolutionary for the specific inhibition of gene expression. However, the application of RNAi has been hampered by the fact that many siRNAs induce dose-dependent unwanted secondary effects. Therefore, new methods to increase inhibition of gene expression with low doses of inhibitors are required. We have tested the combination of RNAi and U1i (U1 small nuclear RNA—snRNA—interference). U1i is based on U1 inhibitors (U1in), U1 snRNA molecules modified to target a pre-mRNA and inhibit its gene expression by blocking nuclear polyadenylation. The combination of RNAi and U1i resulted in stronger inhibition of reporter or endogenous genes than that obtained using either of the techniques alone. The increased inhibition observed is stable over time and allows higher inhibition than the best obtained with either of the inhibitors alone even with decreased doses of the inhibitors. We believe that the combination of RNAi and U1i will be of interest when higher inhibition is required or when potent inhibitors are not available. Also, the combination of these techniques would allow functional inhibition with a decreased dose of inhibitors, avoiding toxicity due to dose-dependent unwanted effects.
INTRODUCTION
Inhibition of gene expression has been successfully applied for functional studies and offers great promise for therapeutic applications. In most laboratories, the expression of the gene of interest is inhibited using RNA interference (RNAi). The inhibitors that mediate RNAi are double-stranded small RNA molecules called small interfering RNAs (siRNAs). For RNAi, exogenous siRNAs are coupled to the RNA-induced silencing complex (RISC) which induces target mRNA cleavage and as a result, target gene expression is inhibited (1). RISC can also load endogenous small non-coding RNAs called microRNAs (miRNAs). miRNAs are transcribed in the nucleus as long primary transcripts or pri-miRNAs which are cleaved into pre-miRNAs, imperfectly paired stem–loop miRNA precursors (2). pre-miRNAs are then exported to the cytoplasm where they bind Dicer, which processes pre-miRNAs into mature double-stranded miRNAs recognized by RISC (3,4). The RISC retains single-stranded mature cellular miRNAs, which can usually bind to their targets with non-perfect complementarity. Binding of the ‘seed’ sequence formed by nucleotides 2–7 of the 5′-end of the miRNA is sufficient for target recognition (5). miRNA binding to the target induces a RISC-mediated translation inhibition and/or mRNA destabilization (6). The cellular silencing machinery can be also used to express siRNAs from exogenous genes. Genes can be designed to transcribe siRNA precursor molecules similar to pre-miRNAs, called small hairpin RNAs (shRNAs) (7). After transcription, shRNAs follow a similar pathway to miRNAs and are loaded into RISC, where they behave akin to synthetic siRNAs leading to target mRNA cleavage.
RNAi is not as specific as originally thought. Under certain circumstances, functional siRNAs can lead to unwanted effects. The three major reasons for this are: (i) some siRNA molecules are sensed by the cell leading to activation of the interferon response (8,9); (ii) overexpression of siRNAs can saturate the cellular silencing machinery which is required to control the expression of many genes involved in essential cellular processes (10); and (iii) many siRNAs are not specific for their target and can act as miRNAs to inhibit the expression of other genes which could be required for proper cell functioning (11,12). As unwanted effects are dose-dependent (11,12), it is essential to develop protocols that improve siRNA performance or allow the efficient dose of siRNA to be reduced to a minimum thus avoiding unwanted effects.
Gene expression can also be inhibited with U1 small nuclear RNA—U1 snRNA—interference (U1i) (13,14). U1 snRNA coupled to U1-70K and other cellular proteins forms a mature nuclear ribonucleoprotein (U1 snRNP), which is a well-studied constitutive splicing factor (15). U1 snRNP functions in splicing by binding the pre-mRNA via a base pairing interaction between nucleotides 2–11 of U1 snRNA and the 5′-splice site sequence. Aside from this splicing function, U1 snRNP can also act as a potent inhibitor of gene expression by inhibiting pre-mRNA 3′-end formation (16). When nt 2–11 of U1 snRNA bind to the 3′-end of a pre-mRNA, U1 snRNP inhibits pre-mRNA polyadenylation. The molecular mechanism that mediates this inhibition has been well-characterized. After U1 snRNP binding to the target pre-mRNA, the U1-70K component of the U1 snRNP directly inhibits polyadenylation and therefore, gene expression (17,18) (Figure 1A). Inhibited pre-mRNA is cleaved at the 3′-end but it is not polyadenylated. Without a polyA tail, the pre-mRNA fails to mature and is rapidly degraded in the nucleus leading to reduced expression.
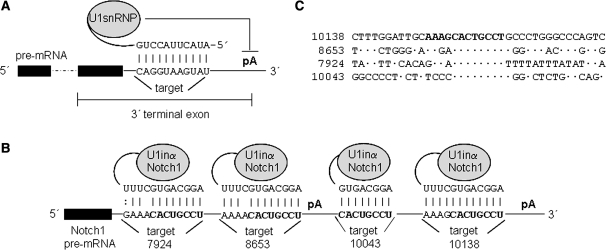
Figure 1.
Schematic of U1i. (A). When the 5′-end of endogenous U1 snRNA base pairs to a target sequence located in the 3′-terminal exon, U1 snRNP inhibits pre-mRNA polyadenylation (pA). Thus, maturation of the pre-mRNA is blocked, mRNA stability, transport to the cytoplasm, and translation are decreased and therefore gene expression is inhibited. 3′-terminal exon sequences are indicated. Intron is depicted with a dashed line. (B and C). The 5′-end of U1 snRNA has been modified to base pair Notch1 pre-mRNA resulting in U1inαNotch1. U1inαNotch1 should bind sequence CACUGCCU located four times in the Notch1 3′-UTR. Two repeats locate at positions 7924 and 8653, upstream of the first polyadenylation site. Two repeats locate at positions 10043 and 10138, upstream of an alternative polyadenylation site. An alignment of the four Notch1 target sites and neighbouring sequences is also shown.
This inhibitory activity of U1 snRNP forms the basis of U1i silencing technology. U1i requires expression of an exogenous 5′-end mutated U1 snRNA designed to base pair to the 3′-terminal exon of a target gene. The U1 snRNA inhibitor (U1in) binds target mRNA and blocks expression by hindering polyadenylation (13,14). Expression of endogenous genes has been inhibited by U1i after transient or stable transfections, thus indicating the good specificity and low toxicity of this technique (14). Expression of multiple U1ins that target different parts of the terminal exon of the same gene results in synergistic enhanced inhibition (14). U1i has been shown to be effective against HIV replication and works in animal models and thus holds great promise for knock-down studies and gene therapy applications [(19), X. Abad and P. Fortes, unpublished data]. The rules that govern U1i were first studied with reporter genes (14). U1ins are expressed from plasmids that include U1 snRNA promoter and terminator sequences. A U1in against an accessible target site located in the 3′-terminal exon of a reporter mRNA, inhibits reporter expression and does not alter the expression of other genes. However, when target sites are located outside the 3′ terminal exon or when they form secondary structures, inhibition is lost (14,17). As RNA secondary structure is difficult to predict, selection of good target sites is a challenge that generally decides the success of the technique.
RNAi and U1i affect target mRNA expression by different mechanisms and in different cell compartments. While U1i affects the nuclear metabolism of the target RNA, RNAi decreases the stability of the RNA in the cytoplasm, where the RISC complex is located. Therefore, both techniques could be combined without increasing the risk of saturation of their silencing machineries. In this study, we show that the combination of RNAi and U1i results in increased inhibition of reporter and endogenous genes. The benefits of the combination have been quantified using luciferase reporters. The results show improved inhibition when shRNAs of different strengths are expressed in combination with U1i inhibitors. This synergism between U1i and RNAi allows the dose of both or either of the inhibitors to be decreased and higher inhibition than when only one of the inhibitors is used to still be obtained. We believe that the combination of RNAi and U1i will allow successful inhibition even when good inhibitors for a specific target have not been identified. Furthermore, this combination will allow the dose of inhibitors to be decreased which may lower the risk of unwanted effects in functional studies and therapeutic applications.
MATERIALS AND METHODS
Cell lines and plasmids
HeLa and 293 cells were obtained from the ATCC and cultured in DMEM medium, supplemented with 10% FBS and 1% penicillin–streptomycin, at 37°C in a 5% CO2 atmosphere. All cell culture reagents were obtained from Gibco BRL/Life Technologies. Plasmid pRL-SV40 (Promega) was used as Renilla luciferase transfection control. pGL3-Promoter (Promega) was used as Firefly luciferase (Photinus pyralis, GenBank Accession No.M15077) expressing plasmid in Figure 3A–C. pNFκβ-Luc (NFκβ 3xLuc; Clontech Co) was used to express Firefly luciferase under NFκβ promoter (Figure 2B). pLuc-U1in (Figures 3B, 4 and 5) was cloned by ligation of base-paired oligonucleotides (Sigma) with the U1inαLuc sequence (Table 1) into the XbaI-NotI site of pGL3-Promoter. A pGem plasmid (Promega) that contains U1 snRNA gene, including promoter and termination sequences in the BamHI site was used to express wild-type U1 snRNA (pGemU1inWT) (14). This plasmid was used as a control for U1i inhibition. Plasmid expressing U1inαLuc has already been described [pGemU1inMut; (17)]. pGemU1inαNotch1, expressing U1inαNotch1, was cloned by ligation of base-paired oligonucleotides with the U1inαNotch1 sequence (Table 1) into the BclI-BglII site of pGemU1inWT. The exact sequence of the 5′-end of U1inαNotch1 is shown in Figure 1B. Ligation of the U1 sequences from pGemU1inWT and pGemU1inαNotch1 digested with EcoRI and HincII into the same sites of pMSCV-Puro (Clontech) resulted in pMSCV-U1inWT and pMSCV-U1inαNoctch, respectively. The MISSION Luciferase shRNA pLKO.1-puro vector was used to deliver all shRNAs targeting Firefly luciferase. shRNAs expressed from this plasmid are transcribed from a U6 promoter. The 5′-end of the shRNA starts with the sense strand and is followed by a CTCGAG loop, the antisense strand and UU. The sense and antisense strands have perfect complementarity and are 21-nt long. Plasmid expressing shαLuc159 is now commercially available (SHC007 Sigma). Plasmid expressing shαNotch1 was cloned by ligation of base-paired oligonucleotides with the shαNotch1 sequence (Table 1) into the HindIII-BglII site of pSuper (7). An irrelevant shRNA sequence was also cloned into the same sites as a negative control. shRNAs expressed from pSuper are transcribed from a H1 promoter, start with the sense strand followed by a TTCAAGAGA loop, the antisense strand and end with UU. The sense and antisense strands have perfect complementarity and are 19-nt long. Ligation of the shαNotch1 gene sequences from pSuper digested with NotI and XhoI and filled in with Klenow into the XhoI site of pIRESNeo (Clontech) resulted in pNeoshαNotch1. To produce pNeoU1in/shαNotch1, pNeoshαNotch1 was digested with BamHI and ligated to U1 sequences digested with BamHI from pGemU1inαNotch1. Irrelevant shRNA sequences and U1WT gene were also cloned into the same sites as negative controls. All clones were verified by sequencing in an ABI Prism 310 genetic analyzer (Applied Biosystems). Plasmid DNA was purified with a Maxiprep kit (Marlingen) before transfection.
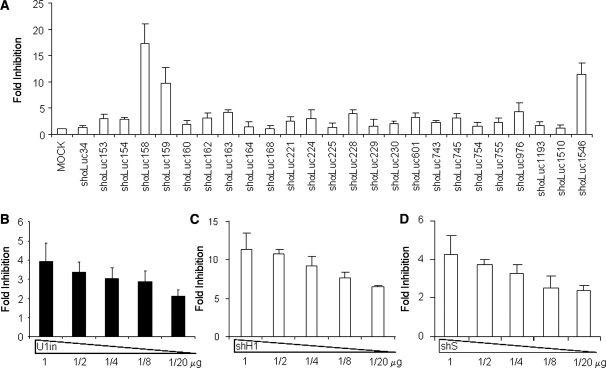
Figure 3.
Analysis of shRNAs and U1in targeting luciferase. (A). Identification of shRNAs targeting Firefly luciferase. Luciferase activity was measured in HeLa cells co-transfected with a plasmid expressing Firefly luciferase and a control plasmid or plasmids expressing the indicated shRNA. The shRNAs are named according to the position of the first nucleotide they bind to in luciferase mRNA. (B–D) Fold inhibition of luciferase activity with decreasing doses of inhibitors. Luciferase activity was measured in HeLa cells co-transfected with a plasmid that expresses Firefly luciferase and a control plasmid or the indicated amount of plasmids expressing luciferase inhibitors (see ‘Materials and Methods’ section for details). Plasmids used express U1inαLuc (B), shαLuc1546 (shH1, C) and shαLuc163 (shS, D). The fold inhibition is indicated for each inhibitor. Error bars are standard deviations of nine different experiments. Note that the scale is different for each figure.
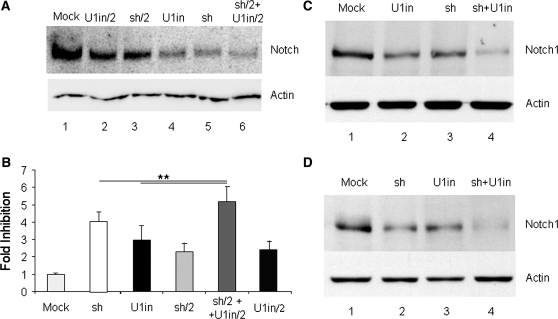
Figure 2.
Inhibition of endogenous Notch1 expression and function with RNAi and U1i. (A). Inhibition of endogenous Notch1 expression. HeLa cells were transfected with a control plasmid (mock), with plasmids expressing U1inαNotch1 (U1in) or shαNotch1 (sh) at the highest dose, or half of the highest dose (X/2) or with the combination of plasmids expressing U1inαNotch1 and siαNotch1 (sh/2 + U1in/2). HeLa cell lysates were collected 3 days after transfection and Notch1 (upper box) and actin (lower box) expression was evaluated in two independent western blots. (B). Functional assay for Notch1 gene silencing. 293 cells were co-transfected with pNFκβ-Luc, Renilla luciferase control and the plasmids described in A. Luciferase activity was evaluated 3 days after transfection and the data obtained were corrected for equal transfection efficiency. Results were plotted to indicate the fold inhibition obtained in each case. Fold inhibition was calculated as the ratio of the luciferase activity obtained in mock transfected cells versus the luciferase activity obtained in cells expressing the indicated inhibitor. Error bars denote standard deviations of three independent experiments. (C and D) Analysis of the silencing of endogenous Notch1 over time. (C) Pools of clones of HeLa cells stably transfected with a control plasmid (mock), or plasmids expressing U1inαNotch1(U1in), shαNotch1 (sh) or both (sh + U1in) were used to evaluate Notch1 (upper box) and actin (lower box) expression by western blot. (D) Representative clones of HeLa cells expressing U1WT or U1inαNotch1 were transfected with a control plasmid (mock and U1in, respectively) or with a plasmid expressing shαNotch1 (sh and sh + U1in, respectively). Stably transfected pools of clones were used to determine Notch1 (upper box) or actin (lower box) expression by western blot.
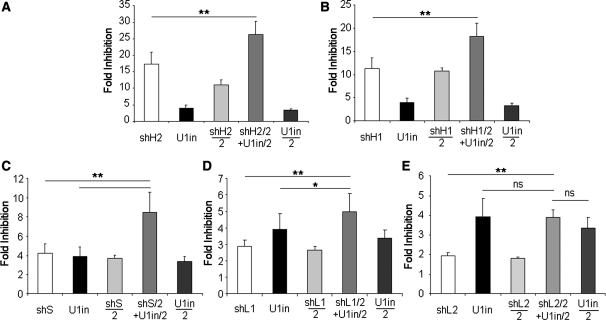
Figure 4.
Increased inhibition is observed when U1in is combined with potent, standard and poor shRNA inhibitors. Luciferase activity was measured in HeLa cells transfected with a plasmid that expresses Firefly luciferase and a control plasmid or plasmids expressing luciferase inhibitors. 1 µg or 1/2 µg (X/2) of plasmid expressing the inhibitor was used for transfection. Inhibitors were U1inαLuc, shH2 (shαLuc158, A), shH1 (shαLuc1546, B), shS (shαLuc163, C), shL1 (shαLuc154, D) and shL2 (shαLuc230, E) alone or a combination of U1inαLuc and shαLuc (shX/2 + U1in/2). The fold inhibition is indicated for each case. Error bars are standard deviations of nine different experiments. Note that the scale is different for each figure.
Figure 5.
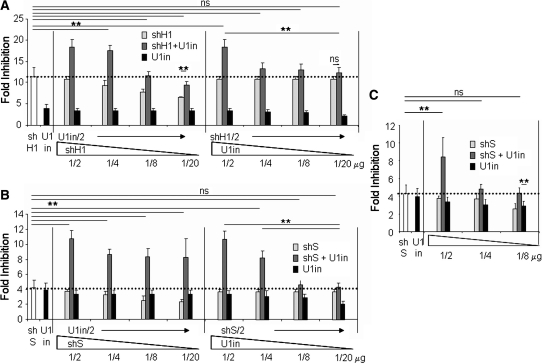
Increased inhibition observed by combination of U1i and RNAi allows the effective dose of inhibitors to be decreased. Luciferase activity was measured in HeLa cells transfected with a plasmid that expresses Firefly luciferase and a control plasmid or the amount of plasmids expressing luciferase inhibitors indicated at the bottom of the graph. Inhibitors were U1inαLuc, and shH1 (A) and shS (B and C) alone or a combination of U1inαLuc and shαLuc. The fold inhibition is indicated for each case. A horizontal dotted line serves to mark the best fold inhibition obtained with the best inhibitor (either a shRNA or a U1in) on its own. Error bars show standard deviations of nine different experiments. Note that the scale is different for each figure.
Table 1.
List of inhibitors used in this study
| Target gene | Name | Target sequence | Position |
|---|---|---|---|
| Notch1 | U1inαNotch1 | CACUGCCU | 7924, 8653, 10 043 and 10 138 |
| shαNotch1 | ACACCAACGUGGUCUUCAA | 4878 | |
| Firefly (P. pyralis) luciferase | U1inαLuc | CCUGCCAUGGAACUA | Exogenous 3′-UTR |
| shαLuc34 | GCGCCAUUCUAUCCGCUGGAA | 34 | |
| shαLuc153 | CACUUACGCUGAGUACUUCGA | 153 | |
| shαLuc154 (L1) | ACUUACGCUGAGUACUUCGAA | 154 | |
| shαLuc158 (H2) | ACGCUGAGUACUUCGAAAUGU | 158 | |
| shαLuc159 | CGCUGAGUACUUCGAAAUGUC | 159 | |
| shαLuc160 | GCUGAGUACUUCGAAAUGUCC | 160 | |
| shαLuc162 | UGAGUACUUCGAAAUGUCCGU | 162 | |
| shαLuc163 (S) | GAGUACUUCGAAAUGUCCGUU | 163 | |
| shαLuc164 | AGUACUUCGAAAUGUCCGUUC | 164 | |
| shαLuc168 | CUUCGAAAUGUCCGUUCGGUU | 168 | |
| shαLuc221 | CAAAUCACAGAAUCGUCGUAU | 221 | |
| shαLuc224 | AUCACAGAAUCGUCGUAUGCA | 224 | |
| shαLuc225 | UCACAGAAUCGUCGUAUGCAG | 225 | |
| shαLuc228 | CAGAAUCGUCGUAUGCAGUGA | 228 | |
| shαLuc229 | AGAAUCGUCGUAUGCAGUGAA | 229 | |
| shαLuc230 (L2) | GAAUCGUCGUAUGCAGUGAAA | 230 | |
| shαLuc601 | UCUACUGGUCUGCCUAAAGGU | 601 | |
| shαLuc743 | GAAUGUUUACUACACUCGGAU | 743 | |
| shαLuc745 | AUGUUUACUACACUCGGAUAU | 745 | |
| shαLuc754 | ACACUCGGAUAUUUGAUAUGU | 754 | |
| shαLuc755 | CACUCGGAUAUUUGAUAUGUG | 755 | |
| shαLuc976 | GCGGUUGCCAAGAGGUUCCAU | 976 | |
| shαLuc1193 | UGUCCGGUUAUGUAAACAAUC | 1193 | |
| shαLuc1510 | AGUCAAGUAACAACCGCGAAA | 1510 | |
| shαLuc1546 (H1) | GUUGUGUUUGUGGACGAAGUA | 1546 |
Position and sequence of the target is also indicated.
Cell transfections
To evaluate Notch1 expression, HeLa cells were transfected with Lipofectamine 2000 (Invitrogen) and 4 μg of the indicated plasmid per 5 × 105 cells, following the recommendations of the supplier. Seventy-four hours after transfection, cells were harvested and western blot analysis was performed. To test the effect of inhibitors on luciferase expression, HeLa or 293 cells were transfected with calcium phosphate as described (20) and were harvested 48-h post-transfection. A total of 150 000 cells were transfected with 0.75 µg of the plasmid expressing Firefly luciferase, 0.25 µg of pRL-SV40 (Promega) and a total of 1 µg of plasmids expressing the inhibitors, the control inhibitor or a combination of both (0.5 µg of plasmid expressing one inhibitor +0.5 µg of plasmid expressing the control). Note that for the combination of RNAi and U1i, 0.5 µg of a plasmid expressing the shRNA inhibitor was mixed with 0.5 µg of a plasmid expressing the U1in. When indicated, a dilution of one or both plasmids expressing the inhibitors was used. In all cases, the total amount of plasmid transfected was constant and transfection efficiency was controlled by Renilla luciferase analysis. The amount of plasmid transfected was chosen because (i) this amount results in a transfection efficiency close to 100% with non-detectable toxicity; (ii) transfection of increasing amounts (from 0.1 to 2 µg) of a luciferase reporter plasmid results in a linear response of luciferase expression; and (iii) transfection of increasing amounts (from 0.05 to 1 µg) of plasmids expressing the inhibitors results in increased inhibition, indicating that the system is not oversaturated (Figure 3B–D). To obtain stably transfected cell lines, HeLa cells were transfected with 2 µg of linear pMSCV-U1inαNoctch, pNeoU1in/shαNotch1 or the corresponding control plasmids and cells were selected for longer than 1 month with 8 µg/ml of puromycin or 1400 µg/ml of G418, respectively. Cells stably expressing pMSCV-U1inαNoctch or pMSCV-U1inWT were also transfected with 2 µg of linear pNeoshαNotch1 or the corresponding control plasmids and cells were selected with G418 as indicated before.
Western blot
Whole cell lysates used for western blotting were prepared by lysis of cells in RIPA buffer, centrifugation for 60 min at 15 000 r.p.m., and collection of the supernatant. Proteins were separated by SDS–PAGE and transferred to an Immobilon-P nylon membrane (Millipore) (21). Notch1 protein was bound by antihNotch-1 antibody (sc-6014-R; Santa Cruz) diluted 1/500 and actin by antiβ-Actin antibody (A2066; Sigma) diluted 1/1000. Horseradish peroxidase-conjugated secondary antibodies (1:5000) were from Cell Signalling (#7074). Western blots were revealed by chemiluminescence (NEL 101001; Perkin Elmer) and quantified with Image Quant TL Software (GE Lifesciences).
Luciferase activity measurements
Renilla and Firefly luciferase activities were measured using the Dual Luciferase System (Promega) as previously described (22) in a Berthold Luminometer (Lumat LB 9507). The values obtained for Firefly luciferase were corrected for equal transfection efficiency with Renilla luciferase activity. The fold inhibition of each inhibitor was calculated as the ratio of the luciferase activity obtained in mock transfected cells versus the luciferase activity obtained in cells expressing the inhibitor studied. Mock transfected cells express a matched reference plasmid used as negative control expressing either U1inWT or the shRNA of irrelevant sequence. By definition, the reference plasmid has an inhibitory activity set to 1.0.
Statistical analysis
Data are expressed as means ± SD. Statistical analysis was performed with Student’s t-test using the SPSS software package (SPSS INC, Chicago, IL, USA). Statistically significant differences (P < 0.05) are indicated with a star.
RESULTS AND DISCUSSION
Combination of RNAi and U1i results in stronger inhibition of the expression of endogenous genes
To test the combination of RNAi and U1i in the inhibition of the expression of a target gene, we first designed siRNAs and U1in inhibitors against an endogenous gene. We decided to target human Notch1, a gene of therapeutic interest (23). Notch1 is a transmembrane receptor involved in transduction of developmental signals in most eukaryotes. Notch1 ligand binding induces the proteolytic cleavage of the intracellular domain of Notch1, which translocates to the nucleus and activates gene expression. Overexpression of the intracellular domain of Notch1 can induce breast cancer and T-cell transformation that results in T-cell acute lymphoblastic leukemias and lymphomas (23,24). Therefore, inhibition of Notch1 could have therapeutic applications for the treatment of several tumours. Aberrant Notch signalling can also trigger signals implicated in promoting autoimmune diseases. However, the cellular response to Notch signalling can be variable depending on the cell type and activation context. Thus, Notch signalling can also induce responses that are critical to negatively regulating immune responses or suppressing carcinogenesis, for instance in neuroendocrine tumours (25,26).
We designed a U1in targeting the sequence CACUGCCU in Notch1 pre-mRNA (Figure 1B, U1inαNotch1, Table 1). We chose this sequence because it is repeated four times in Notch1 3′-terminal exon. Two repeats locate before the first polyadenylation site and the other two before an alternative polyadenylation signal. Homology among the four target sites is not extended further (Figure 1C). We designed four siRNAs targeting Notch1. Three siRNAs were designed following recommendations published by Reynolds et al. (27). These authors list requirements that functional siRNAs must fulfil and describe an algorithm that helps to design potent siRNA inhibitors. According to their results, functional siRNAs have a score >6 out of a maximum of 10. Thus, we designed three siRNAs targeting Notch1 that have a score of 8 or 9. These siRNAs should also be potent inhibitors according to Matveeva et al. [Supplementary Table S1 (28)]. We also used an additional siRNA sequence with a score of 3 as calculated following Reynolds et al. (27), as this has been reported to be an efficient inhibitor of Notch1 expression (29). A pSuper backbone was used to construct plasmids that express the four designed siRNAs (7). When these plasmids were transfected into HeLa cells down-regulation of Notch1 protein was observed with the vector expressing the low score siRNA already published. However, only one of the plasmids expressing a high score siRNA was able to down-regulate Notch1 efficiently as detected by western blot analysis (Table 1, Figure 2A). Transfection of the plasmid expressing U1inαNotch1 into HeLa cells also resulted in down-regulation of Notch1 protein. A 50% reduction of the amount of transfected plasmid expressing either of the inhibitors resulted in a less efficient down-regulation of Notch1 expression. This indicated that inhibition was not saturated in cells transfected with half a dose of the plasmids that express the inhibitors. Quantitation of the western blot indicated that both inhibitors down-regulate Notch1 expression to similar levels under the conditions used (Supplementary Figure S1). Surprisingly, co-transfection of half of the dose of the plasmids expressing the U1inαNotch1 and the shαNotch1 resulted in a strong down-regulation of Notch1. Quantitation of the western blot indicated that the co-expression of U1inαNotch1 and shαNotch1 resulted in stronger inhibition of Notch1 than the expression of the highest dose of either of the inhibitors on their own.
We also wanted to evaluate the functional relevance of Notch1 down-regulation. Notch1 expression increases NFκβ activity by facilitating its nuclear retention (30), resulting in increased expression from NFκβ specific promoters. Thus, 293 cells were transfected with a plasmid that expresses Firefly luciferase under an NFκβ specific promoter (pNFκβ-Luc) and a plasmid that expresses Renilla luciferase independently of Notch1 expression, as an internal control. We also co-transfected a control plasmid, plasmids expressing U1inαNotch1 or shαNotch1 at the highest dose or half dose and the combination of plasmids expressing U1inαNotch1 and shαNotch1. Luciferase activity was evaluated 2 days after transfection and the data obtained were corrected with Renilla luciferase values. Luciferase measurements allowed evaluation of Notch1 function with a wider range of quantification than western blots. The results showed that all inhibitors decreased Firefly luciferase expression (Figure 2B). The fold inhibition was similar in cells transfected with similar doses of plasmids expressing U1inαNotch1 or shαNotch1. As observed before, co-transfection of half of the dose of the plasmids expressing the U1inαNotch1 and the shαNotch1 resulted in the highest fold inhibition.
To address the stability of Notch1 silencing over time we used cells displaying long-term expression of the inhibitors. Two sets of experiments were performed. Notch1 expression was first evaluated in pools of HeLa clones which stably express a control plasmid or plasmids expressing U1inαNotch1, shαNotch1 or both. Stability of expression was achieved as the pools of clones were selected and amplified for longer than 1 month. The results showed that the strongest decrease in Notch1 expression was observed in cells expressing both U1inαNotch1 and shαNotch1 (Figure 2C). Furthermore, we performed a similar experiment with transfections in tandem. We first isolated clones of HeLa cells stably transfected with a plasmid expressing U1inαNotch1 or U1WT, as a control. Then a control clone and a representative clone expressing U1inαNotch1 were transfected with a control plasmid or a plasmid expressing shαNotch1. Pools of clones that express these plasmids stably were selected and amplified for longer than 1 month. Notch1 expression was evaluated by western blot in extracts isolated from these clones. The results showed that the strongest inhibition of Notch1 was observed in cells stably expressing both U1inαNotch1 and shαNotch1 (Figure 2D).
Identification of U1in and siRNA inhibitors targeting luciferase expression
We wanted to evaluate the inhibition obtained by the combination of U1i and RNAi in a more quantitative and direct manner. Measurement of luciferase activity is very sensitive and allows quantification within a broad linear range. Therefore, we decided to design novel inhibitors that target Firefly luciferase based on U1i and RNAi. The siRNAs targeting Firefly luciferase were selected using the algorithm described by the RNAi Consortium (TRC). All possible siRNAs of 21-mer derived from Firefly luciferase mRNA sequence were rewarded for having the weakest base pairing at the 3′-end and the strongest at the 5′-end, for a GC content between 30 and 50% and for the presence of AT base pairs at positions 6–11. Candidates were penalized when they contained a repeat of four bases in a row, an AA start and internal loops. The penalties and rewards are specified at http://www.broad.mit.edu/science/projects/rnai-consortium/trc-shrna-design-process. We then chose and evaluated the 25 best scoring candidates (Table 1). Most of these siRNAs should also be potent inhibitors according to Matveeva et al. [Supplementary Table S1 (28)]. A pLKO.1 backbone was used to construct plasmids that express the 25 siRNAs from a precursor shRNA. These shRNAs were named according to the position of the first nucleotide they bind to in Firefly luciferase mRNA. Three hot spots targeted by the best predicted shRNAs were detected: 9 shRNAs target sequences from nucleotides 153–189, 6 shRNAs nucleotides 221–251 and 4 positions 743–776. The remaining 6 shRNAs base pair to sequences that were far from targets of other selected shRNAs.
To evaluate Firefly luciferase down-regulation by the selected shRNAs, HeLa cells were co-transfected with pGL3-Promoter, which expresses Firefly luciferase, and control plasmids or plasmids expressing each of the shRNAs to be assayed. A plasmid that expresses Renilla luciferase was also co-transfected as an internal control. Cells were collected at 48-h post-transfection and luciferase activity was evaluated in cell extracts. Renilla luciferase activity was similar in all cases, suggesting comparable transfection efficiency in all cases and specificity of the shRNAs for Firefly luciferase. The fold inhibition of each shRNA was calculated as the ratio of the luciferase activity obtained in mock transfected cells versus the luciferase activity obtained in cells expressing the inhibitor studied (Figure 3A). Three shRNAs were very efficient as they showed inhibition of 10–20 fold, which corresponds to a decrease in expression of over 90% (Supplementary Table S1). Three shRNAs were standard as they showed a 3-5 fold inhibition, which represents a 70–80% decrease in expression. Most of the shRNAs tested (19/25) were the least effective as inhibition was <3-fold, a down-regulation <70%. We were surprised by the low success of our shRNAs. These shRNAs should express a functional siRNA core, given that the siRNA sequence has been designed using algorithms built from the analysis of a large number of effective and ineffective siRNAs and a better understanding of the molecular mechanisms that drive RNAi (27,31). Also, our shRNA design parameters, such as stem length, core placement and loop selection have been successfully used by many others (see ‘Materials and Methods’ section for details). We cannot rule out that some genes such as luciferase could be less sensitive to inhibition than others.
Five shRNAs were selected for further studies: two efficient shRNAs, shαLuc1546 and shαLuc158, that target luciferase with high potency and which, for simplicity, were renamed shH1 and shH2, respectively; one standard shRNA, shαLuc163, renamed shS, and two shRNAs with low potency, shαLuc154 and shαLuc230, renamed shL1 and shL2.
Instead of designing a novel U1in targeting Firefly luciferase, we introduced into this gene a target sequence for U1inMut. U1inMut has been previously used to target an exogenous sequence in the 3′-UTR of Renilla luciferase mRNA resulting in a 10–20 fold decrease in Renilla luciferase activity (14). Similarly, U1inMut decreased the expression from Firefly luciferase gene containing the U1inMut target (pLuc-U1in), but the inhibition was only 4-fold (Figure 3B). The difference in the inhibition obtained with Renilla and Firefly luciferases could reflect the effect of target neighbour sequences. We consider that a 4-fold inhibition of luciferase was ideal in our experiments as it would allow a good quantification of increased inhibitions when U1i and RNAi based luciferase inhibitors are combined. In this study, we decided to rename U1inMut as U1inαLuc, for clarity (Table 1).
Fold inhibition of Firefly luciferase expression decreased when the amount of transfected plasmid expressing the shRNAs or U1in targeting Firefly luciferase was decreased (Figures 3B–D). This result was obtained when both efficient (shH1, Figure 3C) or standard (shS, fig 3D) shRNAs were used. This is in line with previous studies that have reported that lower doses of inhibitors have reduced secondary effects but result in weaker inhibition (11,12).
Combination of shRNAs and U1in targeting luciferase results in stronger inhibition of luciferase expression
HeLa cells were co-transfected as before with pLuc-U1in and control plasmids or plasmids expressing the different luciferase inhibitors alone or in combination. Fold inhibition of Firefly luciferase expression in the presence of shRNAs targeting Firefly luciferase was identical when pLuc-U1in or pGL3-Promoter plasmids were used, as both plasmids only differ in the few nucleotides that comprise the U1in target sequence (compare Figures 3 and 4). As indicated above, when we transfected 1/2 µg of the plasmids expressing the inhibitors, the inhibition of Firefly luciferase expression was lower than when 1 µg of these plasmids was used for the transfection (Figure 4). However, when 1/2 µg of the plasmids expressing the shRNAαLuc was co-transfected with 1/2 µg of the plasmid expressing the U1inαLuc, Firefly luciferase expression was most strongly inhibited. The inhibition of luciferase expression when shRNAs and U1in targeting luciferase were combined was higher than when either of the inhibitors was used alone at the highest possible dose in our experimental settings (1 µg). This statistically significant increased inhibition obtained by combining RNAi and U1i was observed with shRNAs that target luciferase with high potency (shH2, Figure 4A and shH1, Figure 4B), with modest potency (shS, Figure 4C) and with low potency (shL1, Figure 4D), but not upon combination of U1in with shL2, which on its own only inhibits <2-fold (Figure 4E). These results suggest that the combination of both techniques could be necessary to achieve functional inhibition of some genes, such as those that are specially difficult to target. Furthermore, the silencing efficiency correlates with the efficiency of the shRNA used. Standard or less-efficient shRNAs combined with U1in can only increase inhibition from 3–4 fold to 5–8 fold (Figures 4C and D). However, combination with U1i can increase silencing efficiency of potent shRNAs from 10–20 fold to 15–30 fold (Figure 4A and B). A synergy index was calculated to evaluate whether the increased inhibition obtained by combination of RNAi and U1i represents a synergic effect (Supplementary Table S2). The synergy index shows that co-expression of U1in and shRNAs of high potency and modest potency results in a synergistic inhibition of luciferase expression.
Increased inhibition was not observed when different shRNAs were combined. Combination of a potent and a standard shRNA (Supplementary Figure S2A), and standard and a less-efficient shRNA (Supplementary Figure S2B) or two less-efficient shRNAs (Supplementary Figure S2C) resulted in fold inhibition similar to that obtained with the more potent inhibitor used in the combination. Calculation of the synergy index shows no synergism in these cases (Supplementary Table S2). This suggests that the parallel application of two small RNAs acting at different positions of a target mRNA is not sufficient to result in synergistic inhibition. Therefore, the synergism that we observe on combining RNAi and U1i is the result of the combination of these two different knock-down mechanisms.
Synergism between RNAi and U1i allows the effective dose of inhibitor to be decreased
Decreasing the dose of inhibitor leads to a decrease in secondary effects but also to a decrease in the inhibition (Figure 3B–D) (11,12). Therefore, it would be interesting to analyse whether strong inhibitions can still be obtained when decreased doses of U1in and shRNA are used in combination. To address this question, we first used decreasing doses of one of the inhibitors while maintaining the amount of the second inhibitor constant. Doses tested were 1/2, 1/4, 1/8 and 1/20 µg of plasmid in the transfection. Initially, we analysed the combination of the potent shH1 with U1inαLuc. When we decreased shH1 maintaining U1inαLuc constant, the synergistic inhibition decreased gradually (Figure 5A, Supplementary Table S2). The combination of 1/2 µg of the plasmid expressing U1inαLuc with 1/8 or 1/20 µg of the plasmid expressing shH1, resulted in inhibitions of luciferase that were not significantly different to those obtained when 1 µg of the plasmid expressing shH1 was transfected alone, but were higher than those obtained when only 1/2 µg of the plasmid expressing shH1 was transfected. When we maintained shH1 constant, a decrease in the plasmid expressing U1inαLuc from 1/2 to 1/4 µg caused the highest decrease in the synergistic inhibition. The combination of 1/2 µg of the plasmid expressing shH1 with 1/20 µg of the plasmid expressing U1inαLuc resulted in a inhibition of luciferase that was not significantly different to that obtained when 1/2 µg of the plasmid expressing shH1 was transfected alone. Therefore, in this case, a decrease in the amount of U1in led to a large reduction in the synergistic effect.
When we combined dilutions of the plasmids expressing the standard shS or U1inαLuc, we also observed that lower doses of either of the inhibitors could be used that still result in potent inhibitions (Figure 5B). However, in this case lowering the dose of shS had less impact on the strength of the synergistic inhibition than when shH1 was used (Supplementary Table S2). The combination of 1/2 µg of the plasmid expressing U1inαLuc with 1/8 or 1/20 µg of the plasmid expressing shS, resulted in inhibitions of luciferase that were significantly higher than those obtained when 1 µg of the plasmid expressing shS was transfected alone. This suggests to us that, as indicated above, the silencing efficiency of the synergy correlates with the efficiency of the shRNA used. Decreasing the dose of plasmid expressing U1inαLuc while maintaining shS constant, resulted in a gradual decrease in the inhibition. The combination of 1/2 µg of plasmid expressing shS with 1/4 µg of plasmid expressing of U1inαLuc, resulted in higher inhibition than that obtained when 1 µg of the plasmid expressing shS or U1inαLuc was transfected alone. Further decreases in plasmid expressing U1inαLuc resulted in inhibitions of luciferase that were not significantly different to those obtained when 1/2 µg of the plasmid expressing shS was transfected alone.
Finally, we tested the effect on inhibition of decreasing the amount of both inhibitors at the same time. When the transfected amount of plasmids that express shS and U1inαLuc was decreased simultaneously, luciferase inhibition decreased very rapidly (Figure 5C). Nevertheless, the combination of 1/8 µg of plasmid expressing shS with 1/8 µg of plasmid expressing U1inαLuc resulted in similar inhibition than that obtained when 1 µg of the plasmid expressing shS or U1inαLuc was transfected alone.
These results show that in general, combination of RNAi and U1i allows the dose of the inhibitors to be decreased and potent inhibition to still be achieved. However, different combinations of shRNAs and U1ins are affected by dilution of the inhibitors in a different manner. Therefore, for each combination of shRNA and U1in we recommend that a careful analysis be made of the inhibition obtained with several doses of inhibitors.
Concluding remarks
Our results show a synergistic effect of the combination of RNAi and U1i, indicating that these mechanisms can work together to produce higher levels of inhibition than those obtained by any of the techniques independently. Further experiments will be required to clarify the exact molecular mechanism underlying this phenomenon. Increased inhibition could result if combination of both methods can bypass a partial saturation obtained only when any of the techniques is used independently. The synergism could be the result of a nuclear decrease in target RNA by U1i coupled to a cytoplasmic decrease of the same target by RNAi. Thus, U1i action could decrease the amount of targetable mRNAs in the cytoplasm. There, it may be easier for RNAi to cleave a lower amount of target. However, RNAi is capable of efficiently inhibiting high concentrations of target mRNAs. RNAi has been described to involve a catalytic reaction where RISC, after target cleavage, is released to act on another target molecule. Thus, a single activated RISC can undergo multiple rounds of target mRNA cleavage (32).
Our work shows that when only low efficient shRNAs or U1in are available that target a specific gene, combination of both inhibitors could result in increased inhibition that could allow proper gene function studies. Also, even when good shRNAs or U1in are available, combination could increase inhibition. This may be of interest when a high inhibition is mandatory, for instance when targeting a replicative viral RNA. Also, this may allow the dose of either of the inhibitors or both inhibitors to be decreased thus, avoiding damaging secondary effects and at the same time adequate levels of inhibition to be obtained. Furthermore, the combination of RNAi with U1i, allows different sequences in a gene of interest, different genes in a given pathway or different viral or cellular genes required for efficient viral production to be targeted. In this last case, the combination of RNAi and U1i could serve to decrease the possibility of viral escape by selection of viruses resistant to a single inhibitor. In conclusion, we believe that the increased inhibition obtained by combination of RNAi and U1i will be of therapeutic interest as this results in an improved protocol for the specific inhibition of gene expression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish ministry of education and science (BIO2006-13225, BIO 2009/09295) and ‘UTE project CIMA’ to (P.F.). Funding for open access charge: Spanish ministry of education and science BIO 2009/09295.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Victor Segura for help in the calculation of synergy indexes, Erkuden Casales for technical assistance, Henry J. George and Paloma Arias for plasmids expressing shRNAs targeting firefly luciferase and Dr Paul Miller for English editorial work.
REFERENCES
- 1.Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 2.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. Embo. J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 5.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 6.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 7.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 8.Bridge AJ, Pebernard S, Ducraux A, Nicoulaz AL, Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat. Genet. 2003;34:263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- 9.Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 10.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 11.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 12.Persengiev SP, Zhu X, Green MR. Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs) RNA. 2004;10:12–18. doi: 10.1261/rna5160904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beckley SA, Liu P, Stover ML, Gunderson SI, Lichtler AC, Rowe DW. Reduction of target gene expression by a modified U1 snRNA. Mol. Cell Biol. 2001;21:2815–2825. doi: 10.1128/MCB.21.8.2815-2825.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fortes P, Cuevas Y, Guan F, Liu P, Pentlicky S, Jung SP, Martinez-Chantar ML, Prieto J, Rowe D, Gunderson SI. Inhibiting expression of specific genes in mammalian cells with 5′ end-mutated U1 small nuclear RNAs targeted to terminal exons of pre-mRNA. Proc. Natl Acad. Sci. USA. 2003;100:8264–8269. doi: 10.1073/pnas.1332669100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Will CL, Luhrmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- 16.Furth PA, Choe WT, Rex JH, Byrne JC, Baker CC. Sequences homologous to 5′ splice sites are required for the inhibitory activity of papillomavirus late 3′ untranslated regions. Mol. Cell Biol. 1994;14:5278–5289. doi: 10.1128/mcb.14.8.5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abad X, Vera M, Jung SP, Oswald E, Romero I, Amin V, Fortes P, Gunderson SI. Requirements for gene silencing mediated by U1 snRNA binding to a target sequence. Nucleic Acids Res. 2008;36:2338–2352. doi: 10.1093/nar/gkn068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gunderson SI, Polycarpou-Schwarz M, Mattaj IW. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol. Cell. 1998;1:255–264. doi: 10.1016/s1097-2765(00)80026-x. [DOI] [PubMed] [Google Scholar]
- 19.Sajic R, Lee K, Asai K, Sakac D, Branch DR, Upton C, Cochrane A. Use of modified U1 snRNAs to inhibit HIV-1 replication. Nucleic Acids Res. 2007;35:247–255. doi: 10.1093/nar/gkl1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narvaiza I, Aparicio O, Vera M, Razquin N, Bortolanza S, Prieto J, Fortes P. Effect of adenovirus-mediated RNA interference on endogenous microRNAs in a mouse model of multidrug resistance protein 2 gene silencing. J. Virol. 2006;80:12236–12247. doi: 10.1128/JVI.01205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vera M, Sobrevals L, Zaratiegui M, Martinez L, Palencia B, Rodriguez CM, Prieto J, Fortes P. Liver transduction with a simian virus 40 vector encoding insulin-like growth factor I reduces hepatic damage and the development of liver cirrhosis. Gene Ther. 2007;14:203–210. doi: 10.1038/sj.gt.3302858. [DOI] [PubMed] [Google Scholar]
- 22.Aparicio O, Razquin N, Zaratiegui M, Narvaiza I, Fortes P. Adenovirus virus-associated RNA is processed to functional interfering RNAs involved in virus production. J. Virol. 2006;80:1376–1384. doi: 10.1128/JVI.80.3.1376-1384.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palomero T, Ferrando A. Oncogenic NOTCH1 control of MYC and PI3K: challenges and opportunities for anti-NOTCH1 therapy in T-cell acute lymphoblastic leukemias and lymphomas. Clin. Cancer Res. 2008;14:5314–5317. doi: 10.1158/1078-0432.CCR-07-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Efstratiadis A, Szabolcs M, Klinakis A. Notch, Myc and breast cancer. Cell Cycle. 2007;6:418–429. doi: 10.4161/cc.6.4.3838. [DOI] [PubMed] [Google Scholar]
- 25.Kunnimalaiyaan M, Chen H. Tumor suppressor role of Notch-1 signaling in neuroendocrine tumors. Oncologist. 2007;12:535–542. doi: 10.1634/theoncologist.12-5-535. [DOI] [PubMed] [Google Scholar]
- 26.Talora C, Campese AF, Bellavia D, Felli MP, Vacca A, Gulino A, Screpanti I. Notch signaling and diseases: an evolutionary journey from a simple beginning to complex outcomes. Biochim. Biophys. Acta. 2008;1782:489–497. doi: 10.1016/j.bbadis.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 27.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 28.Matveeva O, Nechipurenko Y, Rossi L, Moore B, Saetrom P, Ogurtsov AY, Atkins JF, Shabalina SA. Comparison of approaches for rational siRNA design leading to a new efficient and transparent method. Nucleic Acids Res. 2007;35:e63. doi: 10.1093/nar/gkm088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purow BW, Haque RM, Noel MW, Su Q, Burdick MJ, Lee J, Sundaresan T, Pastorino S, Park JK, Mikolaenko I, et al. Expression of Notch-1 and its ligands, Delta-like-1 and Jagged-1, is critical for glioma cell survival and proliferation. Cancer Res. 2005;65:2353–2363. doi: 10.1158/0008-5472.CAN-04-1890. [DOI] [PubMed] [Google Scholar]
- 30.Shin HM, Minter LM, Cho OH, Gottipati S, Fauq AH, Golde TE, Sonenshein GE, Osborne BA. Notch1 augments NF-kappaB activity by facilitating its nuclear retention. Embo. J. 2006;25:129–138. doi: 10.1038/sj.emboj.7600902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boese Q, Leake D, Reynolds A, Read S, Scaringe SA, Marshall WS, Khvorova A. Mechanistic insights aid computational short interfering RNA design. Methods Enzymol. 2005;392:73–96. doi: 10.1016/S0076-6879(04)92005-8. [DOI] [PubMed] [Google Scholar]
- 32.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.