Abstract
RNA editing regulates mitochondrial gene expression in trypanosomatid pathogens by creating functional mRNAs. It is catalyzed by a multi-protein complex (the editosome), and is found to be essential in both insect stage and mammalian blood stream form of Trypanosoma brucei. This particular form of RNA editing is unique to trypanosomatids, and thus provides a suitable drug target in trypanosomatid pathogens. Here, we demonstrate the feasibility of a rapid and sensitive fluorescence-based reporter assay to monitor RNA editing based on ribozyme activity. We could validate our new assay using previously identified inhibitors against the essential RNA editing ligase. The principle advantages of this assay are: (i) the use of non-radioactively labeled materials, (ii) sensitivity afforded by fluorescence instrumentation applicable to high-throughput screening of chemical inhibitors against the essential editosome and (iii) a rapid and convenient ‘mix and measure’ type of assay in low volume with a high signal to noise ratio. This assay should enhance rapid identification and characterization of the editosome inhibitors primarily based on the overall composition of the editosomes from T. brucei. These inhibitors could also be tested against the editosomes from the closely related pathogens including T. cruzi and Leishmania species.
INTRODUCTION
The three major trypanosomatid pathogens; Trypanosoma brucei, T. cruzi, and Leishmania major are responsible for devastating human diseases around the world. They are the causative agents of African sleeping sickness, Chagas disease, and Leishmaniasis, respectively (1,2). Current trypanocidal drugs are found to be generally unsuitable for treatment as they are often toxic, not very effective and can lead to drug resistance (3–6). Therefore, creating a new, effective, and safe drug is essential for the treatment of diseases caused by trypanosomatids. There are several molecular processes that are unique to trypanosomatids. One such process is RNA editing, which regulates parasite gene expression by creating mature functional mRNAs for multiple components of the mitochondrial oxidative phosphorylation system. RNA editing is catalyzed by a large multi-protein complex known as editosome and is a form of post-transcriptional RNA processing by which uridylates (Us) are inserted and deleted in mitochondrial mRNAs as specified by small guide RNAs (gRNAs; 7–9).
Four major enzymatic activities are required for insertion and deletion of Us; (i) endonucleolytic cleavage of pre-edited mRNA at the editing site, (ii) U insertion by terminal uridylate trasferase (TUTase) or (iii) U deletion by Uridylate-specific 3′ exoribonuclease (3′-ExoUase), and (iv) ligation of RNA fragments of the edited products by RNA ligases (10). Purification protocols developed using monoclonal antibodies specific for editosome proteins in combination with column chromatography or a TAP tag; identified 21 proteins in the core complex (11). Knockout or knockdown of some of the editosome proteins results in loss of editosome function and, consequently, in parasite death (12–22), suggesting editing as an essential process and a suitable target for drug development. However, the exact roles of the editosome proteins in RNA editing and the dynamic processing and assembly of the editosome, which might involve interactions among multi-protein complexes and changes in their composition, remain to be determined. Inhibition of different steps of the editing process and subsequent assays on the resultant aberrant products as well as its effects on editosome structure and dynamics should allow resolving some of these remaining questions. To achieve this, a repertoire of inhibitors against different editosome proteins could be very useful. This repertoire will not only give us useful hints about the individual roles of editosome proteins and molecular dynamics of editosome assembly, but also provide us with potential drugs against trypanosomatid pathogens. In order to find such inhibitors we need to develop an assay(s) that can rapidly and accurately monitor the RNA editing process.
Three different biochemical assays have been developed and used to monitor RNA editing activities: (i) full-round in vitro RNA editing assay (23), (ii) pre-cleaved RNA editing assay (24,25) and (iii) a hammerhead ribozyme (HHR)-based assay (26). The first two assays rely on direct visualization of RNA editing product, while the latter uses a HHR and its substrate as a reporter for RNA editing efficiency. One major drawback of the full-round editing assay is its low detection limit (3–5%), while pre-cleaved RNA editing assay bypasses the initial rate limiting step of endonucleolytic cleavage and is useful for examining the U insertion/deletion and RNA ligation catalytic steps of RNA editing. To overcome the low detection limit of full-round editing assay, an in vitro RNA editing assay based on the creation of a HHR was developed (26). This assay entails the conversion of an inactive ribozyme to an active ribozyme, which is specifically edited by the editosome via accurate in vitro deletion editing in which three Us are removed as directed by the appropriate gRNA. The edited functional ribozyme is then used to cleave its targeted RNA substrate. This HHR-mediated assay increased the RNA editing detection limit up to 16.8% (26) .
The above mentioned assays suffer from limitations and drawbacks such as low sensitivity, use of radiolabeled materials and most importantly inapplicability for high-throughput screening.
In this study, we have developed a ‘mix and measure’ HHR-based in vitro reporter assay to monitor RNA editing for rapid identification of the editosome inhibitors. Our assay utilizes a fluorescent resonance energy transfer (FRET) substrate that can monitor full-round deletion RNA editing. We show that this new assay has higher sensitivity compared to previously reported full-round deletion RNA editing assays with a high signal to noise ratio, avoids the use of radiolabel material, and is applicable for high-throughput screening of chemical libraries against the essential editosome proteins. We have also used our assay to confirm the findings of Amaro et al. (27) who have recently reported inhibitors against kinetoplastid RNA editing ligase 1 (KREL1) using a combination of in silico analysis and in vitro adenylation assay. Using our assay, we have shown that the best KREL1 inhibitor reported by Amaro et al. (27) is also capable of inhibiting the full-round deletion RNA editing in the presence of purified editosome, suggesting it as a suitable candidate for development of novel trypanocidal drugs.
MATERIALS AND METHODS
Preparation of RNAs
The pre-edited ribozyme (pre-A6Rbz) for deletion assay, the active ribozyme (A6Rbz), the guide RNA (gA6Rbz) and the guide competitor (gA6Rbz-comp) were transcribed in vitro by T7 polymerase RiboMAX transcription kit (Promega) from synthetic DNA oligonucleotides (pre-A6RBz: 5′-ACATTTGATCTATTGTTTCGTCCTCACGGACTCATCAAAAAGTCACAACTTTCCCTTTCTCTCCTCCCCCTAACCTTTCCCCCTATAGTGAGTCGTATTA-3′; A6Rbz: 5′-ACATTTGATCTATTGTTTCGTCCTCACGGACTCATCAGTCACAACTTTCCCTTTCTCTCCTCCCCCTAACCTTTCCCCCTATAGTGAGTCGTATTA-3′; gA6Rbz: 5′-AAAAAAAAAAAAAAAAATAATTATCATATCACTGTCAAGGGAAAGTTGTGAGGGTGATGAGTCCGTGTATATCCCCCTATAGTGAGTCGTATTA-3′; gA6Rbz-comp: 5′-GGATATACACGGACTCATCACCCTCACAACTTTCCCTTGACAGTGATATGATAATTATTTTTTTTTTTTTTTTTCCCTATAGTGAGTCGTATTA-3′) in combination with a T7 oligonucleotide, where the sequence complementary to the T7 promoter sequence is underlined. All RNAs were purified by electrophoresis through 9% sequencing gels as previously described (28). Unlabeled HHR substrate (5′-GAUCUAUUGUCUCACA-3′) was synthesized by Integrated DNA Technologies (IDT) and the fluorescent labeled substrate, 5′-FAM (6-carboxyfluorescein) –GAUCUAUUGUCUCACA-TAMRA (6-carboxytetramethylrhodamine) −3′ was synthesized and HPLC purified by Eurogentec. Radiolabeling of RNA at the 3′-terminus was performed by [5′-32P]pCp ligation (29). The partial T1 RNA sequencing was performed using the Ambion sequencing kit.
Preparation of mitochondrial extract (editosome) and western blot analyses
Mitochondrial extract from procyclic form of T. brucei 1.7A wild-type cell lines (30) was prepared from 11 × 109 cells (400 ml of 2.7 × 107 cells/ml) by centrifugation in a linear 10–30% (vol/vol) glycerol gradient and fractionated into 21 fractions (500 µl each) as described before (16,31,32,33). From each fraction, 15 µl was separated on 10% SDS–PAGE, blotted onto the PVDF membrane, and probed with four monoclonal antibodies (MAbs) against KREPA1 (1:25 dilution), KREPA2 (1:50 dilution), KREL1 (1:50 dilution) and KREPA3 (1:25 dilution) (34). Goat anti-mouse IgG HRP conjugate (Bio-Rad) was used as the secondary antibody at 1:5000 dilutions in PBST buffer. The western blot was developed by ECL Kit (Amersham).
Editosome complex was also purified by tandem affinity purification from 2 l of TbREL1-TAP expressing T. brucei cells as previously described (31). From each purification method, the fractions were tested for deletion editing activity, and the fractions with the highest activity were used for the experiments. Purified fractions were stored in 15% (vol/vol) glycerol at −80°C between purifications and use in editing assays. The mitochondrial extract and Tap- tag purified editosome concentrations were determined using Bradford assay (BioRad).
Full-round in vitro RNA editing reactions
Assays for RNA editing reactions were performed essentially as described before (26) with some modifications. For radiolabeled full-round deletion RNA editing assays, [32p] pCp-pre-A6Rbz (1 pmol) was initially annealed to gA6Rbz (2.5 pmol) at 70°C and then cooled down at room temperature for 15 min. In the next step, the annealed preA6Rbz and gA6Rbz were added to the editing reaction buffer containing 1× HHE [25 mM HEPES pH 7.9, 10 mM Mg (OAc)2, 50 mM KCL and 1 mM EDTA], 1 mM ATP, 5 mM CaCl2 and 83 ng/µl of Toroula RNA, and 5 µl of editosome purified from mitochondrial preparation was added. Then, RNA editing reactions were incubated at 28°C. The reactions were stopped after 4 h by adding a stop buffer containing 2.5% SDS and 130 mM EDTA. RNA was then extracted by phenol:chloroform:isoamyl alcohol and run on 9% polyacrylamide gel containing 7 M urea for 6–8 h at 55W. Finally, results were visualized by PhosphorImager.
For HHR-based assay, we followed the same protocol as the full-round RNA editing assay except that pre-A6Rbz was not radiolabeled and after 4 h incubation at 28°C, 25 pmol of guide competitor (gA6Comp) was added (10× more than gA6Rbz) and reaction was heated at 95°C for 10 min and cooled down at room temperature for at least 10 min. Then, 2 pmol of [32p] pCp HHR substrate was incubated with editing reaction buffer at 37°C for 45 min. The cleavage product were detected on a 15% polyacrylamide gel using PhosphorImager.
Fluorescent-based RNA editing assays
In this assay, instead of [32p] pCp HHR substrate, 15 pmol of fluorescent-labeled substrate was added to the editing reaction described above. The samples were then transferred to Rotor Gene 3000, where they were incubated for ∼2 h at 37°C. Real-time measurements of the ribozyme activity were recorded at intervals of 1 min. The emission spectra of FAM and TAMRA were 535 nm and 582 nm, respectively, and the excitation wavelength was 470 nm for FAM. The rate of increase of the signal from FAM was used to measure the activity and, thus, the concentration of the edited ribozyme. In order to define the quality of our assay, the Z-factor (35) was calculated for 20 repetitions as:
In which (σ) is the SD of the acquired signal, (μ) is its average, (p) indicates the positive control and (n) indicates the negative control.
KREL1 inhibitors (NCI; 45 207 and 16 209; 27) were dissolved in DMSO (final concentration of <1%) and incubated with editosome for 10 min. We included Triton X-100 (0.1% wt/vol) to prevent compounds from aggregating and non-specific inhibition as previously described (27). IC50 values were calculated and analyzed using the GraphPad Prism 5 software.
RESULTS
Direct visualization of deletion RNA editing of a hammerhead ribozyme and measuring RNA editing efficiency based on HHR reporter substrate
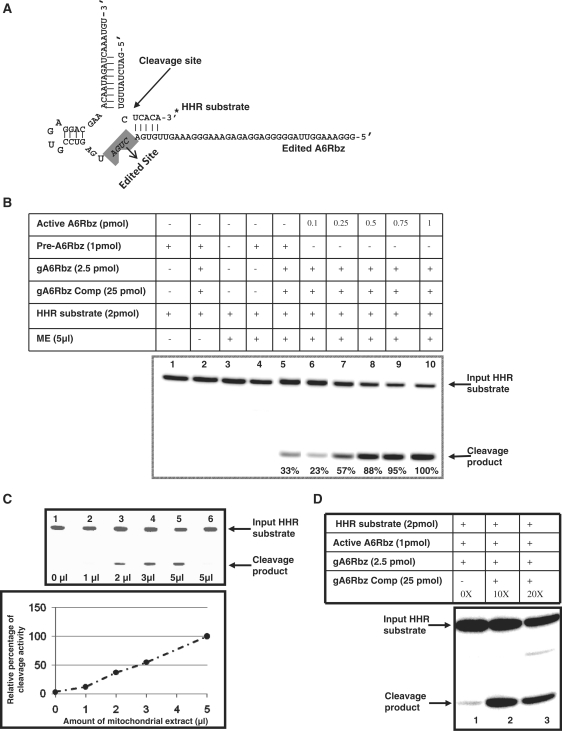
Here, we used the pre-edited HHR which is identical to that used in previous study (26). The pre-edited HHR has three additional Us in its conserved catalytic region that renders the HHR inactive. In the presence of the mitochondrial extract from glycerol gradient, this assay entailed the conversion of the pre-edited HHR to an edited HHR that depends on precise in vitro editing in which three Us are removed as directed by the appropriate gRNA, gA6Rbz (Figure 1A and B). It should be noted that we have performed this assay to monitor and measure the percentage of edited product in the conventional full round RNA editing assay when the editosome complex was purified from glycerol gradient fractionation. The activity peak was observed in fraction 11 of glycerol gradient (data not shown) and we use this fraction for all the experiments in this article in order to have a consistent comparison among different RNA editing assays. The mitochondrial extract concentration for this fraction was 470 µg/ml. Our results indicated that ∼8.8% of input pre-edited HHR was edited by deletion of three Us. In order to accurately report the RNA editing efficiency, HHR substrate cleavage was used as a reporter (Figure 2A). Upon successful completion of full-round RNA editing of HHR, edited functional HHR was able to cleave its radio-labeled substrate which is 16-nt long (Figure 2B, lane 5). We found that gA6Rbz guide RNA had an inhibitory effect on active ribozyme cleavage activity (Figure 2D, lane 1). Figure 2C also shows that the edited HHR cleavage activity is completely abolished in the absence of guide RNA competitor (lane 6). This could possibly be due to absence of enzymes with helicase activity in this fraction. As shown previously (36), different methods of editosome purification lead to pull down of different numbers of editosome proteins; for example, editosome purified from sequential column chromatography contains an enzyme with helicase activity which is assumed to have a role in unwinding the mRNA from the gRNA in the editing process. This activity could be absent from the editosome complex in fraction 11 of glycerol gradient used throughout this article. Hence, to resolve this issue, we added a guide RNA competitor, which was fully complementary to gA6Rbz in order to overcome inhibitory effect of gA6Rbz on HHR activity. We observed that 10× molar excess of guide competitor can rescue the cleavage of HHR substrate by active A6Rbz. On the basis of varying concentration of active A6Rbz (positive control; Figure 2B, lanes 6–10), we have also shown here that the cleavage efficiency of the edited HHR is 33% which is almost two times greater than the 16.8% cleavage efficiency reported previously (26). The in vitro editing activity also showed a linear increase with increasing volume of mitochondrial extract up to 5 µl (Figure 2C). The RNA editing efficiency did not increase using higher volume of mitochondrial extract up to 10 µl (data not shown).
Figure 1.
Full-round HHR-based RNA editing (deletion) assay based on ATPase 6 pre-edited mRNA. (A) The pre-edited ribozyme (pre-A6Rbz) is shown with the gA6Rbz gRNA that specifies the deletion of three Us from the editing site (ES) in the presence of editosome purified from mitochondrial extract (ME). The conserved (5′-CUGA-3′) of A6Rbz in the catalytic core essential for ribozyme activity is highlighted (Edited Site) and the line indicates where the three Us are removed by editing. The 3′ [32p] pCp-label of pre-A6Rbz is indicated by an asterisk. (B) Autoradiogram showing the edited and endonuclease cleavage products (arrows) generated only in the presence of both gA6Rbz and ME. Lane T1 is pre-A6Rbz RNA subjected to partial digestion by RNase T1, which is used as marker. Percentage of RNA editing efficiency in this assay was 8.8% and it was calculated as the percentage of total input pre-A6Rbz.
Figure 2.
Cleavage activity test of HHR. (A) The structure of edited A6Rbz that can cleave the HHR substrate with the cleavage site indicated by an arrow. The 3′ [32P] pCp-label of HHR substrate is indicated by an asterisk. The essential core nucleotides of HHR are highlighted. (B) In the presence of mitochondrial extract, the HHR substrate cleavage can be monitored by edited pre-A6RZ. The cleavage product by edited pre-A6Rbz was generated only in the presence of gA6Rbz and, mitochondrial extract, and gA6Rbz competitor (gA6Rbz Comp; lane 5). There was no cleavage product detected in the absence of either mitochondrial extract (ME) or gA6Rbz gRNA (lanes 2 and 4) and there was no HHR substrate degradation observed in the presence of ME alone (lane 3). Various concentrations of active HHR, A6Rbz, (lanes 6–10) were used to estimate the efficiency of substrate cleavage by pre-A6Rbz upon RNA editing. The percentage of cleavage product was measured as the ratio of density of cleavage product over density of the entire lane. The percent cleavage at the highest concentration of active HHR (1 pmol) was assumed to be 100% and percent cleavage at the lower concentrations (0.1, 0.25, 0.5 and 0.75 pmol) were accordingly normalized to 23%, 57%, 88% and 95%, respectively. (C) The in vitro RNA editing reaction followed by cleavage activity test of HHR using various amounts of ME. The input HHR substrate and cleavage products are indicated by arrows. Lanes 1–5 contain preA6Rbz, gA6Rbz, various amounts of ME as indicated, and gA6Rbz competitor. Lane 6 is same as lane 5 but the gA6Rbz competitor was not added to show the inhibition of HHR substrate cleavage. (D) Inhibitory effect of gA6Rbz on HHR cleavage activity (radiolabel-based assay) in the absence and presence (10× and 20×) of gA6Rbz competitor. In the presence of increasing molar excess of gA6Rbz competitor to gA6Rbz, the A6Rbz cleavage activity was restored.
FRET-based RNA editing assay
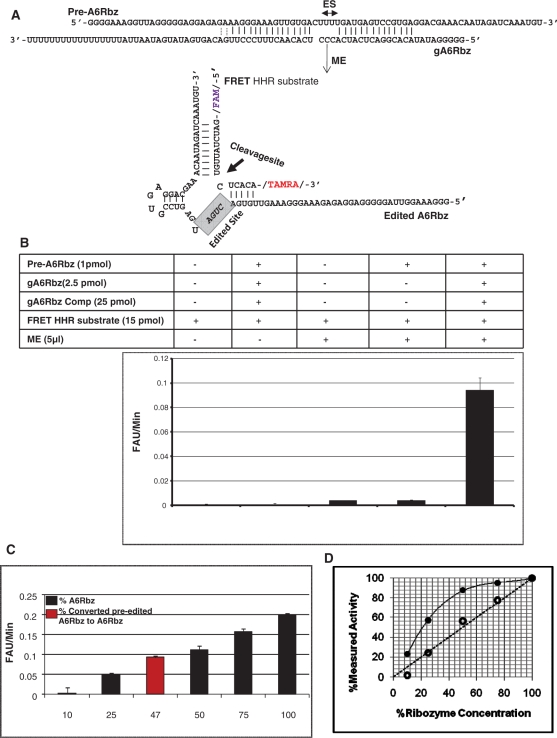
By modifying the previously reported HHR-based radiolabeled in vitro RNA editing assay (26), we created a homogenous fluorescent-based assay that uses a 16-nt-long FRET substrate for real-time determination of HHR activity after completion of full-round RNA editing. The fluorescently labeled substrate contains a fluorescent reporter (FAM) on the 5′-end and a quencher (TAMRA) on the 3′-end. When the two fluorophores are at close physical proximity (10-100 Å), the excited energy state of a donor molecule is transferred non-radioactively to an acceptor molecule, resulting in quenching of the donor fluorescence. On the other hand, when the two fluorophores are separated, a signal is detected upon fluorescence dequenching. Upon RNA editing, the pre-edited inactive ribozyme is converted to the active ribozyme which is capable of cleaving its fluorescently labeled substrate and consequently a signal is detected as a result of cleavage. The complementary paired region between HHR and the HHR substrate is shown in Figure 3A. Cleavage of the substrate by the HHR separates the fluorescent reporter and the quencher, allowing us to monitor, in real time, the cleavage activity of the HHR based on a time-dependent increase of fluorescence. Figure 3B shows that upon completion of full-round RNA editing reaction in the presence of both gA6Rbz and mitochondrial extract, the pre-edited inactive HHR is edited to an active HHR which then cleaves the FRET substrate.
Figure 3.
FRET-based RNA editing assay. (A) Diagram of the pre-edited ribozyme (pre-A6RZ) is shown in association with the gA6RZ gRNA that specifies the deletion of three Us from the editing site (ES). In the presence of functional mitochondrial extract (ME), the pre-A6RZ is edited to active A6RZ that can now cleave the FRET HHR substrate with a fluorescent reporter (FAM) on the 5′-end and a quencher (TAMRA) on the 3′-end. The cleavage site of FRET HHR substrate is indicated by an arrow. (B) FRET-based RNA editing assay. Reactions were performed in a 30µl reaction volume under multiple turn-over conditions. Rotor-Gene 3000TM (CORBETT research) was used as a fluorescent reader. In the graph, y-axis represents the initial velocity of reaction in terms of fluorescent arbitrary unit (FAU) per minute and x-axis indicates the reaction conditions. A high signal-to-noise ratio was measured when both gA6RZ and ME were present (compare complete reaction with other conditions). All reactions containing gA6Rbz competitor (gA6Rbz Comp) were at 10× molar excess to gA6Rbz. The error bars represent the experimental variation (standard deviation) from 20 repetitions and resulting in Z-value of 0.65. (C) Various concentrations of active HHR, A6Rbz, 0.1, 0.25, 0.5, 0.75 and 1 pmol corresponding to 10%, 25%, 50%, 75% and 100% RNA editing was compared to efficiency of edited pre-A6Rbz FRET-based RNA editing assay. The red column indicates that nearly 47% of inactive HHR was edited to active HHR in the presence of gA6Rbz gRNA and ME. (D) FRET-based assay provides a linear measure of ribozyme activity. Close circles represent the measured ribozyme activity based on radiolabeled assay, whereas open circles indicate the measured ribozyme activity based on FRET assay. The dotted line represents a perfectly theoretical linear assay.
Z-factor is most commonly calculated to examine the performance and quality of high throughput screening assays. This factor is a statistical parameter which takes into account the signal to background or signal-to-noise ratio in such assays (35). In order to evaluate the power of our method in discriminating between reactions in which the RNA editing happens and reactions where no editing occurs, we calculated the Z-factor from 20 repetitions of each of the positive and negative reactions (Z: 0.65; Figure 3B). This indicates a high sensitivity and specificity for our FRET-based assay in measuring editing efficiency. In order to accurately measure the edited HHR cleavage activity, the cleavage product of fluorescent substrate was determined at varying concentrations of active HHR (A6Rbz positive control) in the same reaction condition (Figure 3C). Result of this experiment was used as a standard to measure the efficiency of our FRET based assay. We have measured that ∼47% of inactive HHR was converted to active HHR as a result of successful RNA editing. This suggests that the FRET-based assay has ∼14% higher efficiency and, therefore, ∼1.5 times as sensitive as the HHR radiolabeled assay (47% versus 33%). The higher sensitivity of our fluorescent assay in comparison to the radiolabeled HHR-mediated cleavage assay can be attributed to the higher concentration of the substrate. Unlike the radiolabeled substrate, the fluorescent substrate can be used and detected at high concentrations, allowing a more accurate measurement. More importantly, we can measure the velocity of the reaction using the fluorescent substrate, whereas when using the radiolabeled substrates we can only measure the end-point concentration of the cleavage product; the former provides a more accurate and linear estimate of the concentration of active ribozyme (Figure 3D).
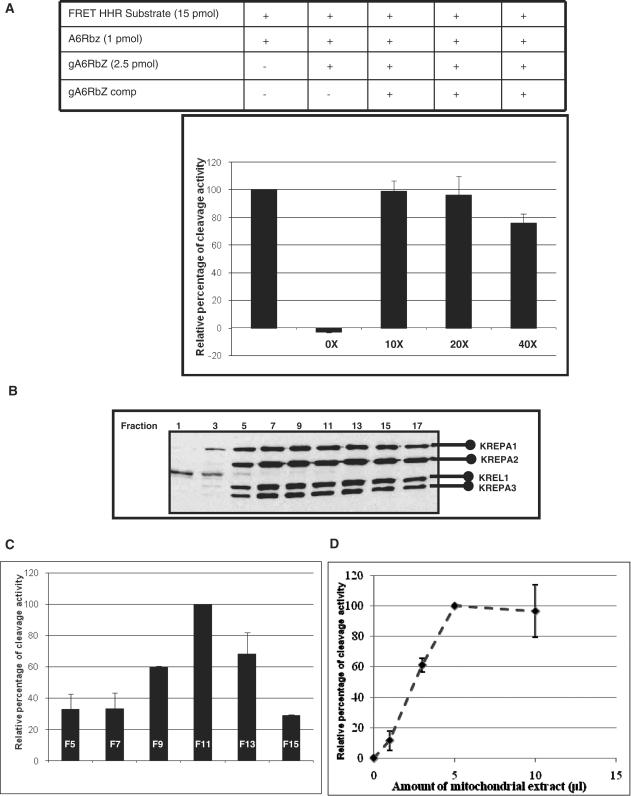
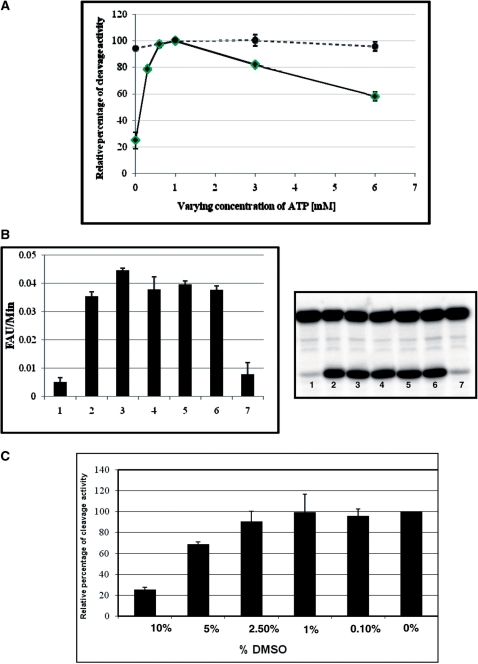
We have also examined various factors which influence our fluorescent-based assay. First, we have shown that gA6Rbz has inhibitory effect on HHR cleavage activity; due to the tight association of the guide RNA and A6Rbz, the edited active HHR could not bind and cleave the HHR substrate (Figure 4A, column 2). This problem was overcome by adding the gA6Rbz competitor (10 × molar excess of gA6Rbz concentration; Figure 4A, column 3). Further increasing gA6Rbz competitor concentration had less or similar effect on cleavage activity (Figure 4A, columns 4 and 5). Second, we have tested the RNA editing activity of mitochondrial extract from different fractions of glycerol gradient (Figure 4B). We have found that fractions 9–13 (Figure 4C), corresponding to 20S editosome, were able to edit inactive HHR to active HHR, but the highest edited active HHR resulted from fraction 11 of glycerol gradient; henceforth, we have used this fraction for our FRET-based assay. Third, the in vitro editing activity showed a linear increase with increasing concentration of mitochondrial extract up to 5 µl (Figure 4D). We have also investigated the effect of the ATP concentration on the RNA editing activity. The mitochondrial extract of T. brucei displays maximum editing activity at ATP concentration up to 1 mM. The RNA editing efficiency does not plateau with higher ATP concentrations, but rather is repressed by further increasing levels of ATP concentration (Figure 5A). In order to determine the potential effect of ATP addition on chelating and lowering the concentration of free Mg2+ required for ribozyme function, we varied the amount of ATP in the presence of the active ribozyme and 10 mM Mg2+ (the concentration used throughout this study). Increasing concentration of ATP up to 6 mM did not inhibit the ribozyme function, indicating that the Mg2+-chelating activity of ATP does not lower the Mg2+ concentration to a level that would inhibit ribozyme cleavage activity (Figure 5A, dotted line). One other likely explanation for this suppression is the inhibitory effect of ATP addition on RNA editing ligase activity. This would be consistent with an earlier observation in bacteriophage T4 RNA ligase 2 that the ATP addition results in the accumulation of the adenylated RNA ligase and RNA-adenylate intermediate and thus suppression of the RNA ligase activity (37). We, however, cannot rule out the possibility of Mg2+ chelation by higher concentrations of ATP and its effect on the RNA editing ligase activity. Since the optimum concentration of Mg2+ for in vitro RNA editing has been established at 10 mM (10), we used this concentration in this study, although increasing the Mg2+ and ATP concentrations simultaneously could increase the editing efficiency.
Figure 4.
Cleavage of FRET HHR substrate with varying concentrations of gA6Rbz competitor and varying amounts of mitochondrial extract. (A) Inhibitory effect of gA6Rbz on HHR cleavage activity (FRET-based assay). Comparison of cleavage with active HHR (A6Rbz) using various concentrations of gA6Rbz competitor (gA6Rbz comp). In the absence of gA6Rbz comp (0×), the signal from cleavage of the substrate is inhibited. In the presence of increasing molar excess of gA6Rbz comp to gA6Rbz (10×, 20×, 40×), the A6Rbz activity was restored. (B) Western analysis of mitochondrial extract fractionated on a 10–30% glycerol gradient. The four editosome proteins; KREPA1, KREPA2, KREPA3 and KREL1 for which monoclonal antibodies are available are indicated. (C) Fractions from (B) were assayed for editing by the FRET-based RNA editing assay. Maximal RNA editing activity was detected in fractions 9–13 (F9–F13) peaking in fraction 11 (F11). The y-axis represents the relative percentages of the cleavage activity for edited HHR in each experiment. (D) Effect of varying concentrations of fraction 11 from (C) on FRET-based in vitro RNA editing assay.
Figure 5.
Effects of ATP and DMSO on RNA editing. (A) FRET-based RNA editing assay (solid line) in the absence (0 mM) and presence (0.3, 0.6, 1, 3 and 6 mM) of ATP, or active ribozyme (dotted line) in the absence (0 mM) and presence (1, 3 and 6 mM) of ATP. The relative percentage of editing is expressed as the percentage of the cleavage activity for edited HHR in each experiment. (B) ATP dependence. Right panel indicates the cleavage of radiolabeled HHR substrate in the absence of ATP (lane 1), in the presence of 1 mM ATP (lane 2), in the presence of both 1 mM ATP and 1 mM dATP (lane 3; 1 mM ATP and 1 mM dATP were incubated with mitochondrial extract, lane 4; 1 mM ATP and 1 mM dATP were added to the reaction buffer, lane 5; 1 mM dATP was incubated with mitochondrial extract separately, lane 6; 1 mM ATP was incubated with mitochondrial extract separately), and in the presence of 1 mM dATP and absence of 1 mM ATP (lane 7), left panel was identical to right panel except FRET substrate substituted for radiolabel substrate. (C) FRET-based RNA editing assay in the absence (0 mM) and presence of varying concentrations of DMSO (0.1–10%). The relative percentage of editing is expressed as the percentage of total cleavage in the absence of DMSO.
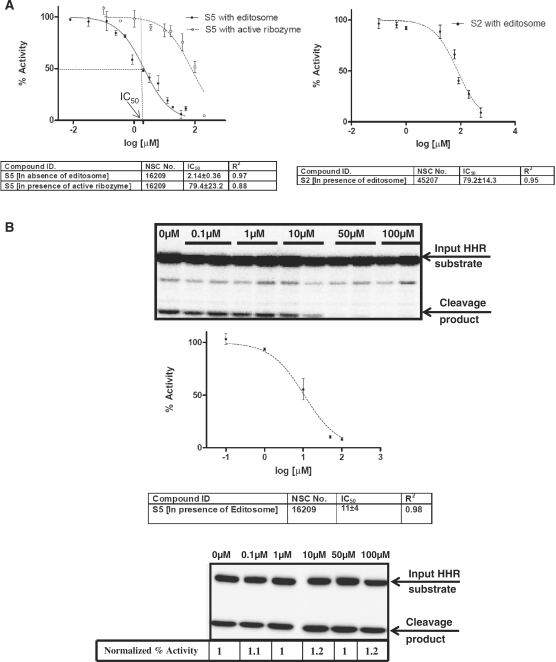
dATP was used here as a possible competitive inhibitor of ATP in the presence and absence of ATP, however no inhibitory effect was detected using this ATP analogue (Figure 5B and C). We have also tested the effect of dimethylsulfoxide (DMSO; a commonly used solvent for chemical compounds) on editing, measured by our fluorescence-based assay. The mitochondrial extract was incubated with different concentrations of DMSO (0–10%). We found that at concentrations up to 1% DMSO did not affect the enzyme activity. However, 5% and 10% DMSO concentrations significantly reduced the editosome activity (Figure 5C). In order to validate our assay with known inhibitors of RNA editing enzymes, we have tested two compounds against KREL1 protein (S2 and S5) reported by Amaro et al. (27). They have shown that S5 had significant inhibitory effect on the adenylation step of RNA editing ligase activity (IC50: 1.01 ± 0.16 μM), whereas S2 did not perturb this step at 10µM concentration. Confirming their findings, here we showed the effect of these two compounds on FRET-based full-round deletion assay. Figure 6A shows the dose–response curves for these two compounds, indicating IC50 values of 2.14 ± 0.36 μM (Figure 6A, left panel) and 79.2 ± 14.3 μM (Figure 6A, right panel) for S5 and S2, respectively. The editosome complex used in this assay was purified by using TAP-tag (324 µg/ml), based on the tagged RNA editing ligase as previously described (31). In order to address the possibility of non-specific inhibitory effect of these compounds on the structure/function of the ribozyme reporter rather than the editing reaction, we included a control in which the effect of S5 was tested on the active A6Rbz/gA6Rbz in the absence of editosome (Figure 6A, ‘S5 with active ribozyme’). The IC50 value of this reaction is 79.4 ± 23.2 μM suggesting that the inhibition is specific to the editing reaction. Both S2 and S5 are dye-like compounds, thus, inhibition observed at highest concentration of these drugs in our fluorescent-based assay is due to the quenching of fluorescent signal by these compounds. Therefore, at higher concentrations of the drug (after ∼50 µM), the IC50 values measured for S2 and active ribozyme are not reliable. We confirmed the specificity of inhibition by S5 in the FRET-based assay using the radiolabeled full-round deletion HHR-based assay. In this assay, higher concentrations of the drug inhibits the editing reaction and therefore cleavage of HHR substrate (Figure 6B, top panel), while the same concentrations cannot inhibit the activity of active A6Rbz in the absence of TAP-tagged purified editosome (Figure 6B, bottom panel). IC50 value of the editing reaction is 11 ± 4 μM as measured using the radio-labeled assay (Figure 6B, top panel). The different IC50 values observed using S5 in radiolabeled and FRET-based assays can be explained since unlike the FRET-based assay, the radiolabeled assay does not provide a linear measure of ribozyme activity (Figure 3D). Thus, the calculated IC50 value from radiolabeled-based assay may not accurately represent the actual IC50 value.
Figure 6.
Effects of known inhibitors of RNA ligase on editosome. (A) Dose–response curves for S5 in the presence of editosome (solid line, close circle, left panel) and in the absence of editosome in the presence of active ribozyme (dashed line, open circle, left panel), and S2 in the presence of editosome (dashed line, close circle, right panel). IC50 and R2-values are reported based on three independent FRET-based full-round editing assays using editosome purified from cells expressing TbREL1-TAP. The IC50 ranges are reported with 95% confidence level. The concentration of S5 that results in 50% inhibition of maximal activity (IC50) is indicated in left panel. (B) The effect of S5 on RNA editing using radiolabel-based HHR assay. Drug concentrations in micromolars have been indicated. The dose–response curve obtained from duplicates for S5 in presence of tap-tag purified editosome has been shown. The IC50 range is determined based on 95% confidence level. The bottom panel shows the effect of drug on active A6Rbz/gA6Rbz in the absence of editosome. The percent cleavage activity (normalized percentage activity) of active ribozyme in the absence of S5 was assumed to be 1 and percent cleavage at the higher concentrations of S5 (0.1, 1, 10, 50 and 100 µM) was accordingly normalized.
DISCUSSION
Here, a convenient in vitro fluorescent-based reporter assay has been developed using an existing in vitro RNA editing assay that is based on HHR activity (26). This fluorescent-based reporter assay contains a 16-nt long FRET substrate labeled with two separate fluorophores, referred to as FAM (fluorescent reporter), and TAMRA (fluorescent quencher). Theoretically, when FAM and TAMRA are physically close to each other, the energy shifts from FAM to TAMRA and results in de-excitation of FAM and excitation of TAMRA, while when they get separated from each other, FAM absorbs light and excited state of this fluorescent reporter is generated (38). Our fluorescent-based assay requires the editing of an inactive pre-edited HHR into an active HHR which is able to cleave its fluorescence-labeled substrate; hence much higher signal compared to background can be detected as a result of cleavage. The editing of HHR requires editosome purified from T. brucei. In order to develop a highly robust and efficient fluorescent-based RNA editing assay, we looked for the most active and pure editosome preparation. Greater activity of editosome can increase the yield of edited HHR and cleavage of its fluorescently labeled substrate and thus higher sensitivity. The editosome preparation from whole cell mitochondrial extract by glycerol gradient fractionation showed a high level of activity. One important advantage of using crude cellular fractions is that the conditions are closer to the inhibitor to be discovered using living cells. On the other hand, a possible drawback of less pure editosome preparation is potentially higher level of false negatives that may influence the high-throughput screening assay. Hence, while testing chemical compounds reported by Amaro et al. (27), we have used a more pure editosome preparation obtained by the TAP-tag method. While they have reported the inhibitory activity of these compounds only on KREL1 adenylation, we further demonstrate that their best drug-like inhibitor, S5, identified by virtual screening against KREL1 and confirmed by adenylation assay is also able to inhibit full-round deletion RNA editing. Virtual screening of chemical libraries against essential protein components of editosome might lead to discovery of drug like inhibitors but this approach is limited by the unavailability of crystal structures of most editosome proteins. Therefore, our assay can be a powerful tool for identifying inhibitors against other essential components of editosome.
The inhibitory effect of gRNA on the HHR activity is an important factor to consider. In RNA editing assays, the gRNA concentration is always in molar excess of pre-edited mRNA, hence during RNA editing assay majority of the mRNAs (edited/pre-edited) form a duplex with gRNA. This causes a problem while monitoring editing in real time using fluorescent based assay, since the edited HHR is not free to cleave its substrate. We have been able to overcome this problem by heat denaturation of the mRNA/gRNA duplex in the presence of gRNA competitor (in 10× molar excess) which could help to unwind the mRNA/gRNA duplex.
A recent publication describes an RNA aptamer-based assay to monitor the editing reaction based on electrochemiluminescent signal. In this assay, the RNA editing of an aptamer causes a conformational change that result in the activation of the streptavidin-binding region of the aptamer, leading to a measurable electrochemiluminescent signal. In contrast, our assay is based on the enzymatic activity of the editing substrate, i.e. the hammer-head ribozyme. This FRET-based ribozyme assay, when compared to the aptamer-based assay, shows a considerably higher signal to background ratio (20-fold selectivity; 39). Also, our FRET-based assay is a simple ‘mix and measure’ homogeneous assay that is much simpler and less time-consuming compared to the aptamer-based assay, particularly because it involves no washing steps. Most importantly, the real-time monitoring of cleavage activity of HHR yields a series of time points with any desired resolution, whereas the aptamer-based assay relies on endpoint signal. This confers a higher accuracy in measuring the amount of edited product (active HHR).
Taken together, we have successfully adapted our ribozyme-based assay for use in high-throughput molecular screening against the whole editosome. A Z-value of 0.65 for full-round deletion RNA editing based on A6Rbz indicates that this assay is suitable for high-throughput screening of chemical compounds against editing machinery. It can be estimated that preparation of mitochondrial extract from 40 l of T. brucei culture using 2 ml of pooled fractions (9–12) of glycerol gradient can yield enough materials for screening 40 000 compounds in search of inhibitors of editing reaction. We calculated the Z-value of 0.6 (data not shown) using this pooled fraction which is slightly lower than the Z-value calculated for the fraction with highest editing activity but still in a desirable range for high-throughput screening. We have also shown that the fluorescent based RNA editing assay demonstrates a greater sensitivity; up to 5-fold more than conventional radiolabel based assay. Furthermore, other major advantages of this method are that the use of radioactive material can be avoided and time-consuming gel electrophoresis can be circumvented. We are currently designing a truncated HHR that can be adapted for pre-cleaved RNA editing assays which are particularly useful in examining the U insertion, U deletion, and ligation catalytic steps of RNA editing. FRET based pre-cleaved RNA editing assays using truncated version of HHR will also serve as a secondary screen to validate the specificity of the inhibitory compounds.
Furthermore, RNA editing is not limited to the insertion and deletion of uridines in mitochondrial encoded RNAs of kinetoplastid, as other types of RNA editing have been described which result in non-encoded adenosines, guanosines, cytidines and inosines by substitution type of RNA editing. The latter type of RNA editing has been observed in chloroplast-encoded RNAs of plants (40) as well as nucleus-encoded RNAs of mammals (41). Our fluorescent-based HHR assay can also be adapted to monitor such processes in order to elucidate the mechanisms involved in this type of RNA editing process using cell-free extracts.
FUNDING
Le Fonds Quebecois de la recherche sur la nature et les technologies (FQRNT; for Centre for Host-Parasite Interactions and research at Institute of Parasitology); National Institutes of Health (NIH # 1R21NS057046-01 to R.S.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Hamed Shateri Najafabadi for critical reviewing of the article and analysis of the data, and also Smriti Kala for critical comments on the article. Monoclonal antibodies against KREPA1, A2, A3 and KREL1 were kindly provided by Dr Kenneth Stuart (Seattle Biomedical Research Institute).
REFERENCES
- 1.Vickerman K, Coombs GH. Protozoan paradigms for cell biology. J. Cell Sci. 1999;112(Pt 17):2797–2798. doi: 10.1242/jcs.112.17.2797. [DOI] [PubMed] [Google Scholar]
- 2.Stuart K, Brun R, Croft S, Fairlamb A, Gurtler RE, McKerrow J, Reed S, Tarleton R. Kinetoplastids: related protozoan pathogens, different diseases. J. Clin. Invest. 2008;118:1301–1310. doi: 10.1172/JCI33945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Legros D, Ollivier G, Gastellu-Etchegorry M, Paquet C, Burri C, Jannin J, Buscher P. Treatment of human African trypanosomiasis–present situation and needs for research and development. Lancet Infect. Dis. 2002;2:437–440. doi: 10.1016/s1473-3099(02)00321-3. [DOI] [PubMed] [Google Scholar]
- 4.Denise H, Barrett MP. Uptake and mode of action of drugs used against sleeping sickness. Biochem. Pharmacol. 2001;61:1–5. doi: 10.1016/s0006-2952(00)00477-9. [DOI] [PubMed] [Google Scholar]
- 5.Fairlamb AH. Chemotherapy of human African trypanosomiasis: current and future prospects. Trends Parasitol. 2003;19:488–494. doi: 10.1016/j.pt.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 6.Delespaux V, de Koning HP. Drugs and drug resistance in African trypanosomiasis. Drug Resist. Updat. 2007;10:30–50. doi: 10.1016/j.drup.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Panigrahi AK, Ernst NL, Domingo GJ, Fleck M, Salavati R, Stuart KD. Compositionally and functionally distinct editosomes in Trypanosoma brucei. RNA. 2006;12:1038–1049. doi: 10.1261/rna.45506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Spek H, Arts GJ, Zwaal RR, van den Burg J, Sloof P, Benne R. Conserved genes encode guide RNAs in mitochondria of Crithidia fasciculata. EMBO J. 1991;10:1217–1224. doi: 10.1002/j.1460-2075.1991.tb08063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rusche LN, Cruz-Reyes J, Piller KJ, Sollner-Webb B. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J. 1997;16:4069–4081. doi: 10.1093/emboj/16.13.4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seiwert SD, Stuart K. RNA editing: transfer of genetic information from gRNA to precursor mRNA in vitro. Science. 1994;266:114–117. doi: 10.1126/science.7524149. [DOI] [PubMed] [Google Scholar]
- 11.Stuart K, Panigrahi AK. RNA editing: complexity and complications. Mol. Microbiol. 2002;45:591–596. doi: 10.1046/j.1365-2958.2002.03028.x. [DOI] [PubMed] [Google Scholar]
- 12.Drozdz M, Palazzo SS, Salavati R, O'Rear J, Clayton C, Stuart K. TbMP81 is required for RNA editing in Trypanosoma brucei. EMBO J. 2002;21:1791–1799. doi: 10.1093/emboj/21.7.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang CE, O'Hearn SF, Sollner-Webb B. Assembly and function of the RNA editing complex in Trypanosoma brucei requires band III protein. Mol. Cell Biol. 2002;22:3194–3203. doi: 10.1128/MCB.22.9.3194-3203.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Hearn SF, Huang CE, Hemann M, Zhelonkina A, Sollner-Webb B. Trypanosoma brucei RNA editing complex: band II is structurally critical and maintains band V ligase, which is nonessential. Mol. Cell Biol. 2003;23:7909–7919. doi: 10.1128/MCB.23.21.7909-7919.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang B, Ernst NL, Palazzo SS, Panigrahi AK, Salavati R, Stuart K. TbMP44 is essential for RNA editing and structural integrity of the editosome in Trypanosoma brucei. Eukaryot. Cell. 2003;2:578–587. doi: 10.1128/EC.2.3.578-587.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carnes J, Trotter JR, Ernst NL, Steinberg A, Stuart K. An essential RNase III insertion editing endonuclease in Trypanosoma brucei. Proc. Natl Acad. Sci. USA. 2005;102:16614–16619. doi: 10.1073/pnas.0506133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salavati R, Ernst NL, O'Rear J, Gilliam T, Tarun S., Jr, Stuart K. KREPA4, an RNA binding protein essential for editosome integrity and survival of Trypanosoma brucei. RNA. 2006;12:819–831. doi: 10.1261/rna.2244106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Babbarwal VK, Fleck M, Ernst NL, Schnaufer A, Stuart K. An essential role of KREPB4 in RNA editing and structural integrity of the editosome in Trypanosoma brucei. RNA. 2007;13:737–744. doi: 10.1261/rna.327707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carnes J, Trotter JR, Peltan A, Fleck M, Stuart K. RNA editing in Trypanosoma brucei requires three different editosomes. Mol. Cell Biol. 2008;28:122–130. doi: 10.1128/MCB.01374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Law JA, O'Hearn SF, Sollner-Webb B. Trypanosoma brucei RNA editing protein TbMP42 (band VI) is crucial for the endonucleolytic cleavages but not the subsequent steps of U-deletion and U-insertion. RNA. 2008;14:1187–1200. doi: 10.1261/rna.899508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarun SZ, Jr, Schnaufer A, Ernst NL, Proff R, Deng J, Hol W, Stuart K. KREPA6 is an RNA-binding protein essential for editosome integrity and survival of Trypanosoma brucei. RNA. 2008;14:347–358. doi: 10.1261/rna.763308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schnaufer A, Panigrahi AK, Panicucci B, Igo R.P., Jr, Wirtz E, Salavati R, Stuart K. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science. 2001;291:2159–2162. doi: 10.1126/science.1058955. [DOI] [PubMed] [Google Scholar]
- 23.Seiwert SD, Heidmann S, Stuart K. Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell. 1996;84:831–841. doi: 10.1016/s0092-8674(00)81062-4. [DOI] [PubMed] [Google Scholar]
- 24.Igo RP, Jr, Palazzo SS, Burgess ML, Panigrahi AK, Stuart K. Uridylate addition and RNA ligation contribute to the specificity of kinetoplastid insertion RNA editing. Mol. Cell Biol. 2000;20:8447–8457. doi: 10.1128/mcb.20.22.8447-8457.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Igo RP, Jr, Weston DS, Ernst NL, Panigrahi AK, Salavati R, Stuart K. Role of uridylate-specific exoribonuclease activity in Trypanosoma brucei RNA editing. Eukaryot. Cell. 2002;1:112–118. doi: 10.1128/EC.1.1.112-118.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang B, Salavati R, Heidmann S, Stuart K. A hammerhead ribozyme substrate and reporter for in vitro kinetoplastid RNA editing. RNA. 2002;8:548–554. doi: 10.1017/s135583820202962x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amaro RE, Schnaufer A, Interthal H, Hol W, Stuart KD, McCammon JA. Discovery of drug-like inhibitors of an essential RNA-editing ligase in Trypanosoma brucei. Proc. Natl Acad. Sci. USA. 2008;105:17278–17283. doi: 10.1073/pnas.0805820105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Alessio JM. RNA sequencing. In: Rickwood D, Hames BD, editors. Gel Electrophoresis of Nucleic Acids: A Practical Approach. Oxford: IRL Press; 1982. pp. 173–97. [Google Scholar]
- 29.Stuart K, Kable ML, Allen TE, Lawson S. Investigating the mechanism and machinery of RNA editing. Methods. 1998;15:3–14. doi: 10.1006/meth.1998.0601. [DOI] [PubMed] [Google Scholar]
- 30.Stuart K, Gobright E, Jenni L, Milhausen M, Thomashow L, Agabian N. The IsTaR 1 serodeme of Trypanosoma brucei: development of a new serodeme. J. Parasitol. 1984;70:747–754. [PubMed] [Google Scholar]
- 31.Stuart K, Panigrahi AK, Schnaufer A. Identification and characterization of trypanosome RNA-editing complex components. Methods Mol. Biol. 2004;265:273–291. doi: 10.1385/1-59259-775-0:273. [DOI] [PubMed] [Google Scholar]
- 32.Schneider A, Charriere F, Pusnik M, Horn EK. Isolation of mitochondria from procyclic Trypanosoma brucei. Methods Mol. Biol. 2007;372:67–80. doi: 10.1007/978-1-59745-365-3_5. [DOI] [PubMed] [Google Scholar]
- 33.Palazzo SS, Panigrahi AK, Igo RP, Salavati R, Stuart K. Kinetoplastid RNA editing ligases: complex association, characterization, and substrate requirements. Mol. Biochem. Parasitol. 2003;127:161–167. doi: 10.1016/s0166-6851(02)00333-x. [DOI] [PubMed] [Google Scholar]
- 34.Panigrahi AK, Schnaufer A, Carmean N, Igo R.P., Jr, Gygi SP, Ernst NL, Palazzo SS, Weston DS, Aebersold R, Salavati R, et al. Four related proteins of the Trypanosoma brucei RNA editing complex. Mol. Cell Biol. 2001;21:6833–6840. doi: 10.1128/MCB.21.20.6833-6840.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 36.Panigrahi AK, Schnaufer A, Ernst NL, Wang B, Carmean N, Salavati R, Stuart K. Identification of novel components of Trypanosoma brucei editosomes. RNA. 2003;9:484–492. doi: 10.1261/rna.2194603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho CK, Shuman S. Bacteriophage T4 RNA ligase 2 (gp24.1) exemplifies a family of RNA ligases found in all phylogenetic domains. Proc. Natl Acad. Sci. USA. 2002;99:12709–12714. doi: 10.1073/pnas.192184699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jenne A, Hartig JS, Piganeau N, Tauer A, Samarsky DA, Green MR, Davies J, Famulok M. Rapid identification and characterization of hammerhead-ribozyme inhibitors using fluorescence-based technology. Nat. Biotechnol. 2001;19:56–61. doi: 10.1038/83513. [DOI] [PubMed] [Google Scholar]
- 39.Liang S, Connell GJ. An electrochemiluminescent aptamer switch for a high-throughput assay of an RNA editing reaction. RNA. 2009;15:1929–1938. doi: 10.1261/rna.1720209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takenaka M, Brennicke A. In vitro RNA editing in pea mitochondria requires NTP or dNTP, suggesting involvement of an RNA helicase. J. Biol. Chem. 2003;278:47526–47533. doi: 10.1074/jbc.M305341200. [DOI] [PubMed] [Google Scholar]
- 41.Sharma PM, Bowman M, Madden SL, Rauscher F.J., III, Sukumar S. RNA editing in the Wilms' tumor susceptibility gene, WT1. Genes Dev. 1994;8:720–731. doi: 10.1101/gad.8.6.720. [DOI] [PubMed] [Google Scholar]