Abstract
MicroRNAs (miRNAs) are naturally occurring small RNAs that regulate the expression of several genes. MiRNAs’ targeting rules are based on sequence complementarity between their mature products and targeted genes’ mRNAs. Based on our present understanding of those rules, we developed an algorithm to design artificial miRNAs to target simultaneously a set of predetermined genes. To validate in silico our algorithm, we tested different sets of genes known to be targeted by a single miRNA. The algorithm finds the seed of the corresponding miRNA among the solutions, which also include the seeds of new artificial miRNA sequences potentially capable of targeting these genes as well. We also validated the functionality of some artificial miRNAs designed to target simultaneously members of the E2F family. These artificial miRNAs reproduced the effects of E2Fs inhibition in both normal human fibroblasts and prostate cancer cells where they inhibited cell proliferation and induced cellular senescence. We conclude that the current miRNA targeting rules based on the seed sequence work to design multiple-target artificial miRNAs. This approach may find applications in both research and therapeutics.
INTRODUCTION
The lack of good therapeutic strategies to deal with complex diseases is pressuring scientists to rethink the way they approach complex human disorders such as cancer and cardiovascular diseases. Because cancer cell development involves the deregulation of multiple genes (1), it is reasonable to believe that inhibiting a single gene is not the best solution to address the problem. In addition, the specific gene expression signature of different cancer cell types suggests we may need to simultaneously inhibit the expression of multiple genes. SiRNAs are effective at gene silencing, but they are designed to target single mRNAs (2). MiRNAs have naturally evolved to inhibit the expression of several genes (3–5), suggesting that it could be possible to use them as tools to inhibit multiple genes.
MiRNA-guide sequences are about 22-nt long and repress the expression of specific genes by guiding the RNA-silencing complex (RISC) to complementary sequences in messenger RNAs (6,7). The guide sequences are located on one strand of stem–loop precursor miRNAs from which they are obtained by enzymatic processing. MiRNA-guide sequences tightly bind to the Argonaute protein of RISCs through their backbone. It is believed that a fair fraction (∼30%) of miRNAs initiate and stabilize the interaction with their targets by involving their nucleotides in positions 2–8 (the seed) in Watson–Crick base pairs with their target mRNAs (7,8). However, in many cases, the seed nucleotides do not have perfect (Watson–Crick) matches with their targets (9) and base pairing with the rest of the miRNAs may compensate to stabilize the interaction. Nevertheless, we implemented in a computer program the current rules of miRNA-target recognition to design artificial miRNAs against gene sets. As a proof of concept, we also tested successfully a few artificial sequences generated by our program.
MATERIALS AND METHODS
Cells and tissue culture
PC3 were obtained from American Type Culture Collection (ATCC) and cultured in RPM1 (GIBCO) supplemented with 10% FBS (Hyclone). IMR90 cells were obtained from ATCC and cultured in Dulbecco’s modified Eagle medium (DMEM, GIBCO) supplemented with 10% fetal bovine serum (FBS, Hyclone) and 1% penicillin G/streptomycin sulfate (GIBCO).
Small-RNA expression
To express our smart RNAs, we used a strategy developed by Paddison et al. (10) where the sequence of the small RNA of interest is cloned in the miR-30 endogenous miRNA pri-precursor backbone. The goal is to form a stem–loop structure where the mature miR-30 sequence is replaced by the sequence of interest, in our case a smart RNA. To do so a PCR template is synthesized containing the 5′ flanking stem sequence of miR-30, the sequence of the small RNA of interest, the mir-30 loop sequence, the complementary sequence of the small RNA of interest and the 3′ flanking stem sequence of miR-30. The miR-30 flanking sequences and loop allow an efficient maturation and expression of the small RNA of interest. This template is amplified by PCR using universal primers, with restriction sites at the 5′ and 3′ flanking miR-30 precursor sequences. The sequence of the templates and universal primers can be found in the Supplementary Table S1. The PCR product is then digested with EcoRI and XhoI, and ligated in the MLP retroviral vector that contain the extending miR-30 flanking sequence. The MLP vector is described in ref. (41). The miR-20 precursor sequence was also cloned in MLP vector as described in ref. (16). Retroviral-mediated gene transfer was performed using retroviral particles produced in Phoenix packaging cells as previously described (42). IMR90 and PC3 cell lines were then infected with the viruses and selected 48 h with puromycin.
miR-20 detection
miR-20 levels in our different cell lines were measured by qPCR. Total RNA was extract with Trizol reagent (Invitrogen) and 2 µg were reverse transcribed and then amplified with a TaqMan microRNA Assay kit specific for miR-20a (Ambion #000580). The level of miR-20 expression was normalized over the expression of U54.
Luciferase assay
Twenty-four hours before transfection, HeLa cells were plated at 50 000 cells per well in a 24-well plate. The pGL3-control plasmids containing the wild type 3′UTR of E2F1, E2F2 or E2F3 were transfected (50 ng) with pRL-globin (50 ng), and the smart RNAs, miR-20 or hairpin control (250 ng) using Lipofectamine LTX (Invitrogen). The luciferase assay was performed 24 h post-transfection using the dual luciferase reporter assay system (Promega) and firefly luciferase activity was normalized to Renilla luciferase activity for each transfected well. The experiment was conducted three times in triplicate.
Western blot
IMR90 cells were trypsinized and washed one time with PBS. The pellet was resuspended in 100 μl of Laemmli buffer and heated 5 min at 95°C. The proteins were quantified with the Bradford reagent and 20 µg were loaded on a 10% SDS–PAGE and transferred to Immobilon-P membranes (Millipore). The following antibodies were used for western blot: anti-E2F1 KH-20 mouse (1 μg/ml), anti-E2F2 CC11 mouse (1 μg/ml), anti-α-tubulin (B-5-1-2 1:5000 mouse; Abcam ab54945, 1:500), anti-E2F3 (ab54945, 1:500 mouse). Signals were revealed after incubation with anti-mouse secondary antibody (1:1500) coupled to peroxidase (GE Healthcare) by using ECL (GE Healthcare).
Growth curves
Twenty-five thousand cells per well were plated into 12-well plates. At the indicated times, cells were washed with PBS, fixed in 4% formaldehyde, and rinsed with distilled water. Cells were stained with 0.1% crystal violet (Sigma) for 30 min, rinsed extensively, and dried. Cell-associated dye was extracted with 2.0 ml 10% acetic acid. Aliquots were diluted 1:4 with H2O, transferred to 96-well microtiter plates, and the optical density at 590 nm was determined. Values were normalized to the optical density at day 0 for the appropriate cell type. Within an experiment, each point was determined in triplicate.
Senescence-associated β-galactosidase
Senescence-associated β-galactosidase activity was detected as previously described with slight modifications (11). Cells were washed once with PBS (pH 7.2), fixed with 0.5% glutaraldehyde (PBS [pH 7.2]), and washed in PBS (pH 7.2) supplemented with 1 mM MgCl2. Cells were stained in X-gal solution (1 mg/ml X-gal, 0.12 mM K3Fe[CN]6, 0.12 mM K4Fe[CN]6, 1 mM MgCl2 in PBS at pH 6.0) overnight at 37°C . Blue cells were counted in triplicate.
Colony formation assay
A total of 500 PC3 GFP+ cells were plated in a 6-well plate and were allowed to form colonies for 8–10 days. Colonies were fixed with glutaraldehyde, stained with crystal violet and counted manually. Data represent the average of three experiments.
Energy calculations and folding
All ΔG and RNA secondary structure predictions were calculated using the Vienna RNA package (12).
RESULTS AND DISCUSSION
Designing small multiple-target artificial (SMART) RNAs to target multiple genes simultaneously is the reverse of finding miRNA targets. Therefore, we implemented known miRNA targeting rules (13) in a computer program (MultiTar) that starts by searching common heptamers in the set of 3′UTRs of the genes we want to target. These heptamers represent common seed binding sites (SBS) for potential Smart RNAs for these genes. We favor SBS that: (i) create low-energy duplexes with the seed; and (ii) expose globally and locally the SBS in the mRNA structure. Once a set of SBSs have been identified and ranked, we use a Tabu search to determine the sequence of the rest of the miRNA (nucleotides 9–22; see Supplementary Methods).
The Tabu search is a local optimization method. It improves the local search by marking a previously found and potential solution as ‘taboo’ so to avoid visiting that solution again (14). We generate a number of sequences by mutating one nucleotide at a time and compute their average hybridization affinities with the mRNAs. We keep the solution that obtains the best average energy at the end of the search. Known examples suggest that the seed alone often suffices to make a miRNA repress the translation of its mRNA targets (8). However, good complementarity in the 3′ region of a miRNA can sometimes compensate for mismatches in the seed (15). Thus, we assign the final scores based on the full miRNA–mRNA duplex complementarity (positions 1–22).
Because it has been shown that both strands can be recruited in the RISC complex, we maximize the incorporation of the guide strand to limit the non-specific effect of the sense strand. To do so, we avoid high thermodynamic stability at the 5′-end of the guide strand, a condition that favors loading into the RISC complex (16,17). This is accomplished in the program by an optional rule (asymmetry rule) where the thermodynamic stability of the 5′-end of the guide strand can be reduced by decreasing its GC content. For more details about the MultiTar algorithm, see Supplementary Figure S1 and the Supplementary Information. The code of MultiTar is available upon request.
We wanted to test whether our algorithm would find the endogenous miRNAs if we give it the sets of natural genes they target. Given the 3′UTR sequence of the validated targets of miR-20 (18–27), miR-206 (28,29) and miR-21 (30–34), our program identified the seed sequences of the corresponding endogenous miRNAs (Table 1). The thermodynamic stability of the seed and its match for these functional miRNAs was highly variable (Table 1). However, the final seed score that takes into account the accessibility of the seed (see ‘Materials and Methods’ section) was more homogenous, suggesting that parameters other than the simple thermodynamics of base pairing influence the activity of miRNAs. We compared the efficiency of MultiTar to TargetCombo, which takes the intersection of the predictions made by TargetScanS, Miranda and Pictar. We took the top predictions (Table 1) for miR-10a and miR-22. In both cases, MultiTar identified the correct seed sequence of the corresponding endogenous miRNAs. Interestingly, prediction ranks for miR-20 and miR-22 improved as the number of targets increases (Table 1).
Table 1.
Finding endogenous miRNAs
| Input genes | miRNA and rank | Seed ΔG score | Total average seed score |
|---|---|---|---|
| E2F1, E2F2, E2F3, RBL2 | miR20 (66/93) | 0.44 | 0.54 |
| E2F1, E2F2, E2F3, CCND1, p21 | miR-20 (2/8) | 0.44 | 0.52 |
| E2F1, E2F2, E2F3, CCND1, p21, RBL2 | miR-20 (1/2) | 0.44 | 0.56 |
| E2F1, E2F2, E2F3, CCND1, p21, RBL2 PTPPRO | miR-20 (1/3) | 0.44 | 0.55 |
| E2F1, E2F2, E2F3, CCND1, p21, RBL2 NCOA3 | miR-20 (1/1) | 0.44 | 0.56 |
| E2F1,E2F2, E2F3, CCND1, p21, RBL2, PTPRO, NCOA3, HIF1 TGFBR2 | miR-20 (1/1) | 0.44 | 0.55 |
| FSTL1, UTRN, GJA1 | miR-206 (8/41) | 0.34 | 0.62 |
| PDCD4, BCL2, SPROUTY2, MTAP, SOX5 | miR-21 (1/1) | 0.38 | 0.54 |
| PAFAH1B1, CIDSPL, SDC1, CTDSYL, ID4 | miR-10a (3/24) | 0.63 | 0.62 |
| PTEN, NDEL1, IL13RA1 | miR-22 (2/33) | 0.74 | 0.65 |
| PTEN, NDEL1, IL13RA1, PLAG2, PTPN9, Cul3 | miR-22 (1/1) | 0.74 | 0.66 |
The endogenous miRNAs are found given their corresponding target genes. All these targets have perfect seed binding sites. The ranking refers to the scores of the solutions found by the algorithm (ranking/number of solutions). The highest scores are ranked first. The scores are between 0 and 1, where 1 indicates perfect matching (see Supplementary methods for their definition).
Next, we used MultiTar to find Smart RNAs that could target simultaneously E2F1, E2F2 and E2F3. The E2Fs are transcription factors that have redundant function in activating gene expression (35) and on cell cycle progression (36). Simultaneous inactivation of E2F1-3 by a conditional gene targeting approach in mice blocks cell cycle progression. However, cells can proliferate normally with only one of the three E2F1-3 genes (36). We have recently shown that these three genes are targeted by miR-20 (20). MiR-20 is found in the solutions of MultiTar for E2F1-3. Its low scoring rank (66th solution of the program) can be explained by the fact that this miRNA is not optimized for recognition of only these three genes. As seen in Table 1, miR-20 is the second best solution when the three E2Fs are combined with RBL2, another target of miR-20.
We experimentally tested the top solutions obtained with different options of the program (Table 2). MT-E2Fs(1) and MT-E2Fs(2) were the best smart RNAs found when we looked for only one binding site in each E2Fs with a perfect seed match. MT-E2Fs(3) targets two sites with one mismatch allowed in the 3′UTR of each E2F (multiplicity option). In all cases shown in Table 2 we activated the asymmetry rule to facilitate the loading of the guide strand into the RISC.
Table 2.
Smart RNAs against E2F1-3
| Name | Sequence | M | Target site |
||
|---|---|---|---|---|---|
| E2F1 | E2F2 | E2F3 | |||
| miR-20 | UAAAGUGCUUAUAGUGCAGGUAG | NA | 363–395 | 881–913 | 1800–1832 |
| 956–988 | 1495–1527 | ||||
| 3259–3291 | |||||
| MT-E2Fs(1) | UAUCUGACUUACGUGACUGCUU | 1 | 740–772 | 974–1006 | 1188–1220 |
| MT-E2Fs(2) | UUUCCCAAUUUCGCCCGGCCCU | 1 | 707–739 | 2019–2051 | 1838–1870 |
| MT-E2Fs(3) | UAGUGGGGAGGGGGUUUCCGGU | 2 | 93–125 | 2262–2294 | 709–741 |
| 946–978 | 2734–2766 | 1219–1251 | |||
M, multiplicity rule. miRNA seeds are underlined.
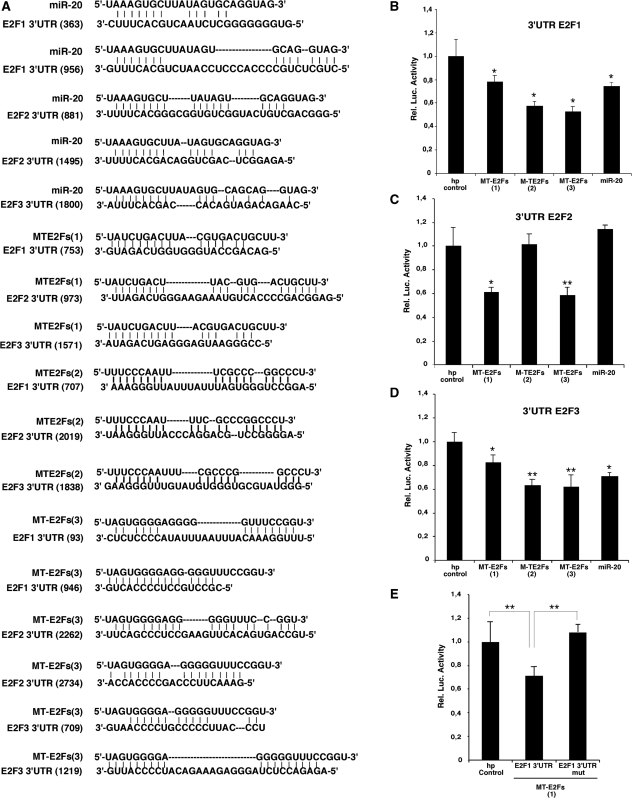
To study the ability of these smart RNAs to repress E2Fs’ activity, we cloned them as DNA cassettes coding for small hairpin RNAs (shRNAs), which upon expression in cells are expected to be processed into mature smart RNAs. The different MT-E2Fs were co-expressed in HeLa cells with a luciferase reporter containing the 3′UTR of E2F1, E2F2 or E2F3 that we used before to test the ability of miR-20 to regulate these mRNAs (20). Base-pairing between these smart RNAs and their target sequences are shown in Figure 1A. When tested with the reporter having the 3′UTR of E2F1, all three smart RNAs reduced its expression similarly to miR-20 (Figure 1B). A similar result was found with the E2F2 reporter. However, MT-E2Fs(2) did not reduce expression of the E2F2 reporter because its binding site is not present in this E2F2-3'UTR reporter sequences (14) (Figure 1C). For the E2F3 3′UTR, the results were identical as for that of E2F1 (Figure 1D). To show that the inhibition induced by our smart RNAs depends on the sites predicted by MultiTar, we mutated the seed match for MT-E2Fs(1) in the 3′UTR of E2F1 for its complementary sequence. For the mutated seed match, we observed luciferase activity up to the control levels (Figure 1E).
Figure 1.
Artificial miRNAs (Smart RNAs) targeting E2F1, E2F2 and E2F3. (A) Base pairing between miR-20, the three MT-E2Fs and the E2Fs binding sites. (B–D) MT-E2Fs target E2F1-3 luciferase reporters. (B) A reporter luciferase gene fused to E2F1 3′UTR. (C) A reporter luciferase gene fused to E2F2 3′UTR. (D) A reporter luciferase gene fused to E2F3 3′UTR. (E) Luciferase activity from HeLa cells co-transfected with MT-E2F(1) and E2F1 3′UTR or a mutant lacking the binding site for MT-E2F1. Paired t-test: *P < 0.05 and **P < 0.02.
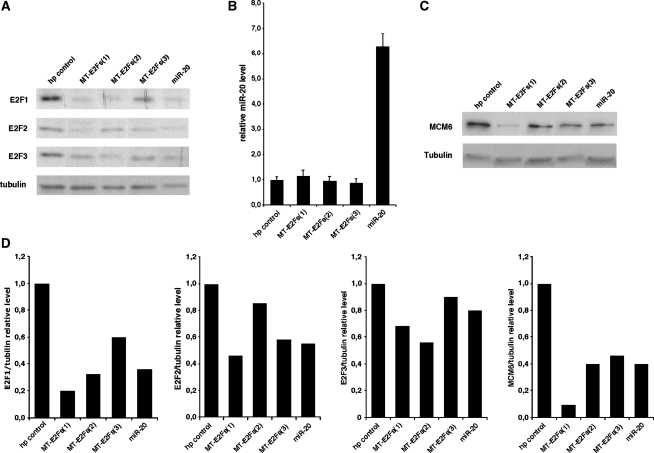
Next we tested the effect of our smart RNAs on endogenous E2F1-3 in normal fibroblasts IMR90 using immunoblots. All three MT-E2Fs reduced E2F1-3 expression as expected (Figure 2A). None of these MT-E2Fs changed the levels of miR-20, the endogenous miRNA known to target E2Fs (Figure 2B). Because the main function of the E2Fs depends on their transcriptional activity, we measured expression of Mcm6, a transcriptional target of the E2Fs required for DNA replication (37). The three MT-E2Fs decrease Mcm6 levels in comparison to the control hairpin (Figure 2C). The decrease was comparable to that induced by miR-20, and was more pronounced for MT-E2Fs(1), which is also the smart RNAs that had the highest inhibitory effect on the E2Fs.
Figure 2.
MT-E2Fs target E2F1-3 endogenous protein. (A) Immunoblots on endogenous E2F1, E2F2 and E2F3 in IMR90 cells expressing the smart RNAs against E2F1-3, control hairpin and miR-20. (B) Measure of the relative miR-20 level by qPCR in the different cell lines expressing the smart RNAs against E2F1-3, control hairpin and miR-20. (C) Mcm6 immunoblots of IMR90 cells expressing the smart RNAs against E2F1-3, a control hairpin and miR-20. (D) Quantification of the immunoblots presented in (A and C) showing the relative level of E2F1, E2F2, E2F3 and MCM6 normalized on the tubulin expression for the different cell lines.
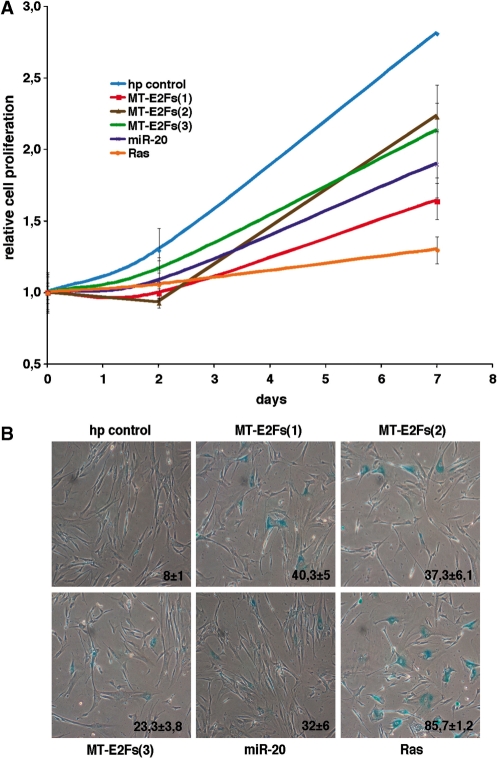
Next, we verified the biological impact of the decreased E2Fs’ activity by our smart RNAs. Blocking E2F activity is expected to arrest cell proliferation, and in some conditions induce cellular senescence (38). We observed a good correlation between the extent of E2Fs decrease, Mcm6 decrease (Figure 2A and C) and that of inhibition of cell proliferation by our MT-E2Fs (Figure 3A). This result suggests that our smart RNAs can efficiently inhibit their three targets and neutralize the redundant function of the three E2Fs in cell cycle progression. Maehara et al. showed that a siRNA against DP1, an essential cofactor for the transcriptional activity of E2F1-3, induced senescence (38). We thus performed a senescence-associated β-galactosidase staining to measure senescence induction in our cells infected with the three smart RNAs, and observed senescence induction for all MT-E2Fs (Figure 3B).
Figure 3.
MT-E2Fs targeting E2F1-3 functions. (A) Growth curves of normal human fibroblasts expressing the smart RNAs against E2F1-3, a control hairpin, miR-20, and RasV12 a positive control for growth arrest and senescence. (B) Senescence-associated β-galactosidase of normal human fibroblasts expressing the smart RNAs against E2F1-3, a control hairpin, miR-20, and RasV12. Cells were stained at day 8 post-selection. Data represent the average and standard deviation of three independent experiments.
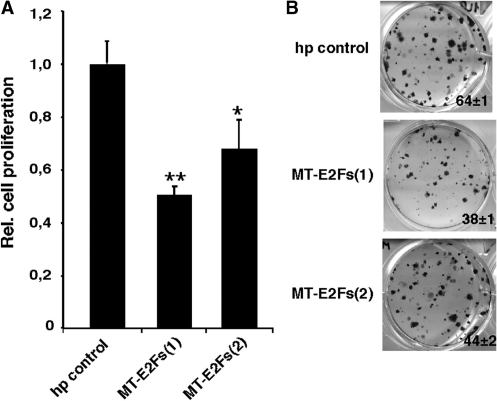
To measure the impact of the inhibition made by our smart RNAs in cancer cells, we infected PC3 prostate cancer cells with our retroviral vectors expressing MT-E2Fs(1) and MT-E2Fs(2). Then, we evaluated cell growth by clonogenic assays, which are commonly used to evaluate anti cancer drug efficacy in culture. Again, our smart RNAs were effective in inhibiting cell proliferation (Figure 4A). We also observed a decrease in the number of colonies formed (Figure 4B).
Figure 4.
Effect of MT-E2Fs(1) and MT-E2Fs(2) on prostate cancer cells. (A) Relative cell growth of PC3 cells expressing MT-E2Fs(1), MT-E2Fs(2) and a hairpin control. (B) Colony formation ability of PC3 cells expressing MT-E2Fs(1), MT-E2Fs(2) and a hairpin control. Paired t-test: *P < 0.05; **P < 0.02.
Our results indicate that we can engineer smart RNAs to inhibit the expression of a pre-determined set of genes. This approach is not limited to sets of genes of the same family. For instance, the 3′UTR of the E2Fs are heterogeneous in length and sequence. A multiple sequence alignment between the three sequences using EMBL Clustal W2 gives distances of 0.48651, 0.49250 and 0.48654 for E2F1, E2F2 and E2F3, respectively. A similar calculation for the unrelated genes CCND1, CDKN1 and RBL2 gave numbers in the same range (0.48734, 0.49699 and 0.47777, respectively)
To estimate the upper bound of the number of genes that can be targeted using the targeting rules employed in MultiTar, we searched for heptamers common to a group of n sequences in a collection of randomly generated sequences of different lengths. For sequences of 3000 nt (our estimate of the average length of a mammalian 3′UTR), we found common heptamers for up to five sequences (Supplementary Figure S2). The probabilities of finding solutions are even higher when one or two mismatches in the seed region are allowed. In 124 validated miRNA targets in TarBase, 20% include one mismatch and 5% two mismatches. In our sets of randomly generated sequences, one or two mismatches allow to find many common heptamers for up to 10 genes (Supplementary Figure S2). Since it is now accepted that miRNAs also target the coding region (39), this means that our approach can find artificial miRNAs for more than 10 genes.
The asymmetry rule can be used to maximize the incorporation of the sequence of interest in the RISC complex but could eliminate some potential solutions. For example, in the in silico validation of MultiTar, this rule prevented us to find the seed sequence of miR-10a, miR-21 and miR-22 given their respective known targets. In fact, these miRNAs contain more than one G or C in their first four nucleotides. It was reported that endogenous miRNAs precursors can express two different miRNAs that are encoded on each strand of the stem (40). Hence, natural miRNAs may sometimes avoid the asymmetry rule. In the case of smart RNAs, it is important to limit the expression of the sense sequence to minimize non-specific effects. To maximize the number of solutions suggested by MultiTar, the asymmetry rule in the program is optional.
The MultiTar strategy provides a novel approach to the problem of inhibiting/repressing multiple genes simultaneously. Alternative approaches include the co-expression of one siRNA per gene. For example, Cheng and colleagues (41) showed an induction of cell death in prostate cancer cells using a cassette expressing six shRNAs. Interestingly, the inhibition of only one gene by the expression of single-gene targeting shRNAs was not enough to induce cell death, suggesting the requirement of multi-targeting. Liu et al. (42) developed an expression cassette based on the miR-17-92 polycistron to express several shRNAs to target HIV mRNAs. These strategies achieved good inhibition of the targets, but necessitated the co-expression of several siRNAs that may increase non-specific effects. In addition, the use of several siRNAs can saturate the RISC complex, triggering toxicity by inhibiting endogenous miRNA functions (43). Another difference between Smart RNAs and siRNAs is the degree of target inhibition. SiRNAs inhibit target gene expression with high efficiency, while Smart RNAs has a microRNA-like effect inhibiting target gene expression by 1.2- to 2-fold. Therefore, Smart RNAs are not substitutes of siRNAs and their power of regulation is based on their effects on multiple targets. For example, it has been shown that the deletion of one of the E2Fs is not sufficient to affect cell proliferation (36). However, the simultaneously moderate inhibition on the three E2Fs induced by our Smart RNAs was enough to decrease cell proliferation.
Smart RNAs, like siRNAs will have off-target effects as first described by Jackson et al. (44). The nature and number of off-target effects will depend on each particular seed sequence. Therefore, as recommended for siRNAs, more than one solution should be tested for each gene set to ensure specificity of any biological effect observed. It is also plausible that some sequences may have more targets than others because they recognize certain repeated patterns in the genome. To minimize as much as possible the non-specific effects of our Smart RNAs, additional bioinformatic tools are required to predict whether other genes could fulfill the recognition criteria for every MulTar solution. The use of the asymmetry rule, or modified small RNAs limiting the expression of the passenger strand or pools of smart RNAs could limit the non-specific off target effects of smart RNAs as well. Nevertheless, our results with MT-E2Fs support the concept of custom design miRNAs. These Smart RNAs with the ability to modulate gene expression patterns may find multiple applications in both research and therapeutics.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Prostate Cancer Canada (to G.F.); Fonds de la Recherche en Santé du Québec (FRSQ; to V.D.G., P.C., G.F.); Canadian Institute of Health and Research (F.M., G.F.). Funding for open access charge: Canadian Prostate Cancer Research Foundation (to G.F.); Canadian Institutes of Health and Research (to F.M., G.F.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Ferbeyre and Major labs for their comments. F.M. is a member of the Robert-Cedergren Centre of the Université de Montréal.
REFERENCES
- 1.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 3.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 4.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 5.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: A comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paddison PJ, Cleary M, Silva JM, Chang K, Sheth N, Sachidanandam R, Hannon GJ. Cloning of short hairpin RNAs for gene knockdown in mammalian cells. Nat. Methods. 2004;1:163–167. doi: 10.1038/nmeth1104-163. [DOI] [PubMed] [Google Scholar]
- 11.Bandyopadhyay D, Gatza C, Donehower LA, Medrano EE. Analysis of cellular senescence in culture in vivo: the senescence-associated beta-galactosidase assay. In: Bonifacino JS, Dasso M, Harford JB, Lippincott-Schwartz J, Yamada KM, editors. Current Protocols in Cell Biology. 2005. Chapter 18, Unit 18.9. [DOI] [PubMed] [Google Scholar]
- 12.Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 14.Cvijovicacute D, Klinowski J. Taboo search: an approach to the multiple minima problem. Science. 1995;267:664–666. doi: 10.1126/science.267.5198.664. [DOI] [PubMed] [Google Scholar]
- 15.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 17.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 18.Hossain A, Kuo MT, Saunders GF. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol. Cell Biol. 2006;26:8191–8201. doi: 10.1128/MCB.00242-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 20.Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G, Chartrand P. An E2F/miR-20a autoregulatory feedback loop. J. Biol. Chem. 2007;282:2135–2143. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- 21.Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J. Biol. Chem. 2007;282:2130–2134. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- 22.Fontana L, Fiori ME, Albini S, Cifaldi L, Giovinazzi S, Forloni M, Boldrini R, Donfrancesco A, Federici V, Giacomini P, et al. Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PloS one. 2008;3:e2236. doi: 10.1371/journal.pone.0002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu Z, Wang C, Wang M, Li Z, Casimiro MC, Liu M, Wu K, Whittle J, Ju X, Hyslop T, et al. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J. Cell Biol. 2008;182:509–517. doi: 10.1083/jcb.200801079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taguchi A, Yanagisawa K, Tanaka M, Cao K, Matsuyama Y, Goto H, Takahashi T. Identification of hypoxia-inducible factor-1 alpha as a novel target for miR-17-92 microRNA cluster. Cancer Res. 2008;68:5540–5545. doi: 10.1158/0008-5472.CAN-07-6460. [DOI] [PubMed] [Google Scholar]
- 25.Xu X, Hong Y, Kong C, Xu L, Tan J, Liang Q, Huang B, Lu J. Protein tyrosine phosphatase receptor-type O (PTPRO) is co-regulated by E2F1 and miR-17-92. FEBS Lett. 2008;582:2850–2856. doi: 10.1016/j.febslet.2008.07.017. [DOI] [PubMed] [Google Scholar]
- 26.Wang Q, Li YC, Wang J, Kong J, Qi Y, Quigg RJ, Li X. miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proc. Natl Acad. Sci. USA. 2008;105:2889–2894. doi: 10.1073/pnas.0800178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenberg MI, Georges SA, Asawachaicharn A, Analau E, Tapscott SJ. MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J. Cell Biol. 2006;175:77–85. doi: 10.1083/jcb.200603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson C, Catoe H, Werner R. MIR-206 regulates connexin43 expression during skeletal muscle development. Nucleic Acids Res. 2006;34:5863–5871. doi: 10.1093/nar/gkl743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 31.Sayed D, Rane S, Lypowy J, He M, Chen IY, Vashistha H, Yan L, Malhotra A, Vatner D, Abdellatif M. MicroRNA-21 targets Sprouty2 and promotes cellular outgrowths. Mol. Biol. Cell. 2008;19:3272–3282. doi: 10.1091/mbc.E08-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y, Liu W, Chao T, Zhang Y, Yan X, Gong Y, Qiang B, Yuan J, Sun M, Peng X. MicroRNA-21 down-regulates the expression of tumor suppressor PDCD4 in human glioblastoma cell T98G. Cancer Lett. 2008;272:197–205. doi: 10.1016/j.canlet.2008.06.034. [DOI] [PubMed] [Google Scholar]
- 33.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 34.Wickramasinghe NS, Manavalan TT, Dougherty SM, Riggs KA, Li Y, Klinge CM. Estradiol downregulates miR-21 expression and increases miR-21 target gene expression in MCF-7 breast cancer cells. Nucleic Acids Res. 2009;37:2584–2595. doi: 10.1093/nar/gkp117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeGregori J, Leone G, Miron A, Jakoi L, Nevins JR. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc. Natl Acad. Sci. USA. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu L, Timmers C, Maiti B, Saavedra HI, Sang L, Chong GT, Nuckolls F, Giangrande P, Wright FA, Field SJ, et al. The E2F1-3 transcription factors are essential for cellular proliferation. Nature. 2001;414:457–462. doi: 10.1038/35106593. [DOI] [PubMed] [Google Scholar]
- 37.Ohtani K, DeGregori J, Leone G, Herendeen DR, Kelly TJ, Nevins JR. Expression of the HsOrc1 gene, a human ORC1 homolog, is regulated by cell proliferation via the E2F transcription factor. Mol. Cell Biol. 1996;16:6977–6984. doi: 10.1128/mcb.16.12.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maehara K, Yamakoshi K, Ohtani N, Kubo Y, Takahashi A, Arase S, Jones N, Hara E. Reduction of total E2F/DP activity induces senescence-like cell cycle arrest in cancer cells lacking functional pRB and p53. J. Cell Biol. 2005;168:553–560. doi: 10.1083/jcb.200411093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ro S, Park C, Young D, Sanders KM, Yan W. Tissue-dependent paired expression of miRNAs. Nucleic Acids Res. 2007;35:5944–5953. doi: 10.1093/nar/gkm641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng TL, Teng CF, Tsai WH, Yeh CW, Wu MP, Hsu HC, Hung CF, Chang WT. Multitarget therapy of malignant cancers by the head-to-tail tandem array multiple shRNAs expression system. Cancer Gene Ther. 2009;16:516–531. doi: 10.1038/cgt.2008.102. [DOI] [PubMed] [Google Scholar]
- 42.Liu YP, Haasnoot J, ter Brake O, Berkhout B, Konstantinova P. Inhibition of HIV-1 by multiple siRNAs expressed from a single microRNA polycistron. Nucleic Acids Res. 2008;36:2811–2824. doi: 10.1093/nar/gkn109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 44.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.