Abstract
Next-generation sequencing has proven an extremely effective technology for molecular counting applications where the number of sequence reads provides a digital readout for RNA-seq, ChIP-seq, Tn-seq and other applications. The extremely large number of sequence reads that can be obtained per run permits the analysis of increasingly complex samples. For lower complexity samples, however, a point of diminishing returns is reached when the number of counts per sequence results in oversampling with no increase in data quality. A solution to making next-generation sequencing as efficient and affordable as possible involves assaying multiple samples in a single run. Here, we report the successful 96-plexing of complex pools of DNA barcoded yeast mutants and show that such ‘Bar-seq’ assessment of these samples is comparable with data provided by barcode microarrays, the current benchmark for this application. The cost reduction and increased throughput permitted by highly multiplexed sequencing will greatly expand the scope of chemogenomics assays and, equally importantly, the approach is suitable for other sequence counting applications that could benefit from massive parallelization.
INTRODUCTION
Next-generation sequencing (NGS) technologies can generate up to several hundred million reads of DNA sequence per lane or slide, and this capacity continues to increase at a rapid pace. This massive capacity has allowed exploration of diverse biological questions (1–4). Although pooled chemogenomic screens of compound–gene interactions in yeast (5–16) and mammalian cells (17,18) are typically assessed using barcode microarrays, counting of individual strains could also be assessed by barcode sequencing. We recently developed such an assay (Bar-seq) to monitor thousands of gene–chemical interactions (19). We now expand upon this proof-of-principle to interrogate 96 samples in parallel, developing the methodology and analytical tools to use NGS to simultaneously monitor several hundred thousand gene–environment interactions using a method that should be readily adaptable to an automated workflow.
Here, we demonstrate successful multiplexing of samples obtained from 96 distinct pooled yeast growth assays, with each sample comprising 6200 uniquely barcoded yeast mutants. This 96-plex experiment represents a 150-fold increase in unique observations over our proof-of-principle assessment, and provides substantial cost reduction/experiment over microarrays. Furthermore, while many aspects of microarray assay costs are fixed, the cost of multiplex barcode sequencing continues to decline as the number of reads per experiment increases. Indeed, this increase in sequencing rate has recently been shown to outpace the rate of Moore’s law (20). To assess the data quality at this level of multiplexing, all 96 samples were also assessed by microarray and we then compared the ability of both platforms to detect specific compound–gene interactions. It is expected that the principle of this 96-fold multiplexing application, with its ability to discriminate many sample types/slide or flow cell can be applied, with modification, to other molecular counting methods such as RNA-seq (21), ChIP-seq (22), promoter assays (23), histone occupancy (24) and Tn-seq (25). To systematically test highly multiplexed Bar-seq, we required a large pool of distinct sequences whose relative abundances could be varied and whose quantities could also be assessed by an orthogonal method. The Yeast Knock Out collection of 6200 Saccharomyces cerevisiae mutants, although designed for testing gene function, provides a suitable test bed for new sequencing methods (19). Each yeast deletion mutant contains three salient features: a dominant drug resistance marker replacing the deleted gene; two unique 20 base molecular barcodes; and universal primers that flank each barcode to allow amplification of all barcodes in a pooled manner using a single set of primers. Pooled competitive growth assays are typically carried out on 6200 mutants, and their relative abundances inferred from the signal from a barcode microarray (5–16). The rapid pace of advance in sequencing depth have led us and others to exploring diverse strategies for multiplexing of samples for NGS samples (19,25–34).
One essential element for multiplexing prior to sequencing is the incorporation (in this instance using modified primers during PCR) of a unique experimental indexing tag (See Supplementary Figure S1 for structure of PCR amplicon). Following PCR, the amplified DNA is purified and quantified, then pooled with amplicons derived from other samples with different indexing tags. The pooled PCR products are then purified from a single lane of a polyacrylamide gel, reducing costs and sample preparation time. Further, combining samples prior to purification reduces potential liquid transfer errors, providing for greater uniformity, and also reducing the number of emulsion PCRs reactions required prior to di-nucleotide sequencing on the SOLiD V3 instrument. In our 20- and 96-plex sequencing runs, two independent reads were obtained for each feature (Supplementary Figure S1): the first sequence read was primed from the P1 adapter sequence, capturing the sequence of the first common primer (U1) and the yeast barcode. The second sequencing read, primed from the IA internal adaptor captures the SOLiD indexing tag used to assign barcode reads to 96 individual sample bins. Data from a real-world 96-plex assay show performance equal to or exceeding both microarray data and published lower complexity multiplexing Bar-seq data. The potential for higher order multiplexing of thousands of samples offers the prospect of greatly reduced cost and such ‘extreme-multiplexing’ will benefit from robust automation to ensure sample uniformity.
METHODS AND MATERIALS
Yeast deletion collection
The yeast deletion collection was obtained from Angela Chu at the Stanford Genome Technology Centre, and stored in YPD-7% dimethyl sulfoxide (DMSO) in −80°C as individual strains in 96-well plates. The plates were thawed and robotically pinned onto YPD agar plates. Cells are grown in 30°C for 2–3 days until colonies form. Slow growing strains were grown separately for 2–3 additional days. All plates were then flooded with 5–7 ml of media, scraped and pooled in YPD-7% DMSO to a final concentration of OD600 = 50, and frozen at −80°C until use, as described by Pierce et al. (11).
Construction of pools with fixed numbers of barcoded strains
A pool of 953 different heterozygous mutants was selected to contain two well-known drug targets as heterozygous deletions. ‘Pool-constant’ was constructed by growing each strain in 100 µl of YPD to saturation in 96-well plates then pooling 20 µl from each well, so that all 953 strains in this pool are at approximately the same abundance. ‘Pool-variable’ consisted of the same 953 strains but in this pool, the number of cells of each strain was varied systematically with one-quarter of the 953 strains added at one of the following ratios (2 : 1 : 0.5 : 0.25) with respect to Pool-constant.
Pooled growth assays
Two deletion pools, a homozygous deletion pool of 5054 strains representing non-essential genes and a heterozygous pool of 1194 strains representing genes essential for viability, were thawed and diluted in YPD to an OD600 of 0.0625. Seven hundred microliters of cultures were grown at 30°C with a chemical inhibitor applied at a dose that produced 10–20% growth inhibition of wild-type. An automated liquid handler was used to maintain logarithmic growth of pools (by dilution), and to collect 0.7 OD600s of heterozygous pool following 20 generations of growth, and 1.4 OD600s of homozygous pool following five generations of growth.
Assessing fitness of barcoded yeast strains by barcode microarray
Except where indicated, pooled assays were performed as described by Pierce et al. (11). Genomic DNA was isolated from cells and barcodes amplified and hybridized to barcode microarrays, where each barcode deletion mutant is represented by 10 hybridization signals (an uptag and downtag for each strain, each present on the array five times). Array signals were quantile normalized such that all tags hybridized with the sample pool had similar distributions. Following normalization, a correction factor was applied to the array data to correct for feature saturation (11) and the fitness of each barcoded deletion strain determined using the uptag barcodes only (to compare to the sequencing samples which contained only uptags as well). Positive fitness defect scores signify a decrease in strain abundance during drug treatment and suggest that the wild-type version of the gene deleted in that strain is required for resistance to that drug or inhibitor.
Assessing fitness of barcoded yeast strains by SOLiD sequencing
DNA was isolated from the deletion pools as described (11). Each 20-mer uptag barcode was amplified with composite primers comprised of the sequences of the common barcode primers and the sequences required for attachment to the SOLiD slide. For the Uptags the following primers were used:
5′-CTGCCCCGGGTTCCTCATTCTCTNNNNNNNN NNCTGCTGTACGGCCAAGGCGGTCGACCTGCAGCGTACG-3′ (Forward) and
5′-CCTCTCTATGGGCAGTCGGTGATGATGTCCACGAGGTCTCT-3′ (Reverse). The 5′ portion (in bold) are the P2 and P1 sequences incorporated into the F and R primer, respectively. The variable sequence (italics) represents the 10-mer indexing tag used for multiplexing. The internal adaptor (IA) sequence (bold italics) is required to sequence the SOLiD multiplexing tags. The 3′ portion (underlined) represents the common primer flanking the uptag barcode and is required to amplify the yeast barcodes. PCR amplification was conducted in 100 μl volumes, using Invitrogen Platinum PCR Supermix (Cat. No. 11306-016) with the following conditions: 95°C/3 min; 25 cycles of 94°C/30 s, 55°C/30 s, 68°C/30 s; followed by 68°C/10 min. PCR product was then purified with Qiagen MinEluteTM 96 UF PCR Purification Kit (Cat. No. 28051). Following PCR purification, DNA was quantified with the Invitrogen Quant-iTTM dsDNA BR Assay Kit (Cat No. Q32853) and then adjusted to a concentration of 10 µg/ml. Equal volumes of normalized DNAs were then pooled. This pooled DNA (20- and 96-plex) consisting of 130-bp PCR products was gel purified from 12% polyacrylamide TBE gels using the crush and soak method (35) followed by ethanol precipitation. Samples were used directly for emulsion PCR and bead enrichment. Each bead on the slide was hybridized twice, first to the P1 primer to sequence the yeast barcode (SOLiD Fragment Library Sequencing kit—Master Mix 50 Cat No. 4406370; SOLiD 3 Instrument Buffer kit Cat No. 4406479) and then to the internal adaptor (IA) to sequence the SOLiD multiplexing tag (SOLiD Fragment Library Sequencing kit Barcode set Cat No. 4406447). The 10-base multiplexing tag allowed postsequencing assignment of each amplicon to a particular experiment. The identity of each bead’s multiplexing tag was determined allowing 0 mismatches. To analyze the Bar-seq data, all counts were quantile normalized such that each experiment had the same count distribution. By analogy with barcode microarray fitness experiments, fitness defect ratios for each strain were calculated and expressed as the log2 ratio (control/treatment).
RESULTS
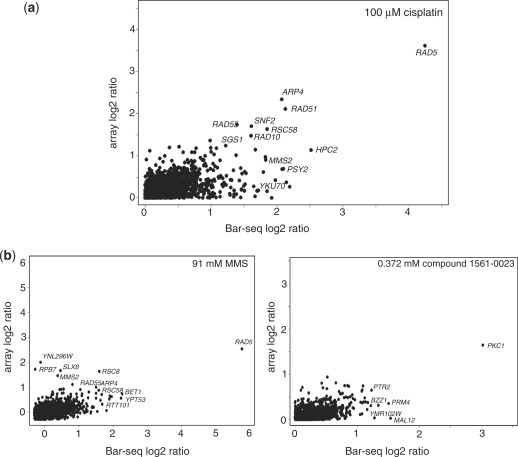
As a proof of principle, we performed a 20-plex analysis of five samples; two untreated controls and three chemically treated samples (four replicates each plus a spike-in control consisting of five unique samples). We found an average correlation of r = 0.989 between raw, unfiltered counts in replicate experiments, indicating the sequencing assay is highly reproducible. The same five samples were hybridized to barcode microarrays. Treatment of the yeast deletion pool with the DNA damaging agent cisplatin resulted in fitness defects in strains deleted for a variety of DNA repair genes, when either sequencing or array were used as the readout [Figure 1a and Supplementary Table S1 (10)]. Treatment of the yeast deletion pool with the drug cervistatin showed a similar concordance between Bar-seq and array readouts. For example, in cervistatin treatment the strain deleted for Hmg1 [the known drug target (19)] scored as one of the top hits in both the microarray and Bar-seq assays (Supplementary Figure S2). The fitness defect ratios across all strains were well correlated (r = 0.739).
Figure 1.
(a) The log2 ratio of control versus treatment for one example from the 20-plex Bar-seq data. The x-axis is the log2 ratio derived from the Bar-seq data, while the y-axis is the log2 ratio derived from barcode microarray data. The profiles are from a 100 µM cisplatin treatment. The black dots represent genes above our threshold; greater than 200 signal intensity in the barcode microarray and greater than 40 barcode sequence counts in the Bar-seq. (b) The log2 ratio of control over treatment for two examples from the 96-plex Bar-seq data. The x-axis is the log2 ratio derived from the Bar-seq data, the y-axis is the log2 ratio derived from barcode microarray data. The left panel shows the result from 91-mM MMS treatment, the right panel shows the profile obtained with an uncharacterized compound 1561-0023 screened at 0.372 mM. The black dots represent genes which were above our threshold; greater than 200 signal intensity in the barcode microarray and greater than 40 barcode sequence counts in Bar-seq.
We next expanded the degree of multiplexing from 20 to 96 individual experiments. The 96-plex experiment includes 12 DMSO-treated vehicle controls, 84 drug-treated samples and 5 spike-in controls. The experimental samples were selected based on the availability of existing hybridization data from our laboratory.
To assess our ability to resolve data from a 96-plex experiment, we first examined the spike-in controls that consisted of a pool of 953 yeast deletion mutants combined at the same relative abundance (Pool-constant). This pool was amplified in four separate PCR reactions, each reaction having a different multiplexing primer with a unique SOLiD indexing tag. Each sample was added to the sequencing reaction at 1 of 4 relative quantities (1×, 2×, 4× or 8×). Comparison between these samples using count ratios (Supplementary Figure S3) indicated that there were sufficient sequencing counts in the 96-plex experiment to identify a 4-fold difference in barcode abundance between two samples. While the counts do not scatter symmetrically around the expected ratio line (likely reflecting slight imprecision at the pooling step) the populations are distinct. Figure 1b shows two samples from the 96-plex that clearly reveal distinct drug targets. Methyl methanesulfonate (MMS), a DNA damaging agent, shows that the Rad5 deletion strain exhibits a prominent fitness defect after analysis by both multiplexed NGS and microarray [Supplementary Table 2 (10)]. Results obtained with a second, previously uncharacterized compound 1561-0023 (from ChemDiv, Inc.) suggest that it may be an inhibitor of Pkc1, a serine/threonine kinase involved in cell wall remodeling during growth (36). We compared the structure of this inhibitor to 150 known Pkc1 inhibitors present in the ‘Chembl’ database (http://www.ebi.ac.uk/chembldb/index.php using ECFP_4 fingerprints to represent the structures and a Tanimoto score cutoff of >0.3). This analysis found no similar structures, suggesting this compound is a potentially novel chemical probe for inhibiting Pkc1.
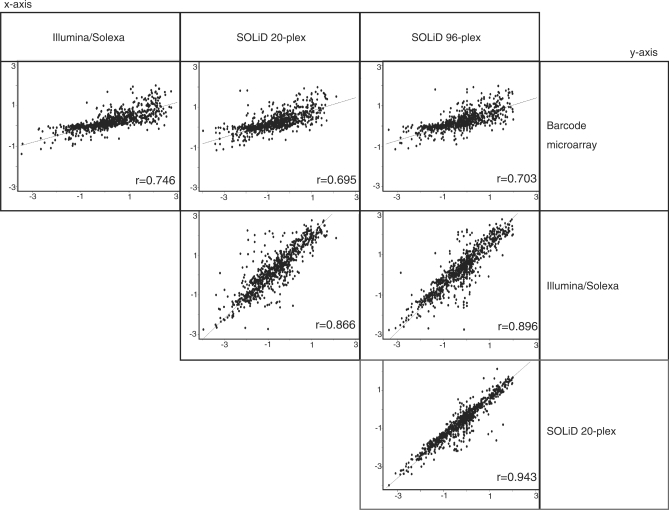
To assess if increasing the degree of multiplexing from 20 to 96 affected either data quality or data reproducibility, we examined the correlation of signal ratios in the spike-in samples that were present in both sequencing runs, using Pool-constant (at 8× abundance) and a second pool of the same 953 strains divided into four levels of abundance relative to Pool-constant (i.e. 2×, 1×, 0.5× and 0.25× of Pool-constant). Ratios were highly correlated between 20- and 96-plex runs (r = 0.943; Figure 2). Since the same pool of 953 strains had previously been assessed with an Illumina Genome Analyzer IIx, we performed a cross-platform comparison. The ratios showed a strong agreement between both methods with correlations of, r = 0.866, 0.896 for the SOLiD 20- and 96-plex, respectively (Figure 2). The ratios calculated by microarray analysis were similar to ratios calculated on either platform at each level of multiplexing (r ranging from 0.695–0.746; Figure 2).
Figure 2.
Scatter plots of the Log2 ratio of the Pool-constant versus the Pool-variable. The data were generated on three different platforms, barcode microarray, Illumina/Solexa, and SOLiD. The SOLiD data is derived from 20- and 96-plex samples. The r-value is indicated for each comparison and the line of best fit is plotted. Data presented were filtered for barcodes that were present in both Illumina/Solexa and SOLiD sequencing counts with counts greater than zero in both platforms.
We also assessed the performance of the SOLiD multiplexing tags used in the 96-plex experiment. Specifically, index tags were first assigned to samples only when there was a perfect match to the expected sequence. We then reanalyzed this data to allow up to two mismatches to the index sequence to ask if additional reads could be mapped, without degrading performance due to misaligned reads. In total, ∼320 million reads were collected in the 96-plex experiment (including spike-in controls, this sample comprises 104 different index sequences). Of these sequences, 64.5% were separated into bins based on a perfect match to the 10-bp index sequence. Of the 96 samples generated from the complete pool, the average bin size was 2.1 million reads. The coefficient of variation across these 96 samples was 19%.
By allowing either one or two mismatches in the index sequence, the total percentage of reads binned increased from 64.5% to 80%. With one mismatch, the percentage increase across the 96 bins ranges between 5.5% and 14.8%, with a further increase of 6.0–17% observed with two mismatches (Supplementary Table S3 and Figure S4).
We examined a number of sequence characteristics in the index sequence to identify any factors that might contribute to the variation in the increased bin sizes after allowing mismatches. The GC content of the 10-base index ranged between 20% and 80% with a median GC content of 50%. No correlation was observed between the GC content and the increased bin sizes after either one or two mismatches. Furthermore, the index sequences with GC content at the extremes (20%, 80%) did not show significant differences in bin increases versus index sequences with balanced GC. Mononucleotide runs of 3 or 4 bases were observed in 31 and 4% of the index sequences, respectively. Index sequences containing these runs did not differ significantly from those that did not have these runs with respect to the increases in bin size after allowance of mismatches. We next examined variation in the dinucleotide pair at the end of the index barcode immediately 3′ to the sequencing primer. Of the 16 possible dinucleotides, 13 were present. No significant differences were seen across these classes. Finally, to determine if any of the index sequences introduced a secondary structure that might affect sequencing accuracy. We determined the free energy (37) of the 70-base sequence common to all reactions, which differed only by the index sequence (sequencing primer/adapter + index + spacer + upstream barcode). Maximal free energies ranged between −11.69 and −5.04 kcal/mol. No correlation was seen between free energy and increases in bin sizes after allowance of mismatches.
A few index barcodes in the 96-plex experiment displayed relatively large increases in bin size after allowance of mismatches compared with most of the sequences. These all appeared in the five samples generated from contrived pools (bins 100–104). After allowance of a single mismatch in the index barcode, bin 100 increased by 53%, followed by a further 24% increase after allowance of two mismatches. After two mismatches, bins 102 and 103 also increased by 32 and 45%, respectively (Supplementary Table S3 and Figure S4). No primary sequence characteristic was observed in these index sequences, which was also not observed in the other index sequences. For these three indexes, the GC content was either 40 or 50%, only a single mononucleotide run of 3 bases was observed, the dinucleotide end sequences were not unique and the free energy was at the less negative end of the range, suggesting less secondary structure interference. Based on this initial analysis of the index sequences, we conclude that despite their sequence differences, their performance for this application is robust.
DISCUSSION
We previously determined, by simulation, that a minimum of 50 counts per barcode was sufficient for accurate assessment of strain abundance in Bar-seq analysis (19). In the 96-plex experiment each barcode was counted, on average, 262 times (ranging from 111 counts to 383 counts/barcode). In contrast, the 20-plex experiment counted each barcode an average of 1224 times. Both the counts between replicates and the relative strain abundance in our spike-in pools are highly correlated between the 20- and 96-plex runs, suggesting that empirically 100–300 counts/barcode is sufficient. Additional studies will be required to determine the actual lower limit of counts at different false discovery thresholds. From a practical perspective, we were able to clearly identify drug targets in many of the experiments in the 96-plex run (Figure 1b), suggesting higher order (>96) multiplexing can be achieved. Fitness defect ratios for all strains/pool were highly correlated across different platforms (SOLiD, Illumina/Solexa and microarray; Figure 2). This matched sequencing array data set of a 20- and 96-plex samples should be useful for assessment of platform-specific performance. Furthermore, given the similarity in experimental design of yeast deletion pool experiments and highly multiplexed shRNA assays in mammalian cells (38), the methods described here should translate well to these more complex pooled assays.
Finally, the data presented in this study may prove useful by providing a large data set to augment computer-aided barcode design efforts (33,39,40), which will become increasingly important as the levels of multiplexing increase further. In summary, we present a robust 96-plex method for sequencing pools of medium complexity (1000–50 000 members/pool) that should be applicable to other sequencing applications.
SUPPLEMENTARY DATA
Supplemental figures are available at NAR Online. Raw data available at: http://chemogenomics.med.utoronto.ca/supplemental/multiplex/.
FUNDING
Funding for open access charge: Canadian Institutes for Health Research (MOP-81340, to G.G, MOP-84305, to C.N.); National Human Genome Research Institute (HG00317-05).
Conflict of interest statement. J.B., A.N.H. and K.M.P. are employees of ABI/Life Technologies. The other authors have declared none.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Alex Robertson and Anjali Shah at ABI/Life Technologies and Malene Urbanus in the HIPHOP lab for helpful comments on the manuscript.
REFERENCES
- 1.Alkan C, Kidd JM, Marques-Bonet T, Aksay G, Antonacci F, Hormozdiari F, Kitzman JO, Baker C, Malig M, Mutlu O, et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat. Genet. 2009;41:1061–1067. doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hillier LW, Marth GT, Quinlan AR, Dooling D, Fewell G, Barnett D, Fox P, Glasscock JI, Hickenbotham M, Huang W, et al. Whole-genome sequencing and variant discovery in C. elegans. Nat. Methods. 2008;5:183–188. doi: 10.1038/nmeth.1179. [DOI] [PubMed] [Google Scholar]
- 4.Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ericson E, Gebbia M, Heisler LE, Wildenhain J, Tyers M, Giaever G, Nislow C. Off-target effects of psychoactive drugs revealed by genome-wide assays in yeast. PLoS Genet. 2008;4:e1000151. doi: 10.1371/journal.pgen.1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giaever G, Flaherty P, Kumm J, Proctor M, Nislow C, Jaramillo DF, Chu AM, Jordan MI, Arkin AP, Davis RW. Chemogenomic profiling: identifying the functional interactions of small molecules in yeast. Proc. Natl Acad. Sci. USA. 2004;101:793–798. doi: 10.1073/pnas.0307490100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giaever G, Shoemaker DD, Jones TW, Liang H, Winzeler EA, Astromoff A, Davis RW. Genomic profiling of drug sensitivities via induced haploinsufficiency. Nat. Genet. 1999;21:278–283. doi: 10.1038/6791. [DOI] [PubMed] [Google Scholar]
- 8.Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D, et al. The chemical genomic potrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho CH, Magtanong L, Barker SL, Gresham D, Nishimura S, Natarajan P, Koh JL, Porter J, Gray CA, Andersen RJ, et al. A molecular barcoded yeast ORF library enables mode-of-action analysis of bioactive compounds. Nat. Biotechnol. 2009;27:369–377. doi: 10.1038/nbt.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee W, St Onge RP, Proctor M, Flaherty P, Jordan MI, Arkin AP, Davis RW, Nislow C, Giaever G. Genome-wide requirements for resistance to functionally distinct DNA-damaging agents. PLoS Genet. 2005;1:e24. doi: 10.1371/journal.pgen.0010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pierce SE, Davis RW, Nislow C, Giaever G. Genome-wide analysis of barcoded Saccharomyces cerevisiae gene-deletion mutants in pooled cultures. Nat. Protoc. 2007;2:2958–2974. doi: 10.1038/nprot.2007.427. [DOI] [PubMed] [Google Scholar]
- 12.Pierce SE, Fung EL, Jaramillo DF, Chu AM, Davis RW, Nislow C, Giaever G. A unique and universal molecular barcode array. Nat. Methods. 2006;3:601–603. doi: 10.1038/nmeth905. [DOI] [PubMed] [Google Scholar]
- 13.St Onge RP, Mani R, Oh J, Proctor M, Fung E, Davis RW, Nislow C, Roth FP, Giaever G. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat. Genet. 2007;39:199–206. doi: 10.1038/ng1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan Z, Costanzo M, Heisler LE, Paw J, Kaper F, Andrews BJ, Boone C, Giaever G, Nislow C. Yeast Barcoders: a chemogenomic application of a universal donor-strain collection carrying bar-code identifiers. Nat. Methods. 2008;5:719–725. doi: 10.1038/nmeth.1231. [DOI] [PubMed] [Google Scholar]
- 15.Parsons AB, Lopez A, Givoni IE, Williams DE, Gray CA, Porter J, Chua G, Sopko R, Brost RL, Ho CH, et al. Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell. 2006;126:611–625. doi: 10.1016/j.cell.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 16.Pan X, Yuan DS, Xiang D, Wang X, Sookhai-Mahadeo S, Bader JS, Hieter P, Spencer F, Boeke JD. A robust toolkit for functional profiling of the yeast genome. Mol. Cell. 2004;16:487–496. doi: 10.1016/j.molcel.2004.09.035. [DOI] [PubMed] [Google Scholar]
- 17.Silva JM, Marran K, Parker JS, Silva J, Golding M, Schlabach MR, Elledge SJ, Hannon GJ, Chang K. Profiling essential genes in human mammary cells by multiplex RNAi screening. Science. 2008;319:617–620. doi: 10.1126/science.1149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 19.Smith AM, Heisler LE, Mellor J, Kaper F, Thompson MJ, Chee M, Roth FP, Giaever G, Nislow C. Quantitative phenotyping via deep barcode sequencing. Genome Res. 2009;19:1836–1842. doi: 10.1101/gr.093955.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Editorial Metagenomics versus Moore's Law. Nat. Methods. 2009;6:623. [Google Scholar]
- 21.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 23.Patwardhan RP, Lee C, Litvin O, Young DL, Pe'er D, Shendure J. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nat. Biotechnol. 2009;27:1173–1175. doi: 10.1038/nbt.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ozsolak F, Song JS, Liu XS, Fisher DE. High-throughput mapping of the chromatin structure of human promoters. Nat. Biotechnol. 2007;25:244–248. doi: 10.1038/nbt1279. [DOI] [PubMed] [Google Scholar]
- 25.van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat. Methods. 2009;6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mamanova L, Coffey AJ, Scott CE, Kozarewa I, Turner EH, Kumar A, Howard E, Shendure J, Turner DJ. Target-enrichment strategies for next-generation sequencing. Nat. Methods. 2010;7:111–118. doi: 10.1038/nmeth.1419. [DOI] [PubMed] [Google Scholar]
- 27.Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lefrancois P, Euskirchen GM, Auerbach RK, Rozowsky J, Gibson T, Yellman CM, Gerstein M, Snyder M. Efficient yeast ChIP-Seq using multiplex short-read DNA sequencing. BMC Genomics. 2009;10:37. doi: 10.1186/1471-2164-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cronn R, Liston A, Parks M, Gernandt DS, Shen R, Mockler T. Multiplex sequencing of plant chloroplast genomes using Solexa sequencing-by-synthesis technology. Nucleic Acids Res. 2008;36:e122. doi: 10.1093/nar/gkn502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porreca GJ, Zhang K, Li JB, Xie B, Austin D, Vassallo SL, LeProust EM, Peck BJ, Emig CJ, Dahl F, et al. Multiplex amplification of large sets of human exons. Nat. Methods. 2007;4:931–936. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- 31.Turner EH, Lee C, Ng SB, Nickerson DA, Shendure J. Massively parallel exon capture and library-free resequencing across 16 genomes. Nat. Methods. 2009;6:315–316. doi: 10.1038/nmeth.f.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Craig DW, Pearson JV, Szelinger S, Sekar A, Redman M, Corneveaux JJ, Pawlowski TL, Laub T, Nunn G, Stephan DA, et al. Identification of genetic variants using bar-coded multiplexed sequencing. Nat. Methods. 2008;5:887–893. doi: 10.1038/nmeth.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat. Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frank DN. BARCRAWL and BARTAB: software tools for the design and implementation of barcoded primers for highly multiplexed DNA sequencing. BMC Bioinformatics. 2009;10:362. doi: 10.1186/1471-2105-10-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Russell DW Cold Spring Harbor Laboratory. Molecular Cloning: A Laboratory Manual. 3rd. edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 2001. [Google Scholar]
- 36.SGD project. Saccharomyces genome database. 2008 Available at www.yeastgenome.org (15 January 2010, date last accessed) [Google Scholar]
- 37.Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33:W577–W581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, Wong KK, Elledge SJ. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parameswaran P, Jalili R, Tao L, Shokralla S, Gharizadeh B, Ronaghi M, Fire AZ. A pyrosequencing-tailored nucleotide barcode design unveils opportunities for large-scale sample multiplexing. Nucleic Acids Res. 2007;35:e130. doi: 10.1093/nar/gkm760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Q, Schlabach MR, Hannon GJ, Elledge SJ. Design of 240,000 orthogonal 25mer DNA barcode probes. Proc. Natl Acad. Sci. USA. 2009 doi: 10.1073/pnas.0812506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.