Abstract
Lysosomal β-hexosaminidase A (Hex A) is essential for the degradation of GM2 gangliosides in the central and peripheral nervous system. Accumulation of GM2 leads to severely debilitating neurodegeneration associated with Tay-Sachs disease (TSD), Sandoff disease (SD) and AB variant. Here, we present the X-ray crystallographic structure of Hex A to 2.8 Å resolution and the structure of Hex A in complex with NAG-thiazoline, (NGT) to 3.25 Å resolution. NGT, a mechanism-based inhibitor, has been shown to act as a chemical chaperone that, to some extent, prevents misfolding of a Hex A mutant associated with adult onset Tay Sachs disease and, as a result, increases the residual activity of Hex A to a level above the critical threshold for disease. The crystal structure of Hex A reveals an αβ heterodimer, with each subunit having a functional active site. Only the α-subunit active site can hydrolyze GM2 gangliosides due to a flexible loop structure that is removed post-translationally from β, and to the presence of αAsn423 and αArg424. The loop structure is involved in binding the GM2 activator protein, while αArg424 is critical for binding the carboxylate group of the N-acetyl-neuraminic acid residue of GM2. The β-subunit lacks these key residues and has βAsp452 and βLeu453 in their place; the β-subunit therefore cleaves only neutral substrates efficiently. Mutations in the α-subunit, associated with TSD, and those in the β-subunit, associated with SD are discussed. The effect of NGT binding in the active site of a mutant Hex A and its effect on protein function is discussed.
Keywords: lysosomal storage disorders, β-hexoasaminidase A, GM2 ganglioside, Tay-Sachs disease, glycoside hydrolase
Introduction
GM2 gangliosidosis is a family of three autosomal recessive, lysosomal storage disorders characterized by the intralysosomal accumulation of the acidic glycolipid GM2 ganglioside, primarily in the brain and peripheral neural tissues.1 Deficiencies of either the α-subunit or the β-subunit of the heterodimeric β-hexosaminidase A (Hex A) protein, or the small monomeric GM2 activator protein (a substrate-specific co-factor for Hex A) leads to the phenotypic neurodegeneration associated with this family of devastating disorders; i.e. Tay-Sachs disease (TSD), Sandhoff disease (SD) and the AB variant form, respectively.2
Hex A is a member of the Family 20 glycoside hydrolases (glycosidase) (EC 3.2.1.52).3 It removes the terminal non-reducing N-acetylgalactosamine (GalNAc) from the GM2 ganglioside. The α-subunit and β-subunit of human Hex A are encoded by the evolutionarily related genes HEXA and HEXB, respectively. The primary sequences of these subunits are approximately 60% identical. The GM2 activator protein (GM2AP) encoded by GM2A, is a lipid transporter that removes GM2 from its membranous environment and presents it to Hex A for hydrolysis.4
Hex A, as well as Hex B (a β-homodimeric Hex isozyme), can carry out the hydrolysis of β-linked GalNAc and/or N-acetylglucosamine (GlcNAc) from substrates, such as the oligosaccharide moieties from proteins and neutral glycolipids, or from certain mucopolysaccharides. The hydrolysis of the GM2 ganglioside, which contains a negatively charged sialic acid group, however, is carried out only by the α-subunit of Hex A.5,6 The specificity of this reaction is made absolute by the mechanism by which the GM2AP–GM2 complex interacts with the Hex A heterodimer.
The GM2 ganglioside, composed of GalNAcβ(1–4)-[NANAα(2–3)-]-Galβ(1–4)-Glc-ceramide, is primarily an intermediate in the synthesis and degradation of the higher brain gangliosides, e.g. GM1 ganglioside. Gangliosides are degraded in the lysosomes in a stepwise manner by interdependent exo-glycosidases. A number of different genetic disorders are the result of a deficiency of one of these exo-glycosidase or its co-factor, which prevents turnover of the remaining macromolecule.7,8 This results in the accumulation of partially degraded glycosphingolipid, e.g. GM2 ganglioside, primarily in neural tissue and resulting in neuro-degeneration.9,10
Mutations in HEXA, HEXB and GM2A genes causing GM2 gangliosidosis have been characterized in detail,2 and include partial gene deletion, splicing mutations, nonsense mutations and missense mutations. These mutations cause defects in transcription, translation, monomer folding and/or dimerization and, more rarely, in the catalytic function of Hex A. Different genotypes result in different clinical phenotypes, which generally correlate biochemically with the amount of residual Hex A activity.11 The most common, severe and fatal form is the acute or infantile onset forms of Tay-Sachs disease (ITSD) or infantile Sandhoff disease (ISD). ITSD and ISD are associated with a total deficiency of Hex A activity. However, in ITSD, total Hex activity is nearly normal due to the stable Hex B isozyme; whereas in ISD, total Hex activity is only ~3% of normal, due to the unstable Hex S (an α-homodimeric Hex isozyme). The less severe late on-set forms of GM2-gangliosidosis, i.e. juvenile/subacute and adult/chronic Tay-Sachs (ATSD), results from mutations that do not completely prevent the formation of catalytically active Hex A; with residual activities ranging from ~1–8% of normal levels. The rare variant AB form of GM2 gangliodosis is due to mutations in the GM2A gene and produces normal levels of both Hex A and Hex B when assayed with simple artificial substrates, but no activity when assayed using GM2 ganglioside as a substrate. In the Ashkenazi Jewish population, the rate of TSD is an astounding 1 in 30. For the general population, the rate is 1 in 300.1
Here, we report the crystallographic structure of the mature lysosomal form of Hex A from human placenta as a native structure to 2.8 Å resolution and co-crystallized with NAG-thiazoline (NGT) to 3.25 Å resolution. NGT is a mechanism-based inhibitor,12 shown to decrease endoplasmic reticulum (ER) retention and hence increase residual Hex A activity ~3-fold in ATSD cells homozygous for the αG269S mutation.13 The native structure reveals the mature heterodimeric, glycosylated α-subunit and β-subunit of Hex A. Two distinct active sites are present in Hex A, one on the α-subunit and one on the β-subunit. In both active sites, a glutamate residue acts as a general acid-base that assists in cleaving the terminal β-linked GalNAc or GlcNAc residues from substrates; whereas an adjacent aspartate residue stabilizes the positively charged oxazolinium intermediate that develops during the substrate-assisted catalytic mechanism carried out by human Hex18,21. In the α-active site, αAsn423 and αArg424 residues promote GM2 binding by interacting favorably with the negatively charged sialic acid residue present on the GM2 oligosaccharide structure. The corresponding residues in the β-subunit active site are βAsp452 and βLeu453, which would be expected to repel the negatively charged sialic acid moiety of GM2. The complex structure of Hex Awith NGTreveals the mechanism by which NGT acts as a chaperone, stabilizing the native conformation of the α-subunit and thereby promoting dimerization and allowing Hex A to exit the ER and to be targeted to the lysosome. These data provide an excellent starting point for therapeutic advancement toward the treatment of late on-set forms of GM2 gangliosidosis through structure-based drug design.
Results and Discussion
Crystallization and overall structure of Hex A
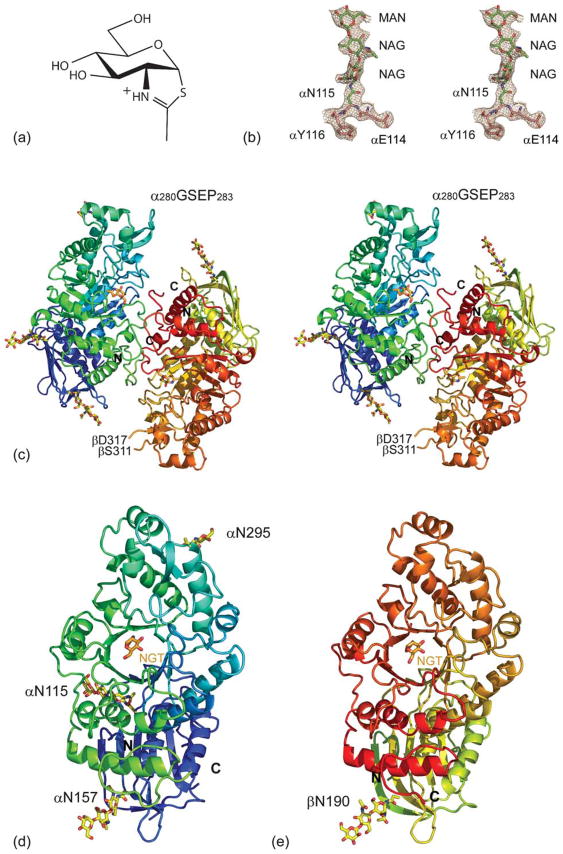
A functionally mature glycosylated form of lysosomal Hex A isolated from human placenta (Mr 112,500) was crystallized in the absence and in the presence of NGT (Figure 1(a)). The glycosylated form was maintained in an attempt to view the carbohydrate moieties involved in mannose-6-phosphate (M6P) receptor interaction(s). Numerous crystals of both native and inhibitor-bound Hex A were subjected to X-ray diffraction, the majority of which diffracted X-rays to approximately 4 Å resolution. In the end, only one native Hex A crystal could be found that diffracted X-rays beyond 3 Å resolution. Native Hex A crystallized in space group C2 with four Hex A heterodimers in the asymmetric unit; a total of 4044 residues are in the asymmetric unit. Well-defined electron density (Figure 1(b)) was obtained from phasing by molecular replacement using the biological dimer of Hex B as the search model. Hex A was built into the experimental electron density at 2.8 Å resolution and refined to an Rwork of 0.26 and an Rfree of 0.28 (Table 1). The biological dimer of Hex A is depicted in Figure 1(c) with the individual α and β-subunits represented in Figure 1(d) and (e), respectively. Hex A co-crystallized with NGT was refined to 3.25 Å resolution (Table 1). As in the native structure, NGT-bound Hex A also crystallized in space group C2 with four molecules in the asymmetric unit. NGT was present in all four α-subunits and four β-subunits in the asymmetric unit Hex A (Figure 1(d) and (e)).
Figure 1.
(a) Hex A structure. Chemical structure of NGT. (b) Stereo view of the 2Fo−Fc map contoured at 1σ on residues αN114 to αY116, including glycosylation at αN115. (c) Stereo view of a ribbon representation for Hex A. The α-subunit N terminus color begins with dark blue and continues to light blue, and then ends with light green at its C terminus. The β-subunit N terminus begins with a greenish yellow color, changing to orange and ending in red at the C terminus. NGT, located at the face of the TIM barrel, is shown in orange. (d) The individual α-subunit and (e) β-subunit are represented as viewed from the dimer interface.
Table 1.
X-ray diffraction data collection and atomic refinement
| A. Crystal information | ||
| Data set | Native | NGT |
| Space group | C2 | C2 |
| Solvent content (%, v/v) | 50.7 | 50.7 |
| Matthew’s coefficienta | 2.5 | 2.5 |
| Molecules/asymmetric unitb | 4 (4044) | 4 (3933) |
| Residues/asymmetric unit | ||
| B. Data collection | ||
| Unit cell dimensions | ||
| a (Å) | 321.1 | 322.2 |
| b (Å) | 110.5 | 109.8 |
| c (Å) | 129.7 | 132.8 |
| β (deg.) | 90.9 | 91.5 |
| Wavelength (Å) | 1.1158 | 1.1271 |
| Resolution range (Å) | 40.00 – 2.80 | 35.0 – 3.25 |
| High-resolution (Å) | 2.90 – 2.80 | 3.37 – 3.25 |
| Total observations | 406,584 | 144,007 |
| Unique reflections | 111,512 | 71,535 |
| 〈I/σI〉c,d | 9.7 (2.0) | 8.7 (1.9) |
| Completeness (%)e | 99.4 (99.7) | 97.8 (97.4) |
| B-value, Wilson plot (Å2) | 75 | 92 |
| Multiplicity | 3.6 | 2.0 |
| Rmergee | 0.089 (0.750) | 0.065 (0.475) |
| C. Refinement | ||
| Rworkf | 0.26 | 0.27 |
| Rfreeg | 0.28 | 0.32 |
| Number of atoms | 32,139 | 32,007 |
| Water | 151 | 11 |
| r.m.s.d from ideal | ||
| Bond lengths (Å) | 0.006 | 0.009 |
| Bond angles (deg.) | 0.818 | 1.09 |
| Ramachandran plot | ||
| Most favored (%)h | 2903 (86.0) | 2884 (85.5) |
| Allowed (%) | 461 (13.7) | 481 (14.3) |
| Generously allowed (%) | 13 (0.4) | 10 (0.3) |
| Disallowed (%) | 0 | 0 |
VM, Å3/Da.
Z, the number of molecules in the unit cell.
Statistics for the highest resolution shell are in parentheses.
〈I/σI〉 is the ratio between the mean intensity and the mean error of the intensity.
Rmerge Σhkl Σ|Ij(hkl)−〈I(hkl)〉|ΣhklΣj j〈I(hkl)〉, with Ij(hkl) representing the intensity of measurement j and 〈I(hkl)〉 is the mean of measurements for the reflection hkl. Although the Rmerge in the outer shell is high, the appropriate resolution limits were deduced from the Wilson plot.
Rwork = Σhkl ||Fobs(hkl)|−Fcalc(hkl)||/Σhkl |Fobs(hkl)|, where Fobs and Fcalc are the observed and calculated structure factors, respectively.
Rfree is calculated in the same manner on 5% of structure factors that were not used in the model refinement.
Numbers in parentheses represent the percentage of residues in each area of the Ramachandran plot.
The four heterodimers in the asymmetric unit of the Hex A crystals are structurally comparable, having an average r.m.s.d. of 0.40 Å for 920±23 matching Cα atoms. (See Supplementary Data for individual r.m.s.d. values for all structural superimpositions.) The NGT-bound Hex A has slightly better structural agreement, with an average r.m.s.d. of 0.31 Å for 961±4 matching Cα atoms. When the four heterodimers from the Hex A structure are superimposed with the four NGT-bound Hex A heterodimers, the average r.m.s.d. is 0.36 Å with 938±20 matching Cα atoms.
The overall structure of the Hex A heterodimer is similar to the structure of the Hex B homodimer, having an average r.m.s.d. of 0.65 Å for 915±8 matching Cα atoms. The NGT-bound Hex A is comparable when superimposed with Hex B, giving an average r.m.s.d. of 0.66 Å for 920±5 matching Cα atoms.
Subunit structure of Hex A
The α-subunit of Hex A is post-translationally cleaved to give the mature form,14 consisting of two polypeptide chains: αLys23 to αGly74, and αThr89 to αGln528 (Figure 1(d)).15 The β-subunit of Hex A is also cleaved post-translationally,16 to give the mature form consisting of three polypeptides: βAla50 to βGly107, βThr122 to βSer311, and βLeu316 to βMet556 (Figure 1(e)).17 With only 60% sequence identity, the structures of the α-subunits and β-subunits are comparable with an r.m.s.d. of 0.71 Å for 460±10 matching Cα atoms when structurally aligned. When the individual α-subunits and β-subunits from the NGT-bound Hex A structure are aligned, the average r.m.s.d. is 0.67 Å for 466±4 matching Cα atoms.
In our X-ray structure of Hex A, both the α-subunit and β-subunit reveal similar topologies. Each subunit consists of two domains. Domain I, residues Leu23 to Pro168 in the α-subunit and βAla50 to βPro201 in the β-subunit, is an N-terminal domain having two parallel α-helices sandwiched between a six-stranded anti-parallel β-sheet and domain II. The function of domain I in Hex A is unknown. Domain II, residues 165 to 529 in the α-subunit and 202 to 556 in the β-subunit, consists of a core TIM barrel fold ((β,α)8-barrel) with a helical insertion, αThr327 to αAsp347 in the α-subunit and βGlu362 to βThr378 in the β-subunit, as well as an extension at the C terminus (Figure 1(d) and (e)).
Important differences exist between the α-subunit and the β-subunit. The α280GSEP283 loop in the α-subunit is post-translationally cleaved in the β-subunit after βSer311 and before βAsp316 (Figure 1(c)). In addition, the α396IPV398 loop found in the α-subunit is not encoded by the HEXB mRNA for the β-subunit. From the structure of Hex B, a model of Hex A was generated, onto which the structure of the GM2A protein was docked.18 The model suggested the necessity for a flexible α280GSEP283 loop in order for the GM2A protein to interact with Hex A. It was demonstrated subsequently through biochemical studies with mutant forms of Hex A in which these loops had been deleted that the flexible α280GSEP283 loop plays the most important role in this interaction.19 Our current Hex A structure is consistent with the biochemical data and confirms the validity of the previous model derived from Hex B.
The active site of Hex A and the proposed mechanism of action
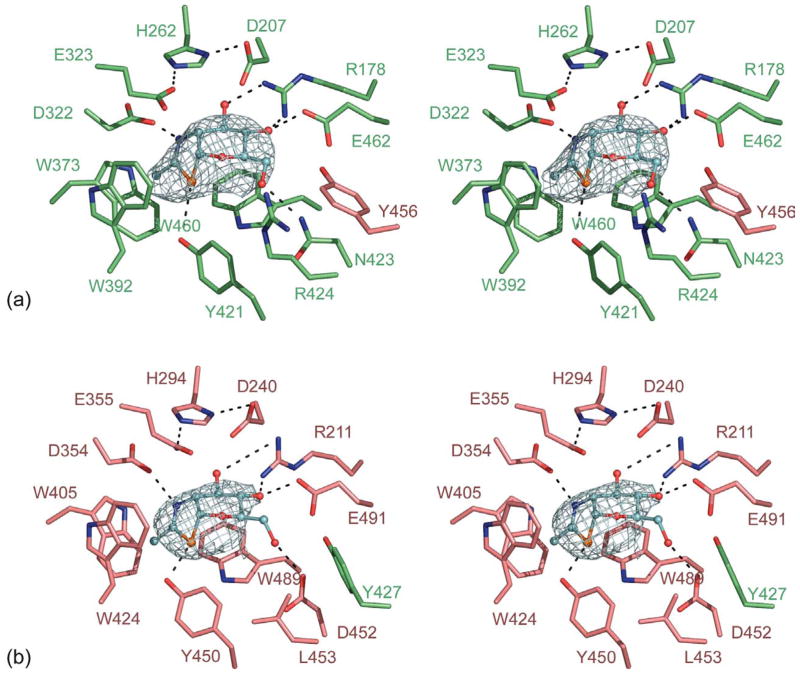
Two active sites are present in the Hex A dimer; one comprising residues from the α-subunit (Figure 2(a)) and a second one from residues of the β-subunit (Figure 2(b)). These active sites are located at the opening of the TIM barrels at the interface between the α and β-subunits. In the α-subunit, NGT is stabilized via hydrogen bonding with αArg178, αGlu462, αAsn423 αTyr421 and αAsp322 (Figure 2(a)). In the β-subunit, NGT forms hydrogen bonds with βArg211, βGlu491, βAsp452, βTyr450, and βAsp354 (Figure 2(b)). There is residue sharing in both active sites: βTyr456 is found in the α-subunit active site, whereas αTyr427 is found in the β-subunit active site. Although not observed in our structure of NGT-bound Hex A, previous analyses of the NGT-bound structure of Hex B solved at 2.5 Å demonstrated that a water molecule along with, βTyr456, stabilizes active site residues αGlu462 and αAsn423 in the α-subunit. These residues participate in hydrogen bonding with NGT bound within the α-subunit. A complementary stabilization takes place within the β-subunit, where αTyr427 hydrogen bonds with water to coordinate βGlu491 and βAsp452 in the active site of the β-subunit. The intimate interactions shared between the two active sites of both Hex A and Hex B suggest that dimerization is essential for activity in each subunit of these isoenzymes. These data are consistent with the lack of any biochemical evidence for the existence of an active α or β-monomeric form of Hex.2
Figure 2.
NGT bound in the active site of Hex A. (a) NGT (shown in blue) bound in the active site of the α-subunit (green) showing a minor contribution of βY456 from the β-subunit (pink). (b) NGT bound in the active site of the β-subunit (pink) showing a minor contribution of αY427 from the α-subunit (green). Unrefined Fo−Fc density shown for NGT is contoured at 2.5σ.
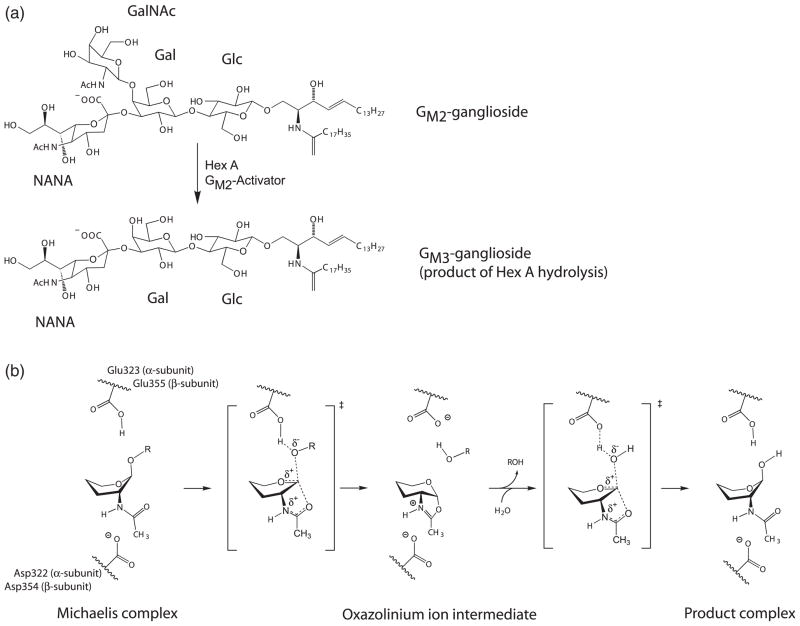
GM2 is presented to Hex A by the GM2 activator protein (GM2AP). Hex A removes the terminal β-linked GalNAc from the GM2 ganglioside to produce the GM3 ganglioside (Figure 3(a)).18 This hydrolysis is catalyzed only by the α-subunit of Hex A. Residues emanating from the C termini of the β-strands comprising the (βα)8 barrel participate in GM2 hydrolysis. From the structures of other Family 20 glycoside hydrolases, it has been demonstrated that Family 20 members use substrate-assisted catalysis with retention of configuration (Figure 3(b)) to remove the terminal β-linked GalNAc and/or GlcNAc residues from their oligosaccharide substrates.13,18,20–22 In Hex A, αGlu323 (α-subunit) and βGlu355 (β-subunit) are the general acid-base residues for protonation of the glycosidic oxygen atom; αAsp322 (α-subunit) and βAsp354 (β-subunit) provide the negatively charged carboxylate groups that stabilize the developing positive charge on the nitrogen atom of the oxazolinium ion during the nucleophilic attack of the N-acetamido oxygen atom on the C1′ of the substrate. In addition, there are strong substrate-orienting effects from the aromatic rings of αTrp373, αTrp392, and αTrp460 in the α-subunit, and βTrp405, βTrp424 and βTrp489 in the β-subunit. Hydrogen bonding from αTyr421 in the α-subunit and βTyr450 in the β-subunit helps to orient the nucleophilic carbonyl oxygen atom as well as to stabilize the oxazolinium ion intermediate (Figure 2). This environment protects the acyl center of the oxazolinium ion from attack and guides an incoming water molecule for the correct attack at the anomeric center of the intermediate to produce a product with net retention of the β-configuration.
Figure 3.
Proposed catalytic mechanism for Hex A. (a) Hydrolysis of the GM2 ganglioside by Hex A results in the loss of GalNAc to produce a GM3 ganglioside. (b) Proposed catalytic mechanism for Hex A showing substrate-assisted catalysis. αGlu323 in the α-subunit and βGlu355 in the β-subunit act as the general base, while αAsp322 in the α-subunit and βAsp354 in the β-subunit act to orient the C2-acetamido group into position for nucleophilic attack and subsequently stabilizes the oxazolinium ion intermediate. The hydroxyl residues and C6 have been removed from the pyranose ring of the substrate for clarity. The exact positions for these groups have not been determined.
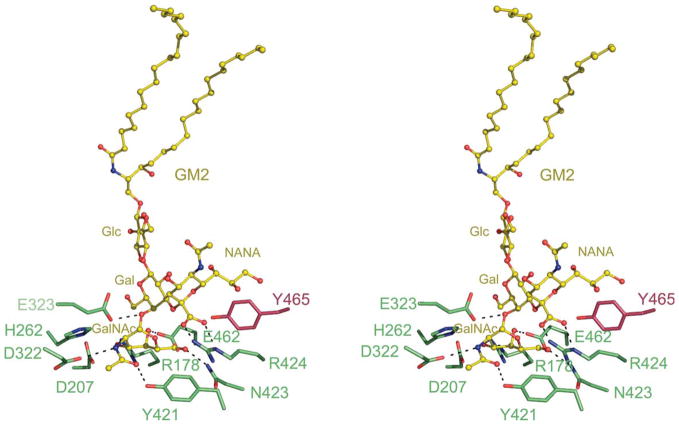
A model of GM2 ganglioside, based on the previously published model,18 was docked onto the α-subunit of Hex A (Figure 4). Only residues interacting with the sugar residues are shown. The remaining HexA residues and GM2AP, which interacts with the acyl chains of the GM2 ganglioside, have been removed for clarity. The only residue that required adjustment in order to accommodate GM2 in the α-subunit active site was αArg424, which was rotated about the Cδ–Cβ torsion angle. The model of GM2 docked into Hex A demonstrates that αArg424 would stabilize the negatively charged carboxylate group of the N-acetylneuraminic acid (NANA) via hydrogen bonding. In addition, it appears that αArg424, which in the unbound structure stacks against αTyr456, moves to stack against αTyr421 in the presence of GM2. Many of the residues in the α and β-active sites are conserved with the exception that αAsn423 and αArg424 in the α-subunit are replaced with βAsp452 and βLeu453 in the β-subunit. The negatively charged carboxylate group of the NANA would be repelled by the carboxylate group of βAsp452 in the β-subunit, making the formation of a productive complex unlikely. Mutagenesis data also support this rationale for Hex A substrate specificity. A double mutant was prepared whereby in Hex B, βAsp452 and βLeu453 were replaced with an Asn and Arg, respectively, in order to mimic the environment of the α-subunit active site.23 Kinetic studies with this Hex B double mutant resulted in a 30-fold increase in the rate of the GM2AP-independent hydrolysis of the negatively charged artificial 6-sulfated substrate 4-methylumbelliferyl-7-(6-sulfo-2-acetamido-2-deoxy)-β-D-glucopyranoside (MUGS) compared with the wild-type Hex B.
Figure 4.
Model of GM2 docked onto the α-subunit active site of Hex A. A model of the GM2 ganglioside (yellow) was docked into the active site of the α-subunit of Hex A based on the model of GM2 bound to the α-subunit active site of Hex B.18 For clarity, only residues interacting with the sugar residues of GM2 are shown. GM2AP, which interacts with the acyl chains of the GM2 ganglioside, has also been removed. αArg424, a positively charged residue unique to the α-subunit of Hex A, is found within hydrogen bonding distance from the negatively charged carboxylate of the NANA group of GM2.
Structural flexibility of Hex A
The fact that four Hex A molecules were found in the asymmetric unit gives us an opportunity to assess the structural variability among the different Hex A molecules and the effect of the NGT binding, a chemical chaperone, on the Hex A structure. There is close structural agreement between all four Hex A dimers in the asymmetric unit for both the native and NGT-bound Hex A. (See Supplementary Data, Table 1.) The structural alignment of the heterodimers of native Hex A gives r.m.s.d. values ranging from 0.36 Å to 0.44 Å, with an average r.m.s.d. of 0.39 Å. When NGT is associated with Hex A, the structural alignment of the heterodimers results in r.m.s.d. values that are somewhat lower, ranging from 0.26 Å to 0.35 Å with the average r.m.s.d. of 0.31 Å. The small deviations from structural superimpositions can be attributed to differences in crystal packing between unbound and NGT-bound Hex A that, interestingly, occur in the α280GSEP283 loop. Flexibility in this loop region is expected for GM2AP docking.
Hex A glycosylation
The α and β-subunits of Hex A display many N-linked glycosyl residues and there are oligosaccharides bound at most of the known glycosylation sites on Hex A (Figure 1(b), (c), (d) and (e)). There are three glycosylation sites on the α-subunit of Hex A: αAsn115, αAsn157 and αAsn295.24 Four glycosylation sites on the β-subunit have been identified: βAsn84, βAsn142, βAsn190, and βAsn327.24,25 The mannose residues of the glycosylated αAsn115, αAsn295 and βAsn84 are preferentially phosphorylated in order to be recognized by the M6P receptor. In our structure, electron density for glycosylation at αAsn115 and αAsn157 was observed in all four α-subunits. In only two α-subunits was electron density for glycosylation seen at αAsn295. In the four β-subunits, all βAsn190 had electron density for glycosylation, whereas electron density for glycosylation was observed in only one βAsn327. No or weak electron density was observed for the remaining glycosylation sites in the β-subunit. In several instances, two N-acetylglucosamine residues followed by mannose are visible.
Characterization of Hex A mutations on the basis of structure
The reduction in the rate of GM2 ganglioside hydrolysis below a surprisingly low critical threshold, estimated to be ~10% of normal,11 leads to its accumulation in the neural tissues and concomitant neurodegeneration. Reduced Hex A activity can occur via numerous types of mutations throughout the HEXA and HEXB genes. A large number of these defects have been identified‡.2 In many cases, genotype can easily be used to predict the ITSD phenotype, e.g. partial gene deletions, mRNA splicing, and nonsense mutations. Missense mutations also play a major role in dysfunctional Hex A and can lead to phenotypes ranging from acute to chronic, making predictions based only on genotype difficult. Interestingly, more disease-associated missense mutations in the α-subunit have been identified than in the β-subunit. This may reflect the lower inherent stability, i.e. greater flexibility of the α-subunit as compared to the β-subunit, which would make the α-subunit more susceptible to destabilizing missense mutations. In vitro mutagenesis and expression experiments that duplicated α-point mutation in the aligned site in the β-subunit support this hypothesis.26
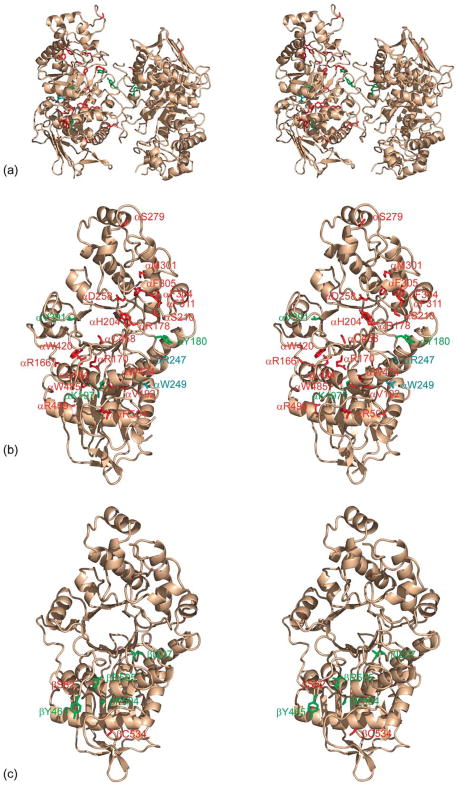
We have mapped all known missense mutations onto the Hex A molecule according to the severity of disease (Figure 5). In the α and β-subunits of Hex A, these include the residues listed in Table 2. Each missense mutation has been colored according to the severity of the GM2 gangliosidosis phenotype, red for acute to subacute, green for chronic and blue for asymptomatic (mutations that lower Hex A activity, but not below the critical threshold needed to prevent storage). The majority of residues involved in acute and chronic TSD are located throughout domain II of the α-subunit, distributed amongst the β-strands and helices comprising the TIM barrel. Notably, only a few mutations are found among residues of the active site.
Figure 5.
Known mutations of Hex A contributing to Tay-Sachs and Sandhoff disease. (a) Stereo view of a ribbon representation of Hex A (wheat), with residues known to disrupt Hex A activity: acute to sub-acute, red; chronic, green; asymptomatic, cyan. (b) A stereo view of the α-subunit of Hex A and residues associated with Tay-Sachs disease. (c) A stereo view of the β-subunit and residues associated with Sandhoff disease.
Table 2.
Missense mutations identified in the α-subunits and β-subunits of Hex A
| Location in α-subunit | Mutation(s) | Structural Phenotype | Cellular phenotype | Clinical phenoype |
|---|---|---|---|---|
| Domain I | L127F,R | Disruption of B-sheet | Na, decreased mRNA | Acute |
| Domain II, at interface between domain I | R166G | Salt bridge lost | Na | Severe subacute |
| Domain II Tim-barrel | R170W,Q | Over packing of residues and disruption of β-sheet | Na | Acute |
| Domain II, active site | R178C,H,L | Over packing of residues disruption of active site | ER retention | Acute |
| Domain II, Tim-barrel | V192L | Over packing of residues and disruption of β-sheet | Targeting mutant, only pro-α generated | Acute |
| Domain II, Tim-barrel | V200M | Over packing of residues and disruption of β-sheet | Targeting mutant, only pro-α generated | Acute |
| Domain II, Tim-barrel | H204R | Over packing of residues and disruption of β-sheet | Na | Acute |
| Domain II, helix at end of Tim-barrel | S210F | Over packing of residues and disruption of β-sheet | Na | Acute |
| Domain II, helix at end of Tim-barrel | F211S | Hydrophobic cavity decreased | Na | Acute |
| Domain II, helix at end of Tim-barrel | D258H | Hydrogen bonding lost and disruption of β-sheet | B1-like activity and targeting mutant | Severe subacute |
| Helical extension of domain II | S279P | Disruption of hydrogen bonding and loop that interacts with the GM2AP | Na | Severe subacute |
| Outer helix of Tim-barrel | M301R | Hydrophobic interaction decreased, buried polar residue | Na | Acute |
| Outer helix of Tim-barrel | F304 | Hydrophobic interaction decreased, buried polar residue | Targeting mutant, only proα generated | Acute |
| Outer helix of Tim-barrel | F305 | Hydrophobic interaction decreased, buried polar residue | Targeting mutant, only proα generated | Acute |
| Domain II, Tim-barrel | G320 | Na | Na | Severe subacute |
| Domain II, Tim-barrel | G321 | Na | Na | Acute |
| Domain II, Tim-barrel | Y420C | Hydrophobic interaction decreased, buried polar residue | No activity, targeting mutation | Acute |
| Domain II, Tim-barrel | G454S,D | Overpacking, backbone strain and disruption of β-sheet | Processing and targeting mutation | Acute |
| Domain II, Tim-barrel | G455R | Overpacking, backbone strain and disruption of β-sheet | Processing and targeting mutation | Acute |
| Domain II, Tim-barrel | C458Y | Overpacking, backbone strain and disruption of β-sheet | No activity | Acute |
| Domain II, Tim-barrel | W474C | Hydrophobic interaction decreased, buried polar residue | Decreased activity, targeting mutation | Acute |
| Domain II, Tim-barrel | E482K | Salt bridge lost | No activity, ER targeting mutation | Acute |
| Domain II, Tim-barrel | L484P | Hydrophobic interaction decreased | No activity, ER targeting mutation | Acute |
| Domain II, Tim-barrel | W485R | Hydrophobic interaction decreased, buried polar residue | No activity, ER targeting mutation | Acute |
| Domain II, Tim-barrel | R499C,H | Salt bridge lost | Decreased activity, targeting mutation | Acute |
| Domain II, Tim-barrel | R504C,H | Salt bridge lost | Decreased activity, targeting mutation | Acute |
| Domain II, Tim-barrel | G250D | Overpacking, buried polar residue | Reduced to Intermediate activity | Subacute |
| Domain II, Tim-barrel | C180H | Decrease hydrophobic interaction | Unstable protein | Chronic |
| Domain II, Tim-barrel | L197T | Salt bridge lost | Na | Chronic |
| Domain II, Tim-barrel | R252H | Hydrogen bonding lost | Na | Chronic |
| Domain II, Tim-barrel | G269S | Overpacking, backbone disortion | Reduced activity | Chronic |
| Domain II, Tim-barrel | V391M | Overpacking | Na | Chronic |
| Domain II, Tim-barrel | R247Y | Salt bridge lost with domain I, Overpacking | Reduced activity, HexA formed but unstable | Asymptomatic |
| Domain II, Tim-barrel | 249 | Salt bridge lost with domain I, Overpacking | Reduced activity, HexA formed but unstable | Asymptomatic |
| Domain II, Tim-barrel | S226F | Overpacking | Na | Uncharacterized |
| Domain II, Tim-barrel | G269D | Overpacking, backbone disortion | Na | Uncharacterized |
| Domain II, Tim-barrel | D314V | Salt bridge lost | Na | Uncharacterized |
| Domain II, extra helix in Tim-barrel | I335F | Overpacking | Na | Uncharacterized |
| Domain I | S62L | No apparent effect | Na | Acute |
| Domain II, Tim-barrel | S255R | Overpacking, buried charged residue | Na | Acute |
| Domain II, Tim-barrel | C534Y | Overpacking, buried hyrophobic residue | Na | Acute |
| Domain II, Tim-barrel | I207V | Decrease in hydrophobic pocket | Neutral polymorphism | Chronic |
| Domain II, in contact with the α-subunit | Y456S | Decrease in hydrophobic pocket | Targeting mutant | Chronic |
| Domain II, Tim-barrel | P504S | Disrupt in backbone | ER retention | Chronic |
| Domain II, Tim-barrel | R505Q | Salt bridge and hydrogen bonding lost | Heat labile HexB | Chronic |
| Domain II, Tim-barrel | A543T | overpacking | Heat labile HexB | Chronic |
Missense mutations identified in the α-subunit and β-subunits of Hex A are listed according to their severity for Tay-Sachs and Sandhoff disease. The color of the text corresponds to the mutations mapped on the structure shown in Figure 6. na: not assessed. References for each individual mutation can be found at http://www.hexdb.mcgill.ca/.2
As noted in Table 2, we have attempted to predict the effect of these missense mutations on the structure of Hex A in addition to listing the cellular phenotype and the severity of disease associated with that mutation. The majority of mutations characterized for Hex A tend to shift the equilibrium from fully folded toward misfolded protein production, leading to the retention of the defective α or β-subunits in the ER and degradation by the ER-associated degradation pathway, ERAD.27 In ERAD, misfolded proteins in the ER are detected by resident ER proteins and undergo retrograde transport back into the cytosol, where they are ubiquitinated and targeted for degradation by the proteasome.28 Indeed, it has been shown for another lysosomal storage disease, GM1 gangliosidosis, that the neuronal cell death associated with lysosomal β-galactosidase deficiency is attributed not to the accumulation of GM1, but to the unfolded protein response that results in the up-regulation of chaperones and apoptotic factors.29 Therefore, characterization of Hex A misfolding is essential to correlate the mutation with the severity of disease.
Because the extent of protein folding and misfolding in the ER for the subunits of Hex A can vary, the classification of misfolded mutants is difficult. Nevertheless, there are two main biochemical phenotypes associated with the destabilizing missense mutations for Hex A. Firstly, the mutations that result in subunits that are completely unable to fold correctly and, because they cannot form heterodimers and exit the ER, none obtains the lysosomal targeting label. These mutant subunits are either not easily detectable because of rapid degradation by the ERAD pathway, or can be extracted from cells only in the presence of detergent, because they form ERAD-resistant aggregates. These aggregates may exacerbate the clinical phenotype.30 The second type of missense mutations are presumably less destabilizing and allow a small proportion of newly synthesized mutant subunits to fold. These properly folded subunits can form heterodimers, can exit the ER and therefore obtain the lysosomal targeting label. In these cases, the levels of residual activity generally correlate with the levels of mature lysosomal protein, indicating that little if any change has occurred in the catalytic capacity of the mutant enzymes. These latter mutations include αTyr180-His, αGly269Ser, βPro504Ser, βArg505Gln and βAla543Ser, resulting in the less severe phenotypes of GM2 gangliosidosis. The reduction in Hex A activity caused by these mutations is a result of the various biochemical consequences of both the position of the mutated residue in the subunit and the degree of conservation of the amino-acid substitution. We see from the overall distribution of the currently characterized Hex A mutations and their biochemical phenotypes that prediction of the severity of a mutation, with the exception of those arising in the active site, would be difficult. Tay-Sachs mutations arising in Hex A that have been studied in more detail are described below, including examples of acute, chronic and asymptomatic phenotypes.
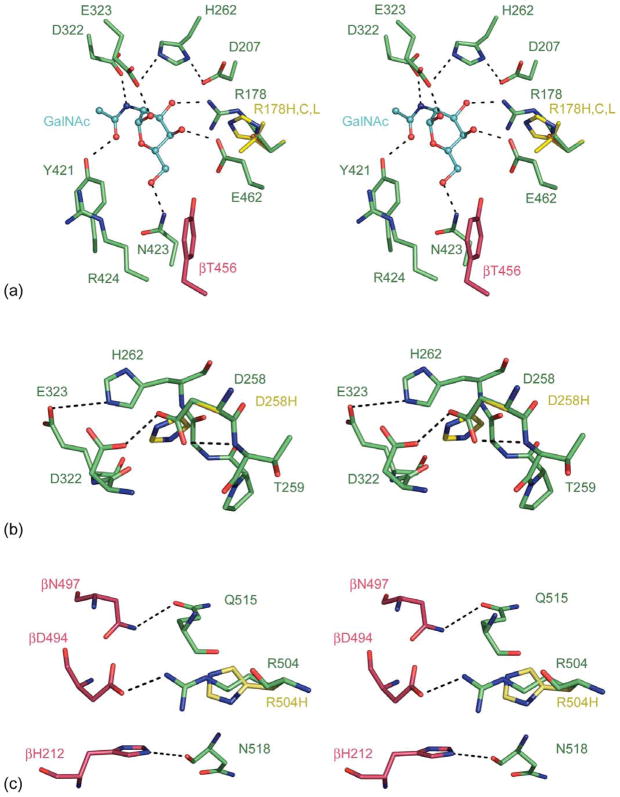
Active site mutation αArg178: acute, B1-variant
Analysis of naturally occurring mutants revealed a rare phenotype of “normally folded but reduced activity” class of active site mutants termed the B1 variant. The B1 variant mutation occurs predominantly at αArg178,31 and results in the formation of a normal Hex A heterodimer that can hydrolyze the common neutral artificial substrate MUG, but is nearly catalytically inactive towards the α-specific substrates MUGS and the GM2 ganglioside.32 With GalNAc from the GM2 substrate docked into the active site of Hex A (Figure 6(a)), we can see that αArg178 is involved in substrate binding by interacting with the 3′ hydroxyl group of the non-reducing βGalNAc. By modeling the αArg178H, αArg178C and αArg178L mutations, we see a disruption in the hydrogen bonding network in the active site that would reduce GM2 binding and severely affect the activity of the mutant α-active site of Hex A. Patients homozygous for αArg178His have a sub-acute phenotype, whereas heterozygotes with a second null allele present with the more severe acute phenotype.33 Other substitutions of the same residue, αArg178Cys or αArg178Leu, result in a more severe phenotype, because of these less conservative substitutions that may also destabilize the α-subunit, as well as decrease its catalytic capacity severely.
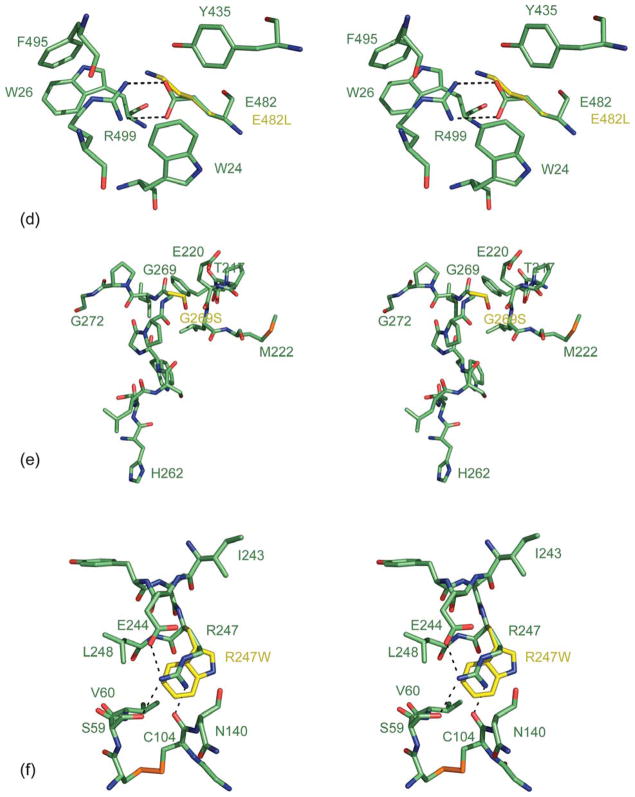
Figure 6.
Model of Hex A mutants. (a) Stereo view representation of the Cα trace for wt-Hex A (α-subunit, green; β-subunit, pink) superimposed with the αArg178H,C,L Hex A substitutions (yellow). GalNAc (cyan) from a GM2 substrate has been docked into the active site. (b) Stereo view representation of the Cα trace for wt-Hex A (α-subunit, green) superimposed with the αAsp258His Hex A mutant (yellow). (c) A stereo view representation of the Cα trace for wt-Hex A (α-subunit, green; β-subunit, pink) superimposed with the αAsp258His substitution (yellow). (d) A stereo view representation of the Cα trace for wt-Hex A (α-subunit, green) superimposed with the αGlu482Lys Hex A substitutions (yellow). (e) A stereo view representation of the Cα trace for wt-Hex A (α-subunit, green) superimposed with the αGly269Ser Hex A substitutions (yellow). (f) A stereo view representation of the Cα trace for wt-Hex A (α-subunit, green) superimposed with the αArg247W Hex A mutant (yellow). Stick representations are shown for residues surrounding substitutions as well as other key residues described in the text.
αAsp258His mutation: severe subacute, B1-variant like
The αAsp258His mutation34 was identified in a TSD patient with a phenotype termed severe subacute. Samples from this patient displayed a higher than expected residual Hex A activity utilizing MUG as a substrate (~15% of normal), but were nearly inactive when the MUGS substrate was used. Thus, biochemically this appeared to be B1-like.35,36 αAsp258 is located adjacent to the active site in the α-subunit. It participates in strong hydrogen bonding with residues αThr259 and αAsp322 (Figure 6(b)), the latter hydrogen bonds with GalNAc, providing a negatively charged carboxylate that stabilizes the developing positive charge on the nitrogen atom of the oxazolinium ion during the nucleophilic attack of the N-acetamido oxygen atom on the C1′ of the substrate. The substitution of αAsp258 for a more bulky His residue would disrupt the coordination of αAsp322 and may displace αGlu323, which acts as the general acid in the GM2 hydrolysis. Consequently, this mutation would be expected to inhibit substrate hydrolysis indirectly by the α-subunit active site of Hex A, as well as destabilize the initial folding of the α-subunit.
αArg504His mutation: subacute
An Arg504His substitution37–39 results in synthesis and proper folding of the α-subunit precursor, such that it was unable to form dimers and thus be transported to the lysosome. This conclusion was based on the observations that no mature (lysosomal) forms of the α-subunit could be detected but treatment of the cells of this patient with NH4Cl could induce the secretion of some inactive, phosphorylated α-monomers. αArg504 is located at the interface between the α and β-subunits and hydrogen bonds with βAsp494 of the β-subunit (Figure 6(c)). This interaction, along with other hydrogen bonding interactions such as αGln515 with βAsn497 and αAsn518 with βHis212 at the center of the subunit interface, plays a role in Hex A dimerization. (See Supplementary Data, Figure 1.) Substitution of αArg504 for a His residue would weaken this interaction at the core of the subunit interface.
αGlu482Lys mutation: acute
This mutation accounts for 2% of cases of TSD found in Moroccan Jews.40 When αGlu482 is substituted for Lys, the protein cannot exit the ER and results in expression of insoluble aggregates. As a result, this single amino acid substitution results in ITSD. αGlu482 is a buried residue that participates in hydrogen bonding with αArg499 (Figure 6(d)). In addition, αGlu482 hydrogen bonds with αTrp26 of domain I. This salt-bridge is surrounded by hydrophobic residues that comprise the interface between domain I and domain II of Hex A. (See Supplementary Data, Figure 2 for interface interactions between domain I and domain II.) Substitution of Lys for αGlu482 would disrupt this hydrogen bonding and may disrupt the interactions between domain I and domain II. Therefore, it is possible that the interaction between domain I and domain II plays a role in protein folding and/or facilitates dimer formation. Currently, the role of domain I in Hex A is unknown.
The most common (chronic) ATSD mutation, αGly269Ser
The αGly269Ser is the predominant mutation found in adult TSD (ATSD).41 Although this mutation is rare in all populations, its expression in Ashkenazi Jews is greatest, as it can pair with one of the two high-frequency ITSD alleles. The overall TSD carrier rate in this population is 1 in 30, with the ATSD allele accounting for ~3% of the total mutant TSD alleles.1 Whether homozygous or heterozygous for this allele, patients present with the ATSD phenotype. This mutation was predicted to disrupt the stability of Hex A, resulting in retention of the majority of the mutant α-subunits in the ER. As a result, these ATSD patients have residual activity between 4% and 8%.13 Modeling the αGly269Ser mutation in Hex A shows that the Cβ of Ser269 would clash with the Cβ of Glu220 (Figure 6(e)). Examination of the region surrounding αGly269 reveals a random coil, while the αGlu220 is found on a 310 coil that appears to be rigidified via hydrogen bonding with neighboring residues. A mutation at αGly269 with a more bulky and polar residue such as Ser would result in a displacement of its randomly coiled backbone. This αGly269Ser mutation is proximal to αHis262, a key residue found in the active site that plays an essential role as a proton donor to αGlu323 and αAsp207 in the active site. Thus, disruption of the backbone from αGly269Ser may result in a movement of αHis262 and may disrupt the coordination of residues in the active site. The fact that NGT can rescue αGly269Ser Hex A from misfolding to some extent,13 indicates that upon NGT binding a slight conformational change may occur in a sufficient manner to enable proper folding and targeting of Hex A from the ER to the Golgi, and ultimately to the lysosomal compartment via the mannose-6-phosphate receptor. Binding of NGT may shift the equilibrium for protein folding toward a more stable conformation that is able to evade the ERAD pathway.
The asymptomatic αArg247Trp and αArg249Trp substitutions in Hex A
Missense αArg247Trp and αArg249Trp substitutions in Hex A have been identified in patients having low levels of Hex A similar to those having TSD, but with no symptom of disease.42 The low levels of Hex A have been attributed to instability of the α-subunit. Using pulse-chase studies, also it has been shown that the rate of conversion from the precursor α-subunit to its mature form is delayed.43 This may reflect an effect on folding or dimerization. Interestingly, once formed, these Hex A mutants are not heat-labile, nor do they have trouble being processed or targeted to the lysosomal compartment. They appear to have normal catalytic capacities. Both residues are located on the surface of Hex A at the interface between domain I and domain II. The αArg247Trp substitution (Figure 6(f)), located in domain II, interacts via hydrogen bonding with αSer59 and αCys104 found in domain I and αGlu244 of domain II of Hex A. (See Supplementary Data, Figure 2 for interface interactions between domain I and domain II.) The αArg249Trp substitution (model not shown), also located in domain II, participates in hydrogen bonding with αArg67 of domain I, as well as αAsp191 and αTyr245 found in domain II. Substitutions of αArg247Trp and αArg249Trp would disrupt these hydrogen bonds and influence the interaction between domains I and II in the α-subunit. As stated above, it is possible that the interaction between domain I and domain II plays a role in protein folding and/or facilitates dimer formation. In addition, it has been shown that single amino acid mutations can result in an increased association of a protein with chaperones leading to their retention in the ER: a D18G transthyretin mutation increases its association with BiP,44 and the human ether-a-go-go (hERG) N470D mutation increases its association with calnexin.45 It is possible that the asymptomatic αArg247Trp and αArg249Trp mutations found on the surface of Hex A may increase its association with ER chaperones leading to retention in the ER.
The determination of the three-dimensional structure of Hex A provides the first glimpse of the molecule responsible for TSD, and provides an opportunity to develop structure-based inhibitors and novel chemical chaperones (CC). Substrate deprivation therapy with N-butyl-DNJ is currently being used to treat Gaucher diseases,46,47 and is being tested for the treatment of ATSD. This drug inhibits the synthesis of glucosylceramide (stored in Gaucher), which is the precursor to neutral and acidic (ganglioside) glycolipid synthesis. N-Butyl-DNJ, however, has a number of unpleasant side-effects, which increase in a dose-dependent manner.48 Thus, more specific drugs such as CC are needed as alternative methods of treatment. Potentially, CC could be used in conjunction with substrate reduction therapy to lower the amount of N-butyl-DNJ needed to treat Gaucher or ATSD. Information obtained from the structure of NGT bound to Hex A is important in understanding the mechanism by which this chemical acts as a chaperone for Hex A protein folding. Primary screening using live cell assays is being developed in order to identify novel CC. Promising candidates can then be docked and hopefully co-crystallized with Hex A. In addition, the three-dimensional-structure of Hex A can be used for in silico docking in order to identify new potential inhibitors or CCs. Moreover, a structure of Hex A will enable structure-based design of new pharmacological chaperones for the treatment of those afflicted with ATSD.
Materials and Methods
Purification and crystallization
Hex A was purified from human placenta as described,49 and was crystallized using the vapor-diffusion method in 13% (w/v) PEG 8000, 0.1 M sodium acetate, 0.2 M thiocyanate (pH 5.5). The protein was used in its mature glycosylated state for all experiments. Initially, small multilayered crystals grew within one week. Macroseeding was used to obtain well-ordered crystals with dimensions of 100 μm × 100 μm×50 μm. Crystals were then soaked briefly in mother liquor containing 20% (v/v) ethylene glycol followed by flash-cooling in liquid nitrogen. NGT-bound HexA crystals were obtained by soaking 5 mM NGT for one to two days into drops containing crystals.
Structure determination and model building
All diffraction data were collected at the Advanced Light Source (ALS) at Lawrence Berkeley National Lab, BL8.3.1 equipped with a Quantum 210 ADSC CCD detector. Intensity data were processed using DENZO and SCALEPACK.50 Phases were calculated using MOLREP for Hex A,51 and PHASER for Hex A-NGT,52 with physiological Hex B (1NOU.pdb) as the search model.
Four Hex A molecules, each consisting of an α-subunit and a β-subunit, were located in the asymmetric unit. The Hex B structure served as a backbone model to facilitate Hex A tracing. The structure was subjected to rigid body refinement followed by successive rounds of restrained refinement using REFMAC5,53 followed by model building using Xfit.54 Both NCS and TLS were used in the refinement to facilitate tracing. Water molecules were placed manually and checked with WATERTIDY in the CCP4 package.55 NCS was omitted for the final round of refinement. The final model has good geometry with no amino acid residue in the disallowed region of the Ramachandran plot, as determined by PROCHECK.56 GM2 and the GM2AP complex structure were docked manually onto the Hex A structure on the basis of the Hex A-NGT structure, and models generated by the Hex B structure.18 Figures were prepared with PYMOL†. The superimposition of similar and related molecules were carried out with ALIGN.57
Protein Data Bank accession number
The atomic coordinates and structure factors have been deposited with RCSB Protein Data Bank as entry pdb 2GJX for native Hex A and 2GKI for NGT-bound Hex A.
Acknowledgments
This work has been supported by the Canadian Institute for Health Research (CIHR) and the Canadian Protein Engineering Network (PENCE). We thank Ernst Bergman, Jonathan Parrish (Alberta Synchrotron Institute, University of Alberta) and James Holden (BL8.3.1, Advanced Light Source, Berkeley National Laboratory) for help with the data collection. X-ray diffraction data were collected at beamline 8.3.1 of the Advanced Light Source (ALS) at Lawrence Berkeley Lab, under an agreement with the Alberta Synchrotron Institute (ASI). The ALS is operated by the Department of Energy and supported by the National Institutes of Health. Beamline 8.3.1 is funded by the National Science Foundation, the University of California and Henry Wheeler. The ASI synchrotron access program is supported by grants from the Alberta Science and Research Authority (ASRA) and the Alberta Heritage Foundation for Medical Research (AHFMR) and Western Economic Diversification (WED), Canada. We thank Amy Leung (Hospital for Sick Children) for purifying the Hex A used in this project. We thank all members of the James laboratory, Dr M. Wang and Dr B. Biswal for their assistance during structure determination and manuscript preparation, Dr J. Parrish for data collection, and Shiraz Khan for his assistance with crystal shipping. B.L.M. was supported by scholarships from the CIHR and AHFMR. M.J.L. is supported by fellowship scholarships with CIHR and AHFMR. M.N.G.J. acknowledges support from the Canada Research Chairs Program.
Abbreviations used
- Hex A
β-hexosaminidase A
- NANA
N-acetyl-neuraminic acid
- NGT
NAG-thiazoline
- NAG
N-acetylglucosamine
- GM2
GalNAc-1,4(NeuAc-2,3)Gal-1,4 Glc-ceramide
- TSD
Tay-Sachs disease
- ATSD
adult/chronic form of Tay-Sachs disease
- ITSD
infantile/acute form of Tay-Sachs disease
- SD
Sandhoff disease
- ISD
infantile/acute form of Sandhoff disease
- ASD
adult/chronic form of Sandhoff disease
- MUG
4-methylumbelliferyl-β-N-acetylglucosaminide
- MUGS
4-methylumbelliferyl-7-(6-sulfo-2-acetamido-2-deoxy)-β-D-glucopyranoside
- GalNAc
N-acetylgalactosamine
- AdNJ
2-acetamido-2-deoxynojirimycin
- DNJ
deoxynojirimycin
- NBDNJ
n-butyl-DNJ
- NGT
N-acetylglucosamine-thiazoline
- ER
endoplasmic reticulum
- WT
wild-type
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jmb.2006.04.004
References
- 1.Gravel RA, Clarke JTR, Kaback MM, Mahuran D, Sandoff K, Suzuki K. The GM2 gangliosidoses. In: Scriver CR, editor. The Metabolic and Molecular Basis of Inherited Diseases. McGraw-Hill: New York; 1995. pp. 2839–2879. [Google Scholar]
- 2.Mahuran DJ. Biochemical consequences of mutations causing the GM2 gangliosidoses. Biochim Biophys Acta. 1999;1455:105–138. doi: 10.1016/s0925-4439(99)00074-5. [DOI] [PubMed] [Google Scholar]
- 3.Henrissat B, Davies G. Structural and sequence-based classification of glycoside hydrolases. Curr Opin Struct Biol. 1997;7:637–644. doi: 10.1016/s0959-440x(97)80072-3. [DOI] [PubMed] [Google Scholar]
- 4.Mahuran DJ. The GM2 activator protein, its roles as a co-factor in GM2 hydrolysis and as a general glycolipid transport protein. Biochim Biophys Acta. 1998;1393:1–18. doi: 10.1016/s0005-2760(98)00057-5. [DOI] [PubMed] [Google Scholar]
- 5.Kresse H, Fuchs W, Glossl J, Holtfrerich D, Gilberg W. Liberation of N-acetylgluco-samine-6-sulfate by human beta-N-acetylhexosaminidase A. J Biol Chem. 1981;256:12926–12932. [PubMed] [Google Scholar]
- 6.Hepbildikler ST, Sandhoff R, Kolzer M, Proia RL, Sandhoff K. Physiological substrates for human lysosomal beta-hexosaminidase S. J Biol Chem. 2002;277:2562–2572. doi: 10.1074/jbc.M105457200. [DOI] [PubMed] [Google Scholar]
- 7.Futerman AH, van Meer G. The cell biology of lysosomal storage disorders. Nature Rev Mol Cell Biol. 2004;5:554–565. doi: 10.1038/nrm1423. [DOI] [PubMed] [Google Scholar]
- 8.Kolter T, Sandhoff K. Principles of lysosomal membrane digestion-stimulation of sphin-golipid degradation by sphingolipid activator proteins and anionic lysosomal lipids. Annu Rev Cell Dev Biol. 2005;21:81–103. doi: 10.1146/annurev.cellbio.21.122303.120013. [DOI] [PubMed] [Google Scholar]
- 9.Itoh H, Tanaka J, Morihana Y, Tamaki T. The fine structure of cytoplasmic inclusions in brain and other visceral organs in Sandhoff disease. Brain Dev. 1984;6:467–474. doi: 10.1016/s0387-7604(84)80029-7. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi T, Goto I, Okada S, Orii T, Ohno K, Nakano T. Accumulation of lysosphingolipids in tissues from patients with GM1 and GM2 gangliosidoses. J Neurochem. 1992;59:1452–1458. doi: 10.1111/j.1471-4159.1992.tb08460.x. [DOI] [PubMed] [Google Scholar]
- 11.Conzelmann E, Sandhoff K. Partial enzyme deficiencies: residual activities and the development of neurological disorders. Dev Neurosci. 1983;6:58–71. doi: 10.1159/000112332. [DOI] [PubMed] [Google Scholar]
- 12.Knapp S, Vocadlo D, Gao Z, Kirk B, Lou J, Withers SG. NAG-thiazoline, an N-acetyl-beta-hexosaminidase inhibitor that implicates aceta-mido participation. J Am Chem Soc. 1996;118:6804–6805. [Google Scholar]
- 13.Tropak MB, Reid SP, Guiral M, Withers SG, Mahuran D. Pharmacological enhancement of beta-hexosaminidase activity in fibroblasts from adult Tay-Sachs and Sandhoff patients. J Biol Chem. 2004;279:13478–13487. doi: 10.1074/jbc.M308523200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Little LE, Lau MML, Quon DVK, Fowler AV, Neufeld EF. Proteolytic processing of the α chain of the lysosomal enzyme β-hexosaminidase, in normal human fibroblasts. J Biol Chem. 1988;263:4288–4292. [PubMed] [Google Scholar]
- 15.Hubbes M, Callahan J, Gravel R, Mahuran D. The amino-terminal sequences in the pro-α and -β polypeptides of human lysosomal β-hexosaminidase A and B are retained in the mature isozymes. FEBS Letters. 1989;249:316–320. doi: 10.1016/0014-5793(89)80649-0. [DOI] [PubMed] [Google Scholar]
- 16.Mahuran DJ, Neote K, Klavins MH, Leung A, Gravel RA. Proteolytic processing of human pro-β hexosaminidase: identification of the internal site of hydrolysis that produces the nonidentical βa and βb polypeptides in the mature β-subunit. J Biol Chem. 1988;263:4612–4618. [PubMed] [Google Scholar]
- 17.Quon DVK, Proia RL, Fowler AV, Bleibaum J, Neufeld EF. Proteolytic processing of the β-subunit of the lysosomal enzyme, β-hexosaminidase, in normal human fibroblasts. J Biol Chem. 1989;264:3380–3384. [PubMed] [Google Scholar]
- 18.Mark BL, Mahuran DJ, Cherney MM, Zhao D, Knapp S, James MN. Crystal structure of human beta-hexosaminidase B: understanding the molecular basis of Sandhoff and Tay-Sachs disease. J Mol Biol. 2003;327:1093–1109. doi: 10.1016/s0022-2836(03)00216-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zarghooni M, Bukovac S, Tropak M, Callahan J, Mahuran D. An alpha-subunit loop structure is required for GM2 activator protein binding by beta-hexosaminidase A. Biochem Biophys Res Commun. 2004;324:1048–1052. doi: 10.1016/j.bbrc.2004.09.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mark BL, Wasney GA, Salo TJ, Khan AR, Cao Z, Robbins PW, et al. Structural and functional characterization of Streptomyces plicatus beta-N-acetylhexosaminidase by comparative molecular modeling and site-directed mutagenesis. J Biol Chem. 1998;273:19618–19624. doi: 10.1074/jbc.273.31.19618. [DOI] [PubMed] [Google Scholar]
- 21.Tews I, Perrakis A, Oppenheim A, Dauter Z, Wilson KS, Vorgias CE. Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease. Nature Struct Biol. 1996;3:638–648. doi: 10.1038/nsb0796-638. [DOI] [PubMed] [Google Scholar]
- 22.Williams SJ, Mark BL, Vocadlo DJ, James MN, Withers SG. Aspartate 313 in the Streptomyces plicatus hexosaminidase plays a critical role in substrate-assisted catalysis by orienting the 2-acetamido group and stabilizing the transition state. J Biol Chem. 2002;277:40055–40065. doi: 10.1074/jbc.M206481200. [DOI] [PubMed] [Google Scholar]
- 23.Sharma R, Bukovac S, Callahan J, Mahuran D. A single site in human beta-hexosaminidase A binds both 6-sulfate-groups on hexosamines and the sialic acid moiety of GM2 ganglioside. Biochim Biophys Acta. 2003;1637:113–118. doi: 10.1016/s0925-4439(02)00221-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sonderfeld-Fresko S, Proia RL. Analysis of the glycosylation and phosphorylation of the lysoso-mal enzyme, β-hexosaminidase B, by site-directed mutagenesis. J Biol Chem. 1989;264:7692–7697. [PubMed] [Google Scholar]
- 25.O’Dowd BF, Cumming DA, Gravel RA, Mahuran D. Oligosaccharide structure and amino acid sequence of the major glycopeptides of mature human beta-hexosaminidase. Biochemistry. 1988;27:5216–5226. doi: 10.1021/bi00414a041. [DOI] [PubMed] [Google Scholar]
- 26.Brown CA, Mahuran DJ. beta-Hexosaminidase isozymes from cells cotransfected with alpha and beta cDNA constructs: analysis of the alpha-subunit missense mutation associated with the adult form of Tay-Sachs disease. Am J Hum Genet. 1993;53:497–508. [PMC free article] [PubMed] [Google Scholar]
- 27.Hampton RY. ER-associated degradation in protein quality control and cellular regulation. Curr Opin Cell Biol. 2002;14:476–482. doi: 10.1016/s0955-0674(02)00358-7. [DOI] [PubMed] [Google Scholar]
- 28.Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: the long road to destruction. Nature Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- 29.Tessitore A, del P Martin M, Sano R, Ma Y, Mann L, Ingrassia A, et al. GM1-ganglioside-mediated activation of the unfolded protein response causes neuronal death in a neurodegenerative gang-liosidosis. Mol Cell. 2004;15:753–766. doi: 10.1016/j.molcel.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 30.Rutishauser J, Spiess M. Endoplasmic reticulum storage diseases. Swiss Med Wkly. 2002;132:211–222. doi: 10.4414/smw.2002.09861. [DOI] [PubMed] [Google Scholar]
- 31.Ohno K, Suzuki K. Mutation in GM2-gangliosidosis B1 variant. J Neurochem. 1988;50:316–318. doi: 10.1111/j.1471-4159.1988.tb13266.x. [DOI] [PubMed] [Google Scholar]
- 32.Hou Y, Vavougios G, Hinek A, Wu KK, Hechtman P, Kaplan F, Mahuran DJ. The Val192Leu mutation in the alpha-subunit of beta-hexosaminidase A is not associated with the B1-variant form of Tay-Sachs disease. Am J Hum Genet. 1996;59:52–58. [PMC free article] [PubMed] [Google Scholar]
- 33.dos Santos MR, Tanaka A, sa Miranda MC, Ribeiro MG, Maia M, Suzuki K. GM2-gangliosidosis B1 variant: analysis of beta-hexosamin-idase alpha gene mutations in 11 patients from a defined region in Portugal. Am J Hum Genet. 1991;49:886–890. [PMC free article] [PubMed] [Google Scholar]
- 34.Bayleran J, Hechtman P, Kolodny E, Kaback M. Tay-Sachs disease with hexosaminidase A: characterization of the defective enzyme in two patients. Am J Hum Genet. 1987;41:532–548. [PMC free article] [PubMed] [Google Scholar]
- 35.Fernandes MJG, Yew S, Leclerc D, Henrissat B, Vorgias CE, Gravel RA, et al. Identification of candidate active site residues in lysosomal beta-hexosaminidase A. J Biol Chem. 1997;272:814–820. doi: 10.1074/jbc.272.2.814. [DOI] [PubMed] [Google Scholar]
- 36.Tse R, Vavougios G, Hou Y, Mahuran DJ. Identification of an active acidic residue in the catalytic site of beta-hexosaminidase. Biochemistry. 1996;35:7599–7607. doi: 10.1021/bi960246+. [DOI] [PubMed] [Google Scholar]
- 37.Paw BH, Moskowitz SM, Uhrhammer N, Wright N, Kaback MM, Neufeld EF. Juvenile GM2 gangliosidosis caused by substitution of histidine for arginine at position 499 or 504 of the alpha-subunit of beta-hexosaminidase. J Biol Chem. 1990;265:9452–9457. [PubMed] [Google Scholar]
- 38.Boustany RM, Tanaka A, Nishimoto J, Suzuki K. Genetic cause of a juvenile form of Tay-Sachs disease in a Lebanese child. Ann Neurol. 1991;29:104–107. doi: 10.1002/ana.410290120. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka A, Sakazaki H, Murakami H, Isshiki G, Suzuki K. JSSIEM meeting. Molecular genetics of Tay-Sachs disease in Japan. J Inherited Metab Dis. 1994;17:593–600. doi: 10.1007/BF00711597. [DOI] [PubMed] [Google Scholar]
- 40.Proia RL, Neufeld EF. Synthesis of beta-hexosaminidase in cell-free translation and in intact fibroblasts: an insoluble precursor alpha chain in a rare form of Tay-Sachs disease. Proc Natl Acad Sci USA. 1982;79:6360–6364. doi: 10.1073/pnas.79.20.6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Navon R, Proia RL. The mutations in Ashkenazi Jews with adult GM2 gangliosidosis, the adult form of Tay-Sachs disease. Science. 1989;243:1471–1474. doi: 10.1126/science.2522679. [DOI] [PubMed] [Google Scholar]
- 42.Cao Z, Natowicz MR, Kaback MM, Lim-Steele JS, Prence EM, Brown D, et al. A second mutation associated with apparent beta-hexosaminidase A pseudodeficiency: identification and frequency estimation. Am J Hum Genet. 1993;53:1198–1205. [PMC free article] [PubMed] [Google Scholar]
- 43.Cao Z, Petroulakis E, Salo T, Triggs-Raine B. Benign HEXA mutations, C739T(R247W) and C745T(R249W), cause beta-hexosaminidase A pseudodeficiency by reducing the alpha-subunit protein levels. J Biol Chem. 1997;272:14975–14982. doi: 10.1074/jbc.272.23.14975. [DOI] [PubMed] [Google Scholar]
- 44.Sorgjerd K, Ghafouri B, Jonsson BH, Kelly JW, Blond SY, Hammarstrom P. Retention of misfolded mutant transthyretin by the chaperone BiP/GRP78 mitigates amyloidogenesis. J Mol Biol. 2006;356:469–482. doi: 10.1016/j.jmb.2005.11.051. [DOI] [PubMed] [Google Scholar]
- 45.Gong Q, Jones MA, Zhou Z. Mechanisms of pharmacological rescue of trafficking defective hERG mutant channels in human long QT syndrome. J Biol Chem. 2006;7:4069–4074. doi: 10.1074/jbc.M511765200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Platt FM, Neises GR, Reinkensmeier G, Townsend MJ, Perry VH, Proia RL, et al. Prevention of lysosomal storage in Tay-Sachs mice treated with N-butyldeoxynojirimycin. Science. 1997;276:428–431. doi: 10.1126/science.276.5311.428. [DOI] [PubMed] [Google Scholar]
- 47.Sango K, Yamanaka S, Hoffmann A, Okuda Y, Grinberg A, Westphal H, et al. Mouse models of Tay-Sachs and Sandhoff diseases differ in neurologic phenotype and ganglioside metabolism. Nature Genet. 1995;11:170–176. doi: 10.1038/ng1095-170. [DOI] [PubMed] [Google Scholar]
- 48.Butters TD, Dwek RA, Platt FM. Imino sugar inhibitors for treating the lysosomal glycosphingolipidoses. Glycobiology. 2005;15:43R–52R. doi: 10.1093/glycob/cwi076. [DOI] [PubMed] [Google Scholar]
- 49.Mahuran D, Lowden JA. The subunit and polypeptide structure of hexosaminidases from human placenta. Can J Biochem. 1980;58:287–294. doi: 10.1139/o80-038. [DOI] [PubMed] [Google Scholar]
- 50.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 51.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Crystallog. 1997;30:1022–1025. [Google Scholar]
- 52.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallog sect D. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 53.Steiner RA, Lebedev AA, Murshudov GN. Fisher’s information in maximum-likelihood macromolecular crystallographic refinement. Acta Crystallog sect D. 2003;59:2114–2124. doi: 10.1107/s0907444903018675. [DOI] [PubMed] [Google Scholar]
- 54.McRee DE. XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 55.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallog sect D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 56.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallog. 1993;26:283–291. [Google Scholar]
- 57.Cohen GH. ALIGN: a program to superimpose protein coordinates, accounting for insertins and deletions. J Appl Crystallog. 1997;30:1160–1161. [Google Scholar]