Abstract
Objectives
CYP2A6 is the main enzyme involved in nicotine metabolism in humans. We have identified a novel allele, CYP2A6*23 (2161C>T, R203C), in individuals of Black-African descent and investigated its impact on enzyme activity and association with smoking status.
Methods
Wildtype and variant enzymes containing amino acid changes R203C (CYP2A6*23), R203S (CYP2A6*16) and V365M (CYP2A6*17) were expressed in Escherichia coli. The effect of CYP2A6*23 in vivo was examined in individuals of Black-African descent given 4 mg oral nicotine.
Results
CYP2A6*23 occurred at an allele frequency of 2.0% in individuals of Black-African descent (N = 560 alleles, 95% confidence interval 0.8%–3.1%) and was not detected in Caucasians (N = 334 alleles), Chinese (N = 288 alleles) or Japanese (N = 104 alleles). In vitro, CYP2A6.23 had greatly reduced activity towards nicotine C-oxidation similar to CYP2A6.17, as well as reduced coumarin 7-hydroxylation. Conversely, CYP2A6.16 did not differ in activity compared to the wildtype enzyme. The trans-3′-hydroxycotinine to cotinine ratio, a phenotypic measure of CYP2A6 activity in vivo, was lower in CYP2A6*1/*23 and CYP2A6*23/*23 individuals(mean adjusted ratio of 0.60, n = 5) compared to CYP2A6*1/*1 individuals (mean adjusted ratio of 1.21, n = 150) (p < 0.04). CYP2A6*23 trended towards a higher allele frequency in nonsmokers (3.1%, N = 9/286 alleles) compared to smokers (0.7%, N = 2/274 alleles) (p = 0.06).
Conclusions
These results suggest the novel CYP2A6*23 allele impairs enzyme function in vitro and in vivo and trends toward an association with lower risk of smoking.
Keywords: CYP2A6, genetic polymorphisms, Black-Africans, nicotine, smoking
Introduction
Nicotine is responsible for the majority of the psychoactive and highly addictive properties of cigarettes [1]. In humans, ~80% of absorbed nicotine is converted into cotinine and the majority of this reaction (~90%) is mediated by the hepatic enzyme cytochrome P450 2A6 (CYP2A6) [2, 3]. Cotinine is further metabolized to trans-3′-hydroxycotinine and this reaction is entirely mediated by CYP2A6 [4]. CYP2A6 is also the main enzyme metabolizing therapeutic compounds such as coumarin [5], tegafur [6], and SM-12502 [7], and it can bioactivate tobacco smoke procarcinogens such as N′-nitrosonornicotine (NNN) and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) [8].
Large interindividual and interethnic differences in nicotine metabolism have been reported in vitro and in vivo (reviewed in [9]). CYP2A6 is highly polymorphic, with 22 numbered alleles and numerous single nucleotide polymorphisms (SNPs) identified so far (http://www.cypalleles.ki.se/cyp2a6.htm). Characterization of CYP2A6 variants is needed because CYP2A6 polymorphisms altering nicotine metabolism may be important determinants of smoking behaviors. Several, but not all, studies have found that individuals with CYP2A6 alleles impairing enzyme activity are less likely to be current smokers [9, 10].
Differences in nicotine metabolism, smoking behaviors and incidence of tobacco-related illnesses have been observed between ethnic groups. Compared to Caucasians, individuals of Black-African descent have a significantly reduced fractional conversion of nicotine to cotinine, reduced metabolic clearance of nicotine to cotinine and higher plasma levels of cotinine for similar cigarette consumption [11, 12]. Together, this suggests reduced nicotine and cotinine metabolism, likely as a result of genetic variations in CYP2A6. Interestingly, populations of Black-African descent are also characterized by unique smoking patterns. Despite having a similar prevalence of current smoking as Caucasians [13], populations of Black-African descent report later ages of smoking initiation [14, 15] and lower cigarette consumption [16, 17]. Furthermore, populations of Black-African descent suffer from disproportionately higher rates of most tobacco-related illnesses [17], such as lung cancer [18, 19], despite their lower levels of smoking.
In this study, we identified a novel CYP2A6 allele (CYP2A6*23) with a SNP at 2161C>T (GenBank accession number NG_000008.7) in exon 4 corresponding to an amino acid change R203C in a population of Black-African descent. We discovered this variant while designing a genotyping assay for the previously identified CYP2A6*16 allele [20], which occurs at the same locus (2161C>A, R203S). Primers were designed for both the C>A and C>T nucleotide change as there had been initial confusion to the specific change found in CYP2A6*16. The effect of CYP2A6*23 on enzyme function was determined in vitro using an Escherichia coli (E. coli) heterologous expression system. We compared it to CYP2A6*16, which encodes a different amino acid change at the same position, and to CYP2A6*17, a previously characterized reduced-activity allele [21]. Furthermore, we examined the effect of CYP2A6*23 on nicotine metabolism in vivo and its association with smoking status.
Methods
Participants
Adults of Black-African descent were recruited from Toronto, Ontario; detailed descriptions of the population and experimental protocol can be found elsewhere [22]. Briefly, male and female current smokers and tried nonsmokers were recruited. Smokers were defined as smoking ≥ 100 lifetime cigarettes and currently smoking at least 5 days of the week while tried nonsmokers had smoked 1–99 lifetime cigarettes but were never regular smokers. Plasma nicotine, cotinine (COT) and trans-3′-hydroxycotinine (3HC) levels were measured following administration of an oral nicotine dose (4 mg capsule). The 3HC/COT ratio, calculated from plasma levels collected at 270 minutes after dosing, has been previously verified as an indicator of CYP2A6 activity [23]. Caucasian (n = 167), Chinese (n = 144) and Japanese (n = 52) individuals recruited for a separate study on CYP2A6 genetic variation and smoking behaviors [10] were also genotyped for CYP2A6*23. The research protocol was approved and monitored by the University of Toronto Ethics Review Office and the Institutional Review Board Services (Aurora, ON).
Genotyping assay
Blood samples were stored at −20°C, and genomic DNA was extracted and stored at −20°C until use (GenElute Mammalian Genomic DNA Kit, Sigma-Aldrich Co., Mississauga, ON). A novel two-step allele specific polymerase chain reaction (PCR) assay was developed for CYP2A6*23. A fragment of CYP2A6 between intron 3 to intron 5 was amplified using the primers: forward (2A6exin3F) 5′–GGC ACT GGC GGT GAG CAG–3′ and reverse (2A6in5R) 5′–GGC CTG TGT CAT CTG CCT–3′. The reaction mixture contained: 50 ng of genomic DNA, 1X Taq buffer with KCl, 200 μM deoxyribonucleoside triphosphates (dNTPs), 1.5 mM MgCl2, 125 nM of each primer, 1.25 U of Taq Polymerase (MBI Fermentas, Burlington, ON) and H2O for a total volume of 25 μl. The reaction conditions were as follows: initial denaturation at 95°C for 1 min., denaturation at 95°C for 15 sec., annealing at 58°C for 30 sec., and extension at 72°C for 2 min. for 30 cycles, followed by final extension step at 72°C for 7 min. (MJ Research PCR Cycler PTC 200; Waltham, MA). The product from this first amplification served as the template for a second allele-specific reaction using the primers: forward (2A6in3F) 5′–CTG CCT CCT GGA ATT CTG AC–3′ and a reverse primer specific to 2161C (2A6ex42161AWR, 5′–GGA AGA TTC CTA GCA TCA TGC G–3′) or to 2161T (2A6ex42161AVR, 5′–GGA AGA TTC CTA GCA TCA TGC A–3′). The reaction mixture contained: 0.8 μl of template, 1X Taq buffer with (NH4)2SO4, 1.0 mM of MgCl2, 75 nM of each primer, 1.25 U of Taq Polymerase and H2O for a total volume of 25 μl. The reaction conditions were: initial denaturation at 95°C for 1 min., denaturation at 95°C for 15 sec., annealing at 57°C for 10 sec., and extension at 72°C for 30 sec. for 20 cycles. The PCR products were separated by electrophoresis on a 1.2% agarose gel stained with ethidium bromide. Subjects had been genotyped for known CYP2A6 variants (CYP2A6*1B,*2, *4A & D, *9, *12, *14, *15, *17, *20,*21, *24, *25, *26, *27, *28, *29).
DNA sequence analyses
We confirmed the detection of CYP2A6*23 by our genotyping assay, and determined its haplotype by sequencing all CYP2A6 exons and exon-intron borders in two heterozygous individuals (CYP2A6*1/*23) and a homozygous individual (CYP2A6*23/*23). A 9.2 kb fragment spanning −1.4 kb upstream and 7.8 kb downstream of the +1 ATG start site of CYP2A6 was amplified from genomic DNA using long-PCR. The primers for long-PCR were: forward (2A65Pr1F) 5′–ACC TAG ACT TAA TCT TCC CGT ATA C–3′ and reverse (2A6R0) 5′–AGG TCA TCT AGA TTT TCT CCT ACA–3′. The product was subcloned into pCR®-XL-TOPO plasmid (TOPO® XL PCR Cloning Kit, Invitrogen Canada Inc., Burlington, ON). DNA sequencing was performed with an ABI 3730XL DNA analyzer at the Centre for Applied Genomics (Toronto, ON)
Construction of CYP2A6 expression plasmids
The bicistronic construct with the full length cDNA of CYP2A6 and human NADPH-CYP450 reductase (hNPR) inserted into the pCW expression vector (8537 bp) [24] was derived from the cognate monocistronic construct prepared by Soucek et al. [25], in which the native CYP2A6 sequence was changed to encode Ala at the second position with only silent nucleotide changes elsewhere in the N-terminus. The SNPs 2161C>T (CYP2A6*23), 2161C>A (CYP2A6*16) and 5065G>A (CYP2A6*17) were introduced into the expression constructs using the QuikChange® II XL Site-Directed Mutagenesis Kits (Stratagene, La Jolla, CA). Primers used in the mutagenesis reactions were: 2161C>T (5′–AGT TCC TGT CAC TGT TGTGCA TGA TGC TAG GAA TC–3′, 5′–GAT TCC TAG CAT CAT GCA CAA CAG TGA CAG GAA CT–3′), 2161C>A (5′–AGT TCC TGT CAC TGT TGAGCA TGA TGC TAG GAA TC–3′, 5′–GAT TCC TAG CAT CAT GCT CAA CAG TGA CAG GAA CT–3′), 5065G>A (5′–ATC CAA AGA TTT GGA GAC ATG ATC CCC ATG AGT TTG G–3′, 5′–CCA AAC TCA TGG GGA TCA TGT CTC CAA ATC TTT GGA T–3′). A negative control was created using BamHI to remove 802 bp from the 5′ end of the CYP2A6 cDNA (total length of 1485 bp). The products were separated by gel electrophoresis and the remaining plasmid (7735 bp) was extracted and re-ligated. The variant constructs were confirmed by sequencing using an ABI 3730XL DNA analyzer at the Centre for Applied Genomics (Toronto, ON).
Expression of CYP2A6 constructs in E. coli
The constructs encoding CYP2A6 and hNPR were expressed in E. coli as described previously [26]. DH5α cells (Invitrogen, Burlington, ON) were transformed with wildtype and variant constructs and a starter culture was grown in Luria-Bertani medium containing ampicillin (100 μg/ml) overnight at 37°C with shaking at 200 rpm. The starter culture was diluted (1:100) in 100 ml of terrific broth containing ampicillin (100 μg/ml), 1.0 mM thiamine, and 0.5 mM δ-aminolevulinic acid. The cultures were incubated at 30°C with shaking at 120 rpm for 4–6 hours, with induction initiated by the addition of 1.0 mM isopropyl β-D-thiogalactopyranoside. The cultures were incubated for a further 19–22 hours at 30°C and shaking at 120 rpm. Membrane fractions were prepared as described in [26] with minor modifications. Briefly, the pelleted bacteria was resuspended in buffer (50 mM Tris-HCl, 0.1 mM EDTA, 0.1 mM dithiothreitol, pH 7.4) and digested with lysozyme (0.25 mg/ml) for 60 min. The spheroplast was pelleted, sonicated, and centrifuged at 12,000 × g for 12 min. The supernatant was centrifuged for 110,000 × g for 90 min, and the pellet containing the membrane fraction was resuspended in 1.15% KCl and stored at −80°C until use. All constructs were initiated for expression concurrently and processed for immunoblotting and activity at the same time. Batch processing was performed to reduce construct to construct differences in degradation.
Immunoblotting
Total amount of membrane protein was measured by the Bradford protein assay (Bio-Rad Labs, Mississauga, ON) and the amount of CYP2A6 protein in the membrane preparations was determined by immunoblotting as previously described [27]. A standard curve was constructed using CYP2A6 expressed by a baculovirus-infected insect cell system (BD Gentest, San Jose, CA). The bacteria membrane preparations and baculovirus-expressed CYP2A6 were serially diluted to establish linear range of detection.
Enzyme assays for nicotine and coumarin
Nicotine C-oxidation and coumarin 7-hydroxylation were determined as previously described [28, 29]. Briefly, the reaction mixture contained 20 nM of CYP2A6, 20 nM of expressed cytochrome b5 (Invitrogen, Burlington, ON), 50 mM Tris-HCl buffer (pH 7.4) and substrate. Two substrate concentrations of nicotine (30 and 300 μM) or coumarin (5 and 50 μM) were initially used to screen for catalytic activity. Subsequently, full kinetic analyses were performed with nicotine, our main substrate of interest, ranging from 1–500 μM. Mouse liver cytosol (1.2 mg protein/ml) was added to the nicotine C-oxidation reaction as a source of aldehyde oxidase; this was added in excess so that CYP-mediated oxidation would be rate-limiting[28, 30]. The mixture was pre-warmed for 2 min. and the reaction was initiated by the addition of 1 mM NADPH. The mixture was incubated at 37°C for 45 min. for nicotine C-oxidation and 30 min. for coumarin 7-hydroxylation, and the reaction was stopped with 4% Na2CO3. The amount of cotinine and trans-3′-hydroxycotinine formed were detected by high-pressure liquid chromatography (HPLC) as previously described [28].
To detect 7-hydroxycoumarin formation, 5 μl of 20% (w/v) of trichloroacetic acid was added to the samples. 10 μl of 4-hydroxycoumarin (1 mg/ml) was also added as an internal standard. The samples were centrifuged at 13,000 rpm for 10 min. and 100 μl of the supernatant was analyzed by HPLC. Coumarin and its metabolite 7-hydroxycoumarin were separated on the ZORBAX SB C18 Column (250 × 4.6 mm I.D.; particle size, 5 μm) from Agilent Technologies Inc. (Mississauga, ON) at a flow-rate of 1 ml/min., with a mobile phase of acetonitrile, water and acetic acid (25 : 75 : 0.1, v/v) and detected at 315 nm. The concentrations of coumarin and 7-hydroxycoumarin were determined from standard curves created by spiking drug-free bacterial membrane preparations with known amounts of coumarin (0.2–10 μg/ml) and 7-hydroxycoumarin (50–1000 ng/ml). The within-day precision were 2.4–12.8% for coumarin and 2.6–8.3% for 7-hydroxycoumarin, and the between-day precision were 4.2–7.7% for coumarin and 5.9–13.7% for 7-hydroxycoumarin.
Bioinformatics
Linkage disequilibrium of CYP2A6*23 to other genotyped CYP2A6 variants was determined using Haploview (version 3.32) [31]. We used SIFT and PMUT, bioinformatics programs that predict whether nonsynonymous SNPs will affect enzyme function or structure based on physicochemical properties and evolutionary conservation of the amino acid changes, to examine how CYP2A6*23 may be affecting enzyme function [32, 33].
Statistics
The catalytic activities of the in vitro expressed wildtype and variant enzymes were compared using one-way ANOVA with the Bonferroni correction for post-hoc analyses. In ethnic groups where the allele frequency of CYP2A6*23 was zero, the score method was used to calculate confidence intervals [34]. A comparison of the CYP2A6*23 allele frequencies between ethnic groups, differences in the distribution of the CYP2A6*23 allele in nonsmokers versus smokers, and the Hardy-Weinberg equilibrium were calculated using Fisher’s Exact Test. Consistent with previous studies, gender [35] and smoking status [36, 37] were found to affect the rate of nicotine metabolism in our population of Black-African descent, as indicated by the 3HC/COT ratio [22]. Thus, the metabolic ratio was adjusted by dividing the value from each individual with the overall mean from their respective group (e.g. female nonsmokers, male nonsmokers, female smokers, male smokers). The effect of genotype on the adjusted 3HC/COT ratio was examined by a two-tailed independent t-test. A multivariate linear regression model was also used to examine the impact of CYP2A6*23 genotypeon the unadjusted 3HC/COT ratio while controlling for the effect of smoking status and gender. The unadjusted 3HC/COT ratio was not normally distributed according to the Kolmogorov-Smirnov test and was log-transformed for the regression model. All statistical analyses were performed using SPSS (version 14.0 for Windows) and GraphPad Prism (version 2.0 for Windows).
Results
CYP2A6*23 was found in populations of Black-African descent
Among the individuals of Black-African descent genotyped for CYP2A6*23 (n = 280), nine heterozygous and one homozygous individuals were found. Thus, CYP2A6*23 occurred at an allele frequency of 2.0% (95% confidence interval 0.8–3.1%, Table 1); genotype frequencies did not deviate from Hardy-Weinberg equilibrium (p = 0.821). Five of the ten individuals with CYP2A6*23 had other CYP2A6 variants (Table 2); CYP2A6*23 was not found in linkage disequilibrium with any of the other CYP2A6 variants genotyped in this population when analyzed with Haploview (version 3.32) [31], and no other nonsynonymous SNPs were found in linkage with CYP2A6*23 in the samples sequenced. CYP2A6*23 was not detected in Caucasian (N = 334 alleles), Chinese (N = 288 alleles) or Japanese individuals (N = 104 alleles) (Table 1). The allele frequency of CYP2A6*23 in individuals of Black-African descent significantly differed from Caucasians and Chinese (p < 0.05, Table 1).
Table 1.
Allele frequency of CYP2A6*23 by ethnicity
| Ethnicity | Allele frequency (%) | Total # of alleles |
95% Confidence Intervals |
p-values a |
|---|---|---|---|---|
| Black-African descent | 2.0 | 560 | 0.8–3.1% | --- |
| Caucasian | 0 | 334 | 0–1.1% | 0.01 |
| Chinese | 0 | 288 | 0–1.3% | 0.02 |
| Japanese | 0 | 104 | 0–3.6% | 0.23 |
The allele frequency of CYP2A6*23 in each ethnic group was compared against the value found in individuals of Black-African descent.
Table 2.
CYP2A6*23 genotype groups and their mean adjusted 3HC/COT ratio
| Allele | Genotype | n | Mean adjusted 3HC/COT | SD | % of wildtype | p-valuesa |
|---|---|---|---|---|---|---|
| *23 | *1/*1 | 150 | 1.210 | 0.634 | 100 | 0.04 |
| *1/*23 | 4 | 0.756 | 0.540 | 62.4 | ||
| *23/*23 | 1 | 0 | 0 | 0 | ||
| *17 | *1/*1 | 150 | 1.210 | 0.634 | 100 | <0.001 |
| *1/*17 | 19 | 0.672 | 0.468 | 55.5 | ||
| *17/*17 | 3 | 0.109 | 0.095 | 9.1 | ||
| More than one variant | *1/*1 | 150 | 1.210 | 0.634 | 100 | 0.02 |
| *17/*23 | 2 | 0.259 | 0.366 | 21.4 | ||
| *20/*23 | 2 | 0.555 | 0.512 | 45.8 | ||
| *9/*23 | 1 | 1.032 | 0 | 85.2 |
The adjusted 3HC/COT ratio was compared between wildtype individuals (CYP2A6*1/*1) to other genotype groups combined using a two-tailed independent t-test.
CYP2A6*23 reduced enzyme activity towards nicotine and coumarin in vitro
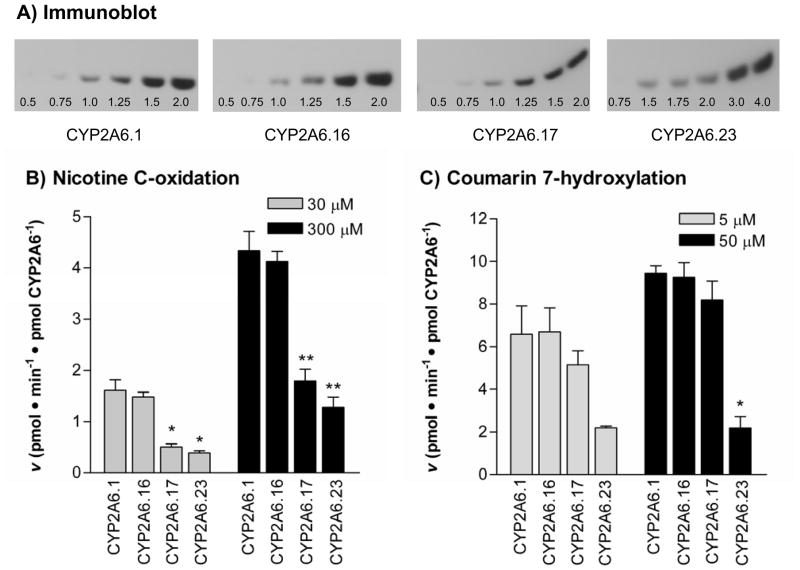
The specific content of CYP2A6 was similar between CYP2A6.1 (0.11 pmol CYP2A6/μg membrane protein), CYP2A6.16 (0.12 pmol CYP2A6/μg of membrane protein) and CYP2A6.17 (0.12 pmol CYP2A6/μg membrane protein), although CYP2A6.23 was expressed at lower levels (0.07 pmol CYP2A6/μg membrane protein) (Fig. 1A). The negative construct did not produce any detectable CYP2A6 protein. Subsequent enzyme assays with the wildtype and variant constructs were performed using equivalent amounts of CYP2A6 (20 nM).
Figure 1. CYP2A6.23 had substantially reduced cata lytic activity towards nicotine and coumarin in vitro.
A) Immunoblot shows the expression of CYP2A6 protein from the wildtype and variant constructs. The amount of total membrane protein loaded is as labeled and ranged from 0.5–2.0 μg for CYP2A6.1, CYP2A6.16 and CYP2A6.17, while CYP2A6.23 was loaded at 0.75–4.0 μg. Detection of CYP2A6 constructs was linear by immunoblotting at lower amounts loaded, and was used to calculate the amount of CYP2A6 detected by comparison to the standard (not shown). B) CYP2A6.17 and CYP2A6.23 have reduced cotinine formation from nicotine (30 and 300 μM) while CYP2A6.16 had similar activity as CYP2A6.1. C) CYP2A6.23 had reduced 7-hydroxycoumarin formation from coumarin (5 and 50 μM) while CYP2A6.16 and CYP2A6.17 had similar activity as CYP2A6.1. No product formation was found using the negative construct. The data is presented as mean ± SEM for nicotine C-oxidation (n = 3) and coumarin 7-hydroxylation (n = 2). * p < 0.01, ** p < 0.001 when compared to CYP2A6.1.
We initially screened the activities of the wildtype and variant constructs towards nicotine and coumarin at two concentrations (Fig. 1B, C). There was a significant difference in activities of the constructs at 30 μM of nicotine (F = 29.0, p < 0.001), 300 μM of nicotine (F = 35.9, p < 0.001), and 50 μM of coumarin (F = 28.3, p < 0.01). Both CYP2A6.23 and CYP2A6.17 had significantly lower activity towards nicotine compared to CYP2A6.1 at 30 μM (p < 0.01) and 300 μM (p < 0.001) of substrate. CYP2A6.23 also had significantly reduced activity towards 50 μM of coumarin (p < 0.01) while CYP2A6.17 retained similar activity as the wildtype enzyme. CYP2A6.16 had similar activities as CYP2A6.1 towards nicotine and coumarin at both concentrations tested (Fig. 1B, C).
We then performed full kinetic analyses on nicotine, our main substrate of interest. CYP2A6.23 and CYP2A6.17 had reduced activity towards nicotine in vitro while CYP2A6.16 had similar activity as CYP2A6.1 (Fig. 2). The apparent Km values did not significantly differ between the wildtype and variant constructs (F = 3.0, p = 0.083), though it trended towards higher values for CYP2A6.17 (p = 0.09, Table 3). However, there was a significant difference in Vmax (F = 35.2, p < 0.001) and Vmax/Km (F = 19.0, p < 0.001) between the constructs. Vmax was significantly lower for CYP2A6.23 (p < 0.001) and CYP2A6.17 (p < 0.01) compared to CYP2A6.1. Likewise, Vmax/Km was significantly lower for CYP2A6.23 (p < 0.001) and CYP2A6.17 (p < 0.01) compared to CYP2A6.1. There was no significant difference in Vmax and Vmax/Km between CYP2A6.23 and CYP2A6.17, or between CYP2A6.16 and CYP2A6.1.
Figure 2. CYP2A6.17 and CYP2A6.23, but not CYP2A6.16, have reduced in vitro nicotine C-oxidation.
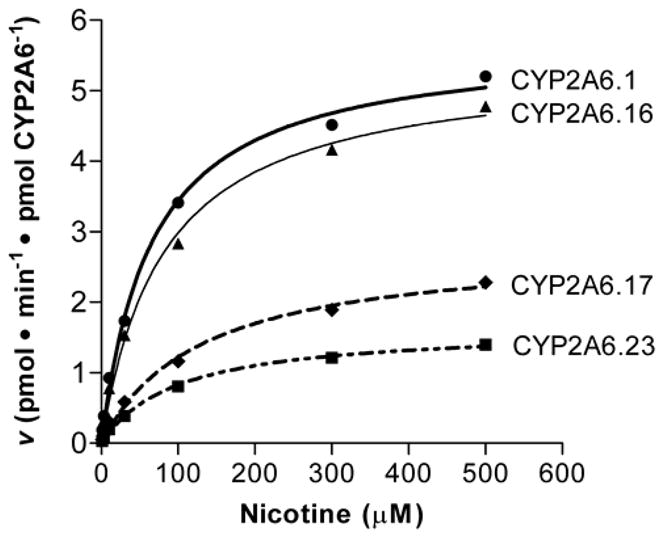
A representative plot is shown, and the curve was fitted to the Michaelis-Menten equation using non-linear regression in GraphPad Prism (version 2.0).
Table 3.
Kinetic parameters of CYP2A6 wildtype and variant constructs for nicotine
| Km (μM)a | Vmax (pmol·min−1·pmol CYP2A6−1) |
Vmax/Km (nL·min−1·pmol CYP2A6−1) |
|
|---|---|---|---|
| CYP2A6.1 | 58.3 ± 6.5 | 5.5 ± 0.5 | 97.3 ± 11.6 |
| CYP2A6.16 | 73.6 ± 6.1 | 5.1 ± 0.2 | 71.4 ± 7.7 |
| CYP2A6.17 | 88.9 ± 6.8 | 2.6 ± 0.2* | 29.6 ± 4.2* |
| CYP2A6.23 | 77.7 ± 10.4 | 1.4 ± 0.2** | 18.4 ± 2.3** |
Kinetic parameters were calculated using non-linear regression in GraphPad Prism (version 2.0). Data is presented as mean ± SEM of parameters calculated from three or four independent experiments.
p < 0.01,
p < 0.001 when compared to CYP2A6.1.
CYP2A6*23 decreased the rate of nicotine metabolism in vivo in a population of Black-African descent
The 3HC/COT is a validated phenotypic measure of CYP2A6 activity and rates of nicotine metabolism [23], with previous studies finding a significant association between the ratio and CYP2A6 genotype [38–40]. Because the 3HC/COT ratio did not differ significantly from CYP2A6*1/*1 individuals in this population of Black-African descent, the wildtype group (*1/*1) included individuals with the CYP2A6*1B allele. The adjusted 3HC/COT ratio was significantly lower in individuals with at least one CYP2A6*23 allele and no other variants (n = 5) in comparison to CYP2A6*1/*1 individuals(n = 150) (p < 0.04, Fig. 3). A gene-dose effect was observed such that CYP2A6*1/*23 heterozygous individuals had ~40% loss in enzyme activity compared to CYP2A6*1/*1 individuals, while one CYP2A6*23/*23 homozygous individual did not produce any 3HC (Fig. 3). A similar impact of CYP2A6*23 was observed in a multivariate linear regression model with the log (3HC/COT) as the dependent variable and including smoking status and gender as predicting variables (R2 = 0.355, p < 0.04). It is notable that the five individuals with CYP2A6*23 in addition to other genetic variants also had significantly lower 3HC/COT ratios compared to the wildtype group (p < 0.02, Table 2).
Figure 3. CYP2A6*23 decreased the rates of nicotine metabolism in vivo, as measured by the 3HC/COT ratio.
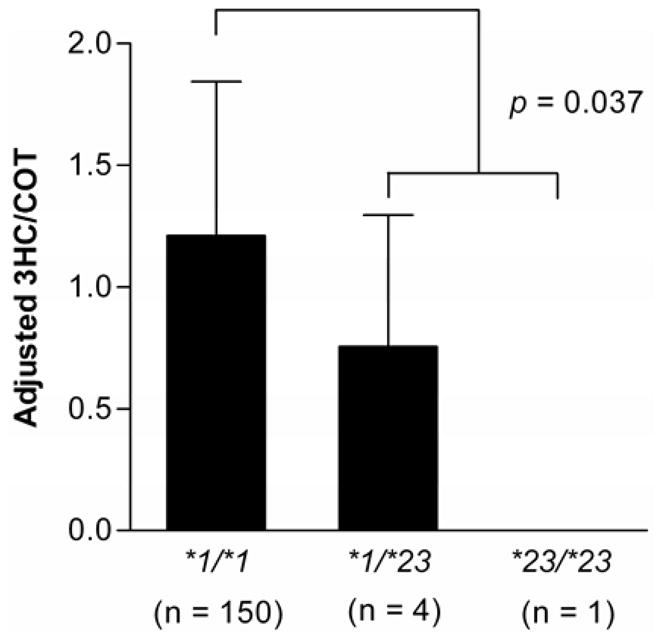
The number of individuals in each genotype group is shown in parentheses on the x-axis. The data is presented as mean ± SD. Individuals with other CYP2A6 genetic variants (CYP2A6*2, *4A & D, *9, *12, *14, *15, *17, *20, *21, *24, *25, *26, *27, *28, *29) were excluded from the wildtype and CYP2A6*23 genotype groups.
CYP2A6*23 was associated with a lower likelihood of being a current adult smoker
CYP2A6*23 trended towards a higher frequency in nonsmokers (3.1%, N = 9/286 alleles) compared to smokers (0.7%, N = 2/274 alleles) (p = 0.06, Fig. 4). Individuals with CYP2A6*23 were approximately 4–5 times less likely to be smokers when all individuals with CYP2A6*23 were taken into account (odds ratio = 0.23). The magnitude of the effect was the same when analyses were restricted to individuals with only CYP2A6*1/*1, CYP2A6*1/*23, and CYP2A6*23/*23 (odds ratio = 0.18, p = 0.11).
Figure 4. Individuals with the CYP2A6*23 allele trended to having a lower likelihood of being current smokers.
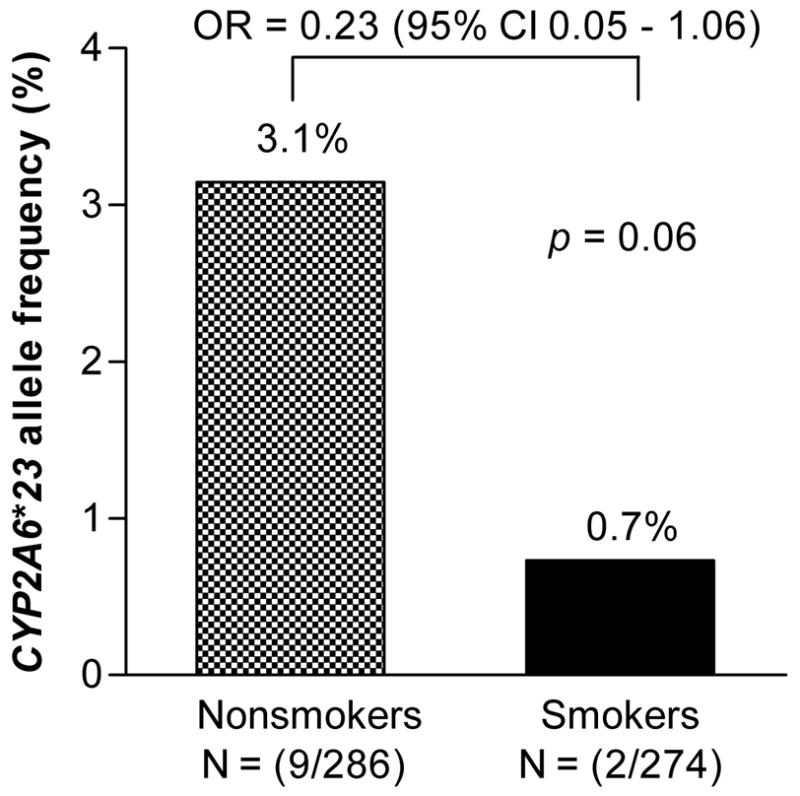
The allele frequency of CYP2A6*23 was calculated from the 280 genotyped individuals in the study, including 143 nonsmokers and 137 smokers.
Discussion
We have identified a novel CYP2A6 allele, CYP2A6*23, which is found in populations of Black-African descent but not in Caucasians, Chinese and Japanese. Two recently described alleles, CYP2A6*17 and CYP2A6*20, have also been identified exclusively in this population [21, 41]. These genetic variants may help explain the reduced rates of nicotine metabolism in individuals of Black-African descent compared to Caucasians [11, 12].
We have demonstrated that CYP2A6*23 (2161C>T, R203C) impairs both nicotine C-oxidation and coumarin 7-hydroxylation in vitro. CYP2A6.23 had a significantly reduced Vmax towards nicotine, and the intrinsic clearance (Vmax/Km) was reduced to 19% of the wildtype enzyme. This is in agreement with the observation that in vivo, the 3HC/COT ratio is reduced in individuals with the CYP2A6*23 allele. CYP2A6*23 may be affecting enzyme function through alteration in substrate binding. Molecular modeling indicates Arg203 may be important in the orientation of Phe209, a residue critical for coumarin binding and possibly involved in nicotine binding [42, 43]. In addition, CYP2A6.23 had a lower level of expression in vitro compared to the other constructs used in this study, which may also contribute to the lower activity observed in vivo.
In contrast to CYP2A6*23 (2161C>T, R203C), CYP2A6*16 (2161C>A, R203S) did not appear to affect enzyme function. Our in vitro data suggests CYP2A6.16 has similar rates of nicotine and coumarin metabolism as the wildtype enzyme. Furthermore, Nakajima et al. recently reported that CYP2A6*16 did not affect rates of nicotine metabolism in vivo [44]. These data suggests the functional impact of CYP2A6*23 (Cys203) differs from that of CYP2A6*16 (Ser203). The wildtype residue (Arg203) is positively charged and hydrophilic while Cys203 is neutral and hydrophobic and Ser203 is neutral and hydrophilic. Furthermore, the bioinformatics program SIFT and PMUT [32, 33] predicted the amino acid change in CYP2A6*23 as not tolerated while CYP2A6*16 was predicted to be benign.
Interestingly, CYP2A6*23 impaired nicotine C-oxidation to at least the same extent as CYP2A6*17 both in vitro and in vivo. Similar to expressed CYP2A6.23, CYP2A6.17 had a significantly reduced Vmax towards nicotine, with Vmax/Km reduced to 30% of the wildtype enzyme. This is in agreement with a previous study where CYP2A6.17 expressed in E. coli did not alter Km but reduced Vmax towards nicotine, with Vmax/Km reduced to 40% of the wildtype construct [21]. In vivo, the 3HC/COT ratio in heterozygous individuals for CYP2A6*17 is reduced to approximately 55% of wildtype activity, while homozygous individuals of CYP2A6*17 had approximately 9% of wildtype activity. This is consistent with previous studies using the COT/NIC ratio as a biomarker [44], and together these data strongly suggest the CYP2A6*17 allele results in reduced nicotine metabolism. We also observed that the impact of CYP2A6.17is substrate-dependent such that it did not differ in coumarin 7-hydroxylation compared to the wildtype enzyme. Accordingly, a previous study found CYP2A6.17 expressed in E. coli had an increased Km but no change in Vmax towards coumarin [21].
A trend was observed where individuals with CYP2A6*23 were less likely to be current adult smokers. Several case-control studies have associated CYP2A6 genetic variations leading to impaired activity with a lower likelihood of smoking [45–47], though negative findings have also been reported [48–50]. A greater understanding of CYP2A6 genetic variation through identification and characterization of novel variants, particularly among different ethnic groups, will allow for better replication of genetic case-control association studies.
In summary, we have discovered a novel CYP2A6 allele, occurring predominantly in a population of Black-African descent, which impairs enzyme activity in vitro and in vivo and may be associated with a lower risk of smoking. An understanding of the genetic factors that contribute to nicotine pharmacokinetics in populations of Black-African descent is important given their unique smoking patterns and higher incidence of tobacco-related illnesses.
Acknowledgments
Funding source: This study was supported by the Centre for Addiction and Mental Health, Canadian Institute for Health Research (CIHR) MOP53248 grant, NSERC CGS-D Postgraduate Scholarship (MKH), CIHR-funded SPICE and TUSP Scholarships (JCM), and a Canada Research Chair in Pharmacogenetics (RFT).
Footnotes
Conflicts of interest: Dr. R.F. Tyndale hold shares in Nicogen Research Inc., a company that is focused on novel smoking cessation treatment approaches. None of the data contained in this manuscript alters or improves any commercial aspect of Nicogen, no Nicogen funds were used in this work, and the manuscript was not reviewed by others affiliated with Nicogen.
References
- 1.Diagnostic and Statistical Manual - Text Revision IV. American Psychiatric Associations; 2000. [Google Scholar]
- 2.Benowitz NL, Jacob P., 3rd Metabolism of nicotine to cotinine studied by a dual stable isotope method. Clinical Pharmacology & Therapeutics. 1994;56:483–493. doi: 10.1038/clpt.1994.169. [DOI] [PubMed] [Google Scholar]
- 3.Messina ES, Tyndale RF, Sellers EM. A Major Role for CYP2A6 in Nicotine C-Oxidation by Human Liver Microsomes. J Pharmacol Exp Ther. 1997;282(3):1608–1614. [PubMed] [Google Scholar]
- 4.Nakajima M, Yamamoto T, Nunoya K, Yokoi T, Nagashima K, Inoue K, et al. Characterization of CYP2A6 involved in 3′-hydroxylation of cotinine in human liver microsomes. J Pharmacol Exp Ther. 1996;277(2):1010–1015. [PubMed] [Google Scholar]
- 5.Pelkonen O, Rautio A, Raunio H, Pasanen M. CYP2A6: a human coumarin 7-hydroxylase. Toxicology. 2000;144(1–3):139. doi: 10.1016/s0300-483x(99)00200-0. [DOI] [PubMed] [Google Scholar]
- 6.Ikeda K, Yoshisue K, Matsushima E, Nagayama S, Kobayashi K, Tyson CA, et al. Bioactivation of Tegafur to 5-Fluorouracil Is Catalyzed by Cytochrome P-450 2A6 in Human Liver Microsomes in Vitro. Clin Cancer Res. 2000;6(11):4409–4415. [PubMed] [Google Scholar]
- 7.Nunoya K, Yokoi Y, Kimura K, Kodama T, Funayama M, Inoue K, et al. (+)-cis-3,5-dimethyl-2-(3-pyridyl) thiazolidin-4-one hydrochloride (SM- 12502) as a novel substrate for cytochrome P450 2A6 in human liver microsomes. J Pharmacol Exp Ther. 1996;277(2):768–774. [PubMed] [Google Scholar]
- 8.Yamazaki H, Inui Y, Yun C-H, Guengerich FP, Shimada T. Cytochrome P450 2E1 and 2A6 enzymes as major catalysts for metabolic activation of N-nitrosodialkylamines and tobacco-related nitrosamines in human liver microsomes. Carcinogenesis. 1992;13(10):1789–1794. doi: 10.1093/carcin/13.10.1789. [DOI] [PubMed] [Google Scholar]
- 9.Malaiyandi V, Sellers EM, Tyndale RF. Implications of CYP2A6 genetic variation for smoking behaviors and nicotine dependence. Clinical Pharmacology & Therapeutics. 2005;77(3):145. doi: 10.1016/j.clpt.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 10.Schoedel KA, Hoffmann EB, Rao Y, Sellers EM, Tyndale RF. Ethnic variation in CYP2A6 and association of genetically slow nicotine metabolism and smoking in adult Caucasians. Pharmacogenetics. 2004;14:615–626. doi: 10.1097/00008571-200409000-00006. [DOI] [PubMed] [Google Scholar]
- 11.Benowitz NL, Perez-Stable EJ, Fong I, Modin G, Herrera B, Jacob P., III Ethnic Differences in N-Glucuronidation of Nicotine and Cotinine. J Pharmacol Exp Ther. 1999;291(3):1196–1203. [PubMed] [Google Scholar]
- 12.Perez-Stable EJ, Herrera B, Jacob P, III, Benowitz NL. Nicotine Metabolism and Intake in Black and White Smokers. JAMA. 1998;280(2):152–156. doi: 10.1001/jama.280.2.152. [DOI] [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention. Cigarette Smoking Among Adults: United States, 2004. Morbidity and Mortality Weekly Report. 2005:1121–1148. [PubMed] [Google Scholar]
- 14.Trinidad DR, Gilpin EA, Lee L, Pierce JP. Do the majority of Asian-American and African-American smokers start as adults? American Journal of Preventive Medicine. 2004;26(2):156. doi: 10.1016/j.amepre.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 15.Trinidad DR, Gilpin EA, Lee L, Pierce JP. Has There Been a Delay in the Age of Regular Smoking Onset Among African Americans? Annals of Behavioral Medicine. 2004;28(3):152–157. doi: 10.1207/s15324796abm2803_2. [DOI] [PubMed] [Google Scholar]
- 16.National Survey on Drug Use and Health. Past Month Cigarette Use among Racial and Ethnic Groups. The NSDUH Report. 2006 [Google Scholar]
- 17.Centers for Disease Control and Prevention; Department of Health and Human Services. Tobacco use among U.S. racial/ethnic minority groups — African-Americans, American-Indians and Alaska Natives, Asian-Americans and Pacific Islanders, and Hispanics: a report of the Surgeon General. Atlanta: 1998. [PubMed] [Google Scholar]
- 18.Haiman CA, Stram DO, Wilkens LR, Pike MC, Kolonel LN, Henderson BE, et al. Ethnic and Racial Differences in the Smoking-Related Risk of Lung Cancer. N Engl J Med. 2006;354(4):333–342. doi: 10.1056/NEJMoa033250. [DOI] [PubMed] [Google Scholar]
- 19.National Center for Chronic Disease Prevention and Health Promotion; Office of Smoking and Health. Tobacco Use Among US Racial/Ethnic Minority Groups—African Americans, American Indians and Alaska Natives, Asian Americans and Pacific Islanders, and Hispanics: A Report of the Surgeon General. Atlanta, GA: 1998. [PubMed] [Google Scholar]
- 20.Kiyotani K, Fujieda M, Yamazaki H, Shimada T, Guengerich FP, Parkinson A, et al. Twenty one novel single nucleotide polymorphisms (SNPs) of the CYP2A6 gene in Japanese and Caucasians. Drug Metab Pharmacokinet. 2002;17(5):482–487. doi: 10.2133/dmpk.17.482. [DOI] [PubMed] [Google Scholar]
- 21.Fukami T, Nakajima M, Yoshida R, Tsuchiya Y, Fujiki Y, Katoh M, et al. A novel polymorphism of human CYP2A6 gene CYP2A6*17 has an amino acid substitution (V365M) that decreases enzymatic activity in vitro and in vivo. Clinical Pharmacology & Therapeutics. 2004;76(6):519. doi: 10.1016/j.clpt.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 22.Mwenifumbo JC, Sellers EM, Tyndale RF. Nicotine metabolism and CYP2A6 activity in a population of black African descent: Impact of gender and light smoking. Drug and Alcohol Dependence. 2007;89(1):24. doi: 10.1016/j.drugalcdep.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 23.Dempsey D, Tutka P, Jacob P, Allen F, Schoedel K, Tyndale RF, et al. Nicotine metabolite ratio as an index of cytochrome P450 2A6 metabolic activity. Clinical Pharmacology & Therapeutics. 2004;76(1):64. doi: 10.1016/j.clpt.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 24.Gillam EMJ, Aguinaldo AMA, Notley LM, Kim D, Mundkowski RG, Volkov AA, et al. Formation of Indigo by Recombinant Mammalian Cytochrome P450. Biochemical and Biophysical Research Communications. 1999;265(2):469. doi: 10.1006/bbrc.1999.1702. [DOI] [PubMed] [Google Scholar]
- 25.Soucek P. Expression of Cytochrome P450 2A6 in Escherichia coli: Purification, Spectral and Catalytic Characterization, and Preparation of Polyclonal Antibodies. Archives of Biochemistry and Biophysics. 1999;370(2):190. doi: 10.1006/abbi.1999.1388. [DOI] [PubMed] [Google Scholar]
- 26.Pritchard MP, McLaughlin L, Friedberg T. Establishment of functional human cytochrome P450 monooxygenase systems in Escherichia coli. In: Phillips IR, Shephard EA, editors. Methods in Molecular Biology, vol. 320: Cytochrome P450 Protocols. 2. Humana Press Inc; Totowa, NJ: 2006. pp. 19–29. [DOI] [PubMed] [Google Scholar]
- 27.Schoedel KA, Sellers EM, Palmour R, Tyndale RF. Down-Regulation of Hepatic Nicotine Metabolism and a CYP2A6-Like Enzyme in African Green Monkeys after Long-Term Nicotine Administration. Mol Pharmacol. 2003;63(1):96–104. doi: 10.1124/mol.63.1.96. [DOI] [PubMed] [Google Scholar]
- 28.Siu ECK, Wildenauer DB, Tyndale RF. Nicotine self-administration in mice is associated with rates of nicotine inactivation by CYP2A5. Psychopharmacology. 2006;184(3–4):401–408. doi: 10.1007/s00213-006-0306-6. [DOI] [PubMed] [Google Scholar]
- 29.Nakajima M, Yamamoto T, Nunoya K, Yokoi T, Nagashima K, Inoue K, et al. Role of human cytochrome P4502A6 in C-oxidation of nicotine. Drug Metabolism and Disposition. 1996;24(11):1212. [PubMed] [Google Scholar]
- 30.Cashman JR, Park SB, Yang ZC, Wrighton SA, Jacob P, Benowitz NL. Metabolism of nicotine by human liver microsomes: stereoselective formation of trans-nicotine N′-oxide. Chem Res Toxicol. 1992;5(5):639–646. doi: 10.1021/tx00029a008. [DOI] [PubMed] [Google Scholar]
- 31.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 32.Ng PC, Henikoff S. Predicting Deleterious Amino Acid Substitutions. Genome Res. 2001;11(5):863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrer-Costa C, Gelpi JL, Zamakola L, Parraga I, de la Cruz X, Orozco M. PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics. 2005;21(14):3176–3178. doi: 10.1093/bioinformatics/bti486. [DOI] [PubMed] [Google Scholar]
- 34.Wilson EB. Probable inference, the law of succession, and statistical inference. Journal of the American Statistical Association. 1927;22:209–212. [Google Scholar]
- 35.Benowitz NL, Lessov-Schlaggar CN, Swan GE, Jacob P., 3rd Female sex and oral contraceptive use accelerate nicotine metabolism. Clin Pharmacol Ther. 2006;79(5):480–488. doi: 10.1016/j.clpt.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 36.Benowitz NL, Jacob P., 3rd Nicotine and cotinine elimination pharmacokinetics in smokers and nonsmokers. Clin Pharmacol Ther. 1993;53(3):316–323. doi: 10.1038/clpt.1993.27. [DOI] [PubMed] [Google Scholar]
- 37.Benowitz NL, Jacob P. Effects of cigarette smoking and carbon monoxide on nicotine and cotinine metabolism[ast] Clin Pharmacol Ther. 2000;67(6):653. doi: 10.1067/mcp.2000.107086. [DOI] [PubMed] [Google Scholar]
- 38.Benowitz NL, Swan GE, Jacob P, Lessov-Schlaggar CN, Tyndale RF. CYP2A6 genotype and the metabolism and disposition kinetics of nicotine[ast] Clin Pharmacol Ther. 2006;80(5):457. doi: 10.1016/j.clpt.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 39.Malaiyandi V, Goodz S, Sellers EM, Tyndale RF. CYP2A6 genotype, phenotype and the use of nicotine metabolites as biomarkers during ad libitum smoking. Cancer Epidemiol Biomarkers Prev. 2006;15(10):1812–1819. doi: 10.1158/1055-9965.EPI-05-0723. [DOI] [PubMed] [Google Scholar]
- 40.Malaiyandi V, Lerman C, Benowitz NL, Jepson C, Patterson F, Tyndale RF. Impact of CYP2A6 genotype on pretreatment smoking behaviour and nicotine levels from and usage of nicotine replacement therapy. Mol Psychiatry. 2006;11(4):400–409. doi: 10.1038/sj.mp.4001794. [DOI] [PubMed] [Google Scholar]
- 41.Fukami T, Nakajima M, Higashi E, Yamanaka H, McLeod HL, Yokoi T. A novel CYP2A6*20 allele found in African-American population produces a truncated protein lacking enzymatic activity. Biochemical Pharmacology. 2005;70(5):801. doi: 10.1016/j.bcp.2005.05.029. [DOI] [PubMed] [Google Scholar]
- 42.Kiyotani K, Fujieda M, Yamazaki H, Shimada T, Guengerich FP, Parkinson A, et al. Drug Metabolism Reviews: Biotransformation and Disposition of Xenobiotics. Maui, Hawaii: Oct 23–27, 2005. Effects of Arg203Ser (CYP2A6*16) on substrate binding of CYP2A6. [Google Scholar]
- 43.Lewis DFV, Dickins M, Lake BG, Eddershaw PJ, Tarbit MH, Goldfarb PS. Molecular modelling of the human cytochrome P450 isoform CYP2A6 and investigations of CYP2A substrate selectivity. Toxicology. 1999;133(1):1. doi: 10.1016/s0300-483x(98)00149-8. [DOI] [PubMed] [Google Scholar]
- 44.Nakajima M, Fukami T, Yamanaka H, Higashi E, Sakai H, Yoshida R, et al. Comprehensive evaluation of variability in nicotine metabolism and CYP2A6 polymorphic alleles in four ethnic populations. Clinical Pharmacology & Therapeutics. 2006;80(3):282. doi: 10.1016/j.clpt.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 45.Schoedel KA, Hoffmann EB, Rao Y, Sellers EM, Tyndale RF. Ethnic variation in CYP2A6 and association of genetically slow nicotine metabolism and smoking in adult Caucasians. Pharmacogenetics. 2004;14:615–626. doi: 10.1097/00008571-200409000-00006. [DOI] [PubMed] [Google Scholar]
- 46.Tyndale RF, Sellers EM. Variable CYP2A6-Mediated Nicotine Metabolism Alters Smoking Behavior and Risk. Drug Metab Dispos. 2001;29(4):548–552. [PubMed] [Google Scholar]
- 47.Pianezza ML, Sellers EM, Tyndale RF. Nicotine metabolism defect reduces smoking. Nature. 1998;393(6687):750. doi: 10.1038/31623. [DOI] [PubMed] [Google Scholar]
- 48.Zhang X, Amemo K, Ameno S, Iwahashi K, Kinoshita H, Kubota T, et al. Lack of association between smoking and CYP2A6 gene polymorphisms in A Japanese population. Nihon Arukoru Yakubutsu Igakkai Zasshi. 2001;36(5):486–490. [PubMed] [Google Scholar]
- 49.Tan W, Chen GF, Xing DY, Song CY, Kadlubar FF, Lin DX. Frequency of CYP2A6 gene deletion and its relation to risk of lung and esophageal cancer in the Chinese population. International Journal of Cancer. 2001;95(2):96–101. doi: 10.1002/1097-0215(20010320)95:2<96::aid-ijc1017>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 50.Ando MHN, Ariyoshi N, Kamataki T, Matsuo K, Ohno Y. Association of CYP2A6 gene deletion with cigarette smoking status in Japanese adults. J Epidemiol. 2003;13(3):176–181. doi: 10.2188/jea.13.176. [DOI] [PMC free article] [PubMed] [Google Scholar]