Abstract
The activities of p53 cover diverse aspects of cell biology, including cell cycle control, apoptosis, metabolism, fertility, differentiation and cellular reprogramming. Although loss of p53 function engenders tumor susceptibility, hyperactivation of p53 is lethal. Therefore, p53 activity must be strictly regulated to maintain normal tissue homeostasis. Critical for the control of p53 function are its two main negative regulators: Mdm2 and Mdmx. Recent reports have provided insight into the complex mechanisms that regulate these two proteins and have revealed novel functions for each. Here, we review and evaluate models of Mdm2- and Mdmx-dependent regulation of p53 activity. Both Mdm2 and Mdmx receive input from numerous signaling pathways and interact with many proteins in addition to p53. Therefore, we also consider roles for Mdm2 and Mdmx in additional cancer-related networks, including Notch signaling and the epithelial-to-mesenchymal transition.
The Mdm2/Mdmx/p53 triumvirate
p53 is a transcription factor that induces expression of numerous downstream targets in response to intra- and extracellular cues. The remarkable evolutionary conservation of p53acrossinvertebrate and vertebrate speciesunder-scores its prominence in many biological processes. Clinical studies and mouse models have shown that p53 is a tumor suppressor that is mutated in ~50% of human cancers, and functionally inactivated in many more. Mdm2 and Mdmx (for mouse double minute, Mdmx is also known as Mdm4) are critical negative regulators of p53, as revealed by models in which deletion of either gene is lethal in a p53-dependent manner (Table 1). Conversely, inactivation of p53 by over-expression of either Mdm2 or Mdmx is oncogenic, as revealed by their frequently elevated levels in human cancers expressing wild type p53 (for a review, see Ref [1]). Mouse models and pre-clinical studies have revealed that inhibiting Mdm2 or Mdmx can have significant therapeutic impact in cancers expressing wild type p53. Thus, understanding the mechanisms that control the function of these oncogenes is important for designing and using future anti-cancer strategies that target the p53 pathway.
Table 1.
Tissue-specific phenotypes of Mdm2 or Mdmx loss.
| Tissues | Genotypea | Phenotype | Embryo | Refs | |
|---|---|---|---|---|---|
| CNS | Nestin-Cre; p53lsl/− | Mdm2−/− | Apoptosis | Lethal | [33] |
| Nestin-Cre | Mdmx−/− | Apoptosis, reduced proliferation | |||
| Mdm2FM/FM | Apoptosis | Lethal | [40] | ||
| MdmxFX/FX | Apoptosis, reduced proliferation | ||||
| p53ER/− | Mdm2−/− | No effect in adult | [36] | ||
| Intestine | Villin-Cre | Mdm2FM/FM | Apoptosis | [38] | |
| Smooth muscle | SM22-CreERT2 | MdmxFX/FX | Apoptosis in proliferative cells | [37] | |
| Mdm2FM/FM MdmxFX/FX |
Apoptosis No effect |
[32] |
|||
| Erythrocyte |
EpoRGFP-Cre/+ |
Mdm2lox/lox Mdmxlox/lox |
Apoptosis Reduced proliferation |
Lethal |
[35] |
| Heart | αMyoHC-Cre | Mdm2FM/− | Apoptosis | Lethal | [34] |
| Thymus | p53-ER | MdmxFX/− | Dilated heart | [39] | |
| Mdm2−/− | Apoptosis | [36] | |||
| Spleen | p53-ER | Mdm2−/− | Apoptosis | [36] | |
| Testis | p53-ER | Mdm2−/− | Apoptosis | [36] | |
| Lung | p53-ERb | Mdm2−/− | No effect | [36] | |
| Kidney | p53-ERb | Mdm2−/− | No effect | [36] | |
| Liver | p53-ERb | Mdm2−/− | No effect | [36] |
Assuming the Cre excises the floxed Mdm2 and Mdmx alleles at similar efficiency, one can conclude that in many cases loss of Mdmx is less severe. However, the possibility remains that Cre-mediated excision is not 100%; therefore, it may be premature to conclude that Mdmx is completely dispensable in all these tissues.
Long term effects of Mdm2 loss could not be studied in these tissues due to premature death.
The degree of sequence similarity between Mdm2 and Mdmx indicates they were retained following duplication from a single ancestral gene. Both proteins bind to p53 via an N-terminal hydrophobic pocket, and this domain contains the highest identity at the amino acid level. The Mdm2 and Mdmx p53-binding domains occlude an N-terminal alpha-helix of p53. This prevents the recruitment of transcriptional co-activators and thereby inhibits p53 transactivation function. This transcriptional antagonism can take place within the nucleus, as Mdm2 and Mdmx have been detected at p53-responsive promoter elements in chromatin [2]. However, Mdm2 and Mdmx are most abundant in the cytosol in many cell lines, suggesting cytoplasmic localization is important for their function [3]. Interestingly, Mdm2 has been reported to promote translocation of p53 from the nucleus to the cytoplasm, and Mdmx may modulate this process [4]. Such a nuclear export mechanism could clearly reduce p53-dependent transcription. p53 function is also inhibited by Mdm2 dependent degradation (see below), which occurs in both the nucleus and cytoplasm, perhaps linked to nuclear export [5]. Conversely, p53 activation can be enhanced by reduced nuclear export and increased nuclear import [6,7].
The role of Mdm2 and Mdmx in p53 post-translational modification
The C termini of both Mdm2 and Mdmx contain a RING (really interesting new gene) domain of the ‘rare’ C2H2C4 type [8]. This nomenclature refers to the order of cysteine and histidine residues within the RING domain, which are required to form a Zn2+ chelating structure important for RING function. In common with many other RING domain proteins, the Mdm2 RING has intrinsic E3 ubiquitin ligase activity, in that it can promote the transfer of ubiquitin molecules from an E2 conjugating enzyme directly to lysine residues of target substrates (Figure 1 and Ref. [9] for review). Despite conservation of the C2H2C4 RING domain, Mdmx shows little intrinsic ubiquitin ligase activity towards p53 [10].
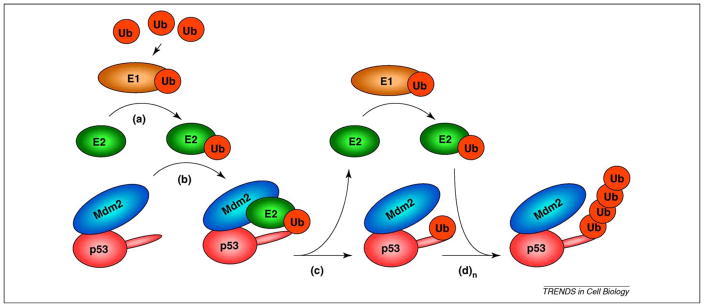
Figure 1.
The E3 ubiquitin ligase function of Mdm2. Free ubiquitin (Ub) is ‘activated’ upon covalent binding to E1 ubiquitin activating enzyme. (a) Ubiquitin is then transferred to the E2 conjugating enzyme active site via a thioester linkage. (b) Mdm2 recruits ubiquitin-loaded E2, bringing it into proximity with p53. (c) Ubiquitin is transferred to p53, and E2 can re-enter the cycle with Ub-charged E1. (d) Ubiquitin-loaded E2 can again be recruited by Mdm2 for (n) cycles, thus extending the ubiquitin chain (n) times. Although mono ubiquitination and polyubiquitination can be performed by the same E2 in vitro, it is unclear whether this is also the case in vivo.
Generally, poly-ubiquitin chains linked via lysine 48 (K48) of ubiquitin trigger protein degradation, whereas monoubiquitylation modulates target protein function. Consistent with this, Mdm2 facilitates the polyubiquitylation and proteasome-dependent degradation of p53 [11]. Thus, Mdm2 negatively regulates p53 by both suppression of transactivation and post-translational destabilization. Under certain conditions, Mdm2 can also promote monoubiquitylation of p53; this does not directly cause p53 degradation, but can induce changes in Mdm2 localization and function (Box 1).
Box 1. Non-degradative functions of Mdm2-dependent ubiquitylation.
Although best known for its role in promoting ubiquitin-dependent proteasome degradation of p53, Mdm2 can also use ubiquitin to modify protein function rather than turnover. For example, Mdm2 can transcriptionally inactivate some p53 target genes by direct ubiquitylation of histones [109]. Additionally, mono-ubiquitination of p53 can facilitate export of p53 from the nucleus to the cytoplasm, thereby inhibiting its role as a transcription factor [110,111]. However, proteasome-dependent degradation of nuclear p53 can also effectively prevent its transactivation function [111,112], suggesting monoubiquitylation of cytoplasmic p53 is required for other processes. Consistent with this, recent data suggest that Mdm2-dependent mono-ubiquitination induces translocation of p53 to the mitochondria [113], where it can activate transcription-independent apoptosis. A p53-dependent pro-apoptotic role for Mdmx at the mitochondria has also been proposed [114], in which Mdmx enhances p53 targeting to the mitochondria. However, whether changes in p53 ubiquitylation status are also involved in Mdmx-dependent mitochondrial p53 translocation was not evaluated. The suggestion that these p53-inhibitory oncogenes also activate p53 is at first glance counterintuitive. Indeed, Mdmx was shown to prevent p53-dependent activation of bax, and consequently apoptosis, in a previous study [115]. Therefore, more rigorous testing of these models is required to firmly establish Mdm2 and Mdmx as positive regulators of p53 activity.
Mdm2 also contributes to negative and positive feedback loops that are built into this system. For example, because Mdm2 itself is a p53 target gene, it is involved in attenuation of the p53 response. However, in addition to degrading p53, Mdm2 also undergoes auto-degradation [12,13] and targets Mdmx for degradation [14–16], which is critical for optimal p53 activation after stress. There is also evidence to suggest that Mdm2 mediates diverse non-ubiquitin modifications of p53 (Box 2).
Box 2. Non-ubiquitin modifications promoted by Mdm2.
In addition to ubiquitin, other small ubiquitin-like (Ubl) molecules can be conjugated to C-terminal lysine residues of p53. SUMO-1 (small ubiquitin-related modifier-1) is reversibly attached to p53 in an enzyme cascade that resembles the ubiquitylation process. This can occur in vitro in the absence of a defined E3 ligase, [116,117], but the PIAS family of E3 ligases was subsequently determined to mediate p53 SUMOylation [118]. Other reports indicate Mdm2 can also fulfill this role [119]. There is some consensus that Arf can stimulate the Mdm2-dependent SUMOylation of both itself and of p53 at K386 [119,120] and that SUMOylation is associated with relocalization of p53 to subnuclear structures [121,122]. However, the biological consequences of Mdm2-dependent SUMOylation are still unclear. Some reports suggest that SUMOylation of p53 increases its transcriptional activity, but this remains controversial [116–118,123].
Mdm2 also promotes the conjugation of NEDD8 (another Ubl molecule), to itself and to p53 [124]. Although Mdm2 ubiquitylates several C-terminal lysines of p53, Mdm2-mediated NEDDylation occurs at a restricted subset of them (K370, K372, K373). Replacing these lysines with arginine leads to increased p53 transcriptional activity in vitro, suggesting that NEDDylation of p53 inhibits its function. The discovery that Mdm2 can facilitate transfer of these Ubl molecules to p53, and to other substrates [125] adds additional layers of complexity to p53 regulation. Determining which of these processes is physiologically relevant is a considerable challenge. Abundance of substrate presumably affects which Ubl modifications are made in vivo. Additionally, some C-terminal lysine residues are subject to multiple types of modification, which complicates the interpretation of mutagenesis studies. For example, K386 can also be ubiquitylated and acetylated [126]. Competition between Mdm2 and other proteins for modification of these sites may therefore contribute to p53 function [127]. Furthermore, mice in which these lysines are mutated to arginine are viable and have a normal lifespan, although p53 regulation is subtly altered in some cell types [128,129]. This suggests that either the lysine residues fine-tune the p53 response or that compensatory mechanisms are present in their absence.
Although Mdm2 can homo-oligomerizea, or hetero-oligomerize with Mdmx via RING/RING interaction, it appears that Mdmx alone is monomeric [17]. Together with the observation that the Mdm2–Mdmx hetero-oligomer is a more effective ligase for p53 in vitro than Mdm2 alone [10], this has led to a model in which the Mdm2–Mdmx ligase complex is more efficient in targeting p53 for ubiquitylation and degradation. Partly, this ‘co-operation model’ has arisen following a growing trend to evaluate Mdm2 and Mdmx function in the context of findings from structure and/or activity relationships of other RING E3 ligases, such as the Brca1 tumor suppressor. Although the Brca1 RING domain is a different (C3HC4) subtype, functional studies have shown that its activity is enhanced via hetero-oligomerization with its non-catalytic partner, Bard1 [18]. In the context of the Brca1–Bard1 heterodimer, E2 binds only to the Brca1 RING, at residues proximal to (but not within) the dimerization interface [19]. This binding mode of E2 s appears to be similar for the Mdm2–Mdmx heterodimer [8,20]. Moreover, the Brca1–Bard1 complex recruits only a subset of E2 enzymes, and only some of these are actually capable of transferring ubiquitin to target lysines [21]. These observations suggest that the Mdm2–Mdmx complex also discriminates when recruiting E2 s. This is important, because it is the E2 enzyme (rather than the E3 scaffold itself) that dictates the type and length of ubiquitin linkage formed [22]. Therefore, determining the identity of E2 s recruited to Mdm2 complexes will inform our interpretation of the biological functions of Mdm2 and Mdmx. Historically, the p53 field has employed the UbcH5 family of E2 s in the in vitro studies of Mdm2-dependent ubiquitylation [23]. In vitro and in vivo evaluation of a subset of E2 enzymes indicates that UbcH5-B and –C contribute to Mdm2-dependent ubiquitylation and degradation of p53 [24]. Interestingly, a large-scale computational and experimental study of interacting E2/E3 pairs has revealed additional E2 s that might contribute to Mdm2 ubiquitin ligase activity [25]. However, some of these E2 s failed to interact with Mdm2 in another high throughput approach [26]. Such discrepancies underscore the technical challenges associated with identification of bona fide E2/E3 pairs, and emphasize the importance of identifying which E2 s are functionally relevant in vivo, and under what conditions.
Recent data suggest that both Mdm2 oligomers and Mdm2–Mdmx oligomers contribute to the regulation of p53 level. Work from both the Vousden and Prives laboratories indicatesthat oligomeric Mdm2is a more effectivep53 ubiquitin ligase than is monomeric Mdm2 [27,28]. Intriguingly, their data show that aromatic residues (Y489 and F490) outside the RING domain at the extreme C-terminus are also critical for Mdm2 ligase activity. It is speculated that these residues act in trans, contacting the RING domain of adjacent Mdm2 molecules to create a scaffold for E2 recruitment. Aromatic residues are conserved in the Mdmx C-terminal tail, and can also act in trans to reactivate E3 ligase function in Mdm2 C-terminal mutants [27,28]. Although not direct proof that Mdm2–Mdmx is the preferred complex for p53 ubiquitylation, this finding underscores the functional importance of hetero-oligomerization. The precise topology and oligomeric status of Mdm2 and Mdmx complexes in vivo remains unclear. However, structural studies of the homo- and heterodimer are consistent with interaction between the C- terminus of one partner and the RING domain of the other [8,20].
All the above studies were performed in the absence of exogenous stress. However, Cheng and colleagues recently found that control of Mdm2 oligomerization is important for p53 stabilization following DNA damage [29]. The group identified several novel sites adjacent to the Mdm2 RING domain that are phosphorylated upon irradiation by damage kinases. The phosphorylation appears to prevent Mdm2 oligomerization, as the Mdm2 RING domain isolated from irradiated cells is unable to interact with non-phosphorylated Mdm2. In cell lines expressing non-phosphorylatable Mdm2 mutants, p53 stabilization and activation after damage is markedly attenuated. Together these data suggest that disruption of Mdm2 oligomers contributes to p53 stabilization. Cheng et al. also show that phosphorylated Mdm2 retains the capacity to bind and degrade Mdmx. This is important, because degradation of Mdmx is required for full activation of p53. Data from an in vivo mouse model also suggest that Mdmx plays a role in control of p53 stability after damage. Phosphorylation of Mdmx at C-terminal serine residues by damage-activated kinases (Ser-341, 367 and 402 in mouse) leads to its Mdm2-dependent degradation. When these serines are mutated to alanine in vivo (Mdmx3SA mice), damage-induced Mdmx degradation is attenuated. Furthermore, p53 stabilization is significantly reduced in Mdmx3SA compared with wild type mice [30]. In Mdmx3SA, Mdm2 remains bound to Mdmx, but degradation of Mdmx is significantly reduced. This would effectively increase the concentration of Mdm2–Mdmx hetero-oligomers that can inactivate p53. Thus, the defect in p53 stabilization in Mdmx3SA mice might be due to the persistence of Mdm2–Mdmx ligase complexes. Together, these results suggest that both disruption of Mdm2 oligomers, and the functional inactivation of Mdm2–Mdmx oligomers via Mdmx degradation, contribute to maximal p53 stabilization after stress (Figure 2). The disruption of Mdm2 oligomers might also contribute to their destabilization following DNA damage [12]. Despite the above findings, the field lacks a complete understanding of the modification status and composition of Mdm2–Mdmx hetero-oligomers. A vexing question suggested by these observations is how phosphorylation of Mdm2 after damage prevents self-oligomerization, while permitting oligomerization with Mdmx since the structures of both RINGs are so similar. The stage is set for exciting new discoveries in this area of Mdm2 and Mdmx research.
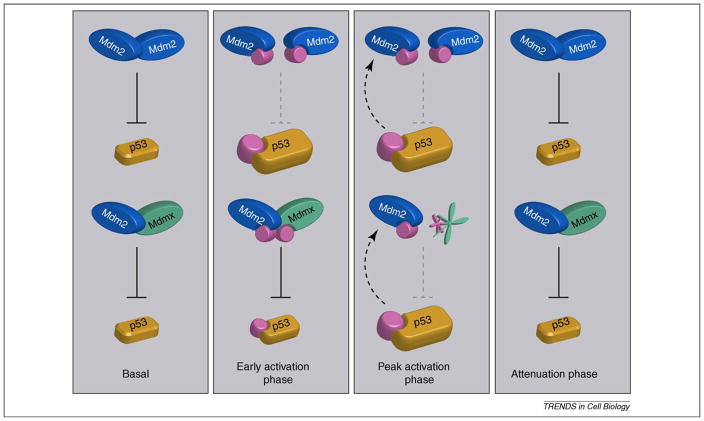
Figure 2.
Control of p53 stability by Mdm2 homo-oligomers and Mdm2–Mdmx hetero-oligomers. Basal p53 levels are regulated by both Mdm2 homo-oligomers and Mdm2/Mdmx hetero-oligomers. In vitro data suggests the hetero-oligomer is a more effective p53 ligase in the absence of stress, and that Mdmx contributes significantly to p53 basal activity, but evidence for the regulatory importance of hetero-oligomers is limited. Immediately following DNA damage, during the early activation phase, Mdm2 is destabilized, and phosphorylated at residues outside the RING domain, destabilizing Mdm2 oligomers, leading to increased p53 levels. At the peak activation phase, Mdm2 degrades itself and Mdmx, which removes Mdm2–Mdm2 and Mdm2–Mdmx oligomers, leading to maximal p53 accumulation. During the activation phase, p53 also transactivates the Mdm2 gene (dashed arrow). The attenuation phase begins when DNA damage signaling abates. Kinase inhibition and phosphatase activation removes the pool of phosphorylated Mdm2 and Mdmx, leading to their stabilization. As a result, the homo- and hetero-oligomers regain p53 ubiquitin ligase activity, reducing p53 to basal levels. Although Mdmx degradation is clearly Mdm2-dependent and therefore requires hetero-oligomerization, the existence and ubiquitin ligase activity of phosphorylated Mdm2–Mdmx hetero-oligomers (early activation phase) is curently speculative. In addition to regulation of Mdm2 and Mdmx, phosphorylation of p53 during the damage response also contributes to p53 activation by decreasing the affinity for negative regulators, and increasing the affinity for transcriptional co-factors.
Recently, a knock-in mouse Mdm2 mutant was described in which one of the critical cysteines in the Mdm2 RING domain was mutated to alanine (C462A) [31]. The homozygous mutation was embryonic lethal in a p53-dependent manner, providing compelling evidence that the Mdm2 RING domain is required for correct regulation of p53 activity in vivo. The authors concluded that loss of Mdm2 E3 ubiquitin ligase activity is likely responsible for the observed phenotype, since the mutant could still bind to p53 and block p53-dependent transactivation. This is an important finding, but it does not directly test whether Mdm2 and Mdmx co-operate as an E3 ligase complex to regulate p53, because the C462A mutation profoundly alters the structure of the RING domain, most likely rendering it unable to bind to Mdmx. Thus, generation and analysis of mutations that block ubiquitin ligase activity but do not profoundly affect RING structure (such as Mdm2 Y489A and Mdmx F488A) are essential for furthering our understanding of the importance of co-operation between Mdm2 and Mdmx in vivo.
Mdm2 and Mdmx seem to contribute to p53 regulation to different extents in different tissues in vivo (Table 1; [32–40]). Clearly in some tissues and cell types [such as the central nervous system (CNS) and proliferative intestinal cells], there is a requirement for both Mdm2 and Mdmx. This lends support to the co-operation model. However, in other cases, such as smooth muscle cells, loss of Mdmx has no phenotype. This might indicate that Mdm2 can effectively antagonize p53 in some tissues. However, since transcription of Mdm2 is increased following loss of Mdmx, this might compensate for sub-optimal ligase activity of Mdm2 oligomers. Importantly, loss of Mdm2 appears to exhibit more profound effects than loss of Mdmx (at least in classically radiosensitive tissues), suggesting that Mdmx cannot compensate for lack of Mdm2 (Table 1). This is probably explained by the increased p53 levels resulting from loss of Mdm2 exceeding the buffering capacity of Mdmx. A general conclusion from the available data is that Mdm2 loss is almost invariably lethal, whereas Mdmx loss can be tolerated in some cases. However, there are no published data addressing the deletion of Mdmx in adult thymus or spleen to compare with Mdm2 knockout in these tissues. Without these data, a direct comparison of the functional redundancy between Mdm2 and Mdmx cannot be made, and the generality of the co-operation model remains to be determined.
It also remains to be seen whether there is a requirement for Mdm2 or Mdmx in other adult tissues. This is difficult to answer using the conditionally active p53 models, since mice die rapidly from the effects of p53 in radiosensitive tissues. Perhaps temporal regulation of Mdm2 or Mdmx knockout in additional tissues such as the liver or kidney may provide some answers. It is quite possible that in some tissues, neither Mdm2 nor Mdmx is required to inhibit p53. Perhaps one of the other E3 ligases reported to control p53 degradation is responsible for its regulation in these tissues [11]. Alternatively, factors that modulate p53 promoter choice or that promote cell survival downstream of p53 activation may be involved.
Regulation of Mdm2 and Mdmx ubiquitylation
Although the importance of Mdm2 and Mdmx in post-translational modification of p53 is well documented, data showing that Mdm2 and Mdmx are themselves extensively modified has recently shed light on the complexity of their regulation, and the signal transduction pathways involving them.
In the absence of exogenous stress, there is balanced turnover of Mdm2, Mdmx and p53. It appears that Mdmx is relatively stable compared with Mdm2 and p53 [41], presumably due to preferential ubiquitylation of the latter proteins. The mechanistic basis for this might be related to the composition of Mdm2 and Mdmx complexes. For example, Mdm2 homo-oligomers are proposed to be relatively unstable, possibly because autoubiquitylation is favored in this context. By contrast, Mdm2’s substrate specificity is switched from itself towards p53 upon Mdmx binding [42]. This switch between target substrate ubiquitylation and autoubiquitylation following changes in binding partner might be a general feature of RING E3 ligases, since ligand binding to IAP (a member of the RING E3 inhibitor of apoptosis proteins) stimulates IAP auto-destruction [43]. Other ligases are also proposed to target p53 for ubiquitin dependent proteolysis under certain circumstances, as recently reviewed by Lee and Gu [44]. In addition to modulation of substrate choice, deubiquitylation is also used to stabilize Mdm2, Mdmx and p53. For example, HAUSP is a deubiquitylase (DUB) that binds to and stabilizes Mdm2 and Mdmx preferentially under normal growth conditions [45,46]. This might explain why Mdm2-bound Mdmx is not constitutively degraded in the absence of stress. In addition, HAUSP-dependent stabilization of Mdm2 and Mdmx might enhance degradation of p53 by the Mdm2–Mdmx heterodimer. More recently a second DUB, Usp2a, has been reported to deubiquitylate both Mdm2 and Mdmx [47,48] and lead to p53 destabilization. Thus, targeting DUBs might be of potential therapeutic benefit for p53 reactivation in tumors. However, most DUBs, including HAUSP, have additional cellular targets, raising the possibility that DUB inhibition could lead to mechanism-based toxicity in normal tissues.
Regulation of Mdm2 and Mdmx phosphorylation
In addition to ubiquitylation, Mdm2 and Mdmx are also subjected to phosphorylation at several sites. A bewildering array of phosphorylation-induced changes has been reported (Figure 3), but the biological effects predictably segregate into those that activate or inhibit p53 (eg [49,50]). Owing to the combinatorial nature of phosphorylation, it is extremely challenging to determine the precise physiological impact of single phospho-mutants. However, some general conclusions can be drawn from the literature. To date, two classes of kinases are known to mediate Mdm2 and Mdmx phosphorylations: those activated by DNA damage, and others involved in mediation of cell growth and survival signals.
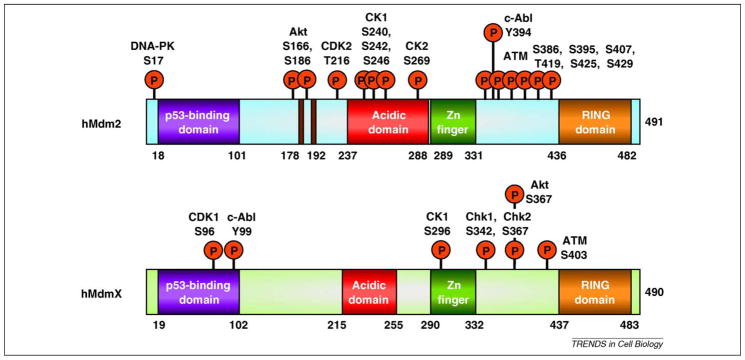
Figure 3.
Phosphorylation of Mdm2 and Mdmx by cellular kinases. Both Mdm2 and Mdmx are extensively phosphorylated by kinases of different classes. These include damage-induced kinases ATM, Chk1, Chk2, DNA-PK and c-Abl, and proliferation/survival kinases including Akt, CK-1 and -2, CDK-1 and -2.
Genotoxic stress triggers a signaling cascade of multiple nuclear kinases (including ataxia telangiectasia mutated, checkpoint kinase-1 and -2, and DNA-dependent protein kinase) that is required to inhibit replication and cell cycle progression. If damage is repaired, these kinases are inactivated and proliferation is resumed. Otherwise, these kinases can contribute to permanent cell cycle arrest or apoptosis. Given the critical role of p53 in these latter processes, it is not surprising that p53 and its negative regulators are targets of damage-activated kinases [51]. DNA damage-induced phosphorylation of N-terminal residues of Mdm2 at Ser17 by DNA-PK [52] or Mdmx at Tyr99 by c-abl [53] have been reported to disrupt binding to p53 in vitro. These modifications might contribute in part to activation of p53-dependent transcription. Additionally, phosphorylation of Mdm2 and Mdmx at serine residues close to the RING domain by ATM or ATM-dependent kinases leads to accelerated auto-degradation of both Mdm2 [54] and Mdmx [41,55–58], with consequent p53 accumulation and activation. Therefore, following a stress, the ligase function of Mdm2 must somehow be switched from p53 towards itself and Mdmx. In vitro data have indicated that DNA damage mediated phosphorylation of multiple serines in Mdm2 and Mdmx (Ser395 and 407 in Mdm2 and Ser342, -367 and -403 in Mdmx) reduced their affinity for HAUSP [46], which effectively increases the rate of Mdm2 and Mdmx ubiquitylation and destabilization. Together with the observation that ATM can inhibit Mdm2 oligomerization and p53 degradation (see above and [29]) these data indicate phosphorylation is an important regulator of E3 ligase function and Mdm2/Mdmx stability [9].
A corollary from the above is that reversal of Mdm2 and Mdmx phosphorylation should inhibit p53 function, and studies of the Wip1 phosphatase suggest this is the case [59,60]. Following resolution of a DNA damage response, the modifications added in response to damage signals must be removed to enable p53 activity to be attenuated. As a p53-inducible protein, Wip1 plays a key role in the p53-dependent DNA damage response [61]. In addition to dephosphorylating p53, Wip1 can specifically dephosphorylate Mdm2 at Ser395 and Mdmx at Ser403 and indirectly at Ser342 and Ser367, which increases their stability in order to inhibit p53 function [59,60].
Not all phosphorylation events lead to enhanced Mdm2 and Mdmx degradation. For example Akt, a pro-survival kinase, has been implicated in the stabilization of both proteins. Following growth factor stimulation, Akt phosphorylates Mdm2 at Ser166 and Ser186 [62], which increases Mdm2 half-life and stimulates Mdm2-dependent p53 degradation. Akt also stimulates Mdm2 translocation to the nucleus, which can contribute to reduced p53-dependent transcription [62,63]. Interestingly, one of the damage kinase target residues (Ser367) of Mdmx is also phosphorylated by Akt [65]. Furthermore, DNA damage and Akt-induced Ser367 phosphorylation both stimulate binding of Mdmx to 14-3-3 proteins [57,64]. However, in contrast to DNA damage, 14-3-3 binding induced by Akt leads to Mdmx stabilization and p53 inactivation.
Various isoforms of CK1 can also phosphorylate both Mdm2 and Mdmx. For example, CK1-δ phosphorylation of the Mdm2 acidic domain blocks Mdm2-dependent p53 degradation [65]. CK1-δ can also phosphorylate Thr18 of p53, which may reduce the affinity of Mdm2 for p53 [66]. Together these data indicate CK1-δ inhibition of Mdm2 contributes to p53 activation. By contrast, CK1-α increases the interaction between Mdmx and p53, which should inactivate p53 [67]. Glycogen synthase kinase-3 (GSK3) also appears to inhibit p53 by enhancing its Mdm2-dependent degradation [68]. These data show that both Mdm2 and Mdmx are critical nodes of multiple cell signaling pathways (Figure 4).
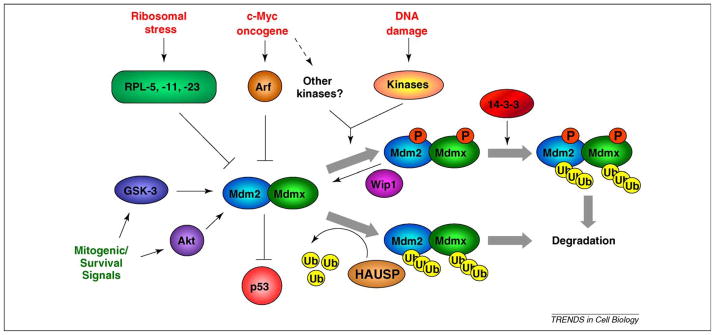
Figure 4.
Multiple cell signaling cascades converge on Mdm2 and Mdmx. p53 activating stresses (red print) can signal through damage-dependent and independent pathways to activate p53. Ribosomal stress proceeds via the release of ribosomal proteins that inhibit Mdm2 ubiquitin ligase activity and stabilize p53. Oncogenes such as c-Myc can engage damage-independent pathways such as the Arf tumor suppressor in order to activate p53, or may induce DNA damage and other kinases, which then phosphorylate either Mdm2 or Mdmx. Following genotoxic stress, multiple damage-activated kinases phosphorylate Mdm2 and Mdmx and inhibit their ligase activity. This can be via increased ubiquitylation and degradation of Mdm2, and increased ubiquitylation and degradation of Mdmx, which requires binding to 14-3-3 proteins. Following resolution of a DNA damage response, the Wip1 phosphatase can dephosphorylate Mdm2 and Mdmx, leading to their stabilization. In non-stressed cells, the levels of Mdm2 and Mdmx ubiquitylation are in part controlled by the deubiquitylase HAUSP, which removes ubiquitin from each protein, leading to their stabilization. Kinases associated with proliferation and survival (green print) can also phosphorylate Mdm2 and enhance its p53 inhibitory function.
The stoichiometric balance between Mdm2, Mdmx and p53 is crucial for the p53 response in vivo
Early models for p53 activation suggested that the disruption of the Mdm2–p53 interaction following DNA damage induced-phosphorylation of p53 at Ser15 and Ser20 was required for stabilization and activation of p53. However, biochemical analysis indicates that these phosphorylation events do not completely prevent Mdm2 binding, and mutation at these phosphorylation sites does not abrogate p53 stabilization after DNA damage in vivo (see [69] for review). Moreover, mouse studies show that blocking p53 phosphorylation at corresponding serine residues only caused modest defects in p53-mediated apoptosis and tumor suppression function [70–73]. Thus, other mechanisms must operate in order to achieve full p53 activation. Indeed, a crucial determinant appears to be the molecular ratio of Mdm2 and Mdmx to p53.
In vitro and in vivo studies show that damage-induced modifications of Mdm2 and Mdmx lead to their destabilization and degradation, reducing their ability to antagonize p53 [49,50]. The increasing ratio between p53 and its negative regulators, concomitant with the onset of p53 transcriptional activity, suggests that damage-induced modifications contribute to p53 activation by changing the stoichiometry of the pathway, and altering the affinity of Mdm2 and Mdmx for p53 [3]. In vivo studies further demonstrate that subtle changes in the stoichiometry of p53 and its negative regulators can profoundly affect p53 function in tumor suppression. For example, a two-fold increase in Mdm2 expression due to a polymorphism in the Mdm2 gene reduces p53 activation enough to significantly increase the risk and decrease the age of onset for hormone-dependent breast cancer in women [74,75]. Perturbing the stoichiometry of the p53 pathway by altering Mdmx regulation also has profound effects in vivo. In Mdmx3SA mice, (see above) there is an approximately 50% reduction of basal p53 activity prior to genotoxic stress [30]. Interestingly, the fold induction of p53 gene targets after DNA damage was similar between Mdmx3SA and wild type mice, but due to reduced basal p53 activity, the absolute transcript levels of p53 target genes ( p21 and puma) were lower in Mdmx3SA mice. This renders the mice remarkably radio-resistant, as they survive an ordinarily lethal dose of ionizing radiation. However, the attenuated p53 activation in Mdmx3SA mice predisposes them to c-Myc-induced tumorigenesis due to deregulated cell proliferation. Previous studies have shown that c-Myc-induced lymphoma was accelerated in transgenic mice overexpressing Mdm2 [76], but was delayed in Mdm2 or Mdmx heterozygous mice [77]. More recently, Murphy et al. demonstrated that low levels of deregulated c-Myc are sufficient to promote cell proliferation and tumorigenesis without engaging the Arf/p53 pathway or triggering apoptosis [78]. Together these studies suggest that the threshold for p53 activation is the critical determinant of tumorigenesis, and that this can be altered by modulating Mdm2 and Mdmx abundance.
In addition to performing tumor surveillance, p53 is also involved in other biological functions. Recently we, and others, showed that p53 activation contributed to the low frequency of generating induced pluripotent stem cells (iPS) during cell reprogramming [79,80]. Knocking down p53 by shRNA increased the efficiency of cell reprogramming. A higher efficiency can be achieved using Mdmx3SA fibroblasts, once again illustrating the crucial function of post-translational modification for fine-tuning p53 activity in biological systems.
Broadening the horizons: novel roles for Mdm2 and Mdmx?
Genetic analyses demonstrate that the major cellular target (at least in development) for both Mdm2 and Mdmx is p53 [49]. However, both Mdm2 and Mdmx interact with many other proteins, including those involved in development, morphogenesis and tumorigenesis (Table 2; [81–92]). Functional characterization of these interactions might provide new links back to p53, but might also reveal p53-independent roles for Mdm2 and Mdmx.
Table 2.
Other cellular factors that interact with Mdm2 or Mdmx.
| Binding cofactor | Mdm2 | Mdmx |
|---|---|---|
| Arf | Arf binds to and sequesters Mdm2 in the nucleolus, which leads to increased p53 levels and activity [81]. | Mdmx can prevent Arf-induced p53 activation and growth arrest. However, whether Mdmx binds to Arf directly remains controversial [83,86, 89]. |
| Arf also promotes sumoylation of Mdm2, which might contribute to the ability of Arf to relocalize Mdm2 to the nucleolus [120]. | ||
| p300 | p300 binds to both Mdm2 and p53 to form a ternary complex and has been shown to enhance Mdm2-mediated ubiquitination and degradation of p53 [87]. | Mdmx can inhibit p53 acetylation induced by endogenous and ectopically expressed p300 [84]. |
| Structural studies indicate that phosphorylation of the p53 transactivation domain increases the affinity of p53 for p300 and decreases binding to Mdm2, which may contribute to the stabilization and activation of p53 [90]. | Mdmx may also exert p53-independent functions, as it can compete with Smad3/4 transcription factors for binding to p300 [85]. | |
| Ribosomal proteins | Nucleolar stress induced by serum starvation or non-genotoxic doses of Actinomycin D and 5-Fluorouracil promotes the binding of ribosomal proteins RPL5, -11, -23 and RPS-7 to Mdm2. This blocks Mdm2 E3 ligase function and increases p53 activity. Conversely, Mdm2 ubiquitylates and degrades RPL-26, leading to decreased p53 mRNA translation [82]. | In contrast to Mdm2, Mdmx is not reported to bind directly to any ribosomal proteins. Rather, ribosomal stress can induce Mdm2-dependent Mdmx degradation in the absence of DNA damage, and overexpression of Mdmx can protect against agents that induce ribosomal stress [88]. |
For example, Notch1 and its negative regulator Numb are both implicated in the regulation of Mdm2 activity. Numb was recently reported to bind and inhibit Mdm2, leading to stabilization of p53, which is consistent with reports of a Numb tumor suppressor function [93]. By contrast, Notch1 can indirectly stimulate Mdm2 ubiquitin ligase activity, thereby inhibiting p53 function [94]. This is in agreement with the role of Notch1 as an oncogene. Determining whether Mdm2 or Mdmx also modulate other aspects of physiological Notch signaling (for example during embryogenesis) is a daunting task, because the pathways are complex [95,96].
The epithelial-to-mesenchymal transition (EMT) is a process required during normal embryonic morphogenesis, but acquisition of EMT-like features increases the metastatic potential of tumor cells. During EMT, the Slug protein is upregulated and represses expression of E-cadherin, the major component of adherens junctions. This effectively ‘loosens’ cell-cell contacts and increases cell motility. In tumor cells, upregulation of Slug and decreased E-cadherin promotes invasiveness of tumor cells as well as acquisition of a ‘cancer stem- cell’ like phenotype and resistance to some chemotherapeutic agents [97]. Mdm2 has been implicated in the ubiquitin-dependent degradation of E-cadherin, and there is a negative correlation between levels of Mdm2 and E-cadherin in primary tissue from metastatic breast cancers [98]. Elevated Mdm2 expression is also associated with increased risk of metastasis in prostate cancer [99]. Together these data suggest that high Mdm2 levels might be correlated with a metastatic phenotype. However, metastases were not reported in transgenic mice overexpressing Mdm2 [100]. Furthermore, Mdm2 was recently reported to downregulate Slug and increase E-cadherin in cell culture, which would presumably prevent invasiveness [101]. Although the picture is not yet clear, models to test the role of Mdm2 in invasion and metastasis are warranted, particularly as targeting these pathologies may be critical for the treatment of many cancers. Adding to the complexity, Mdm2 ubiquitin ligase activity can perform non-degradative functions that either inhibit or activate p53 function (Box 1).
Concluding remarks and future directions
Mdm2 ubiquitin ligase activity is critical for its regulation of p53, yet similar to other RING E3 ligases, details of its cognate E2 and additional substrates in vivo remain important questions to address. Identification of physiological E2 enzymes will provide greater mechanistic insight into Mdm2-dependent ubiquitylation. Similarly, identification of physiological substrates (the above-mentioned notwithstanding) might reveal novel functions for Mdm2.
The majority of in vivo studies to date show that Mdmx loss or overexpression has a milder phenotype when compared with Mdm2. Perhaps this is in part due to the (apparent) lack of ubiquitin ligase activity in Mdmx. Is it really just a stripped down version of Mdm2, or might it have unique cellular functions? Again, studies to define the Mdmx interactome will be invaluable. The generation of additional tissue-specific or conditional Mdmx knockouts in the adult mouse will also provide some of the answers.
Research into the molecular mechanisms by which Mdm2 and Mdmx inhibit p53 has indicated potential therapeutic strategies. For example, small molecule antagonism of the Mdm2–p53 interaction promotes p53-dependent tumor regression in murine models [102,103]. Although the Mdmx–p53 interaction is sufficiently different to limit the efficacy of Mdm2 antagonists [104], structural data will inform medicinal chemistry efforts to develop Mdmx-specific drugs. Additionally, the realization that the Mdm2–Mdmx heterodimer is an important p53 inhibitory complex might provide an additional target for p53 activation. However, as both Mdm2 and Mdmx are required for normal tissue function, the challenge will be to provide maximal therapeutic benefit with minimal mechanism-based toxicity.
Finally, it is unlikely that p53 was selected for as a tumor suppressor in humans, as the frequency of cancer onset is low during peak reproductive years, and increases sharply in old age [105]. There is increasing evidence that in addition to the ‘classical’ function of p53 in the DNA damage response, this versatile protein might be involved in other essential physiological processes, including stem cell homeostasis and fertility [106–108]. As these novel p53 functions are investigated, it will be exciting to determine whether and how Mdm2 and Mdmx are also involved.
Acknowledgments
Research in the Wahl laboratory is funded in part by NIH grant CA061449 awarded to GMW.
Footnotes
In vivo, the exact number of subunits for Mdm complexes is unknown. For clarity, we refer to cell-based complexes as oligomers. Reference to dimers is reserved for structural studies, where the stoichiometry can be precisely determined.
References
- 1.Marine JC, et al. Keeping p53 in check: essential and synergistic functions of Mdm2 and Mdm4. Cell Death Differ. 2006;13:927–934. doi: 10.1038/sj.cdd.4401912. [DOI] [PubMed] [Google Scholar]
- 2.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang YV, et al. Quantitative analyses reveal the importance of regulated Hdmx degradation for p53 activation. Proc Natl Acad Sci U S A. 2007;104:12365–12370. doi: 10.1073/pnas.0701497104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohtsubo C, et al. Cytoplasmic tethering is involved in synergistic inhibition of p53 by Mdmx and Mdm2. Cancer Sci. 2009;100:1291–1299. doi: 10.1111/j.1349-7006.2009.01180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moll UM, Petrenko O. The MDM2-p53 interaction. Mol Cancer Res. 2003;1:1001–1008. [PubMed] [Google Scholar]
- 6.Stommel JM, et al. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marchenko ND, et al. Stress-mediated nuclear stabilization of p53 is regulated by ubiquitination and importin-alpha3 binding. Cell Death Differ. 2009;17:255–267. doi: 10.1038/cdd.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kostic M, et al. Solution structure of the Hdm2 C2H2C4 RING, a domain critical for ubiquitination of p53. J Mol Biol. 2006;363:433–450. doi: 10.1016/j.jmb.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 9.Deshaies RJ, Joazeiro CAP. RING Domain E3 Ubiquitin Ligases. Annual Review of Biochemistry. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 10.Linares LK, et al. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc Natl Acad Sci U S A. 2003;100:12009–12014. doi: 10.1073/pnas.2030930100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell. 2006;21:307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stommel JM, Wahl GM. Accelerated MDM2 auto-degradation induced by DNA-damage kinases is required for p53 activation. EMBO J. 2004;23:1547–1556. doi: 10.1038/sj.emboj.7600145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang S, et al. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 14.de Graaf P, et al. Hdmx protein stability is regulated by the ubiquitin ligase activity of Mdm2. J Biol Chem. 2003;278:38315–38324. doi: 10.1074/jbc.M213034200. [DOI] [PubMed] [Google Scholar]
- 15.Kawai H, et al. DNA damage-induced MDMX degradation is mediated by MDM2. J Biol Chem. 2003;278:45946–45953. doi: 10.1074/jbc.M308295200. [DOI] [PubMed] [Google Scholar]
- 16.Pan Y, Chen J. MDM2 promotes ubiquitination and degradation of MDMX. Mol Cell Biol. 2003;23:5113–5121. doi: 10.1128/MCB.23.15.5113-5121.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanimura S, et al. MDM2 interacts with MDMX through their RING finger domains. FEBS Lett. 1999;447:5–9. doi: 10.1016/s0014-5793(99)00254-9. [DOI] [PubMed] [Google Scholar]
- 18.Hashizume R, et al. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J Biol Chem. 2001;276:14537–14540. doi: 10.1074/jbc.C000881200. [DOI] [PubMed] [Google Scholar]
- 19.Brzovic PS, et al. Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitinligase complex. Proc Natl Acad Sci U S A. 2003;100:5646–5651. doi: 10.1073/pnas.0836054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linke K, et al. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ. 2008;15:841–848. doi: 10.1038/sj.cdd.4402309. [DOI] [PubMed] [Google Scholar]
- 21.Christensen DE, et al. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat Struct Mol Biol. 2007;14:941–948. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- 22.Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Honda R, et al. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 24.Saville MK, et al. Regulation of p53 by the Ubiquitin-conjugating Enzymes UbcH5B/C in Vivo. J Biol Chem. 2004;279:42169–42181. doi: 10.1074/jbc.M403362200. [DOI] [PubMed] [Google Scholar]
- 25.van Wijk SJ, et al. A comprehensive framework of E2-RING E3 interactions of the human ubiquitin-proteasome system. Mol Syst Biol. 2009;5:295. doi: 10.1038/msb.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markson G, et al. Analysis of the human E2 ubiquitin conjugating enzyme protein interaction network. Genome Res. 2009;19:1905–1911. doi: 10.1101/gr.093963.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uldrijan S, et al. An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. EMBO J. 2007;26:102–112. doi: 10.1038/sj.emboj.7601469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poyurovsky MV, et al. The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J. 2007;26:90–101. doi: 10.1038/sj.emboj.7601465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng Q, et al. ATM activates p53 by regulating MDM2 oligomerization and E3 processivity. EMBO J. 2009;28:3857–3867. doi: 10.1038/emboj.2009.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang YV, et al. Increased Radioresistance and Accelerated B Cell Lymphomas in Mice with Mdmx Mutations that Prevent Modifications by DNA-Damage-Activated Kinases. Cancer Cell. 2009;16:33 –43. doi: 10.1016/j.ccr.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Itahana K, et al. Targeted inactivation of Mdm2 RING finger E3 ubiquitin ligase activity in the mouse reveals mechanistic insights into p53 regulation. Cancer Cell. 2007;12:355–366. doi: 10.1016/j.ccr.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 32.Boesten LS, et al. Mdm2, but not Mdm4, protects terminally differentiated smooth muscle cells from p53-mediated caspase-3-independent cell death. Cell Death Differ. 2006;13:2089–2098. doi: 10.1038/sj.cdd.4401973. [DOI] [PubMed] [Google Scholar]
- 33.Francoz S, et al. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc Natl Acad Sci U S A. 2006;103:3232–3237. doi: 10.1073/pnas.0508476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grier JD, et al. Tissue-specific differences of p53 inhibition by Mdm2 and Mdm4. Mol Cell Biol. 2006;26:192–198. doi: 10.1128/MCB.26.1.192-198.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maetens M, et al. Distinct roles of Mdm2 and Mdm4 in red cell production. Blood. 2007;109:2630–2633. doi: 10.1182/blood-2006-03-013656. [DOI] [PubMed] [Google Scholar]
- 36.Ringshausen I, et al. Mdm2 is critically and continuously required to suppress lethal p53 activity in vivo. Cancer Cell. 2006;10:501–514. doi: 10.1016/j.ccr.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 37.Valentin-Vega YA, et al. Mdm4 loss in the intestinal epithelium leads to compartmentalized cell death but no tissue abnormalities. Differentiation. 2009;77:442–449. doi: 10.1016/j.diff.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valentin-Vega YA, et al. The intestinal epithelium compensates for p53-mediated cell death and guarantees organismal survival. Cell Death Differ. 2008;15:1772–1781. doi: 10.1038/cdd.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiong S, et al. Loss of Mdm4 results in p53-dependent dilated cardiomyopathy. Circulation. 2007;115:2925–2930. doi: 10.1161/CIRCULATIONAHA.107.689901. [DOI] [PubMed] [Google Scholar]
- 40.Xiong S, et al. Synergistic roles of Mdm2 and Mdm4 for p53 inhibition in central nervous system development. Proc Natl Acad Sci U S A. 2006;103:3226–3231. doi: 10.1073/pnas.0508500103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pereg Y, et al. Phosphorylation of Hdmx mediates its Hdm2-and ATM-dependent degradation in response to DNA damage. Proc Natl Acad Sci U S A. 2005;102:5056–5061. doi: 10.1073/pnas.0408595102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okamoto K, et al. Mdmx enhances p53 ubiquitination by altering the substrate preference of the Mdm2 ubiquitin ligase. FEBS Lett. 2009;583:2710–2714. doi: 10.1016/j.febslet.2009.07.021. [DOI] [PubMed] [Google Scholar]
- 43.Varfolomeev E, et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell. 2007;131:669–681. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 44.Lee JT, Gu W. The multiple levels of regulation by p53 ubiquitination. Cell Death Differ. 2010;17:86–92. doi: 10.1038/cdd.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li M, et al. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell. 2004;13:879–886. doi: 10.1016/s1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- 46.Meulmeester E, et al. Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol Cell. 2005;18:565–576. doi: 10.1016/j.molcel.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 47.Allende-Vega N, et al. MdmX is a substrate for the deubiquitinating enzyme USP2a. Oncogene. 2010;29:432–441. doi: 10.1038/onc.2009.330. [DOI] [PubMed] [Google Scholar]
- 48.Stevenson LF, et al. The deubiquitinating enzyme USP2a regulates the p53 pathway by targeting Mdm2. EMBO J. 2007;26:976–986. doi: 10.1038/sj.emboj.7601567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marine JC, et al. MDMX: from bench to bedside. J Cell Sci. 2007;120:371–378. doi: 10.1242/jcs.03362. [DOI] [PubMed] [Google Scholar]
- 50.Meek DW, Knippschild U. Posttranslational Modification of MDM2. Molecular Cancer Research. 2003;1:1017–1026. [PubMed] [Google Scholar]
- 51.Roos WP, Kaina B. DNA damage-induced cell death by apoptosis. Trends Mol Med. 2006;12:440–450. doi: 10.1016/j.molmed.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 52.Mayo LD, et al. Mdm-2 Phosphorylation by DNA-dependent Protein Kinase Prevents Interaction with p53. Cancer Res. 1997;57:5013–5016. [PubMed] [Google Scholar]
- 53.Zuckerman V, et al. c-Abl Phosphorylates Hdmx and Regulates Its Interaction with p53. Journal of Biological Chemistry. 2009;284:4031–4039. doi: 10.1074/jbc.M809211200. [DOI] [PubMed] [Google Scholar]
- 54.Maya R, et al. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev. 2001;15:1067–1077. doi: 10.1101/gad.886901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen L, et al. ATM and Chk2-dependent phosphorylation of MDMX contribute to p53 activation after DNA damage. EMBO J. 2005;24:3411–3422. doi: 10.1038/sj.emboj.7600812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.LeBron C, et al. Regulation of MDMX nuclear import and degradation by Chk2 and 14-3-3. EMBO J. 2006;25:1196–1206. doi: 10.1038/sj.emboj.7601032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Okamoto K, et al. DNA Damage-Induced Phosphorylation of MdmX at Serine 367 Activates p53 by Targeting MdmX for Mdm2-Dependent Degradation. Mol Cell Biol. 2005;25:9608–9620. doi: 10.1128/MCB.25.21.9608-9620.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pereg Y, et al. Differential Roles of ATM- and Chk2-Mediated Phosphorylations of Hdmx in Response to DNA Damage. Mol Cell Biol. 2006;26:6819–6831. doi: 10.1128/MCB.00562-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang X, et al. Phosphorylation and Degradation of MdmX Is Inhibited by Wip1 Phosphatase in the DNA Damage Response. Cancer Res. 2009;69:7960–7968. doi: 10.1158/0008-5472.CAN-09-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu X, et al. The Wip1 phosphatase and Mdm2: cracking the “Wip” on p53 stability. Cell Cycle. 2008;7:164–168. doi: 10.4161/cc.7.2.5299. [DOI] [PubMed] [Google Scholar]
- 61.Lu X, et al. PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes Dev. 2005;19:1162–1174. doi: 10.1101/gad.1291305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mayo LD, Donner DB. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc Natl Acad Sci U S A. 2001;98:11598–11603. doi: 10.1073/pnas.181181198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moumen A, et al. Met acts on Mdm2 via mTOR to signal cell survival during development. Development. 2007;134:1443–1451. doi: 10.1242/dev.02820. [DOI] [PubMed] [Google Scholar]
- 64.Lopez-Pajares V, et al. Phosphorylation of MDMX Mediated by Akt Leads to Stabilization and Induces 14-3-3 Binding. J Biol Chem. 2008;283:13707–13713. doi: 10.1074/jbc.M710030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Winter M, et al. Protein kinase CK1delta phosphorylates key sites in the acidic domain of murine double-minute clone 2 protein (MDM2) that regulate p53 turnover. Biochemistry. 2004;43:16356–16364. doi: 10.1021/bi0489255. [DOI] [PubMed] [Google Scholar]
- 66.Sakaguchi K, et al. Damage-mediated phosphorylation of human p53 threonine 18 through a cascade mediated by a casein 1-like kinase. Effect on Mdm2 binding J Biol Chem. 2000;275:9278–9283. doi: 10.1074/jbc.275.13.9278. [DOI] [PubMed] [Google Scholar]
- 67.Chen L, et al. Regulation of p53-MDMX interaction by casein kinase 1 alpha. Mol Cell Biol. 2005;25:6509–6520. doi: 10.1128/MCB.25.15.6509-6520.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kulikov R, et al. Glycogen synthase kinase 3-dependent phosphorylation of Mdm2 regulates p53 abundance. Mol Cell Biol. 2005;25:7170–7180. doi: 10.1128/MCB.25.16.7170-7180.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 70.Chao C, et al. Ser18 and 23 phosphorylation is required for p53-dependent apoptosis and tumor suppression. EMBO J. 2006;25:2615–2622. doi: 10.1038/sj.emboj.7601167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.MacPherson D, et al. Defective apoptosis and B-cell lymphomas in mice with p53 point mutation at Ser 23. EMBO J. 2004;23:3689–3699. doi: 10.1038/sj.emboj.7600363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sluss HK, et al. Phosphorylation of Serine 18 Regulates Distinct p53 Functions in Mice. Mol Cell Biol. 2004;24:976–984. doi: 10.1128/MCB.24.3.976-984.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu Z, et al. Mutation of Mouse p53 Ser23 and the Response to DNA Damage. Mol Cell Biol. 2002;22:2441–2449. doi: 10.1128/MCB.22.8.2441-2449.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bond GL, et al. MDM2 SNP309 accelerates tumor formation in a gender-specific and hormone-dependent manner. Cancer Res. 2006;66:5104–5110. doi: 10.1158/0008-5472.CAN-06-0180. [DOI] [PubMed] [Google Scholar]
- 75.Bond GL, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 76.Wang P, et al. Elevated Mdm2 expression induces chromosomal instability and confers a survival and growth advantage to B cells. Oncogene. 2008;27:1590–1598. doi: 10.1038/sj.onc.1210788. [DOI] [PubMed] [Google Scholar]
- 77.Terzian T, et al. Haploinsufficiency of Mdm2 and Mdm4 in tumorigenesis and development. Mol Cell Biol. 2007;27:5479–5485. doi: 10.1128/MCB.00555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Murphy DJ, et al. Distinct Thresholds Govern Myc’s Biological Output In Vivo. Cancer Cell. 2008;14:447–457. doi: 10.1016/j.ccr.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kawamura T, et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Krizhanovsky V, Lowe SW. Stem cells: The promises and perils of p53. Nature. 2009;460:1085–1086. doi: 10.1038/4601085a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weber JD, et al. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- 82.Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16:369–377. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang X, et al. MDM2 and MDMX can interact differently with ARF and members of the p53 family. FEBS Lett. 2001;490:202–208. doi: 10.1016/s0014-5793(01)02124-x. [DOI] [PubMed] [Google Scholar]
- 84.Sabbatini P, McCormick F. MDMX inhibits the p300/CBP-mediated acetylation of p53. DNA Cell Biol. 2002;21:519–525. doi: 10.1089/104454902320219077. [DOI] [PubMed] [Google Scholar]
- 85.Kadakia M, et al. MdmX inhibits Smad transactivation. Oncogene. 2002;21:8776–8785. doi: 10.1038/sj.onc.1205993. [DOI] [PubMed] [Google Scholar]
- 86.Jackson MW, et al. MdmX binding to ARF affects Mdm2 protein stability and p53 transactivation. J Biol Chem. 2001;276:25336–25341. doi: 10.1074/jbc.M010685200. [DOI] [PubMed] [Google Scholar]
- 87.Grossman SR, et al. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol Cell. 1998;2:405–415. doi: 10.1016/s1097-2765(00)80140-9. [DOI] [PubMed] [Google Scholar]
- 88.Gilkes DM, et al. MDMX regulation of p53 response to ribosomal stress. EMBO J. 2006;25:5614–5625. doi: 10.1038/sj.emboj.7601424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ghosh M, et al. MdmX inhibits ARF mediated Mdm2 sumoylation. Cell Cycle. 2005;4:604–608. [PubMed] [Google Scholar]
- 90.Ferreon JC, et al. Cooperative regulation of p53 by modulation of ternary complex formation with CBP/p300 and HDM2. Proc Natl Acad Sci U S A. 2009;106:6591–6596. doi: 10.1073/pnas.0811023106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Marine JC, Lozano G. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ. 2010;17:93–102. doi: 10.1038/cdd.2009.68. [DOI] [PubMed] [Google Scholar]
- 92.Prives C, White E. Does control of mutant p53 by Mdm2 complicate cancer therapy? Genes & Development. 2008;22:1259–1264. doi: 10.1101/gad.1680508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Colaluca IN, et al. NUMB controls p53 tumour suppressor activity. Nature. 2008;451:76–80. doi: 10.1038/nature06412. [DOI] [PubMed] [Google Scholar]
- 94.Beverly LJ, et al. Suppression of p53 by Notch in lymphomagenesis: implications for initiation and regression. Cancer Res. 2005;65:7159–7168. doi: 10.1158/0008-5472.CAN-05-1664. [DOI] [PubMed] [Google Scholar]
- 95.Dotto GP. Crosstalk of Notch with p53 and p63 in cancer growth control. Nat Rev Cancer. 2009;9:587–595. doi: 10.1038/nrc2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 98.Yang J-Y, et al. MDM2 Promotes Cell Motility and Invasiveness by Regulating E-Cadherin Degradation. Mol Cell Biol. 2006;26:7269–7282. doi: 10.1128/MCB.00172-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Khor LY, et al. MDM2 and Ki-67 predict for distant metastasis and mortality in men treated with radiotherapy and androgen deprivation for prostate cancer: RTOG 92-02. J Clin Oncol. 2009;27:3177–3184. doi: 10.1200/JCO.2008.19.8267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jones SN, et al. Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proc Natl Acad Sci U S A. 1998;95:15608–15612. doi: 10.1073/pnas.95.26.15608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang S-P, et al. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nat Cell Biol. 2009;11:694–704. doi: 10.1038/ncb1875. [DOI] [PubMed] [Google Scholar]
- 102.Shangary S, et al. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc Natl Acad Sci U S A. 2008;105:3933–3938. doi: 10.1073/pnas.0708917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 104.Wade M, Wahl GM. Targeting Mdm2 and Mdmx in cancer therapy: better living through medicinal chemistry? Mol Cancer Res. 2009;7:1–11. doi: 10.1158/1541-7786.MCR-08-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Aranda-Anzaldo A, Dent MAR. Reassessing the role of p53 in cancer and ageing from an evolutionary perspective. Mechanisms of Ageing and Development. 2007;128:293–302. doi: 10.1016/j.mad.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 106.Kang HJ, et al. Single-nucleotide polymorphisms in the p53 pathway regulate fertility in humans. Proc Natl Acad Sci U S A. 2009;106:9761–9766. doi: 10.1073/pnas.0904280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pearson BJ, Sanchez Alvarado A. A planarian p53 homolog regulates proliferation and self-renewal in adult stem cell lineages. Development. 2010;137:213–221. doi: 10.1242/dev.044297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lu W-J, et al. p53 ancestry: gazing through an evolutionary lens. Nat Rev Cancer. 2009;9:758–762. doi: 10.1038/nrc2732. [DOI] [PubMed] [Google Scholar]
- 109.Minsky N, Oren M. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol Cell. 2004;16:631–639. doi: 10.1016/j.molcel.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 110.Carter S, et al. C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nat Cell Biol. 2007;9:428–435. doi: 10.1038/ncb1562. [DOI] [PubMed] [Google Scholar]
- 111.Li M, et al. Mono- Versus Polyubiquitination: Differential Control of p53 Fate by Mdm2. Science. 2003;302:1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- 112.Joseph TW, et al. Nuclear and cytoplasmic degradation of endogenous p53 and HDM2 occurs during down-regulation of the p53 response after multiple types of DNA damage. Faseb J. 2003;17:1622–1630. doi: 10.1096/fj.02-0931com. [DOI] [PubMed] [Google Scholar]
- 113.Marchenko ND, et al. Monoubiquitylation promotes mitochondrial p53 translocation. EMBO J. 2007;26:923–934. doi: 10.1038/sj.emboj.7601560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mancini F, et al. MDM4 (MDMX) localizes at the mitochondria and facilitates the p53-mediated intrinsic-apoptotic pathway. EMBO J. 2009;28:1926–1939. doi: 10.1038/emboj.2009.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wade M, et al. BH3 activation blocks Hdmx suppression of apoptosis and co-operates with Nutlin to induce cell death. Cell Cycle. 2008;7:1973–1982. doi: 10.4161/cc.7.13.6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gostissa M, et al. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 1999;18:6462–6471. doi: 10.1093/emboj/18.22.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rodriguez MS, et al. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 1999;18:6455–6461. doi: 10.1093/emboj/18.22.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Stehmeier P, Muller S. Regulation of p53 family members by the ubiquitin-like SUMO system. DNA Repair. 2009;8:491–498. doi: 10.1016/j.dnarep.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 119.Chen L, Chen J. MDM2-ARF complex regulates p53 sumoylation. Oncogene. 2003;22:5348–5357. doi: 10.1038/sj.onc.1206851. [DOI] [PubMed] [Google Scholar]
- 120.Xirodimas DP, et al. P14ARF promotes accumulation of SUMO-1 conjugated (H)Mdm2. FEBS Lett. 2002;528:207–211. doi: 10.1016/s0014-5793(02)03310-0. [DOI] [PubMed] [Google Scholar]
- 121.Di Ventura B, et al. Reconstitution of Mdm2-dependent post-translational modifications of p53 in yeast. PLoS ONE. 2008;3:e1507. doi: 10.1371/journal.pone.0001507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mauri F, et al. Modification of Drosophila p53 by SUMO Modulates Its Transactivation and Pro-apoptotic Functions. J Biol Chem. 2008;283:20848–20856. doi: 10.1074/jbc.M710186200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kwek SS, Tyner DJ, Shen AL, Gudkov AVZ. Functional analysis and intracellular localization of p53 modified by SUMO-1. Oncogene. 2001;20:2587–2599. doi: 10.1038/sj.onc.1204362. [DOI] [PubMed] [Google Scholar]
- 124.Xirodimas DP, et al. Mdm2-Mediated NEDD8 Conjugation of p53 Inhibits Its Transcriptional Activity. Cell. 2004;118:83–97. doi: 10.1016/j.cell.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 125.Sundqvist A, et al. Regulation of nucleolar signalling to p53 through NEDDylation of L11. EMBO Rep. 2009;10:1132–1139. doi: 10.1038/embor.2009.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tang Y, et al. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wu SY, Chiang CM. Crosstalk between sumoylation and acetylation regulates p53-dependent chromatin transcription and DNA binding. EMBO J. 2009;28:1246–1259. doi: 10.1038/emboj.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Krummel KA, et al. The C-terminal lysines fine-tune p53 stress responses in a mouse model, but are not required for stability control or transactivation. Proc Natl Acad Sci U S A. 2005;102:10188–10193. doi: 10.1073/pnas.0503068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Feng L, et al. Functional analysis of the roles of posttranslational modifications at the p53 C terminus in regulating p53 stability and activity. Mol Cell Biol. 2005;25:5389–5395. doi: 10.1128/MCB.25.13.5389-5395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]