Abstract
Clathrin triskelions can assemble into lattices of different shapes, sizes and symmetries. For many years, the structures of clathrin lattices have been studied by single particle cryo-electron microscopy, which probed the architecture of the D6 hexagonal barrel clathrin coat at the molecular level. By introducing additional image processing steps we have recently produced a density map for the D6 barrel clathrin coat at subnanometer resolution, enabling us to generate an atomic model for this lattice [Fotin, A., Cheng, Y., Sliz, P., Grigorief, N., Harrison, S.C., Kirchhausen, T., Walz, T., 2004. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature 432, 573–579]. We describe in detail here the image processing steps that we have added to produce a density map at this high resolution. These procedures should be generally applicable and may thus help determine the structures of other large protein assemblies to higher resolution by single particle cryo-electron microscopy.
Keywords: Clathrin coats, Clathrin cages, Cryo-electron microscopy, Single particle reconstruction, Endocytosis
1. Introduction
Clathrin-coated vesicles, found in all eukaryotic cells from yeast to human, transport lipids and selected cargo molecules, such as transferrin receptors and low-density lipoprotein receptors, from the plasma membrane to early endosomes. They also carry cargo molecules between the trans-Golgi network and endosomes. In synapses, clathrin-coated vesicles are involved in the recycling of synaptic vesicles and the uptake of neurotransmitters (reviewed in Brodsky et al., 2001; Kirchhausen, 2000; Sudhof, 2004). The building block of clathrin coats is a trimer, referred to as a triskelion, which has three elongated legs radiating from a central hub (Kirchhausen and Harrison, 1981; Ungewickell and Branton, 1981). The clathrin coat has a striking molecular organization: the clathrin legs intertwine so that the triskelions form lattices with mostly pentagonal and hexagonal facets. Assembly of clathrin coats on the membrane accompanies introduction of membrane curvature, and eventually the clathrin coat encapsulates a lipid vesicle (Pearse and Crowther, 1987; Ehrlich et al., 2004). Since clathrin triskelions can form lattices of different shapes and sizes, clathrin coats can accommodate a large range of vesicle sizes and hence a large range of cargo proteins. For a detailed review on clathrin structure and function, see Kirchhausen (2000).
Images of negatively stained clathrin-coated vesicles purified from pig brain first revealed the arrangement of the clathrin triskelions in coats of various designs (Crowther et al., 1976). In particular, several lattices with high symmetry were identified, including what we now designate as the small “mini-coat” with tetrahedral symmetry, the “hexagonal barrel” with D6 symmetry, and the “soccer ball” with icosahedral symmetry (Crowther et al., 1976). A less common lattice design, the “tennis ball”, has the same size and number of hexagons and pentagons as the D6 barrel but only D2 symmetry (Crowther et al., 1976).
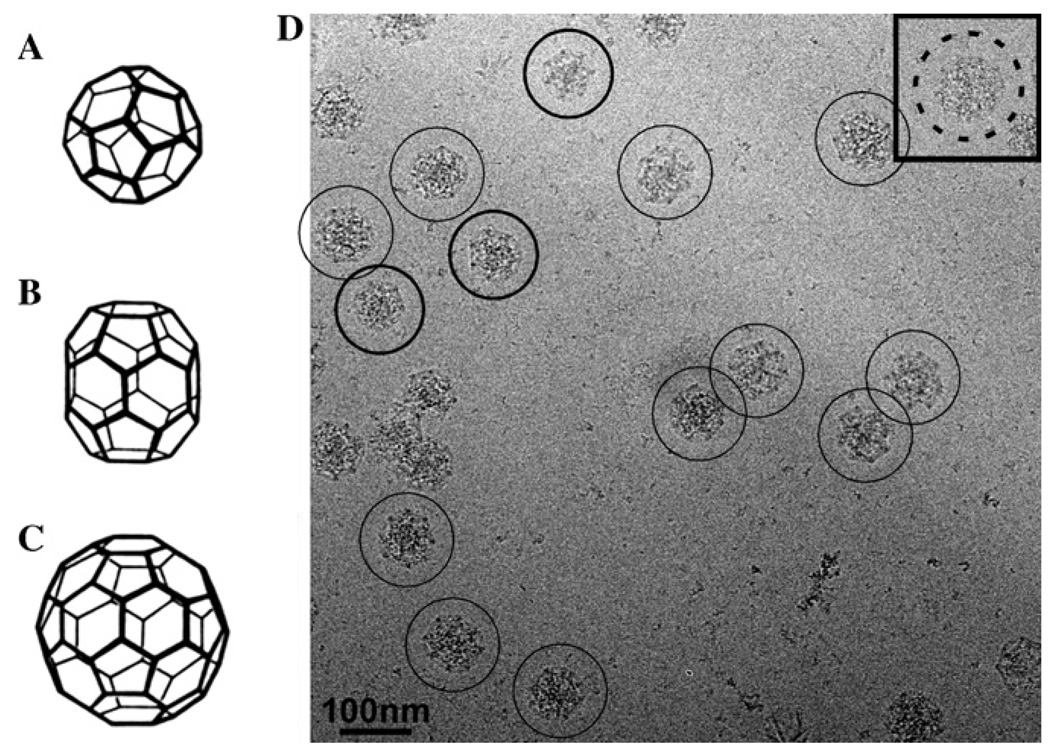
Clathrin coats without membrane vesicles can be assembled in vitro from purified clathrin and adaptor protein (AP) complexes, such as AP-2, resulting in a mixture of coats of different sizes and lattices. Typical coat designs include the tetrahedral “mini coat” (Fig. 1A), the hexagonal D6 “barrel” (Fig. 1B), and the icosahedral “soccer ball” (Fig. 1C) (Crowther et al., 1976). In vitro assembled clathrin coats were indeed used to calculate the first three-dimensional (3D) reconstruction of a D6 barrel clathrin coat using images obtained by cryo-electron microscopy (cryo-EM) of frozen hydrated specimens (Vigers et al., 1986). Although at a relatively modest resolution, the resulting density map confirmed the previously proposed architecture of the D6 barrel, with hexagons at the top and bottom of the coat, each surrounded by six pentagons and joined together by an equatorial ring of six hexagons (Crowther et al., 1976; Vigers et al., 1986). Because of their high symmetry and rigidity, in vitro assembled D6 barrels were also used for further single particle cryo-EM studies, which eventually produced a structure at a resolution of 21 Å (Smith et al., 1998). This density map confirmed the predicted packing of the clathrin triskelions in the lattice. Further analysis of this density map, in light of the crystal structure of the N-terminal domain of the clathrin heavy chain (ter Haar et al., 1998), further revealed how the clathrin triskelions interact with each other and how the individual domains are oriented within the lattice (Musacchio et al., 1999).
Fig. 1.
(A–C) Schematic drawing of clathrin coats: (A) tetrahedral coat (mini-coat), (B) hexagonal D6 barrel coat, and (C) icosahedral coat (soccer ball). (D) Cryo-electron micrograph of in vitro assembled clathrin coats in vitrified ice; mini-coats (thick circle), D6 barrel coats (thin circle) and inset: soccer ball (dashed circle).
Since the determination of the 21 Å resolution structure of the D6 clathrin coat (Smith et al., 1998), the instrumentation, image processing algorithms and software packages used in single particle cryo-EM have advanced considerably. In several cases the structures of protein complexes have now been determined to subnanometer resolution, revealing individual α-helices (e.g., Cheng et al., 2004; Ludtke et al., 2004). Determining the structures of clathrin coats to a similarly high resolution remained difficult, however, mainly because in vitro assembled clathrin coats consist of a mixture of different sizes and lattice designs. Structural flexibility, even within a coat population of the same lattice design, posed further difficulties for obtaining higher resolution reconstructions, and may have been the reason why most of the recent density maps of clathrin coats also have a resolution of less than 21 Å (Heymann et al., 2005; Smith et al., 2004).
We recently reported a 3D reconstruction of the D6 barrel clathrin coat at a nominal resolution of 7.9 Å (Fotin et al., 2004). Based on this subnanometer resolution density map and the crystal structures of a proximal region and the N-terminal domain of the clathrin heavy chain, we proposed a Cα model for a complete D6 barrel clathrin coat. Among the reasons for the higher resolution of our clathrin coat reconstructions are an optimized procedure for the assembly of clathrin coats, the collection of high quality images using a microscope equipped with a field emission electron source, and the use of a new refinement algorithm implemented in the program FREALIGN (Stewart and Grigorief, 2004). In addition, we have made modifications to the image processing procedures. In particular, we have implemented strategies to select only clathrin coats with near-perfect symmetry, to improve the alignment of the particle images by masking out unstructured regions from the reference density map, and to apply “non-crystallo-graphic” averaging. Because these strategies should also be applicable to the study of other large protein assemblies using single particle cryo-EM, we describe in this manuscript the details of the procedures we used to improve the resolution of the 3D reconstruction of the clathrin D6 coat.
2. Materials and methods
2.1. Clathrin coats preparation
Clathrin coats (without associated light chains) were prepared as described in the supplementary information of Fotin et al. (2004).
2.2. Cryo-electron microscopy and image processing
To ensure the structural integrity of the clathrin coats, only freshly in vitro assembled clathrin coats were used for data collection. The sample was diluted with coats assembly buffer (Matsui and Kirchhausen, 1990) to a final concentration of ~0.05 mg/ml before plunge freezing. 2.5 µl of diluted sample were applied to a glow-discharged Quantifoil holey carbon grid (Quantifoil Micro Tools GmbH, Germany), blotted with filter paper, and plunged into liquid ethane at −180 °C using a Reichert KF-80 quick freezing apparatus.
Frozen hydrated clathrin coats were imaged using a Tecnai F20 electron microscope equipped with a field-emission electron source (FEI Company, USA) and operated at an acceleration voltage of 200 kV. Micrographs at a defocus ranging from −2 to −5 µm were recorded at a nominal magnification of 50,000× on Kodak SO-163 film following strict low-dose procedures. The negatives were developed for 12 minutes with full-strength Kodak D-19 developer at 20 °C. Micrographs free of obvious drift were digitized with a Zeiss SCAI scanner using a step size of 7 µm. The digitized micrographs were binned over 6 × 6 pixels for initial image processing, re-binned over 3 × 3 pixels during initial refinement cycles, and over 2 × 2 pixels in the final stages of refinement to give a final pixel size on the specimen level of 2.8 Å.
The program CTFFIND3 (Mindell and Grigorieff, 2003) was used to determine the defocus values for the digitized micrographs. The clathrin coats were selected interactively using Ximdisp, the display program associated with the MRC program suite (Crowther et al., 1996). The particle images were subjected to several rounds of multi-reference alignment, multi-variate statistic analysis and classification using the IMAGIC V software package (van Heel et al., 1996) following established procedures (van Heel et al., 2000). Unique class averages were used to generate an initial 3D reconstruction of the D6 barrel clathrin coat by angular reconstitution (van Heel, 1987). No further refinement of the initial 3D reconstruction was carried out with IMAGIC V. Instead, the program FREALIGN (Stewart and Grigorieff, 2004) was used to refine further both the Euler angles and the in-plane shifts, to correct for the contrast transfer function (CTF), and to calculate further 3D reconstructions. The resolutions cited in this paper are the nominal resolutions based on the Fourier shell correlation (FSC) = 0.143 criterion (Rosenthal and Henderson, 2003). The FSC curves were calculated without removing the unstructured density in the center of the coat. The surface-rendered views of the density maps were produced with the program Chimera (Pettersen et al., 2004).
3. Results
3.1. Particle selection
Thon rings (Thon, 1966) are easily seen in a laser diffractometer and are therefore commonly used to assess the quality of micrographs, i.e., to identify images that contain high-resolution information and are free of drift either due to specimen charging or specimen movement. This selection procedure did not work well for our micrographs of vitrified clathrin coats, because the signal from the vitreous ice layer is very weak and there were too few particles in each micrograph to generate clearly visible Thon rings, especially at higher resolution. Therefore, we could only discard micrographs that were so strongly affected by drift that it could clearly be seen at rather low resolution. Further micrograph selection was therefore carried out once the particles were selected and boxed out from the digitized micrographs.
Among different lattice designs present in the in vitro assembled coats, the hexagonal D6 barrel has been the target of almost all cryo-EM studies (Fotin et al., 2004; Heymann et al., 2005; Smith et al., 1998, 2004; Vigers et al., 1986). By using stringent relatively high salt assembly conditions, D6 barrels containing AP-2 can be made the most abundant clathrin coat in the preparation, but the formation of other clathrin coats, such as mini coats and soccer balls, cannot be completely prevented. A typical image of in vitro assembled clathrin coats embedded in vitreous ice shows the presence of these three clathrin lattices (marked by different circles in Fig. 1D). Since the different types of clathrin coats have different sizes, as a first particle selection criterion, only particles with a size consistent with that of a hexagonal D6 barrel were interactively selected from the digitized micrographs. Viewed along the 6-fold axis, the size of the D6 barrel is similar to that of the mini coat. The two coats can be distinguished, however, because the D6 barrel seen in this orientation displays sharp straight edges while mini coats appear round. Using this rough size selection criterion, 9426 particles of D6 barrel coats were manually selected from 350 digitized micrographs.
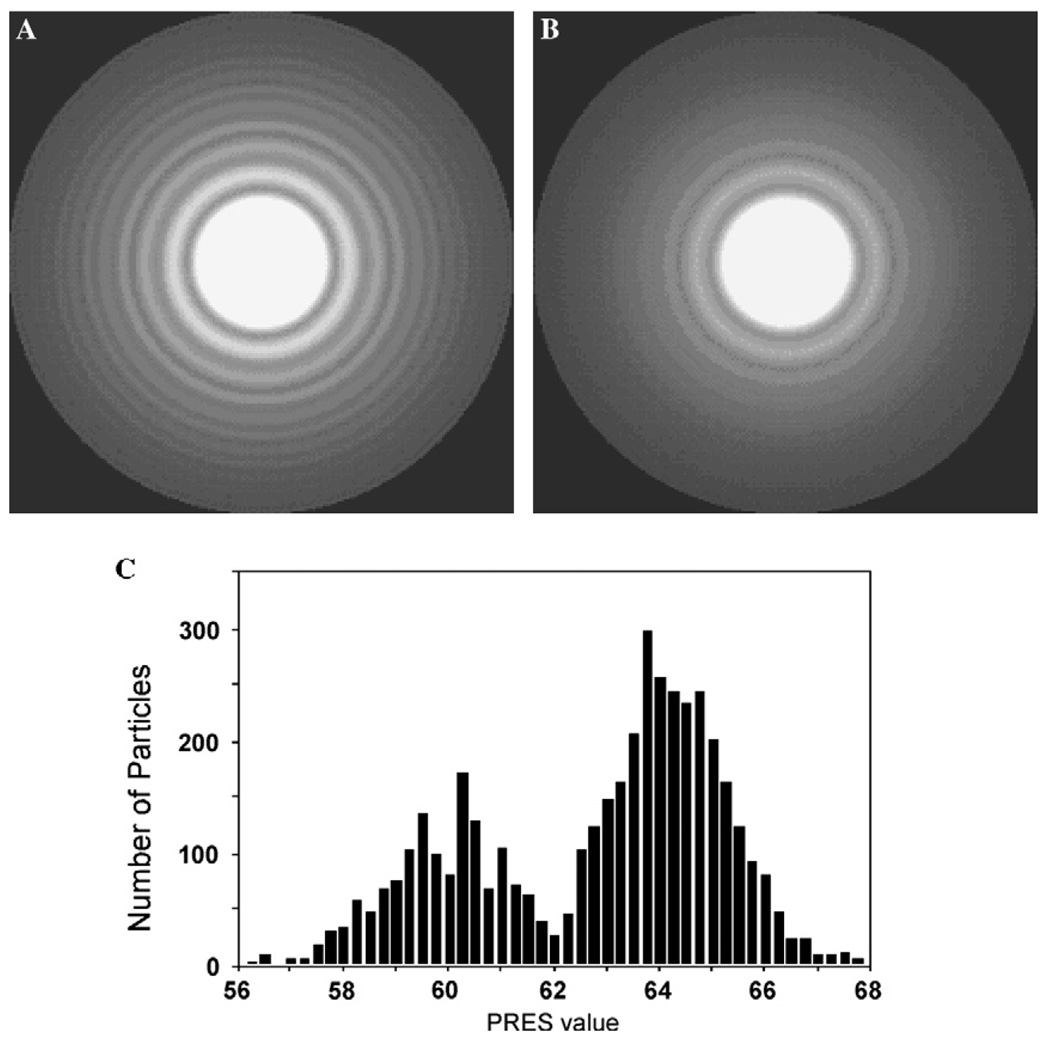
In a next step to assess the quality of the micrographs, we calculated rotationally averaged Fourier power spectra for all the boxed-out particles from the individual micrographs and obtained an average. Good micrographs recorded with an electron microscope equipped with a field emission electron source should show Thon rings that extend towards the edge of the power spectrum (Fig. 2A). By contrast, Thon rings from poor micrographs will only extend to a relatively low resolution (Fig. 2B). The loss of Thon rings in the high-resolution range can be due to a number of factors, such as beam-induced specimen movement, severe astigmatism of the objective lens and local tilt of the specimen. We did not differentiate among these causes and simply discarded all micrographs for which the summed rotationally averaged Fourier power spectra of the particles did not show Thon rings extending to 1/10 Å−1. Based on this criterion, particles from 125 of the initial 350 digitized micrographs were excluded from further processing. A similar procedure to select high quality micrographs has been used in other laboratories (e.g., Saad et al., 2001). We found that there was a direct correlation between the visibility of the Thon rings in the sum of the rotationally averaged Fourier power spectra and the correlation coefficient determined by CTFFIND3 (Mindell and Grigorief, 2003), which is a program that finds the defocus values of a micrograph by fitting simulated Fourier power spectra to the experimental one. The correlation coefficient between the simulated and the experimental Fourier power spectrum provides a quality estimate for the defocus determination. The micrographs we selected in this step all had correlation coefficients higher than 0.2.
Fig. 2.
Rotationally averaged summation of Fourier power spectra of all boxed out particles from (A) a “good” micrograph and (B) a micrograph with limited resolution. The outer ring corresponds to a spatial frequency of 1/10 Å−1. (C) Distribution of PRES values of aligned clathrin particles. Peak I and peak II include 1460 and 3790 particles, respectively. The particles in peak I have lower PRES values and are therefore more similar to the reference model.
The remaining particles were subjected to 10 cycles of iterative multi-reference alignment, multivariate statistic analysis and classification, following a standard protocol (van Heel et al., 2000). Particles corresponding to class averages that did not show well-defined features or did not correspond to the size of D6 barrels were excluded from further image processing. After this second particle selection step, only 5250 particles remained in the data set, about half of the initially selected particles.
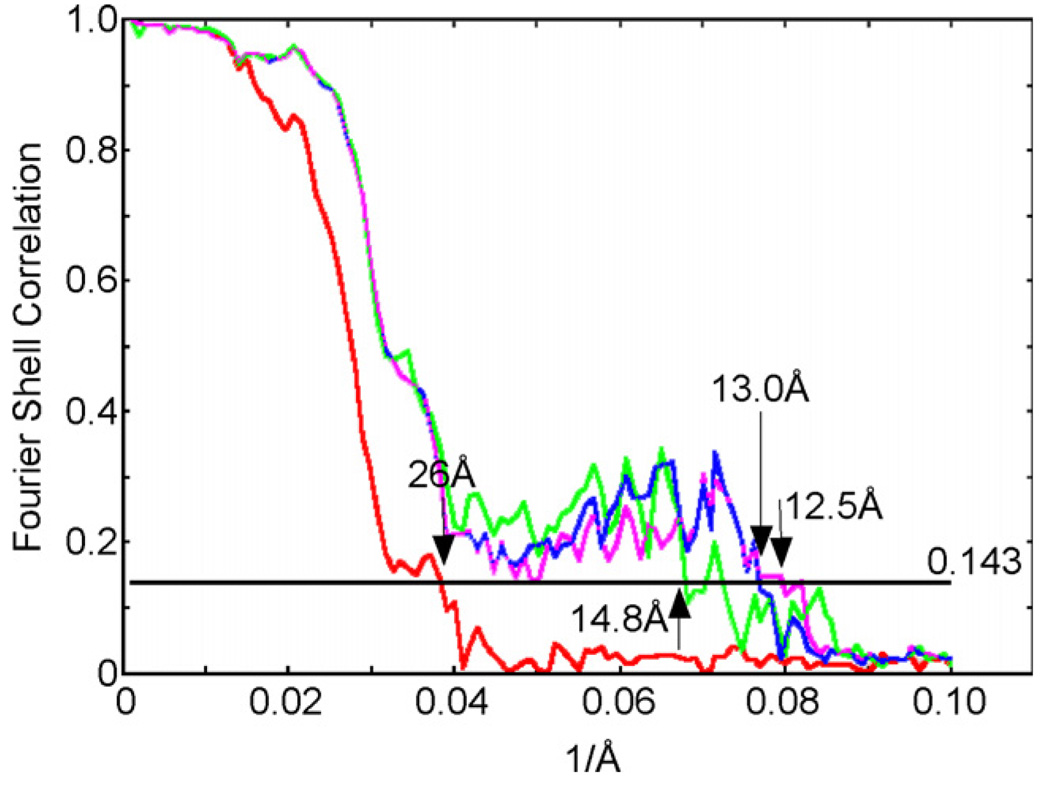
We selected the unique class averages and used them to reconstruct an initial 3D density map of the D6 barrel clathrin coat using the angular reconstitution procedure implemented in the IMAGIC V software package (van Heel et al., 1996). The initial 3D reconstruction at a nominal resolution of 42 Å (not shown) revealed all the structural features of a D6 barrel clathrin coat expected from previously determined structures (e.g., Smith et al., 1998). Further refinement of the density map was performed using FREALIGN (Stewart and Grigorieff, 2004). The weighted correlation coefficient (named PRES in FREALIGN which is calculated as the inverse cosine of the correlation coefficient) between the particle images and the 3D reference structure is one of the parameters listed in the FREALIGN output file. Since a lower PRES value indicates a better match between the particle images and the reference structure, we used this parameter, together with the FSC curve, to follow the progress of the refinement of the orientational parameters. After 20 iterative refinement cycles using only relatively low resolution information in the range of 800–40 Å, we displayed the distribution of particles in PRES bins (Fig. 2C). The histogram shows a clearly bimodal distribution of the particles with a boundary around a PRES value of 62°. We removed the particles from the data set that had PRES values higher than 62°, corresponding to the second peak in the histogram, and calculated a 3D reconstruction with the remaining 1450 particles with PRES values lower than 62°. The features of the clathrin legs in the resulting 3D reconstruction were much better defined than in the one calculated from the whole dataset of 5250 particles. The distribution of particle images into the two peaks had no correlation with the orientation of the particles. The 1450 “high quality” particles provided all the views needed to define the D6 asymmetric unit (see Supplementary Fig. 1d in Fotin et al., 2004) and were used for further refinement cycles. The 3D reconstruction after these refinement cycles reached a resolution of 14.8 Å (Fig. 3, green FSC curve). For comparison, the remaining 3,800 particles from the second peak were also used for a separate refinement, but the resulting 3D reconstruction never exceeded a resolution of 26 Å (Fig. 3, red FSC curve).
Fig. 3.
The FSC curves correspond to 3D reconstructions calculated from the particles of the second PRES peak (red), from the particles of first peak (green), after removing the central unstructured density from the reference map (blue) and after applying a tight mask to the reference map (purple). The FSC curves were calculated without removing the unstructured density in the center of the coat.
3.2. Removing unstructured density from the 3D reference map
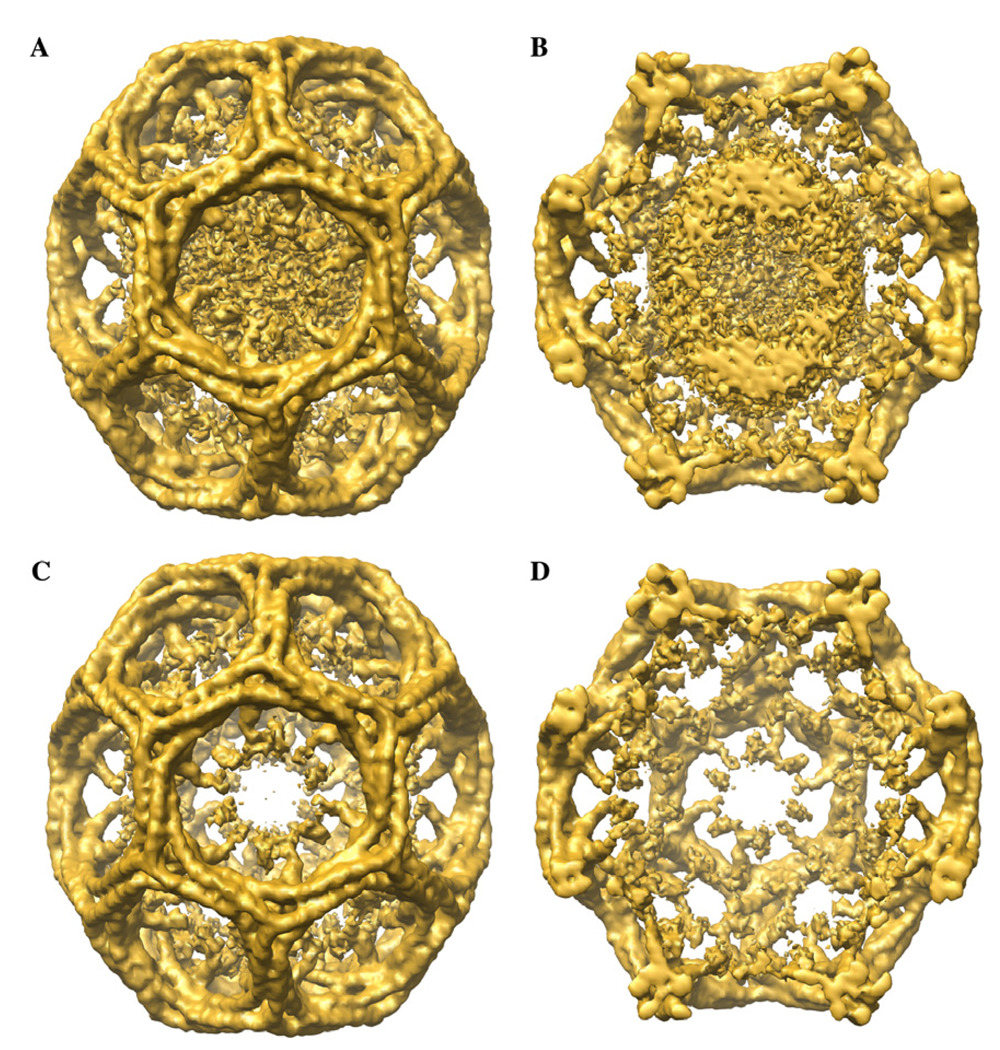
All previously published 3D reconstructions of D6 barrel clathrin coats showed a layer of unstructured density beneath the N-terminal domains of the clathrin legs (Smith et al., 1998, 2004). This layer of density probably represents the AP-2 complexes, for which the position, and perhaps the stoichiometry, is likely to vary from coat to coat. Since their arrangements do not follow the D6 symmetry of the clathrin lattices, D6 symmetrization of the 3D reconstruction causes the densities for the AP-2 complexes to be smeared out into a featureless layer of density. Our symmetrized 3D reconstruction obtained with FREALIGN also contains this layer of unstructured density. Fig. 4A shows a 3D map of the D6 barrel clathrin coat at a resolution of 12.5 Å, and removal of the front half of the map reveals that the center of the 3D reconstruction is almost completely filled with unstructured density (Fig. 4B). In the iterative refinement process, the 3D reconstruction obtained from each cycle is used as the reference 3D map for the next cycle, to which all the raw images are aligned. The presence of a central unstructured density in the 3D reference map is likely to cause misalignment of individual particle images, an effect that will limit the resolution. To prevent its influence on the alignment of the particle images, particularly in the high-resolution range, we applied a soft-edged mask (Gaussian mask with a fall-off of 6 pixels) to the reference map to remove the central unstructured density (Fig. 3C and D). This procedure further improved the resolution of the 3D reconstruction to 13 Å (Fig. 3, blue FSC curve).
Fig. 4.
The central density of the 3D reference model corresponding to an unstructured AP-2 protein shell was masked out during the refinement procedure. (A) Unmodified density map of the reference model and (C) density map of the reference model after removal of the central density. (B) and (D) Same density maps as in (A) and (C) but with the front half removed to show the interior of the density maps.
As the refinement proceeded to high resolution, we also applied a tight mask to the reference map by setting the FREALIGN parameter XSTD to 2.0. This mask removes noise densities in the solvent area. This procedure is equivalent to solvent flattening, which has already been used in other studies (Yonekura and Toyoshima, 2000). At this stage, our 3D reconstruction of the D6 barrel clathrin coat had a nominal resolution of 12.5Å (Fig. 3, purple FSC curve).
3.3. Local symmetry density averaging
The last step used to improve the resolution of the 3D reconstruction of the clathrin coat was to apply local symmetry density averaging. The D6 barrel clathrin coat is formed by 36 triskelions, comprising a total of 108 clathrin molecules. The D6 symmetry imposed during the refinement procedure only results, however, in a 12-fold averaging. Each asymmetric unit thus contains nine independent clathrin molecules and these D6 symmetry-independent molecules can be averaged to improve further the signal-to-noise ratio of the 3D reconstruction. The procedure used for local symmetry density averaging has been described in detail before (Fotin et al., 2004). This final step resulted in subnanometer resolution of our 3D reconstruction of the clathrin coat (7.9 Å, Supplementary Fig. 1e in Fortin et al., 2004).
4. Discussion
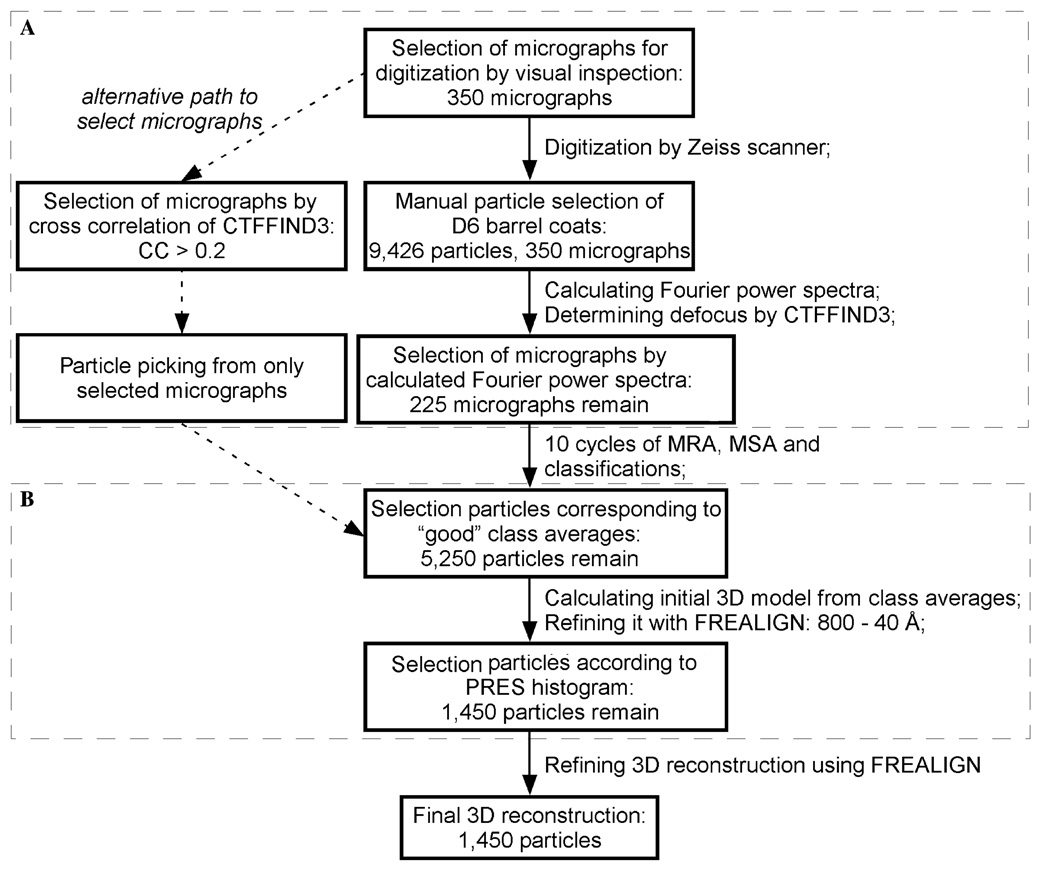
To visualize high-resolution features in single particle cryo-EM reconstructions, it is necessary to boost the signal-to-noise ratio of the reconstructions. This task is commonly accomplished by averaging thousands of individual particle images. It is, however, not only the number of particle images that will determine what resolution the resulting 3D reconstruction will have but also the quality of those images. We therefore attempted to select only the very best particle images for inclusion in the structure determination. Our selection was based on two types of criteria, as shown in the low-chart in Fig. 5.
Fig. 5.
Flow chart of particle selections. Part A concerns the selection of good micrographs and part B shows the selection for undistorted particles.
The first criterion concerned the quality of the individual micrographs (part A in Fig. 5). While this can often simply be assessed by inspection of the micrographs in a laser difractometer, our images of vitrified clathrin coats did not produce sufficient signal to see the Thon rings towards high resolution. Therefore, we judged the quality of individual micrographs based on the Thon rings seen in the sum of the rotationally averaged Fourier power spectra from all particles selected from a given micrograph, similar to a previously described method (Saad et al., 2001). This method requires that particles be selected from all micrographs, even from those that are of poor quality and will be discarded at a later stage. This is a significant drawback considering that particle picking, either manually done or using automated procedures, is a tedious and time-consuming process. We found, however, that there is a correlation between our micrograph selection based on the averaged Fourier power spectra of the particles and the correlation coefficient output by CTFFIND3. In our case, all selected micrographs had a correlation coefficient ≥ 0.2, while micrographs with a correlation coefficient of less than 0.2 were discarded. Therefore, it seems possible to select high quality micrographs based on the correlation coefficient from CTFFIND3, so that only the particles from “good” images are picked. The cut-off correlation coefficient value may differ slightly for different specimens. In the case of human transferrin receptor–transferrin complex (Cheng et al., 2004), we used a cut-off value of 0.18.
Our second selection criterion focused on the structural heterogeneity of the particles (part B in Fig. 5). In vitro assembled clathrin coats are large protein assemblies displaying both structural heterogeneity, such as different lattice designs, and structural flexibility due to their large size and hollow interior. Selecting particles with as little deviation from the D6 barrel structure as possible should therefore help significantly in resolving high-resolution features in 3D reconstructions. As we used FREALIGN to align the raw images to reference models, we could use the PRES value of the individual particle images as a means to identify and discard imperfect particles from the data set. Such similarity criteria between particle images and a reference model to remove “bad” particles is commonly used, and was also used, for example, to select the “best” 9 particles to calculate a 3D reconstruction of the icosahedral catalytic core of the pyruvate dehydrogenase complex from Bacillus stearothermophilus (Borgnia et al., 2004). Since the reference model improves from iteration to iteration, the similarity between the particle images and the reference model is assessed in each cycle. During the refinement of the density map towards higher resolution, the selection of particles that pass the selection criterion may change. In the case of our D6 barrel clathrin coat, the imperfections of the clathrin coats were most readily detected during refinement at low resolution, between 800 Å and 40 Å, probably because in this low-resolution range raw images of the clathrin coats have clearly defined features that are not contaminated by high frequency noise. We therefore permanently removed the particles corresponding to the second peak in the PRES histogram (Fig. 2) after only 20 refinement cycles. In refinement cycles including higher resolution information, the difference between perfect and imperfect coats becomes smaller due to the contaminating noise. Although we did not track the change in PRES values for individual particles during refinement cycles, this observation argues that it becomes less efficient to sort out the perfect D6 barrel coats once the refinement has proceeded to a higher resolution range.
The bimodal distribution of particles in the PRES histogram may not be typical, as we did not observe the same distribution for the tetrahedral coats. One reason might be the smaller size of the tetrahedral mini-coats, which could limit the structural flexibility in this lattice design. Permanently removing bad particles from the data set at an early stage of the refinement process based on the PRES value should improve the reconstruction, regardless of whether there is a bimodal distribution of the particles in the PRES histogram or not. If there is no obvious separation between “good” and “bad” particles in the PRES histogram, the best cut-off value has to be experimentally determined. In the case of the tetrahedral mini-coats, we simply used the same PRES cut-off criterion as in the D6 barrel coats.
3D reconstructions obtained by single particle averaging often show featureless densities for unstructured domains. For example, the RNA of Sindbis virus appears as an unstructured density in 3D reconstructions with imposed icosahedral symmetry since the RNA in the core is not packed with icosahedral symmetry (Zhang et al., 2002). Similarly, AP-2 complexes in in vitro assembled clathrin coats do not follow the symmetry of the clathrin lattice. In our D6-symmetrized reconstructions of the hexagonal barrel, the densities for the AP-2 complexes thus appear as a layer of unstructured density beneath the N-terminal domains of the clathrin molecules. This unstructured density will contaminate, especially at high-resolution spatial frequency, the signal from the ordered structural features of the 3D reference map, which grows weaker towards higher resolution. Removing featureless density prior to calculating FSC curves thus helps to more accurately estimate the resolution of 3D reconstructions. It is more important, however, to remove the featureless density from the reference map also during the refinement process itself. This eliminates its influence on the alignment of the particle images and thus produces better orientational parameters and a better map. Masking has to be done carefully to avoid the introduction of spurious features in the reconstruction that could influence the FSC or could be taken as real features in the structure. In particular, a smooth edge has to be used and the mask has to be sufficiently generous to make sure it does not affect density of the structure itself. In our masking procedure, we used a Gaussian mask.
All the image processing steps described here were necessary to determine the 3D structure of the D6 barrel clathrin coat to subnanometer resolution. We applied the same procedures to the clathrin D6 barrel coat with associated light chains and obtained a density map with a nominal resolution of 8.2 Å. We believe that similar procedures could be applied to other types of large protein assemblies, such as viruses, to obtain 3D reconstructions at higher resolutions.
Acknowledgments
We thank Werner Boll and Iris Rapoport for help in the purification of clathrin and adaptors. This work was supported by National Institutes of Health Grant GM62580 (to David DeRosier) and GM36548 (T.K). The molecular EM facility at Harvard Medical School was established by a generous donation from the Giovanni Armenise Harvard Center for Structural Biology.
References
- Borgnia MJ, Shi D, Zhang P, Milne JL. Visualization of alpha-helical features in a density map constructed using 9 molecular images of the 1.8 MDa icosahedral core of pyruvate dehydrogenase. J. Struct. Biol. 2004;17:136–145. doi: 10.1016/j.jsb.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Brodsky FM, Chen CY, Knuehl C, Towler MC, Wakeham DE. Biological basket weaving: formation and function of clathrin-coated vesicles. Annu. Rev. Cell Dev. Biol. 2001;17:517–568. doi: 10.1146/annurev.cellbio.17.1.517. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Zak O, Aisen P, Harrison SC, Walz T. Structure of the human transferrin receptor–transferrin complex. Cell. 2004;116:565–576. doi: 10.1016/s0092-8674(04)00130-8. [DOI] [PubMed] [Google Scholar]
- Crowther RA, Finch JT, Pearse BM. On the structure of coated vesicles. J. Mol. Biol. 1976;103:785–798. doi: 10.1016/0022-2836(76)90209-6. [DOI] [PubMed] [Google Scholar]
- Crowther RA, Henderson R, Smith JM. MRC image processing programs. J. Struct. Biol. 1996;116:9–16. doi: 10.1006/jsbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- Ehrlich M, Boll W, Van Oijen A, Hariharan R, Chandran K, Nibert ML, Kirchhausen T. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell. 2004;118:591–605. doi: 10.1016/j.cell.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Fotin A, Cheng Y, Sliz P, Grigorieff N, Harrison SC, Kirchhausen T, Walz T. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature. 2004;432:573–579. doi: 10.1038/nature03079. [DOI] [PubMed] [Google Scholar]
- Heymann JB, Iwasaki K, Yim YI, Cheng N, Belnap DM, Greene LE, Eisenberg E, Steven AC. Visualization of the binding of Hsc70 ATPase to clathrin baskets: implications for an uncoating mechanism. J. Biol. Chem. 2005;280:7156–7161. doi: 10.1074/jbc.M411712200. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T. Clathrin. Annu. Rev. Biochem. 2000;69:699–727. doi: 10.1146/annurev.biochem.69.1.699. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T, Harrison SC. Protein organization in clathrin trimers. Cell. 1981;23:755–761. doi: 10.1016/0092-8674(81)90439-6. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Chen DH, Song JL, Chuang DT, Chiu W. Seeing GroEL at 6 A resolution by single particle electron cryomicroscopy. Structure (Camb.) 2004;12:1129–1136. doi: 10.1016/j.str.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Matsui W, Kirchhausen T. Stabilization of clathrin coats by the core of the clathrin-associated protein complex AP-2. Biochemistry. 1990;29:10791–10798. doi: 10.1021/bi00500a011. [DOI] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Musacchio A, Smith CJ, Roseman AM, Harrison SC, Kirchhausen T, Pearse BM. Functional organization of clathrin in coats: combining electron cryomicroscopy and X-ray crystallography. Mol. Cell. 1999;3:761–770. doi: 10.1016/s1097-2765(01)80008-3. [DOI] [PubMed] [Google Scholar]
- Pearse BM, Crowther RA. Structure and assembly of coated vesicles. Annu. Rev. Biophys. Biophys. Chem. 1987;16:49–68. doi: 10.1146/annurev.bb.16.060187.000405. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Saad A, Ludtke SJ, Jakana J, Rixon FJ, Tsuruta H, Chiu W. Fourier amplitude decay of electron cryomicroscopic images of single particles and effects on structure determination. J. Struct. Biol. 2001;133:32–42. doi: 10.1006/jsbi.2001.4330. [DOI] [PubMed] [Google Scholar]
- Smith CJ, Grigorieff N, Pearse BM. Clathrin coats at 21 A resolution: a cellular assembly designed to recycle multiple membrane receptors. EMBO J. 1998;17:4943–4953. doi: 10.1093/emboj/17.17.4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CJ, Dafforn TR, Kent H, Sims CA, Khubchandani-Aswani K, Zhang L, Saibil HR, Pearse BM. Location of auxilin within a clathrin cage. J. Mol. Biol. 2004;336:461–471. doi: 10.1016/j.jmb.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Stewart A, Grigorief N. Noise bias in the refinement of structures derived from single particles. Ultramicroscopy. 2004;102:67–84. doi: 10.1016/j.ultramic.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Sudhof TC. The synaptic vesicle cycle. Annu. Rev. Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- ter Haar E, Musacchio A, Harrison SC, Kirchhausen T. Atomic structure of clathrin: a beta propeller terminal domain joins an alpha zigzag linker. Cell. 1998;95:563–573. doi: 10.1016/s0092-8674(00)81623-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon F. Zur Defokussierungsäbhängigkeit des Phasenkontrastes bei der elektronenmikroskopischen Abbildung. Z. Naturforsch. 1966;21a:476–478. [Google Scholar]
- Ungewickell E, Branton D. Assembly units of clathrin coats. Nature. 1981;289:420–422. doi: 10.1038/289420a0. [DOI] [PubMed] [Google Scholar]
- van Heel M. Angular reconstitution: a posteriori assignment of projection directions for 3D reconstruction. Ultramicroscopy. 1987;21:111–123. doi: 10.1016/0304-3991(87)90078-7. [DOI] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J. Struct. Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- van Heel M, Gowen B, Matadeen R, Orlova EV, Finn R, Pape T, Cohen D, Stark H, Schmidt R, Schatz M, Patwardhan A. Single-particle electron cryo-microscopy: towards atomic resolution. Quart. Rev. Biophys. 2000;33:307–369. doi: 10.1017/s0033583500003644. [DOI] [PubMed] [Google Scholar]
- Vigers GP, Crowther RA, Pearse BM. Three-dimensional structure of clathrin cages in ice. EMBO J. 1986;5:529–534. doi: 10.1002/j.1460-2075.1986.tb04242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura K, Toyoshima C. Structure determination of tubular crystals of membrane proteins. III. Solvent lattening. Ultramicroscopy. 2000;84:29–45. doi: 10.1016/s0304-3991(00)00008-5. [DOI] [PubMed] [Google Scholar]
- Zhang W, Mukhopadhyay S, Pletnev SV, Baker TS, Kuhn RJ, Rossmann MG. Placement of the structural proteins in Sindbis virus. J. Virol. 2002;76:11645–11658. doi: 10.1128/JVI.76.22.11645-11658.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]