Abstract
New Zealand identified its first pandemic H1N1 influenza cases in late April 2009, immediately prior to the historical start of the New Zealand influenza season. Both pandemic and oseltamivir-resistant seasonal H1N1 viruses cocirculated in the population for a period of time. Thus, concerns were raised about the possibility of reassortment events between the two strains. An RT-PCR–based genotyping assay was developed so that H1N1 influenza coinfections and reassortants could be detected quickly. The assay differentiated effectively the seasonal and pandemic strains. It also confirmed the identification of the first reported coinfection of pandemic and seasonal H1N1 strains during the 2009 Southern Hemisphere influenza season in New Zealand.
Keywords: pandemic influenza, seasonal influenza, genotyping PCR, reassortant
1. Introduction
During March 2009, the Centers for Disease Control and Prevention (CDC) reported an increase in influenza-like illness in Mexico (2009a). The etiologic agent was identified subsequently as a novel H1N1 influenza virus. This new influenza virus arose through the reassortment of a North American triple-reassortant swine influenza virus and a Eurasian swine influenza virus (Smith et al., 2009). The virus, pandemic A/H1N1 2009, spread quickly throughout the world giving rise to a new influenza pandemic that persisted during the course of the 2009 Southern Hemisphere influenza season.
In New Zealand, the first pandemic A/H1N1 2009 influenza cases were confirmed in late April, immediately prior to the historical start of the New Zealand influenza season. Initially, both the pandemic A/H1N1 2009 and seasonal H1N1 viruses cocirculated in the population, but by early July, the pandemic virus was the predominant circulating influenza virus (CDC, 2009b). Despite widespread circulation and unlike their seasonal H1N1 counterparts, the pandemic A/H1N1 2009 viruses isolated in New Zealand remained antigenically stable and oseltamivir sensitive, (Hall et al., 2009). The finding that both seasonal and pandemic H1N1 viruses had cocirculated did, however, raise concerns that reassortment could lead to an oseltamivir-resistant pandemic strain. The aim of the present study was to develop a molecular assay capable of rapid identification and genotyping of seasonal-pandemic H1N1 reassortants.
2. Materials and methods
2.1 Clinical material
Clinical samples were obtained from influenza-like illness cases, which were defined as an acute respiratory tract infection characterized by an abrupt onset of at least two of the following symptoms: fever, chills, headache, or myalgia (2009b). Nasopharyngeal or throat swabs were collected in New Zealand as part of a 2009 national surveillance program. All samples were screened for influenza A, pandemic A/H1N1 2009, and seasonal H1N1 by real-time RT-PCR following the World Health Organization’s recommended protocols. For the present study, a confirmed seasonal H1N1–positive (A/New Zealand/3362/2009, VIR-3362), a pandemic A/H1N1 2009–positive (A/New Zealand/2047/2009, VIR-2047), and a double-positive (A/New Zealand/891/2009, VIR-891) specimen (Peacey et al., unpublished results) were used (real-time PCR–positive samples, data not shown).
2.2 Virus isolation and RNA extraction
Clinical specimens were passaged three times in Madin-Darby canine kidney sialyltransferase-1 (MDCK-SIAT1) cells before use in the present study to allow for a suitable volume for assay development. Briefly, influenza viruses were isolated from the clinical specimens on MDCK-SIAT1 cells (Matrosovich et al., 2003), grown in DMEM-SF12 (Gibco, Grand Island, NY, USA) with 2% fetal calf serum (Gibco), L-Glutamine (Gibco), penicillin and streptomycin (Invitrogen, Carlsbad, CA, USA), gentamicin (Pfizer, New York, NY, USA), and geneticin (Sigma-Aldrich, St. Louis, MO, USA). TPCK trypsin (1.6μL/mL; Sigma-Aldrich) was added to MDCK-SIAT1 serum-free medium prior to sample inoculation. RNA was extracted from culture supernatant with the ZR Viral RNA Kit (Zymo Research, Orange, CA, USA), according to the manufacturer’s instructions. RNA was eluted into 50 μL nuclease-free water.
2.3 RT-PCR assay design
RT-PCR assays were designed so that each viral gene segment could be subtyped as either seasonal H1N1 or pandemic A/H1N1 2009. RT-PCRs were performed in 50 μL final volume with the one-step SuperScript® III Taq Polymerase kit (Invitrogen). Cycling reactions were performed in a GeneAmp PCR System 9700 thermocycler (Applied Biosystems, Foster City, CA, USA) as follows: 50 °C for 30 min and 95 °C for 2 min; followed by 40 cycles of 95 °C for 30 s, 57 °C for 30 s, and 68 °C for 3 min; and a final extension at 68 °C for 7 min. PCR amplicons were analyzed in a 2% SeaKem LE agarose gel (Lonza, Rockland, ME, USA) using 0.5× TBE (Tris, Boric Acid, EDTA; Invitrogen) as electrophoresis running buffer and stained with gel red (Biotium, Hayward, CA, USA).
2.4 Oseltamivir resistance test
Surveillance for oseltamivir resistance in pandemic A/H1N1 2009 viruses in New Zealand was carried out using a fluorometric neuraminidase inhibition assay on viral isolates maintained in culture as previously described (Hall et al., 2009; Hurt et al., 2004). VIR-2047 was sensitive to oseltamivir, but VIR-891 was resistant (data not shown).
2.5 Confirmatory DNA sequencing
All amplicons were sequenced to reconfirm the H1N1 subtype. Sequencing was conducted using a BigDye® Terminator v3.1 cycle sequencing kit (Applied Biosystems, Nieuwerkerk, NL) on a capillary sequencer (model 3100 avant, Applied Biosystems,) using the PCR primers as sequencing primers. Sequences were analyzed using BioEdit (Hall, 1999) and ClustalW (Chenna et al., 2003).
3. Results
Differential PCR primers were designed for each of the eight influenza A virus gene segments to identify reassortants between seasonal and pandemic H1N1 viruses. To achieve this, multiple sequence alignments were made with contemporary seasonal H1N1 viruses and pandemic A/H1N1 2009 viruses. Regions of sequence identity within virus strains were selected for primer design. Table 1 summarizes the primers designed for each gene segment, their concentrations, and the anticipated amplicon sizes (140–500 bp with at least a 120-bp difference between the seasonal and pandemic amplicons).
Table 1.
PCR primers and conditions for genotyping of H1N1 influenza viruses
| Gene | Strain specificity | Primer sequences1 | Primer concentrations specificities2 (per template) | Amplicons (bp) |
|---|---|---|---|---|
| PB2 | seasonal H1N1 | PB2-seas-300-316F: 5′ CAGAAATGGACCAGTGG 3′ | - | seasonal: 440 |
| PB2-seas-722-740R: 5′ CCTGGAGTGTACATCTGCT 3′ | ||||
| pandemic H1N1 | PB2-pan-1133-52F: 5′ CCAGGAGATTGATCCAGTTG 3′ | - | pandemic: 230 | |
| PB2-pan-1339-59R: 5′ GGATTCAATTCCCCAGTTCTG 3′ | ||||
| PB1 | seasonal and pandemic H1N1 (multiplex) | PB1-seas-1072-93F: 5′ GAAAGCAAGAGTATGAAACTGA 3′ | seasonal: 160 | |
| PB1-seas-1208-29R: 5′ CCCATCATCATYCCAGGACTCA 3′ | pandemic: 310 | |||
| PB1-pan-1112-32F: 5′ AAATGCTAGCAAGCATTGACC 3′ | ||||
| PB1pand1401-21R: 5′ CCCACTAACTTGCAGGTCCTG 3′ | ||||
| PA | seasonal and pandemic H1N1 (multiplex) | PA-409-32F: 5′ AAAATWAAATCTGAGAAGACACAC 3′ | 600 nmol/L of PA-F, 200 nmol/L of PA-seas-R, and 400 nmol/L of PA-pan-R | seasonal: 260 |
| PA-seas-654-72R: 5′ GGAGAAGTTCGGCGGAAGG 3′ | pandemic: 570 | |||
| PA-pan-962-84R: 5′ TTTCTCATGTGGTTTGACTATGT 3′ | ||||
| HA | seasonal and pandemic H1N1 | HA-40-59F: 5′ GACACTGTWGACACAGTACT 3′ | seasonal: 200 | |
| HA-580-97R: 5′ CATATGCATCTGCATTCT 3′ | pandemic: 560 | |||
| NP | seasonal H1N1 | NP-seas-1110-30F: 5′ ATGGATGCTATTGTGTCAAG 3′ | seasonal: 200 | |
| NP-seas-1288-1307R: 5′ CTTCCCTCTGTATTCCCAGA 3′ | ||||
| pandemic H1N1 | NP-pan-921-38F: 5′ CCAAAACAGCCAAGTGGT 3′ | pandemic: 320 | ||
| NP-pan-1222-39R: 5′ TGAGAATGTAGGCTGCAC 3′ | ||||
| NA | seasonal and pandemic H1N1 (multiplex) | NA-seas-226-43F: 5′ GCTGGAGAGGACAAAACG 3′ | 200 nmol/L of each of the 4 primers | seasonal: 430 |
| NA-seas-639-57R: 5′ CCAACTTTTTATGGTTCCA 3′ | pandemic: 210 | |||
| NA-pan-226-43F: 5′ GCTGGACAGTCAGTGGTT 3′ | ||||
| NA-pan-414-35R: 5′ GGAATGTTTGTCATTTAGCAAG 3′ | ||||
| M | seasonal and pandemic H1N1 (multiplex) | M-181-98F: 5′ GGATTTGTGTTCACGCTC 3′ | seasonal: 140 | |
| M-seas-300-19R: 5′ TCTCCCTCTTAAGCTTTCGA 3′ | ||||
| M-pan-484-504F: 5′ CACAGACAAATGGCTACTACC 3′ | pandemic: 310 | |||
| M-pan-769-90R: 5′ CCCAATGATATTTGCTGCAATG 3′ | ||||
| NS | seasonal H1N1 | NS-seas-47-64F: 5′ GGCATGTCCGCAAACAAG 3′ | seasonal: 300 | |
| NS-seas-335-52R: 5′ TGACACAAAGAGGGCCAG 3′ | ||||
| pandemic H1N1 | NS-pan-417-39F: 5′ CCGATTAGAGACCTTGATACTAC 3′ | pandemic: 200 | ||
| NS-pan-593-613R: 5′ TTCTCCAAGCGAATCTCTGTA 3′ | ||||
The primers nt coordinates are indicated in the primers names (H3 numbering for HA)
Unless specified, 400 nmol/L of each primer was added for each template.
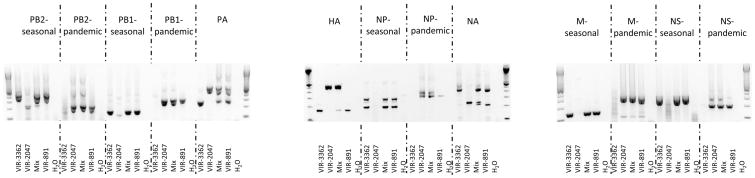
The genotyping assay was able to differentiate between seasonal and pandemic H1N1 viruses that circulated in New Zealand in 2009 (Fig. 1). A specific band at the expected size (Table 1) was observed for the seasonal virus (VIR-3362) and the pandemic virus (VIR-2047) for each of the eight viral gene segments (Fig. 1). To simulate coinfection, the RNA of VIR-3362 and VIR-2047 were mixed in a 1:1 ratio, and the genotyping assay was conducted. As expected, double–positive amplicons of the expected sizes were detected for each gene segment, indicating the presence of RNA from both viruses (Fig. 1). A sample grown from a suspected natural seasonal–pandemic H1N1 coinfection (VIR-891) also displayed the same pattern as that of the mixed RNA, i.e., both the seasonal H1N1 and the pandemic H1N1 PCR bands were observed on the agarose gel (Fig. 1). VIR-891 was, therefore, confirmed as coinfected with the two H1N1 strains. This is one of the first eleven reported cases of coinfection with seasonal and pandemic H1N1 influenza viruses (Peacey et al., unpublished observation). Each amplicon was sequenced, and the accuracy of the genotyping assay was confirmed: either seasonal or pandemic H1N1 sequences were obtained as expected (data not shown).
Fig. 1.
Typical agarose gel electrophoresis pattern of RT-PCR products from seasonal H1N1 (VIR-3362), pandemic A/H1N1 2009 (VIR-2047), seasonal H1N1 + pandemic A/H1N1 2009 mixed template (Mix), natural coinfected template (VIR-891), and water (H2O) as a negative control, as indicated under each gel lane. Indicated above the agarose gel picture are the genes targeted by each RT-PCR and, where appropriate, whether seasonal- or pandemic-specific primers were used. The extreme left and right lanes of each gel contain a 100-bp DNA ladder (Invitrogen).
4. Discussion
The emergence of a new influenza pandemic has raised a number of questions relating to its epidemiology and evolution. Although the pandemic strain has remained stable antigenically, and the reports of oseltamivir resistance have been relatively rare, there is concern that the pandemic strain may acquire further traits by reassortment with seasonal strains. However, the capacity of the pandemic and seasonal strains to reassort and produce viable viral progeny remains unknown.
Reassortment compatibility studies can be conducted in laboratory settings, but examining natural human coinfections is the preferable approach. During the 2009 influenza season, seasonal and pandemic H1N1 strains cocirculated in New Zealand, thereby raising the possibility of detecting such coinfections. This scenario is of particular concern (as compared to seasonal H3N2–pandemic A/H1N1 2009 coinfections) because a recombinant pandemic virus containing a contemporary seasonal H1N1 neuraminidase gene that imparts oseltamivir resistance could be generated. No natural coinfections or reassortment events involving pandemic A/H1N1 2009 viruses have been described to date, possibly due to the lack of significant levels of cocirculating pandemic A/H1N1 2009 with other influenza viruses in many countries where the emergence of the pandemic strain coincided with the disappearance of seasonal strains. In contrast, starting the week of June 8, 2009, 14% of the influenza-positive samples collected in New Zealand were confirmed as pandemic A/H1N1 2009; this number increased to 80% during the week of June 29, 2009. Both viruses cocirculated in the country throughout the winter (CDC, 2009b).
Reassortment events between seasonal influenza viruses have been reported previously, though they have appeared relatively infrequently in France in 2002–2003 (Al Faress et al., 2008), in Cambodia in 2005–2008 (Fourment et al., 2009), and in Thailand in 2006–2008 (Bai et al., 2009). Natural coinfections of influenza are also reported rarely (Eshaghi et al., 2009; Falchi et al., 2008; Takao et al., 2005; Toda et al., 2006). However coinfection events may have been largely underreported and/or underdetected, as suggested recently by Ghedin et al. (2009), with the characterization of clinical specimens from the USA and New Zealand and by Furuse et al. (2009) in a study of Japanese clinical specimens. Nevertheless, once identified, viral populations from coinfections need to be genotyped to detect possible reassortants. In response to this need, a genotyping RT-PCR assay was developed to distinguish between the eight gene segments of pandemic H1N1 virus and those of seasonal H1N1 viruses. This assay allowed the identification of each gene segment as either seasonal H1N1- or pandemic A/H1N1 2009–derived and detected both viruses in an artificially mixed RNA sample and in a naturally coinfected human sample.
This eight-gene–based genotyping assay confirmed the coinfection of one patient with seasonal H1N1–pandemic A/H1N1 2009 during the 2009 influenza season in New Zealand. Although this screen was not done under conditions that made it possible to determine individual viral genotypes (i.e., the screen was conducted on a viral pool as proof of concept for this assay), it did prove that coinfections are possible and that clinical samples should be screened for both pandemic and seasonal influenza viruses. The current study also highlights the possibility of detecting a false oseltamivir-resistant pandemic A/H1N1 2009 strain such as specimen VIR-891; coinfection of seasonal H1N1 and pandemic A/H1N1 2009 may explain why a pandemic A/H1N1 2009 PCR-positive sample is oseltamivir resistant.
The novel genotyping assay described here relies on the present genetic stability of the pandemic A/H1N1 2009. The assay was validated with clinical specimens: 11 seasonal H1N1 and 10 pandemic A/H1N1 2009 positive swabs were genotyped successfully. Changes in the primary sequence of the virus must be monitored closely, as selective pressure on the virus increases in the future. The assay conditions are a compromise between the optimized RT-PCR chemistries and cycling programs for each gene segment, so that all genes of a virus can be genotyped quickly during a single RT-PCR run. Therefore, uniplex and multiplex RT-PCRs have been selected for different gene segments. This compromise also causes the appearance of several nonspecific bands in Figure 1, mainly for PB2 seasonal (unspecific band at 220 bp for a pandemic template), NP seasonal (nonspecific band at 250 bp with a seasonal template), and M (at 180 bp for all H1N1 templates). These nonspecific bands do not compromise the genotyping and disappear if more stringent RT-PCR conditions are chosen. The eight-gene–genotyping assay was also tested with Northern American H1N1 strains, and it genotyped efficiently all genes of A/Memphis/13/09 (2008–09 season), A/New Jersey/15/07 (2006–07 season), and A/California/04/09 and A/Tennessee/560/09 (pandemic A/H1N1 2009). Most of the older H1N1 strains tested were also genotyped easily. However, in some runs, cross-reactive pandemic bands were observed for the NP, NS, and PB2 genes in A/Memphis/15/00, A/Memphis/7/01, A/Memphis/13/06, A/Memphis/2/07, and/or A/Memphis/7/08 (data not shown). The assay is, therefore, suitable to screen only recent (2008–2009 season onwards) human seasonal H1N1 strains.
5. Conclusions
In conclusion, an RT-PCR–based, eight-gene–genotyping assay was developed to detect in a rapid manner H1N1 influenza coinfections and reassortants. The assay identified effectively a natural coinfection of pandemic and seasonal H1N1 strains during the 2009 Southern Hemisphere influenza season in New Zealand.
Acknowledgments
This work was supported by Contract No. HHSN266200700005C from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services. We thank Dr. Natalia Ilyushina for technical support, Dr. Adrianus C.M. Boon for reviewing critically the manuscript, and Dr. Angela McArthur for editorial assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al Faress S, Ferraris O, Moules V, Valette M, Hay A, Lina B. Identification and characterization of a late AH1N2 human reassortant in France during the 2002–2003 influenza season. Virus Res. 2008;132:33–41. doi: 10.1016/j.virusres.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Bai GR, Chittaganpitch M, Kanai Y, Li YG, Auwanit W, Ikuta K, Sawanpanyalert P. Amantadine- and oseltamivir-resistant variants of influenza A viruses in Thailand. Biochem Biophys Res Commun. 2009;390:897–901. doi: 10.1016/j.bbrc.2009.10.071. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) Outbreak of swine-origin influenza A (H1N1) virus infection - Mexico, March-April 2009. MMWR Morb Mortal Wkly Rep. 2009a;58:467–470. [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) Surveillance for the 2009 pandemic influenza A (H1N1) virus and seasonal influenza viruses - New Zealand, 2009. MMWR Morb Mortal Wkly Rep. 2009b;58:918–921. [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshaghi A, Blair J, Burton L, Choi KW, De Lima C, Duncan C, Guyard C, Higgins R, Lombos E, Low DE, Mazzulli T, Drews SJ. Characterization of an influenza A and influenza B co-infection of a patient in a long-term care facility with co-circulating influenza A and influenza B. Int J Infect Dis. 2009;13:e127–e128. doi: 10.1016/j.ijid.2008.06.024. [DOI] [PubMed] [Google Scholar]
- Falchi A, Arena C, Andreoletti L, Jacques J, Leveque N, Blanchon T, Lina B, Turbelin C, Dorleans Y, Flahault A, Amoros JP, Spadoni G, Agostini F, Varesi L. Dual infections by influenza A/H3N2 and B viruses and by influenza A/H3N2 and A/H1N1 viruses during winter 2007, Corsica Island, France. J Clin, Virol. 2008;41:148–151. doi: 10.1016/j.jcv.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Fourment M, Mardy S, Channa M, Buchy P. Evidence for persistence of and antiviral resistance and reassortment events in seasonal influenza virus strains circulating in Cambodia. J Clin, Microbiol. 2010;48:295–297. doi: 10.1128/JCM.01687-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse Y, Suzuki A, Kishi M, Nukiwa N, Shimizu M, Sawayama R, Fuji N, Oshitani H. Occurrence of mixed populations of influenza A viruses that can be maintained through transmission n a single host and potential for reassortment. J Clin Microbiol. 2010;48:369–374. doi: 10.1128/JCM.01795-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghedin E, Fitch A, Boyne A, Griesemer S, DePasse J, Bera J, Zhang X, Halpin RA, Smit M, Jennings L, St George K, Holmes EC, Spiro DJ. Mixed infection and the genesis of influenza virus diversity. J Virol. 2009;83:8832–8841. doi: 10.1128/JVI.00773-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall RJ, Peacey MP, Ralston JC, Bocacao J, Ziki M, Gunn W, Quirk A, Huang QS. Pandemic influenza A(H1N1)v viruses currently circulating in New Zealand are sensitive to oseltamivir. Euro Surveill. 2009;14:19282. doi: 10.2807/ese.14.30.19282-en. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hurt AC, Barr IG, Hartel G, Hampson AW. Susceptibility of human influenza viruses from Australasia and South East Asia to the neuraminidase inhibitors zanamivir and oseltamivir. Antiviral Res. 2004;62:37–45. doi: 10.1016/j.antiviral.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Matrosovich M, Matrosovich T, Carr J, Roberts NA, Klenk HD. Overexpression of the alpha-2,6-sialyltransferase in MDCK cells increases influenza virus sensitivity to neuraminidase inhibitors. J Virol. 2003;77:8418–8425. doi: 10.1128/JVI.77.15.8418-8425.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacey M, Hall RJ, Sonnberg S, Ducatez MFSP, Nicole M, Ralston JC, Bandaranayake DVH, Webby RJ, Huang QS. Natural co-infections of pandemic swine-origin H1N1 (2009) and seasonal H1N1 in six human patients - first report. (Unpublished results) [Google Scholar]
- Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JS, Guan Y, Rambaut A. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459:1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- Takao S, Hara M, Kakuta O, Shimazu Y, Kuwayama M, Fukuda S, Miyazaki K. Eleven cases of co-infection with influenza type A and type B suspected by use of a rapid diagnostic kit and confirmed by RT-PCR and virus isolation. Kansenshogaku Zasshi. 2005;79:877–886. doi: 10.11150/kansenshogakuzasshi1970.79.877. [DOI] [PubMed] [Google Scholar]
- Toda S, Okamoto R, Nishida T, Nakao T, Yoshikawa M, Suzuki E, Miyamura S. Isolation of influenza A/H3 and B viruses from an influenza patient: confirmation of co-infection by two influenza viruses. Jpn J Infect Dis. 2006;59:142–143. [PubMed] [Google Scholar]