Abstract
The envelope (Env) glycoproteins of human immunodeficiency virus (HIV-1) mediate viral entry and are also the primary target of neutralizing antibodies. The gp160 envelope glycoprotein precursor undergoes proteolytic cleavage in the Golgi complex to produce the gp120 exterior glycoprotein and the gp41 transmembrane glycoprotein, which remain associated non-covalently in the trimeric Env complex. Monomeric soluble gp120 has been used extensively to investigate conformational states, structure, antigenicity and immunogenicity of the HIV-1 Env glycoproteins. Expression of gp120 alone (without gp41) leads to the accumulation not only of monomeric gp120 but also an aberrant dimeric form. The gp120 dimers were sensitive to reducing agents. The formation of gp120 dimers was disrupted by a single amino acid change in the inner domain, and was reduced by removal of the V1/V2 variable loops or the N and C termini. Epitopes on the gp120 inner domain and the chemokine receptor-binding surface were altered or occluded by gp120 dimerization. Awareness of the existence and properties of gp120 dimers should assist interpretation of studies of this key viral protein.
Keywords: HIV-1, virus entry, CD4, envelope glycoprotein, conformational change
1. Introduction
The envelope (Env) glycoproteins of human immunodeficiency virus (HIV-1) mediate viral entry and are also the primary target of neutralizing antibodies. Following synthesis in the endoplasmic reticulum, post-translational events including oligomerization, disulfide bond formation, and glycosylation occur (Allan et al., 1985; Robey et al., 1985). The gp160 Env glycoprotein precursor is transported through the Golgi complex, where additional carbohydrate modifications and proteolytic cleavage take place. The resulting mature Env glycoproteins, gp120 (SU) and gp41 (TM), constitute a trimeric complex that is transported to the surface of infected cells and is anchored on the virion surface by the membrane-spanning segments of gp41 (Chan et al., 1997; Farzan et al., 1998; Weissenhorn et al., 1997; Zhu et al., 2003). The gp120 exterior Env glycoprotein is retained on the trimer via labile, noncovalent interactions with the gp41 ectodomain (Helseth et al., 1991). The gp120 glycoprotein binds the initial receptor, CD4 (Dalgleish et al., 1984; Klatzmann et al., 1984). CD4 binding triggers conformational changes in gp120 that promote its interaction with one of the chemokine receptors, CCR5 or CXCR4, and that eventually result in the fusion of the viral and target cell membranes (Chan et al., 1997; Lu et al., 1995; Weissenhorn et al., 1997; Alkhatib et al., 1996; Choe et al., 1996; Deng et al., 1996; Doranz et al., 1996; Dragic et al., 1996; Feng et al., 1996; Trkola et al., 1996; Wu et al., 1996).
Soluble forms of the HIV-1 envelope glycoproteins have been produced for structural and biochemical studies. For example, cleavage-defective gp140 glycoproteins consist of the complete gp120 and the gp41 ectodomain lacking the transmembrane anchor and cytoplasmic tail (Earl et al., 1990). It has been previously shown by sucrose gradient sedimentation and/or chemical cross-linking that mammalian-cell-expressed gp160 and soluble gp140 (sgp140) glycoproteins exist as a mixture of dimers and higher-order oligomers (Center et al., 2000; Doms et al., 1991; Earl et al., 1990; Schawaller et al., 1989). Soluble gp140 glycoproteins have been shown to be cleaved at a low level into gp120 glycoproteins, some which exist as disulfide-linked dimers (Hallenberger et al., 1993). Soluble gp120 expressed in the absence of gp41 was also found to contain a fraction of disulfide-linked dimers (Center et al., 2000). Dimerization of gp120 was more efficient when the variable loop 2 (V2) region of gp120 was intact (Center et al., 2000).
This manuscript reports that expression of gp120 in the absence of gp41 results in the formation of stable dimers within the cells and that these dimers are ultimately secreted into the supernatant along with monomeric gp120. These dimers represent a substantial amount of the overall secreted gp120. Changes in a single residue located within the gp120 inner domain can modulate the extent of dimer formation. Removal of the conformationally flexible major variable loops, particularly the V1/V2 loop, and/or the N and C termini also decreased the proportion of dimers formed. Several conformational epitopes on the monomeric gp120 are either disrupted or not accessible on dimeric gp120. An awareness of these differences is important for structural, biochemical and biophysical studies. These results suggest methods to separate these two forms of gp120 to allow their independent analysis.
2. Materials and Methods
2.1 Cell lines
293T human embryonic kidney, Cf2Th canine thymocytes (American Type Culture Collection) and TZM-bl (NIH AIDS Research and Reference Reagent Program) cells lines were grown at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (Sigma, St Louis, MO, USA) and 100 μg/ml of penicillin-streptomycin (Mediatech, Inc., Manassas, VA, USA).
2.2 Site-directed mutagenesis
Mutations (H66A, L111A, S375W, L111A/S375W, C54A, C205A) were introduced into the pSVIIIenv plasmid expressing the HIV-1YU2 gp120 glycoprotein using the QuikChange II XL site-directed mutagenesis protocol (Stratagene, Cedar Creek, TX, USA). The codon-optimized pcDNA3.1-HIV-1YU2 gp120 vector has been previously described (Yang et al., 2004). The presence of the desired mutations was confirmed by DNA sequencing. All residues are numbered according to those of the prototypic HXBc2 sequence, as per current convention (Korber, 1998). The mutants are designated with the amino acid residue to the right of the number substituted for the amino acid residue to the left of the number.
The codon-optimized pcDNA3.1-HIV-1YU2 44-492 expressor plasmid contains the signal peptide of the T-cell surface CD5 glycoprotein, with the following junctional sequence at the N-terminus: ...SVLA 44VWKE... (leader sequence underlined) and a stop codon at position 493.
The codon-optimized pcDNA3.1-HIV-1YU2 ΔV1/V2 expression construct was made by replacing the sequence encoding ...124PLCVTLNCTDLRN...VPIDNASYRLISCNT198... from the V1/V2 loop with a sequence encoding a GG linker in the codon-optimized HIV-1YU2 gp120 expression construct.
The codon-optimized pcDNA3.1-HIV-1YU2 ΔV3 expression construct was made by replacing the sequence encoding ...302NTRKSINIGPGRALYTTGEII323... from the V3 loop with a sequence encoding a GGSGSG linker in the codon-optimized HIV-1YU2 gp120 expression construct.
In the ΔV1/V2/V3 and 44-492 ΔV1/V2/V3 proteins, the truncations described above were present in combination.
2.3 Immunoprecipitation of envelope glycoproteins
For pulse-labeling experiments, 2 × 106 293T cells were cotransfected by the calcium phosphate method with pLTR-Tat, a plasmid expressing the HIV-1 Tat protein and the pSVIIIenv plasmid expressing the HIV-1YU2 envelope glycoproteins. Beginning one day after transfection, the cells were metabolically labeled for 16 hours with 100 μCi/mL [35S]-methionine/cysteine ([35S] protein labeling mix; Perkin-Elmer, Waltham, MA, USA) in Dulbecco's modified Eagle's medium lacking methionine and cysteine and supplemented with 5% dialyzed fetal bovine serum. For pulse-chase analysis, cells were metabolically labeled for 2 hours with 100 μCi/mL [35S]-methionine/cysteine in Dulbecco's modified Eagle's medium (DMEM) lacking methionine and cysteine and supplemented with 5% dialyzed fetal bovine serum; the cells were then chased for different time intervals in DMEM containing excess unlabeled methionine and cysteine. Cells were subsequently lysed in RIPA buffer (140 nM NaCl, 8 mM Na2HPO4, 2 mM NaH2PO4, 1% NP40, 0.05% sodium dodecyl sulfate (SDS)).
Precipitation of radiolabeled HIV-1 envelope glycoproteins from cell lysates or medium was performed with a mixture of sera from HIV-1-infected individuals. Alternatively, the radiolabeled gp120 envelope glycoprotein in the medium was precipitated with various amounts of anti-gp120 monoclonal antibodies or the recombinant CD4-Ig protein for 1 hour at 37°C in the presence of 70 μl of 10% Protein A-Sepharose (American BioSciences, Blauvelt, NY, USA).
2.4 CCR5 binding
To assess CCR5-binding ability, normalized amounts of radiolabeled gp120 envelope glycoproteins from transfected 293T cell supernatants were incubated in the presence or absence of 400 nM sCD4 for 1 hour at 37°C. This concentration of sCD4 greatly exceeds the Kd and therefore most of the monomeric and dimeric gp120 would likely be complexed to sCD4. The gp120-sCD4 mixtures were then incubated with 2 × 106 Cf2Th-CCR5 cells for 2 hours at 37°C. The cells were washed twice with PBS prior to lysis in RIPA buffer. The bound gp120 glycoproteins in the cell lysates were immunoprecipitated with a mixture of sera from HIV-1-infected individuals.
All precipitated proteins were boiled for 5 min before being analyzed on NuPAGE Novex Bis-Tris polyacrylamide gels (Invitrogen, Carlsbad, CA, USA), in absence or presence of 1.25% of β-mercaptoethanol (BioRad, Hercules, CA, USA) followed by autoradiography and quantification with a PhosphorImager (Molecular Dynamics, Pittsburgh, PA, USA).
3. Results
3.1 Expression of gp120 from 293T cells results in a mixture of monomeric and dimeric gp120
Numerous investigators studying HIV-1 Env glycoprotein structure and antigenicity have utilized a secreted recombinant form of the gp120 protein lacking gp41 sequences (Kang et al., 1994; Kwong et al., 2000; Kwong et al., 1998; Moore et al., 1994). The expression of gp120 alone is efficient and results in the secretion of high levels of the gp120 glycoprotein into the culture medium, facilitating purification. Interestingly, HIV-1 soluble gp120 glycoprotein overexpressed in 293T cells migrated at the expected size on SDS-polyacrylamide gels under reducing conditions (Figure 1A). However, under non-reducing conditions, an additional band appeared with a relative mobility above 150 kD, suggesting the presence of an oligomeric form of gp120.
Figure 1. Expression of gp120 in 293T cells results in a mixture of monomeric and dimeric gp120.
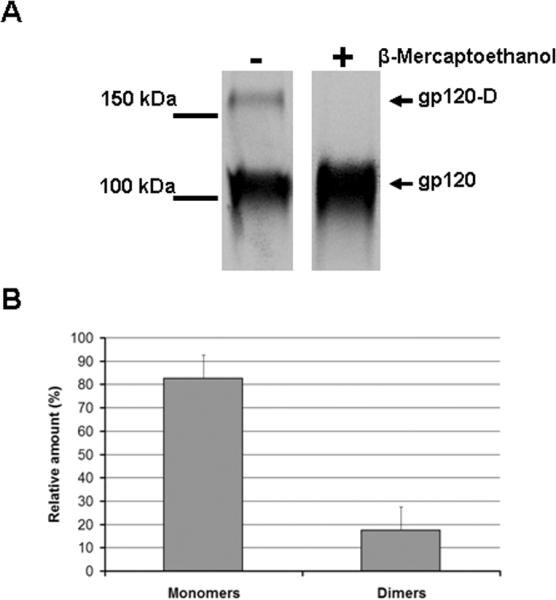
(A) A transfected 293T cell supernatant containing radiolabeled wild-type gp120 was incubated with a serum mixture from HIV-1-infected individuals for two hours at 37°C. Precipitates were analyzed by SDS-PAGE in the presence or absence of 5 mM β-mercaptoethanol followed by autoradiography/ densitometry. The result shown is representative of those obtained in at least three independent experiments. The gp120 dimer is designated gp120-D. (B) The bands presented in (A) were quantified by densitometry. Results are expressed as the percentage of each different form (monomers or dimers) relative to the total amount of gp120 (monomers + dimers). Data shown represent the means +/- SEM of two independent experiments.
To determine the oligomeric state of the gp120 glycoprotein in the 293T cell supernatant, the two bands observed under non-reducing conditions were analyzed by mass spectrometry. Of note, the name applied to gp120 reflects its relative mobility on SDS-polyacrylamide gels rather than its true mass. A mass between 89-92 kD was previously determined for monomeric gp120 by scanning transmission electron microscopy, mass spectrometry and gel filtration (Center et al., 2000; Thomas et al., 1991). Consistent with this analysis, the fast-migrating gp120 band had an estimated mass of 91.1 kD. The mass of the slow-migrating band was estimated to be 181.4 kD, indicating that this band represented a dimer of gp120 molecules. Quantification of the dimeric and monomeric forms of gp120 indicated that dimeric gp120 represented 15-20 % of the overall mixture (Figure 1B).
3.2 Dimeric gp120 is poorly recognized by CD4-induced antibodies
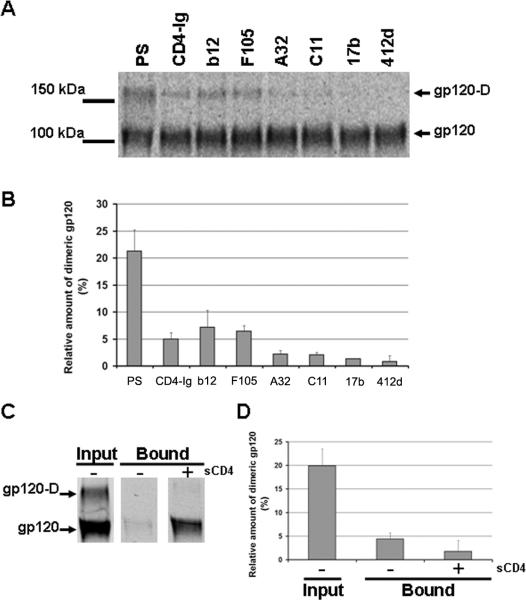
The appearance of dimeric gp120 on SDS-polyacrylamide gels was sensitive to β-mercaptoethanol treatment (reducing conditions) (Figure 1A above). As the non-reduced samples were boiled prior to loading, a treatment that denatures HIV-1 gp120 (McDougal et al., 1986), this observation suggests that dimer formation involves intermolecular disulfide bonds. As unliganded HIV-1 gp120 normally samples multiple conformations (Myszka et al., 2000; Kwong et al., 2002), such disulfide bonds might limit the conformational flexibility of gp120 and the access of particular antibodies to their epitopes. To test this possibility, secreted gp120 was immunoprecipitated with a panel of monoclonal antibodies that recognize different gp120 conformations, as well as with CD4-Ig (Figure 2A). CD4-Ig is a fusion protein in which the N-terminal two domains of CD4 are linked to the Fc component of human immunoglobulin G (Chowdhury et al., 1991). Monoclonal antibodies b12 and F105 recognize epitopes overlapping the CD4-binding site (CD4BS epitopes) (Thali et al., 1991; Zhou et al., 2007), whereas A32 and C11 recognize the gp120 inner domain (Boots et al., 1997; Moore and Sodroski, 1996; Wyatt et al., 1995). The binding of CD4-induced (CD4i) antibodies, which preferentially recognize the CD4-bound conformation (Trkola et al., 1996; Wu et al., 1996; Thali et al., 1993), was also tested. Only CD4-Ig and CD4-binding site (CD4BS) antibodies precipitated modest amounts of dimeric gp120, whereas antibodies directed against the gp120 inner domain or CD4i antibodies barely recognized the gp120 dimers (Figure 2A and B). These observations suggest that dimerization may occlude or disrupt the inner domain and/or the co-receptor binding site.
Figure 2. Dimeric gp120 is poorly recognized by CD4-induced antibodies and CCR5.
(A) A 293T cell supernatant containing radiolabeled wild-type gp120 was incubated with a polyclonal mixture of sera from HIV-1-infected individuals (PS) or 13 nM of monoclonal antibodies or CD4-Ig for two hours at 37°C. Precipitates were analyzed by SDS-PAGE without β-mercaptoethanol followed by autoradiography/densitometry. The result shown is representative of those obtained in two independent experiments. The gp120 dimer is designated gp120-D. (B) The gp120 bands detected in (A) were quantified by densitometry. Results are expressed as the percentage of dimeric gp120 relative to the total amount of gp120 (monomers + dimers). Data shown represent the means +/-SEM of two independent experiments. (C) Radiolabeled HIV-1YU2 gp120 glycoproteins in 293T cell supernatants were incubated in the absence or presence of 200 nM sCD4 prior to addition to Cf2Th cells expressing CCR5. After two hours at 37°C, the amount of input and bound gp120 was determined by immunoprecipitation with PS. Samples were analyzed by SDS-PAGE under non-reducing conditions. The gp120 dimer is designated gp120-D. (D) The gp120 bands detected in the experiment shown in (C) were quantified by densitometry. The percentage of dimeric gp120 in each sample relative to the total gp120 is shown. The data represent the means +/- SEM of two independent experiments.
To compare the ability of the two forms of gp120 to bind CCR5, monomeric and dimeric gp120 with or without a soluble form of CD4 (sCD4) were incubated CCR5-expressing cells (Figure 2C). In the absence of sCD4, only a small fraction of monomeric gp120 bound to CCR5-expressing cells; an even lower amount of dimeric gp120 bound CCR5 (Figures 2C and 2D). Importantly, whereas addition of sCD4 enhanced by more than ten-fold the binding of monomeric gp120 to CCR5, it had only a modest effect on the binding of the dimeric form. Altogether, these data suggest that dimerization disrupts or occludes gp120 structures on the inner domain and near the CCR5-binding site.
3.3 A residue in the inner domain affects dimer formation
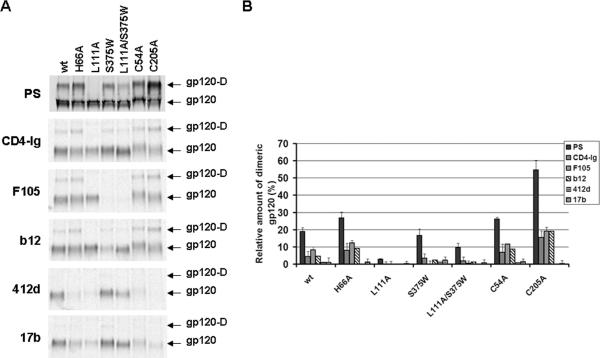
The possibility that gp120 inner domain changes, reported to alter the spontaneously sampling of the CD4-bound conformation (Kassa et al., 2009a,b; Finzi et al., 2010), may affect dimer formation was also addressed. As previously reported (Finzi et al., 2010), gp120 variants H66A and L111A exhibited slight decreases in recognition by CD4-Ig and substantial decreases in recognition by CD4i antibodies (Figure 3). The H66A change slightly increased the amount of dimers in the supernatant relative to the amount of monomers. By contrast, the replacement of leucine 111 with alanine almost completely abrogated dimer formation, implicating the gp120 inner domain in modulating the formation of secreted dimers.
Figure 3. Recognition of gp120 variants by CD4 and monoclonal antibodies.
(A) Comparable amounts of radiolabeled wild-type (WT) and mutant gp120 glycoproteins in transfected 293T cell supernatants were incubated with a polyclonal mixture of sera from HIV-1-infected individuals (PS), CD4-Ig, or different monoclonal antibodies (13 nM) for two hours at 37°C. Precipitates were analyzed by SDS-PAGE without β-mercaptoethanol followed by autoradiography/ densitometry. The results shown are representative of those obtained in two independent experiments. The gp120 dimer is designated gp120-D. (B) The gp120 bands observed in (A) were quantified by densitometry. Results are expressed as the percentage of dimeric gp120 relative to the total amount of gp120 (monomers + dimers). Data shown represent the means +/- SEM of two independent experiments.
The possibility that gp120 changes known to favor the spontaneous sampling of the CD4-bound conformation may affect dimer formation was also addressed. Serine 375 flanks the Phe 43 cavity; substitution of a tryptophan residue for serine 375 fills the Phe 43 cavity with the indole ring (Zhou et al., 2007). As a result, the S375W mutant favors conformation(s) closer to that of the CD4-bound state (Xiang et al., 2002). As previously reported (Xiang et al., 2002; Finzi et al., 2010), relative to wild-type gp120, the S375W mutant was recognized better by CD4-Ig and CD4i antibodies and less efficiently by CD4BS antibodies. However, the S375W change did not modify the relative amount of gp120 dimers. Nevertheless, when combined with the L111A change, the S375W change partially restored dimer formation. These results suggest that dimer formation can be modulated by the different conformations adopted by the gp120 glycoprotein.
An analysis of the gp120 dimers by mass spectrometry suggested that cysteines 54 and 205 in the inner domain may be involved in disulfide bonding during gp120 dimer formation (not shown). The individual replacement of these residues by alanine resulted in an enhancement of the relative amount of dimers (Figure 3). These results suggest that an unpaired cysteine residue in the gp120 inner domain can promote dimer formation. The C54A and C205A mutants were recognized by CD4-Ig and CD4BS antibodies, but not by CD4i antibodies. When cysteines 54 and 205 were simultaneously changed to alanine, the resulting mutant gp120 (C54A/C205A) still formed dimers (data not shown). Thus, at least in the absence of cysteines 54 and 205, other cysteines can contribute to the formation of disulfide bonds between secreted gp120 monomers.
To determine whether the dimers in the secreted gp120 preparations would return to a native conformation after reduction, supernatants of gp120-expressing 293T cells were treated with 0.5 and 5 mM β-mercaptoethanol, followed by dialysis. Although the amount of gp120 dimer in the preparation (as assessed by recognition by polyclonal sera from HIV-1-infected individuals) was reduced by β-mercaptoethanol treatment, the recognition of the total amount of gp120 in the preparation by the 17b antibody was also greatly reduced by the treatment (data not shown). Thus, β-mercaptoethanol treatment, although able to oxidize the disulfide bond(s) stabilizing the dimers, also resulted in significant disruption of the native conformation of gp120.
3.4 The gp120 dimers form intracellularly
To determine if the inter-gp120 contacts leading to dimer formation were made within the cell prior to secretion, transfected 293T cells were metabolically labeled for 2 hours and then chased for different time intervals in DMEM containing excess unlabeled methionine and cysteine. As shown in Figure 4, dimers were observed intracellularly from the first time point. Over time, a decrease in both monomeric and dimeric gp120 was observed within the cells, with a concomitant increase of both forms in the supernatant. Therefore, dimer formation occurs early in the process of gp120 folding inside the cell.
Figure 4. Intracellular gp120 dimer formation.
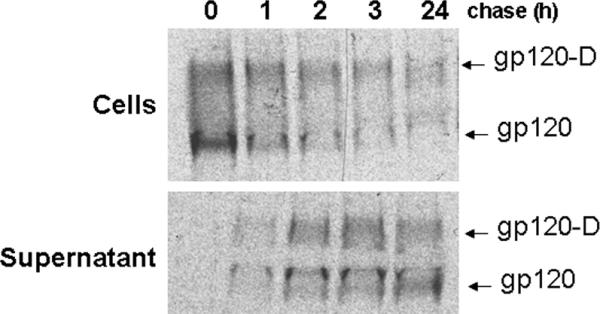
Transfected 293T cells were metabolically labeled for 2 hours and then chased for different time intervals in DMEM containing excess unlabeled methionine and cysteine. Cell lysates and supernatants were incubated with a mixture of sera from HIV-1-infected individuals. Precipitates were analyzed by SDS-PAGE without β-mercaptoethanol followed by autoradiography/ densitometry. The results shown are representative of those obtained in two independent experiments. The gp120 dimers are designated gp120-D.
3.5 The variable V1/V2/V3 loops and the N- and C-termini contribute to dimer formation
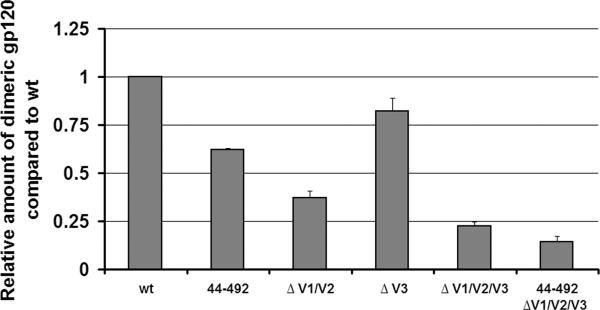
To identify gp120 regions that influence dimer formation, the N- and C-termini (residues 1-44 and 492-511) and the V1/V2 and V3 variable loops were deleted alone or in combination. The V1/V2 variable loops apparently make a significant contribution to dimer formation, as their removal decreased dimer formation by more than 60%. Deletion of the V3 loop had a smaller effect, with a 20% decrease in dimer formation. Finally, removal of the N- and C-termini decreased dimer formation by almost 40%. Importantly, these effects were additive, as a mutant lacking the V1/V2 and V3 variable loops and the N- and C- termini exhibited a decrease in dimer formation of more than 80%. Thus, different regions of the gp120 glycoprotein contribute to dimer formation.
4. Discussion
It has been shown previously that expression of the full-length HIV-1 gp160 envelope glycoprotein can result in the formation of oligomeric complexes, some of which were apparently native and others apparently aberrant (Owens and Compans, 1990). The aberrant oligomers were found to contain disulfide bonds between the individual gp160 subunits (Owens and Compans, 1990). Likewise, overexpression of the soluble HIV-1 gp140 glycoprotein in cells infected by recombinant vaccinia viruses resulted in disulfide-linked dimers of gp140 and cleaved gp120 (Hallenberger et al., 1993). This manuscript reports that over-expression of soluble HIV-1 gp120 also results in a substantial amount of dimer formation. The gp120 dimers appear to be stabilized by intermolecular disulfide bonds, as β-mercaptoethanol treatment disrupted the gel-stable dimers, whereas boiling in SDS sample buffer did not. Analysis of cell lysates and supernatants following pulse-labeling demonstrated that gp120 dimers formed intracellularly.
Interestingly, gp120 dimers were recognized efficiently by patient serum and, to a lesser extent, by CD4 and CD4-binding site (CD4BS) antibodies. Thus, the gp120 region in the vicinity of the CD4-binding site is still exposed to some extent in the context of gp120 dimers. The transitions of unliganded gp120 into the particular conformations recognized by CD4 and the CD4BS antibodies apparently can occur in the gp120 dimers (Myszka et al., 2000; Kwong et al., 2002). By contrast, gp120 dimers did not efficiently bind to cells expressing the CCR5 coreceptor and were poorly recognized by CD4i antibodies. Whereas addition of sCD4 increased the binding of monomeric gp120 to CCR5, this was not the case for dimeric gp120. Thus, one or more elements of the gp120 CCR5-binding region is apparently occluded and/or disrupted in the dimers. Finally, gp120 dimers were poorly recognized by two antibodies directed against the inner domain, suggesting that elements of the inner domain may likewise be occluded or disrupted by dimer association.
The results presented in this study implicate the gp120 inner domain, which interacts with the gp41 ectodomain in the assembled HIV-1 envelope glycoprotein trimer, in the formation of aberrant dimers. Mass spectrometric analysis of the gp120 dimers suggests that cysteines 54 and 205, which normally form an internal disulfide bond in the gp120 inner domain, may be involved in the disulfide bond linking the gp120 dimers. Alteration of these cysteines increased dimer formation, perhaps as a result of positioning an unpaired cysteine at the dimer interface. However, when both cysteines 54 and 205 were changed to alanine, disulfide-linked dimers of gp120 still formed. Thus, at least in this context, other cysteines can participate in the formation of the disulfide bond(s) that stabilize the gp120 dimers. For example, cysteine residues located in the V1/V2 stem-loop structure may participate in intermolecular disulfide bonds during the formation of dimers, as the deletion of this structure significantly reduced the appearance of gp120 dimers. This is consistent with a previous report suggesting the involvement of the V2 loop in the formation of gp120 dimers (Center et al., 2000).
It has been reported recently that the transition from the unliganded to the CD4-bound state is regulated by two potentially flexible topological layers (“Layers 1 and 2”) in the gp120 inner domain (Finzi et al., 2010). The interaction between these layers contributes to the ability of HIV-1 gp120 to sample the CD4-bound conformation. The alteration of one Layer 2 residue, leucine 111, involved in the Layer 1-Layer 2 interaction decreased dramatically dimer formation, whereas changes in Layer 1 residues (histidine 66 and tryptophan 69) that also contribute to the Layer 1-Layer 2 interaction did not. Thus, the inner domain Layer 1-Layer 2 interaction per se is not required for gp120 dimer formation. However, leucine 111 is involved in a network of interactions that stabilize gp120 association with the unliganded trimer (Finzi et al., 2010; Xiang et al., 2010). This network involves gp120 regions that are also involved in the transition to a conformational state that is competent for chemokine receptor binding. The gp120 conformation that binds CCR5 and that is recognized by CD4i antibodies is very sensitive to disruption (Thali, 1993). Indeed, inner domain alterations involving leucine 111 have been shown to decrease the binding of CCR5 and CD4i antibodies (Finzi et al., 2010). Taken together, these results suggest that inner domain interactions are involved in dimer formation, which results in disruption and/or occlusion of the gp120 regions involved in coreceptor binding. Perhaps some of the hydrophobic interactions that normally exist between gp120 and gp41 in the Env glycoprotein trimer contribute to the interactions that promote gp120 dimer formation.
5. Conclusions
In summary, this manuscript reports that expression of gp120 in the absence of gp41 results in the formation of stable dimers within the cells; these disulfide-linked dimers are then secreted into the supernatant. Dimers represent a substantial fraction of the overall secreted gp120 and exhibit differences in the conformation and/or accessibility of certain surfaces compared with monomeric gp120. Consequently, awareness of the secreted gp120 dimers is important for interpreting biochemical, biophysical and antigenic analyses of secreted gp120 glycoproteins. Therefore, samples should be analyzed under non-reducing conditions by SDS-PAGE when assessing different gp120 conformations, in an effort to distinguish between native monomeric gp120 and aberrant disulfide-linked dimers. For some studies, purification of the native monomeric gp120 glycoprotein is desirable. Reduction and dialysis proved damaging to the native conformation of a significant fraction of the gp120 preparation. Instead, immunoaffinity purification using CD4i antibodies, which do not recognize the dimers, represents an attractive option for obtaining native gp120 monomers.
Figure 5. Effect of deletion of the gp120 N/C-termini and variable loops on dimer formation.
293T cells were transfected with plasmids expressing wild-type (wt) HIV-1YU2 gp120, the 44-492 gp120, or gp120 protein with deletions of the V1/V2 and/or V3 variable loops. Comparable amounts of radiolabeled wild-type (wt) and mutant gp120 glycoproteins in transfected 293T cell supernatants were incubated with a polyclonal mixture of sera from HIV-1-infected individuals for two hours at 37°C. Precipitates were analyzed by SDS-PAGE without β-mercaptoethanol followed by autoradiography/ densitometry. The results shown are representative of those obtained in four independent experiments and are normalized to the amount of dimer observed for wt gp120.
Acknowledgements
The authors would like to thank Ms. Yvette McLaughlin and Ms. Elizabeth Carpelan for manuscript preparation. This work was supported by grants from the National Institutes of Health (AI24755, GM56550 and AI67854), by the International AIDS Vaccine Initiative, and by the late William F. McCarty-Cooper. The authors have no conflicts of interest to report.
Abbreviations
- Env
envelope
- sCD4
soluble CD4
- CD4i antibody
CD4-induced antibody
- CD4BS antibody
CD4-binding site antibody
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–8. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- Allan JS, Coligan JE, Barin F, McLane MF, Sodroski JG, Rosen CA, Haseltine WA, Lee TH, Essex M. Major glycoprotein antigens that induce antibodies in AIDS patients are encoded by HTLV-III. Science. 1985;228:1091–4. doi: 10.1126/science.2986290. [DOI] [PubMed] [Google Scholar]
- Boots LJ, McKenna PM, Arnold BA, Keller PM, Gorny MK, Zolla-Pazner S, Robinson JE, Conley AJ. Anti-human immunodeficiency virus type 1 human monoclonal antibodies that bind discontinuous epitopes in the viral glycoproteins can identify mimotopes from recombinant phage peptide display libraries. AIDS Res Hum Retroviruses. 1997;13:1549–59. doi: 10.1089/aid.1997.13.1549. [DOI] [PubMed] [Google Scholar]
- Center RJ, Earl PL, Lebowitz J, Schuck P, Moss B. The human immunodeficiency virus type 1 gp120 V2 domain mediates gp41-independent intersubunit contacts. J Virol. 2000;74:4448–55. doi: 10.1128/jvi.74.10.4448-4455.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–73. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–48. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- Chowdhury IH, Koyanagi Y, Takamatsu K, Yoshida O, Kobayashi S, Yamamoto N. Evaluation of anti-human immunodeficiency virus effect of recombinant CD4-immunoglobulin in vitro: a good candidate for AIDS treatment. Med Microbiol Immunol. 1991;180:183–92. doi: 10.1007/BF00215247. [DOI] [PubMed] [Google Scholar]
- Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF, Weiss RA. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–7. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton RE, Hill CM, Davis CB, Peiper SC, Schall TJ, Littman DR, Landau NR. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–6. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- Doms RW, Earl PL, Moss B. The assembly of the HIV-1 env glycoprotein into dimers and tetramers. Adv Exp Med Biol. 1991;300:203–19. doi: 10.1007/978-1-4684-5976-0_13. discussion 220-1. [DOI] [PubMed] [Google Scholar]
- Doranz BJ, Rucker J, Yi Y, Smyth RJ, Samson M, Peiper SC, Parmentier M, Collman RG, Doms RW. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–58. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, Cayanan C, Maddon PJ, Koup RA, Moore JP, Paxton WA. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–73. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- Earl PL, Doms RW, Moss B. Oligomeric structure of the human immunodeficiency virus type 1 envelope glycoprotein. Proc Natl Acad Sci U S A. 1990;87:648–52. doi: 10.1073/pnas.87.2.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzan M, Choe H, Desjardins E, Sun Y, Kuhn J, Cao J, Archambault D, Kolchinsky P, Koch M, Wyatt R, Sodroski J. Stabilization of human immunodeficiency virus type 1 envelope glycoprotein trimers by disulfide bonds introduced into the gp41 glycoprotein ectodomain. J Virol. 1998;72:7620–5. doi: 10.1128/jvi.72.9.7620-7625.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–7. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- Finzi A, Xiang S-H, Pacheco B, Wang L, Haight J, Kassa A, Danek B, Pancera M, Kwong PD, Sodroski J. Topological layers in the HIV-1 gp120 inner domain regulate gp41 interaction and CD4-triggered conformational transitions. Molecular Cell. 2010;37:656–67. doi: 10.1016/j.molcel.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallenberger S, Tucker SP, Owens RJ, Bernstein HB, Compans RW. Secretion of a truncated form of the human immunodeficiency virus type 1 envelope glycoprotein. Virology. 1993;193:510–4. doi: 10.1006/viro.1993.1156. [DOI] [PubMed] [Google Scholar]
- Helseth E, Olshevsky U, Furman C, Sodroski J. Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J Virol. 1991;65:2119–23. doi: 10.1128/jvi.65.4.2119-2123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang CY, Hariharan K, Nara PL, Sodroski J, Moore JP. Immunization with a soluble CD4-gp120 complex preferentially induces neutralizing anti-human immunodeficiency virus type 1 antibodies directed to conformation-dependent epitopes of gp120. J Virol. 1994;68:5854–62. doi: 10.1128/jvi.68.9.5854-5862.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassa A, Finzi A, Pancera M, Courter JR, Smith AB, 3rd, Sodroski J. Identification of a Human Immunodeficiency Virus (HIV-1) Envelope Glycoprotein Variant Resistant to Cold Inactivation. J Virol. 2009a doi: 10.1128/JVI.02110-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassa A, Madani N, Schon A, Haim H, Finzi A, Xiang SH, Wang L, Princiotto A, Pancera M, Courter J, Smith AB, 3rd, Freire E, Kwong PD, Sodroski J. Transitions to and from the CD4-bound conformation are modulated by a single-residue change in the human immunodeficiency virus type 1 gp120 inner domain. J Virol. 2009b;83:8364–78. doi: 10.1128/JVI.00594-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman JC, Montagnier L. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature. 1984;312:767–8. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- Korber B, Foley BT, Kuiken C, Pillai SK, Sodroski JG. Numbering Positions in HIV Relative to HXB2CG. Human Retroviruses and AIDS. 1998:III, 102–111. [Google Scholar]
- Kwong PD, Wyatt R, Majeed S, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure. 2000;8:1329–39. doi: 10.1016/s0969-2126(00)00547-5. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–59. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt S, Majeed S, Steenbeke T, Venturi M, Chaiken I, Fung M, Katinger H, Parren P, Robinson J, Wang L, Burton DR, Freire E, Wyatt R, Sodroski J, Hendrickson WA, Arthos J. HIV-1 evades neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–82. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- Lu M, Blacklow SC, Kim PS. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat Struct Biol. 1995;2:1075–82. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- McDougal JS, Kennedy MS, Sligh JM, Cort SP, Mawle A, Nicholson JKA. Binding of HTLV-III/LAV to T4+ T cells by a complex of the 110K viral protein and the T4 molecules. Science. 1986;231:382–5. doi: 10.1126/science.3001934. [DOI] [PubMed] [Google Scholar]
- Moore JP, Sattentau QJ, Wyatt R, Sodroski J. Probing the structure of the human immunodeficiency virus surface glycoprotein gp120 with a panel of monoclonal antibodies. J Virol. 1994;68:469–84. doi: 10.1128/jvi.68.1.469-484.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JP, Sodroski J. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J Virol. 1996;70:1863–72. doi: 10.1128/jvi.70.3.1863-1872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myszka D, Sweet RW, Hensley P, Brigham-Burke M, Kwong PD, Hendrickson WA, Wyatt R, Sodroski J, Doyle M. Energetics of the HIV gp120-CD4 binding reaction. Proc Natl Acad Sci USA. 2000;97:9026–31. doi: 10.1073/pnas.97.16.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens RJ, Compans RW. The human immunodeficiency virus type 1 envelope glycoprotein precursor acquires aberrant intermolecular disulfide bonds that may prevent normal proteolytic processing. Virology. 1990;179:827–33. doi: 10.1016/0042-6822(90)90151-g. [DOI] [PubMed] [Google Scholar]
- Robey WG, Safai B, Oroszlan S, Arthur LO, Gonda MA, Gallo RC, Fischinger PJ. Characterization of envelope and core structural gene products of HTLV-III with sera from AIDS patients. Science. 1985;228:593–5. doi: 10.1126/science.2984774. [DOI] [PubMed] [Google Scholar]
- Schawaller M, Smith GE, Skehel JJ, Wiley DC. Studies with crosslinking reagents on the oligomeric structure of the env glycoprotein of HIV. Virology. 1989;172:367–9. doi: 10.1016/0042-6822(89)90142-6. [DOI] [PubMed] [Google Scholar]
- Thali M, Olshevsky U, Furman C, Gabuzda D, Posner M, Sodroski J. Characterization of a discontinuous human immunodeficiency virus type 1 gp120 epitope recognized by a broadly reactive neutralizing human monoclonal antibody. J Virol. 1991;65:6188–93. doi: 10.1128/jvi.65.11.6188-6193.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DJ, Wall JS, Hainfeld JF, Kaczorek M, Booy FP, Trus BL, Eiserling FA, Steven AC. gp160, the envelope glycoprotein of human immunodeficiency virus type 1, is a dimer of 125-kilodalton subunits stabilized through interactions between their gp41 domains. J Virol. 1991;65:3797–803. doi: 10.1128/jvi.65.7.3797-3803.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Dragic T, Arthos J, Binley JM, Olson WC, Allaway GP, Cheng-Mayer C, Robinson J, Maddon PJ, Moore JP. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–7. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–30. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- Wu L, Gerard NP, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso AA, Desjardins E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–83. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- Wyatt R, Moore J, Accola M, Desjardins E, Robinson J, Sodroski J. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J Virol. 1995;69:5723–33. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang SH, Kwong PD, Gupta R, Rizzuto CD, Casper DJ, Wyatt R, Wang L, Hendrickson WA, Doyle ML, Sodroski J. Mutagenic stabilization and/or disruption of a CD4-bound state reveals distinct conformations of the human immunodeficiency virus type 1 gp120 envelope glycoprotein. J Virol. 2002;76:9888–99. doi: 10.1128/JVI.76.19.9888-9899.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Tomov V, Kurteva S, Wang L, Ren X, Gorny MK, Zolla-Pazner S, Sodroski J. Characterization of the outer domain of the gp120 glycoprotein from human immunodeficiency virus type 1. J Virol. 2004;78:12975–86. doi: 10.1128/JVI.78.23.12975-12986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–7. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Chertova E, Bess J, Jr., Lifson JD, Arthur LO, Liu J, Taylor KA, Roux KH. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc Natl Acad Sci U S A. 2003;100:15812–7. doi: 10.1073/pnas.2634931100. [DOI] [PMC free article] [PubMed] [Google Scholar]