Abstract
Amelogenins are the dominant proteins present in ameloblasts during the early stages of enamel biomineralization, making up >90% of the matrix protein. Along with the full-length protein there are several splice-variant isoforms of amelogenin present including LRAP (Leucine-Rich Amelogenin Protein), a protein that consists of the first 33 and the last 26 residues of full-length amelogenin. Using solution-state NMR spectroscopy we have assigned the 1H-15N HSQC spectrum of murine LRAP (rp(H)LRAP) in 2% acetic acid at pH 3.0 by making extensive use of previous chemical shift assignments for full-length murine amelogenin (rp(H)M180). This correlation was possible because LRAP, like the full-length protein, is intrinsically disordered under these solution conditions. The major difference between the 1H-15N HSQC spectra of rp(H)M180 and rp(H)LRAP was an additional set of amide resonances for each of the seven non-proline residues between S12* and Y12 at the N-terminus of rp(H)LRAP indicating that the N-terminal region of LRAP exists in two different conformations. Analysis of the proline carbon chemical shifts suggest that the molecular basis for the two states is not a cis-trans isomerization of one or more of the proline residues in the N-terminal region. Starting from 2% acetic acid, where rp(H)LRAP was monomeric in solution, NaCl addition effected residue specific changes in molecular dynamics manifested by the reduction in intensity and disappearance of 1H-15N HSQC cross peaks. As observed for the full length protein, these perturbations may signal early events governing supramolecular self-assembly of rp(H)LRAP into nanospheres. However, the different pattern of 1H-15N HSQC cross peak perturbation between rp(H)LRAP and rp(H)M180 in high salt suggest that the termini may behave differently in their respective nanospheres, and perhaps, these differences contribute to the cell signaling properties attributable to LRAP but not the full-length protein.
Keywords: LRAP, intrinsic disorder, nanosphere self-assembly, amelogenesis, enamel, NMR spectroscopy, dynamic light scattering, biomineralization, cell signaling
1. Introduction
The 1-2 mm of dental enamel on the outer surface of the tooth is the hardest tissue in the bodies of vertebrates [1]. Its strength is necessary because the primary function of teeth is to masticate food in the bacteria-filled environment of the mouth and enamel cannot undergo self-repair or remodeling. The robust mechanical properties of enamel are due to a combination of high mineral content and unique three-dimensional architecture [2]. Ninety-five percent of mature enamel consists of long and narrow crystals of carbonated hydroxyapatite packed into parallel arrays, called enamel rods or prisms, that are intricately interwoven into an unique lattice [3, 4].
While there is little, if any, protein in mature dental enamel, the nucleation, growth, and organization of enamel (amelogenesis) occurs in a protein-filled extracellular matrix composed predominantly of one protein, amelogenin [4-8]. The importance of amelogenin in the biomineralization process is demonstrated by amelogenin-null mice and specific mutations to amelogenin that both result in dramatic enamel phenotypes similar to amelogenesis imperfecta [4], a group of genetic disorders that results in malformed enamal [9, 10].
A key component in directing control of enamel crystallites appears to be amelogenin's ability to self-assemble into a unique quaternary structure, 40 - 100 nm in diameter, called a nanosphere. Such nanospheres have been observed both in vivo [7] and in vitro [6] and mutants of amelogenin that disrupt nanosphere formation in vitro [11] are observed to cause malformed enamel in knock-in mice in vivo [12] supporting the importance of this quaternary structure in enamel biomineralization. It is hypothesized that nanosphere assembly occurs through the progressive association of amelogenin molecules with monomers first associating into dimer and trimer building blocks [13, 14]. The self-association is a complex process and depends on the interplay between protein concentration and the many properties of the solution (ionic strength, pH, solutes, and temperature) [14-17] that, in turn, affect the physical properties of the protein (hydrophobicity and electrostatics) [4, 18, 19].
Yeast two-hybrid assays [20] suggest that the regions of the protein essential in nanosphere self-assembly are near the N- and C-termini, M1-M42 and S157-K173, respectively. Further evidence for the importance of the amino- and carboxy-terminal regions in nanosphere formation is the high conservation of these regions among species [21] and solution NMR studies tracking the self-assembly process [22]. It has been postulated that these termini are exposed on the surface of amelogenin nanopheres [6, 12, 16, 20], increasing the nanosphere's solubility and enhancing its interactions with calcium phosphate [23]. Support for these hypotheses is the demonstration that removing the C-terminus results in a reduced affinity for hydroxyapatite and a reduction in the ability to inhibit crystal growth [24, 25].
The 59-residue amelogenin LRAP (Leucine-Rich Amelogenin Protein), composed of the first 33 and last 26 residues of full length amelogenin, is a naturally occurring result of alternative splicing of the primary transcript [10]. The biological role of LRAP in enamel formation has not been established, however, it has been proposed to be involved in both biomineralization [26] and cell signaling events [27-29]. Evidence for a role in biomineralizaton includes the observation that LRAP shares many biological properties with full-length amelogenin including the ability to form nanospheres at high protein concentrations [19, 30] and associate intimately with the surface of hydroxyapatite [31, 32]. Furthermore, while LRAP cannot completely rescue the amelogenin null phenotype [33], enamel crystals generated in the presence of LRAP are more organized [34] and another study using explant mouse molars and amelogenin knock-out mice showed increased enamel width in the presence of LRAP [35]. Evidence for its role in cell signaling are observations that show LRAP enhances osteoblastic cell differentiation in mouse embryonic stem cells [29] and other cell types [27, 28]. Unfortunately, there is little detailed structural information available for LRAP that may shed light into its potential biological roles.
To better understand the structure of LRAP in solution, we have used solution NMR spectroscopy and dynamic light scattering [36] to probe the behavior of the mouse splice-variant of amelogenin, rp(H)LRAP [37], as a function of increasing ionic strength using the salt sodium chloride. To conduct these studies we assigned the 1H, 13C, and 15N chemical shifts for rp(H)LRAP under acidic conditions, where it behaves as a monomer in solution, with the assistance of the chemical shift assignments for the full-length protein [38]. Consequently, it was also possible to identify the specific residues involved in the early stages of nanosphere assembly in rp(H)LRAP by following perturbations to their amide chemical shifts in the 1H-15N HSQC spectrum as a function of NaCl concentration. Such perturbations, typically chemical shift and/or intensity reductions to the backbone 1HN and 15N resonances [22, 39], reflect changes in the chemical environment of nuclei at interaction interfaces. By comparing the spectral properties of rp(H)LRAP with rp(H)M180 [17], under conditions where they behave as monomers in solution (2% acetic acid, pH 3) and large molecule weight complexes (2% acetic acid plus excess NaCl, pH 3.0), it may be possible to extract insights into structural differences that may account for some of the functional differences between the two proteins.
2. Materials and methods
All chemicals and enzymes were purchased from the Sigma Chemical Company (St. Louis, MI) except when indicated.
2.1 Protein expression and purification
Recombinant splice-variant mouse leucine-rich amelogenin protein (rp(H)LRAP) [40] was expressed and purified from Escherichia coli BL21(DE3) cells using methods previously described for recombinant amelogenin [38] and exchanged into the final NMR buffer (2% CD3CO2D, 5% D2O/95% H2O, pH 3.0) [17] used as the starting point for the NaCl titration study and chemical shift assignments. The protein was prepared 13C- and 15N-labelled with a 12-residue, unremovable, N-terminal histidine tag (MRGSHHHHHHGS-) to assist protein purification. The residues for the N-terminal affinity tag are numbered 1 thro,ugh 12 with an asterisk and the rp(H)LRAP residues are numbered sequentially using the numbering scheme from rp(H)M180 (P2-P33 and P166-D180) to facilitate comparison with the full-length protein.
2.2 NMR data collection and processing
The NMR data on the double-labeled sample (0.6 mM) was collected at 293 K using Varian Inova-600, and –750 spectrometers equipped with triple resonance probes, cyroprobes (Inova-600), and pulse field gradients. Pulse sequences for the two- and three-dimensional experiments were from Protein Pack. Three-dimensional HNCACB and CCC-TOCSY data were collected to verify amide chemical shift assignments based primarily on comparison to the 1H-15N HSQC spectrum of previously assigned rp(H)M180. For the NaCl titration study, a stock solution of 1 M NaCl was prepared in NMR buffer and the pH measured to assure that there was no significant variation from the starting condition. Aliquots of the sodium chloride solutions were added directly to the rp(H)LRAP sample (0.6 mM (260 μL)) and following gentle agitation a high resolution 1H-15N HSQC spectrum (20°C) was immediately acquired at a 1H resonance frequency of 600 MHz. Spectra were recorded in the absence of salt and at NaCl:protein molar ratios of 120, 240, 420, 540, 1050 and 1260 to one. At the end of the titration, due to dilution, the final concentration of rp(H)LRAP was 0.35 mM with 440 mM NaCl in a final volume of 470 μL. All NMR data was processed using Felix2007 (Felix NMR, Inc, San Diego, CA) and Sparky [41] software and the 1H, 13C, and 15N chemical shifts were referenced to DSS (DSS = 0 ppm) using indirect methods [42].
2.3 Dynamic light scattering (DLS)
The DLS measurements were obtained at 25°C on the rp(H)LRAP NMR sample at the final NaCl titration point using a Brookhaven Instruments 90 Plus (Brookhaven Instruments Corporation, Holtsville, NY) equipped with a 657 nm 35 mW laser. Data was also collected in 2% acetic acid, pH 3.0, on a synthetic, unlabelled murine LRAP sample that lacked the 12-residue N-terminal tag (synthesized and purified by the University of Texas, Protein Chemistry Technology Center). Protein solutions were analyzed in triplicate using a 90° scattering angle with acquisition times of 1 to 3 minutes. Standard NIST traceable polystyrene 92 nm ± 3.7 nm latex standards and a blank, 0.02 μm filtered DI ultrapure water (VWR International, Wester Chester, PA) were run as standards. The autocorrelation functions were deconvoluted to obtain size distributions using both the non-negatively constrained least squares fit (multiple pass NNLS).
3. Results and discussion
3.1 Amide chemical shift assignments
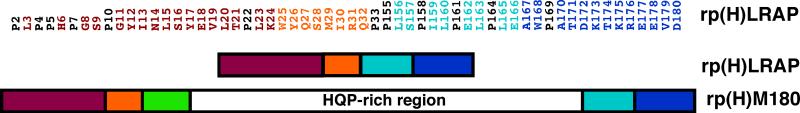
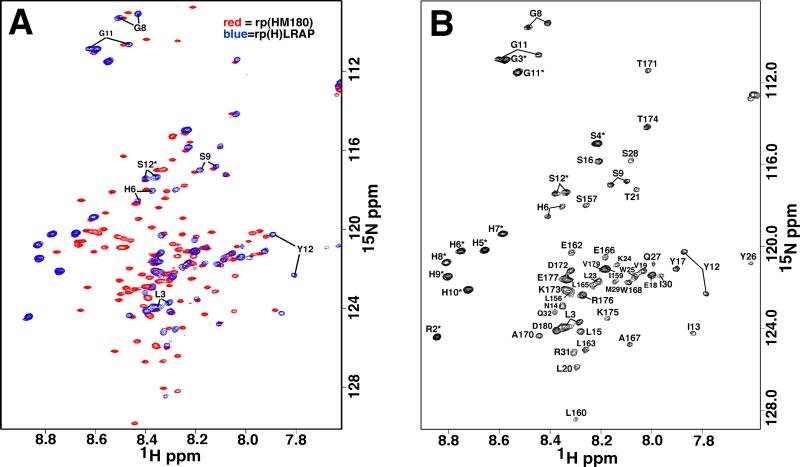
As shown in Figure 1, the amino acid sequence of rp(H)LRAP is identical to that of rp(H)M180 except for the absence of the lectin-like binding tri-tyrosine domain (green) and the central HQP-rich region (white), residues Y34-Q165. Because rp(H)M180 itself is intrinsically disordered in 2% acetic acid at pH 3.0 [38], it was anticipated that removing the central region to generate rp(H)LRAP would have little effect on the remaining amide chemical shifts in the splice-variant under the same solution conditions. Such an observation was previously observed with N- and C-terminal truncated constructs of another intrinsically disordered protein, human high mobility group protein A (HMGA1) [43]. This turned out to be true for amelogenin as well with the 1H-15N HSQC spectrum of rp(H)LRAP overlaying well with the 1H-15N HSQC spectrum of rp(H)M180 as shown in Figure 2A. However, where the truncated constructs of HMGA1 were used to assist the amide chemical shift assignment of full-length HMGA1 [43], here we used the assignments of full-length rp(H)M180 to assist in the assignment of the smaller rp(H)LRAP. Most of the amide resonances of rp(H)LRAP were directly assigned by comparison with the 1H-15N HSQC spectrum of rp(H)M180. To verify these amide assignments, the corresponding side chain carbon chemical shifts were checked in 3D HNCACB, and CCC-TOCSY spectra. The completely assigned 1H-15N HSQC spectrum for rp(H)LRAP is shown in Figure 2B.
Fig. 1.
Amino acid sequence of full-length amelogenin rp(H)M180 and the natural splice variant rp(H)LRAP. Underneath the sequence for rp(H)LRAP is a schematic illustration of identified regions in the proteins [19]. The N-terminal may be divided into three components: magenta = protein-protein interaction region, orange = linker region, and green = lectin-like binding tri-tyrosine domain. The C-terminal contains two regions: cyan = hydrophobic segment cleaved by enamelysin, and blue = C-terminal mineral-binding domain. The central majority of protein is enriched in the amino acids P, L, H, and Q and called the HQP-rich region. The two proteins are identical except for the absence of the lectin-like binding tri-tyrosine domain (green) and HQP-rich region in rp(H)LRAP.
Fig. 2.
A) Overlay of the 1H-15N HSQC spectrum of rp(H)M180 (red) and rp(H)LRAP (blue) under similar conditions (2% acetic acid, pH 3.0, 20°C) recorded at a 1H resonance frequency of 750 (rp(H)LRAP) and 900 (rp(H)M180) MHz. B) Assignment of the amide resonance in the 1H-15N HSQC spectra of rp(H) LRAP. In both figures the solid lines identify the two sets of cross peaks observed for the amide resonances between S12* and Y12.
While the overlap of the 1H-15N HSQC spectra of rp(H)M180 with rp(H)LRAP strongly suggest that both proteins are similarly disordered in solution, there was one noticeable difference between the 1H-15N HSQC spectra of both proteins. As shown by the connecting solid lines in Figure 2, there is a duplication of the amide cross peaks for all the non-proline residues between S12* and Y12 in the 1H-15N HSQC spectrum of rp(H)LRAP. Heating the protein to 50°C failed to collapse these duplicate resonances into a single set, suggesting the observation is not due to a self-association equilibrium (eg: monomer □ dimer). Such a duplication of resonances was not observed in the full-length protein (rp(H)M180) and suggests that the N-terminus of rp(H)LRAP is in slow exchange (>ms) between two different conformations [22, 44]. A duplication of a subset of four amide cross peaks was reported for full-length porcine amelogenin although this was for residues in the HQP-rich region that are not present in the LRAP splice-variant [45]. The presented explanation for the duplicate set of cross peak in porcine amelogenin was a cis-trans conformational exchange of proline residues near these duplicate resonances. Evidence for the cis-trans conformational exchange was the chemical shift difference between the 13Cβ and 13Cγ proline carbons (Δβγ = σ13Cβ – σ13Cγ) [46]. Typically, Δβγ = 4.5 ± 1.2 ppm and 9.6 ± 1.3 ppm for the trans-proline and cis-proline conformations, respectively. As shown in Table 1, the Δβγ values for 10 of the 11 proline residues in rp(H)LRAP are characteristic of the trans-proline conformation (values for P33 could not be obtained because the residue C-terminal to it is P155). Indeed, the absolute chemical shift values for all the proline 13Cβ and 13Cγ resonances are characteristic of the trans-proline conformation [46]. Hence, unless it is a cis-trans conformational exchange at P33 or P4, the only prolines for which chemical shift assignments could not be made, then the conformational exchange that gives rise to the duplication of the resonances between S12* and Y12 must be due to something else. While the reason for this conformational exchange is not known, its absence at the N-terminus of full-length amelogenin (Figure 2A) may be one of the physical properties responsible for some of the subtle biochemical differences between LRAP and the full-length protein such as the cell signaling capacity associated with rp(H)LRAP but not rp(H)M180. Further investigations at more physiologically relevant conditions together with repeated observations in the corresponding protein from different species are necessary to further evaluate such structure-function correlations.
Table 1.
Carbon chemical shifts of the proline 13Cβ and 13Cγ atoms of rp(H)LRAP in 2% acetic acid, pH 3.0 (20°C)1.
| Proline Residue | 13Cβ | 13Cγ | Δ β γ |
|---|---|---|---|
| P2 | 32.0 | 27.1 | 4.9 |
| P2 | 31.8 | 27.1 | 4.7 |
| P4 | ---- | ---- | ---- |
| P5 | 31.9 | 27.1 | 4.8 |
| P5 | 31.8 | 27.1 | 4.7 |
| P7 | 31.8 | 27.3 | 4.5 |
| P7 | 31.8 | 27.1 | 4.7 |
| P10 | 31.8 | 27.1 | 4.7 |
| P10 | 31.8 | 27.1 | 4.7 |
| P22 | 31.8 | 27.2 | 4.6 |
| P33 | ---- | ---- | ---- |
| P155 | 31.8 | 27.0 | 4.8 |
| P158 | overlap | 27.1 | ---- |
| P161 | 31.8 | 27.1 | 4.7 |
| P164 | 31.8 | 27.2 | 4.6 |
| P169 | 31.9 | 27.2 | 4.7 |
Where values are provided for two proline residues, the upper value is for the downfield amide resonance of the duplicate pair.
3.2 Sodium chloride titrations
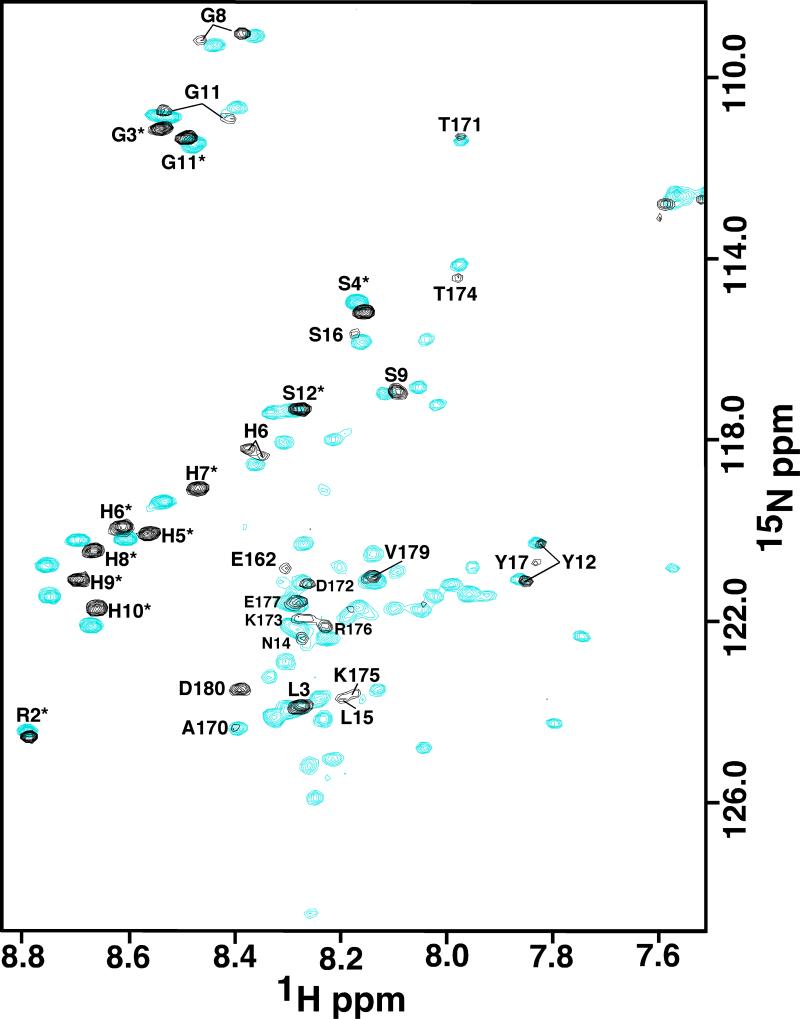
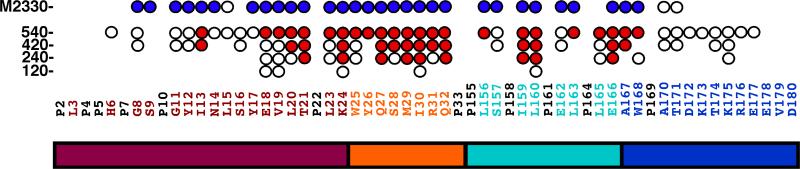
Previously, it was shown that adding increasing amounts of NaCl or CaCl2 to rp(H)M180 effected a stepwise self-assembly of amelogenin into larger units [17]. This transition was monitored by the disappearance of amide cross peaks in the 1H-15N HSQC of rp(H)M180 as a function of increasing salt concentration. Because LRAP has also been observed to form nanospheres in solution [19, 30], the same NMR titration experiment was performed with rp(H)LRAP and NaCl to determine if LRAP behaved similarly with increasing salt concentration. Figure 3 is an overlay of the 1H-15N HSQC spectra of rp(H)LRAP in the absence (cyan) and in the presence of 540:1 molar excess NaCl (black). As observed with rp(H)M180, a large number of the amide cross peaks in the 1H-15N HSQC spectrum of rp(H)LRAP disappear in the presence of excess NaCl. For the few cross peaks that remain, most show slight chemical shift perturbation, although these perturbations are not significant given the narrow chemical shift scale, approximately one ppm, in the proton dimension. Increasing the salt concentration further, to a 1260:1 molar ratio, had little further effect on the 1H-15N spectrum (data not shown). Because the 1H-15N HSQC spectrum of rp(H)LRAP has been assigned in the absence of salt (Figure 2) and the chemical shift perturbations to the spectra following the addition of salt are small, it is possible to track the sequential disappearance of amide cross peaks as a function of the molar ratio of salt present and these assignments are shown in Figure 3 and summarized in Figure 4. As described for rp(H)M180, the disappearance of amide cross peaks in the 1H-15N HSQC spectrum at high salt concentrations likely reflects restricted motion at a protein-protein interface [17].
Fig. 3.
Overlay of the 1H-15N HSQC spectrum of rp(H)LRAP in the absence of salt (cyan) and in the presence of a 540:1 molar ratio of NaCl:rp(H)LRAP. The tentative assignments of the amide resonances still remaining at the high salt concentration are shown.
Fig. 4.
Summary of the results of the titration of NaCl into a solution of rp(H)LRAP in 2% acetic acid. The amide resonances that start to disappear or completely disappear in the 1H-15N HSQC spectrum of 15N-labelled rp(H)LRAP are indicated by open and red circles, respectively. The NaCl:rp(H)LRAP molar ratios are indicated on the left. The top row (M2330) is the previously published results for rp(H)M180 at a NaCl:rp(H)M180 molar ratio of 2330:1 with the amide resonances that have begun to disappear or completely disappear in the 1H-15N HSQC indicated by open and blue circles, respectively [17]. Underneath is a schematic illustration of the various regions of LRAP [19] (magenta = protein-protein interaction region, orange = linker, cyan = hydrophobic segment cleaved by enamelysin, blue = hydrophilic C-terminal mineral-binding domain).
The disappearing amide cross peaks in the 1H-15N HSQC spectrum of rp(H)LRAP can be understood in relation to the solution-state NMR timescale of motion [22, 44]. In the fast NMR timescale (<μs) nuclei undergo rapid changes in their local chemical environment and generate observed resonances that represent the weighted average of all chemical environments sampled by the nuclei. In the slow NMR timescale (>ms) nuclei undergo slow changes in their local chemical environment and generate observed resonances for each chemical environment sampled. For example, the two set of resonances observed for a subset of residues near the N-terminus of rp(H)LRAP suggest that the rate of exchange between these two states is greater than milliseconds. If there are significantly more than two different chemical environments in slow exchange it may not be possible to detect any of these resonances above the noise background. In between these two extremes of motion is the intermediate NMR timescale (ms to μs) where no resonances are observed because the resonances have broadened beyond detection [44, 47]. Except for the region between S12* and Y12, a single set of sharp, assignable resonances was observed in the 1H-15N HSQC spectrum for rp(H)LRAP at pH 3.0 in 2% acetic acid, indicating that the majority of the molecule was in fast exchange on the NMR timescale (<μs). For residues between S12* and Y12 the set of two sharp assignable resonances for each amide in this region indicates that this part of the molecule is in slow exchange between two different states. Relatively narrow line shapes for all the cross peaks in the 1H-15N HSQC spectrum of rp(H)LRAP (Figure 2B) and dynamic light scattering data (Table 2) are consistent with a monomer or dimer in solution. Upon the addition of NaCl to rp(H)LRAP, amide resonances disappeared and the line widths of those that remained became very broad (Figure 3). The broadening of amide resonances are indicative of the formation of large molecular weight complexes and is corroborated by dynamic light scattering data, shown in Table 2, showing the physical size of the species in solution quadrupled in the presence of a high concentration of NaCl. The disappearance of amide resonances is indicative of a shift from a state of fast motion to one of intermediate motion or slower. There are many possible explanations for the observed shift in dynamics including restricted motion at a protein-protein or protein-ligand (Na+) interface, an intermediate rate of chemical exchange between the free and associated states, or a salt induced change in the local monomer conformation. However, as argued for rp(H)M180 [17], protein-protein interaction is the explanation most consistent with our data. Regardless of the physical cause, in many proteins motion in the intermediate timescale range is frequently associated with regions involved in binding and catalysis [22, 44, 48].
Table 2.
Salt dependence of the diameter of porcine LRAP as determined by dynamic light scattering.1
| sample condition | smallest observed diameter rp(H)LRAP (nm) |
|---|---|
| No salt, pH 3.0 | 52 |
| 100 mM NaCl, pH 3.0 | 20 |
All samples contained 2% acetic acid.
Synthethic unlabeled LRAP without the 8-residue N-terminal tag.
In full-length murine amelogenin a stepwise disappearance of the amide resonances was observed upon increasing the NaCl concentration, beginning near the N-terminus (Y12-I51) followed by a region near the C-terminus (L141-T171) [17]. As shown in Figure 4 there was no such stepwise disappearance of amide resonance in rp(H)LRAP upon increasing the NaCl concentration. Instead, amide resonances begin to disappear in the central region (T21-E166) first with the effect extending outward from this area with increasing salt. Note that the changes appeared to plateau at a Na+:protein molar ratio of 540:1 as no significant changes were observed upon increasing the Na+:protein molar ratio to 1260:1 (date not shown). A plateau was also observed with rp(H)M180, however, this was not reached until a Na+:protein molar ratio of 2330:1 was attained. At the titration plateau for both rp(H)LRAP and rp(H)M180 there appears to be some difference in the dynamics at the termini between the two proteins in high salt as reflected by the greater number of amide resonances gone completely in the N-terminal region (P2-Y17) of rp(H)M180 (seven blue closed circles) relative to rp(H)LRAP (one red closed circle). At the C-terminal region (A170-D180) the dynamic difference is reversed with fewer residues perturbed (open circles) at the C-terminal region of rp(H)M180 (two) relative to rp(H)LRAP (eight). Note that for rp(H)LRAP the duplicate cross peaks for S12*, L3, and S9 collapsed to one cross peak with H6 almost fully collapsed at a Na+:protein molar ratio of 540:1. On the other hand, the amide resonances for G8, G11, and Y12 are still duplicated at this salt concentration, suggesting slow exchange between two conformations is still occurring at the N-terminus in high salt. Perhaps the biophysical cause of the slow exchange between two conformations in rp(H)LRAP, that was not observed in rp(H)M180, may also be responsible for the differences in dynamics between rp(H)LRAP and rp(H)M180 observed in high salt.
As illustrated in Figure 2B, the 1H-15N HSQC spectrum of rp(H)LRAP is well resolved in 2% acetic acid. This is in sharp contrast to the only other published 1H-15N HSQC spectrum for LRAP, the human sequence, collected in 10 mM acetate at pH 4.0 [49]. This difference in spectral quality exists even though our 1H-15N HSQC spectrum for rp(H)LRAP was recorded at a slightly lower field strength (1H resonance frequency of 750 MHz versus 800 MHz) and much higher solution concentrations (0.6 mM versus 0.040 mM). While pH is a well documented initiator of amelogenin nanosphere formation [4], little change in the spectral features of the 1H-15N HSQC spectrum of rp(H)M180 was observed between pH 3.0 to 5.0 when the pH was adjusted with acetic acid [17]. However, when the pH of rp(H)M180 was adjusted with NaOH, changes similar to those observed in the salt titration experiments were observed in the 1H-15N HSQC spectrum of rp(H)M180. Consequently, one of the explanations for the poor spectral qualities of the previously reported 1H-15N HSQC spectrum of human LRAP may be related to the manner by which the pH was adjusted, or, perhaps some residual salt still present in the protein. Alternatively, human LRAP may be more sensitive to self-aggregation as a function of pH than murine LRAP.
3.3 Biological implications
The NMR data presented here suggest that, like the full-length protein, high salt effects a transition from a small molecular weight species (monomers/dimers) to a larger one (nanospheres). Such an interpretation follows the current amelogenin nanosphere assembly paradigm where dimers and trimers form first followed by the assembly of hexamers that progressively congregate to form nanospheres [14]. With rp(H)LRAP, dimer assembly is first initiated at the center of LRAP (T21-E166) followed by a greater involvement of the N- and C-terminal regions as the ionic strength is increased. This is different from full-length amelogenin where there appeared to be a stepwise formation of dimers beginning near the N-terminal (Y12-I51) followed by a region at the C-terminal (L141-T171) [17].
Intermolecular association of hydrophophic regions in proteins is believed to be one of the driving forces responsible for protein association/aggregation [50]. Increasing the ionic strength of the solution generally increases the tendency for such regions to interact [51]. Because there is significant sequence conservation of amelogenin among different species and, aside from the acidic C-teminal tail, the full-length amelogenin sequence is rich in hydrophobic residues [17, 21], it has been suggested that intermolecular hydrophobic interactions play a functional role in the self-assembly of amelogenin in the high ionic strength environment of ameloblasts [14, 16]. Indeed, towards the latter stages of calcium hydroxyapatite crystal growth the hydrophobic environment within nanospheres is believed to be crucial for proper enamel formation [19, 30]. The difference between the amino acid sequence of rp(H)LRAP and rp(H)M180 is the loss of 101 hydrophobic residues in the HQP-rich region and lectin-like binding tri-tyrosine domain (Figure 1). Despite the loss of these hydrophobic residues, nanospheres still formed and a much lower molar ratio of Na+:protein was required to reach a plateau for rp(H)LRAP (540) than for rp(H)M180 (2330). At the plateau there were differences in dynamics between the two proteins at both termini suggesting that the termini of rp(H)LRAP and rp(H)M180 behaved differently in nanosphere assemblies. Regardless of the salt dependent oligomeric state of the proteins, a structural difference was also observed between the two proteins as the N-terminal region of rp(H)LRAP adopted two slowly (>ms) interconverting conformations as a monomer or a nanosphere. Perhaps these differences in biophysical properties, especially the structural differences near the N-terminal, have biological functional significance given the cell signaling properties attributed to LRAP but not the full-length protein.
Acknowledgements
This research was supported by NIH-NIDCR Grant DE-015347 and internal Laboratory Directed Research Development (LDRD) funds. The research was performed at the Pacific Northwest National Laboratory (PNNL), a facility operated by Battelle for the U.S. Department of Energy, and at the W.R. Wiley Environmental Molecular Sciences Laboratory (EMSL), a national scientific user facility sponsored by the U.S. DOE Biological and Environmental Research program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ten Cate AR. Oral histology: developement, structure, and function. 4th ed. Mosby; St. Louis: 1994. [Google Scholar]
- 2.White SN, Luo W, Paine ML, Fong H, Sarikaya M, Snead ML. Biological organization of hydroxyapatite crystallites into a fibrous continuum toughens and controls anisotrophy in human enamel. J. Dent. Res. 2001;80:321–326. doi: 10.1177/00220345010800010501. [DOI] [PubMed] [Google Scholar]
- 3.Hunter G. Interfacial aspects of biomineralization. Curr. Opin. Mat. Sci. 1996;1:430–435. [Google Scholar]
- 4.Margolis HC, Beniash E, Fowler CE. Role of macromoleculare assembly of enamel matrix proteins in enamel formation. Crit. Rev. Oral Biol. Med. 2006;85:775–793. doi: 10.1177/154405910608500902. [DOI] [PubMed] [Google Scholar]
- 5.Termine JD, Belcourt AB, Christner PJ, Conn KM, Nylen MU. Properties of dissociatively extracted fetal tooth matrix proteins. J. Biol. Chem. 1990;255:9760–9768. [PubMed] [Google Scholar]
- 6.Fincham AG, Moradian-Oldak J, Simmer JP, Sarte P, Lau EC, Diekwisch T, Slavkin HC. Self-assembly of a recombinant amelogenin protein generates supramolecular structures. J. Struct. Biol. 1994;112:103–109. doi: 10.1006/jsbi.1994.1011. [DOI] [PubMed] [Google Scholar]
- 7.Fincham AG, Moradian-Oldak J, Diekwisch TGH, Lyaruu DM, Wright JT, P.B., Slavkin HC. Evidence for amelogenein“nanospheres” as functional components of secretory-stage enamel matrix. J. Struct. Biol. 1995;115:50–59. doi: 10.1006/jsbi.1995.1029. [DOI] [PubMed] [Google Scholar]
- 8.Uchida T, Tanabe T, Fukae M, Shimizu M, Yamada M, Miake K, Kobayashi S. Immunochemical and immunohistochemical studies, using antisera against porcine 25 kDa amelogenin, 89 kDa enamelin and the 12-17 kDa nonamelogenins, on immature enamel of the pig and rat. Histochemistry. 1991;96:129–138. doi: 10.1007/BF00315983. [DOI] [PubMed] [Google Scholar]
- 9.Ravassipour D, Hart PS, Hart TC, Ritter AV, Yamauchi M, Gibson C, Wright JT. Unique enamel phenotype associated with amelogenin gene (AMELX) codon 41 point mutation. J. Dent. Res. 2000;79:1476–1481. doi: 10.1177/00220345000790070801. [DOI] [PubMed] [Google Scholar]
- 10.Gibson CW, Yaun ZA, Hall B, Longenecker G, Chen EH, Thyagarajan T, Sreenath T, Wright JT, Decker S, Piddington R, Harrison G, Kulkarni AB. Amelogenin-deficient mice display an amelogenesis imperfecta type phenotype. J. Biol. Chem. 2001;276:31871. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- 11.Moradian-Oldak J, Paine ML, Lei YP, Fincham AG, Snead ML. Self-assembly properties of recombinant engineered amelogenin proteins analyzed by dynamic light scattering and atomic force microscopy. J. Struct. Biol. 2000;131:27–37. doi: 10.1006/jsbi.2000.4237. [DOI] [PubMed] [Google Scholar]
- 12.Paine ML, White SN, Luo W, Fong H, Sarikaya M, Snead ML. Regulated gene expression dictates enamel structure and tooth function. Matrix Biol. 2001;20:273–292. doi: 10.1016/s0945-053x(01)00153-6. [DOI] [PubMed] [Google Scholar]
- 13.Wen HB, Fincham AG, Moradian-Oldak J. Progressive accretion of amelgenin molecules during nanosphere assembly revealed by atomic force microscopy. Matrix Biol. 2001;20:387–395. doi: 10.1016/s0945-053x(01)00144-5. [DOI] [PubMed] [Google Scholar]
- 14.Du C, Falini G, Fermani S, Abbott C, Moradian-Oldak J. Supramolecular assembly of amelogenin nanospheres into birefringent microribbons. Science. 2005;307:1450–1454. doi: 10.1126/science.1105675. [DOI] [PubMed] [Google Scholar]
- 15.Moradian-Oldak J, Simmer JP, Lau EC, Sarte PE, Slavkin HC, Fincham AG. Detection of monodisperse aggregates of a recombinant amelogenin by dynamic light scattering. Biopolymers. 1994;34:1339–1347. doi: 10.1002/bip.360341006. [DOI] [PubMed] [Google Scholar]
- 16.Moradian-Oldak J, Leung W, Fincham AG. Temperature and pH-dependence of amelogin self-assembly: A particle size distribution study. J. Struct. Biol. 1998;122:320–327. doi: 10.1006/jsbi.1998.4008. [DOI] [PubMed] [Google Scholar]
- 17.Buchko GW, Tarasevich BJ, Bekhazi J, Snead ML, Shaw WJ. A solution NMR investigation into the early events of amelogenin nanosphere self-assembly initiated with sodium chloride or calcium chloride. Biochemistry. 2008;47:6571–6582. doi: 10.1021/bi8018288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dempsey CE. The actions of melittin on membranes. Biochim. Biophys. Acta. 1990;1031:143–161. doi: 10.1016/0304-4157(90)90006-x. [DOI] [PubMed] [Google Scholar]
- 19.Moradian-Oldak J. Amelogenins: assembly, processing and control of crystal morphology. Matrix Biol. 2001;20:293–305. doi: 10.1016/s0945-053x(01)00154-8. [DOI] [PubMed] [Google Scholar]
- 20.Paine ML, Snead ML. Protein interactions during assembly of the enamel organic extracellular matrix. J. Bone Min. Res. 1997;12:221–227. doi: 10.1359/jbmr.1997.12.2.221. [DOI] [PubMed] [Google Scholar]
- 21.Toyosawa S, O'hUigin F, Figueroa F, Tichy H, Klein J. Identification and characterization of amelogenin genes in monotremes, reptiles, and amphibians. Proc Natl Acad Sci USA. 1998;95:13056–13061. doi: 10.1073/pnas.95.22.13056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buchko GW, McAteer K, Wallace SS, Kennedy MA. Solution-state NMR investigation of DNA binding interactions in Escherichia coli formamidopyrimidine-DNA glycosylase (Fpg): a dynamic description of the DNA/protein interface. DNA Repair. 2005;4:327–339. doi: 10.1016/j.dnarep.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 23.Moradian-Oldak J, Tan J, Fincham AG. Interaction of emelogenin with hydroxyapatite cyrstals: an adherence effect through emelogenin self-association. Biopolymers. 1998;46:225–238. doi: 10.1002/(SICI)1097-0282(19981005)46:4<225::AID-BIP4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 24.Aoba T, Moreno EC. The enamel fluid in the early secretory stage of porcine amelogenesis: chemical composition and saturation with respect to enamel mineral. Calcif. Tissue Int. 1987;41:86–94. doi: 10.1007/BF02555250. [DOI] [PubMed] [Google Scholar]
- 25.Moradian-Oldak J, Bouropoulos N, Wang L, Gharakhanian N. Analysis of the self-assembly and apetite binding properties of amelogenin proteins lacking the hydrophilic C-terminal. Matrix Biol. 2002;21:197–205. doi: 10.1016/s0945-053x(01)00190-1. [DOI] [PubMed] [Google Scholar]
- 26.Gibson CW. The amelogenin “enamel proteins” and cells in the periodontium. Cells Tissues Organs. 2008;18:345–360. doi: 10.1615/critreveukargeneexpr.v18.i4.30. [DOI] [PubMed] [Google Scholar]
- 27.Veis A, Tompkinds K, Alvares K, Wei K, Wang L, Wang S, Brown AG, Jengh S-M, Healy KE. Specific amelogenin gene splice products have signaling effects on cells in culture and in implants in vivo. J. Biol. Chem. 2000;275:41263–41272. doi: 10.1074/jbc.M002308200. [DOI] [PubMed] [Google Scholar]
- 28.Boabaid F, Gibson CW, Kuehl MA, Berry JE, Snead ML, Nociti FHJ, Katchburian E, Sommerman MJ. Leucine-rich amelogenin peptide: a candidate signalling molecule during cementogenesis. J. Periodontol. 2004;75:1126–1136. doi: 10.1902/jop.2004.75.8.1126. [DOI] [PubMed] [Google Scholar]
- 29.Warotayanont R, Zhu D, Snead ML, Zhou Y. Leucine-rich amelogenin peptide induces osteogenesis in mouse embryonic stem cells. Biochem. Biophys. Res. Commum. 2008;367:1–6. doi: 10.1016/j.bbrc.2007.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Habelitz S, Kullar A, Marshall SJ, DenBesten PK, Balooch M, Marshall GW, Li W. Amelogenin-guided crystal growth on fluoroapatite glass-ceramics. J. Dent. Res. 2004;83:698–702. doi: 10.1177/154405910408300908. [DOI] [PubMed] [Google Scholar]
- 31.Shaw WJ, Ferris K. Structure, orientation, and dynamics of the C-terminal hexapeptide of LRAP determined using solid-state NMR. J. Phys. Chem. B. 2008;112:16975–16981. doi: 10.1021/jp808012g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaw WJ, Ferris K, Tarasevich BJ, Larson JL. The structure and orientation of the C-terminus of LRAP. Biophysical J. 2008;94:3247–3257. doi: 10.1529/biophysj.107.119636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen EH, Yuan Z-A, Wright JT, Hong SP, Li Y, Collier PM, Hall B, D'Angelo M, Decker S, Piddington R, Abrams WR, Kulkarni AB, Gibson CW. The small bovine amelogenin LRAP fails to rescue the amelogenin null phenotype. Calcif. Tissue Int. 2003;73:487–495. doi: 10.1007/s00223-002-0036-7. [DOI] [PubMed] [Google Scholar]
- 34.Gibson CW, Li Y, Daly B, Suggs C, Yuan Z-A, Fong H, Simmons D, Aragon M, Kulkarni AB, Wright JT. The leucine-rich amelogenin peptide alters the amelogenin null enamel phenotype. Cells Tissues Organs. 2008;189:169–174. doi: 10.1159/000151384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ravindranath RMH, Devarajan A, Jr PB. Enamel formation in vitro in mouse molar explants exposed to emelogenin polypeptides ATMP and LRAP on enamel development. Arch. Oral Biol. 2007;52:1161–1171. doi: 10.1016/j.archoralbio.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 36.Goto Y, Kogure E, Takagi T, Aimoto S, Aoba T. Molecular conformation of porcine amelogenin in solution: three folding units at the N-terminal, central, and C-termininal regions. J. Biochem. (Toyko) 1993;113:55–60. doi: 10.1093/oxfordjournals.jbchem.a124003. [DOI] [PubMed] [Google Scholar]
- 37.Snead ML, Lau EC, Zeichner-David M, Fincham AG, Woo SL, Slavkin HC. DNA sequence for cloned cDNA for murine amelogenin reveals the amino acid sequence for enamel-specific proteins. Biochem. Biophys. Res. Commum. 1986;129:812–818. doi: 10.1016/0006-291x(85)91964-3. [DOI] [PubMed] [Google Scholar]
- 38.Buchko GW, Bekhazi J, Cort JR, Valentine NB, Snead ML, Shaw WJ. 1H, 13C and 15N Resonance assignments of murine amelogenin, an enamel biomineralization protein. Biomol. NMR Assign. 2008;2:89–91. doi: 10.1007/s12104-008-9092-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zuiderweg ERP. Mapping protein-protein interactions in solution using NMR spectroscopy. Biochemistry. 2002;41:1–7. doi: 10.1021/bi011870b. [DOI] [PubMed] [Google Scholar]
- 40.Simmer JP, Lau EC, Hu CC, Bringas P, Santos V, Aoba T, Lacey M, Nelson D, Zeichner-David M, Snead ML, Slavkin HC, Fincham AG. Isolation and characterization of a mouse amelogenin expressed in Escherichia coli. Calcif. Tissue Int. 1994;54:312–319. doi: 10.1007/BF00295956. [DOI] [PubMed] [Google Scholar]
- 41.Goddard TD, Kneller DG. Sparky 3. University of California; San Francisco: [Google Scholar]
- 42.Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. 1H, 13C and 15N Chemical shift referencing in biomolecular NMR. J. Biomol. NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 43.Buchko GW, Ni S, Lourette NM, Reeves R, Kennedy MA. NMR resonance assignments of the high mobility group protein HMGA1. J. Biomol. NMR. 2007;38:185. doi: 10.1007/s10858-006-9116-8. [DOI] [PubMed] [Google Scholar]
- 44.Ishima R, Torchia DA. Protein dynamics from NMR. Nature Struct. Biol. 2000;7:740–743. doi: 10.1038/78963. [DOI] [PubMed] [Google Scholar]
- 45.Delak K, Harcup C, Lakshminarayanan R, Sun Z, Fan Y, Moradian-Oldak J, Evans JS. The tooth enamel protein, procine amelogenin, is an intrinsically disordered protein with an extended molecular configuration in the monomeric form. Biochemistry. 2009;48:2272–2281. doi: 10.1021/bi802175a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schubert M, Labudde D, Oschkinat H, Schmieder P. A software tool for prediction of Xaa-Pro peptide bond conformations in proteins based on 13C chemical shift statistics. J. Biomol. NMR. 2002;24:149–154. doi: 10.1023/a:1020997118364. [DOI] [PubMed] [Google Scholar]
- 47.Lee D, Hilty C, Wider G, Wuthrich K. Effective rotational correlation times of proteins from NMR relaxation interference. J. Magn. Reson. 2006;178:72–76. doi: 10.1016/j.jmr.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 48.Mulder FAA, Hon G, Muhandiram DR, Dahlquist FW, Kay LE. Flexibility and ligand exchange in a buried cavity mutant of T4 lysozyme studied by multinuclear NMR. Biochemistry. 2000;39:12614–12622. doi: 10.1021/bi001351t. [DOI] [PubMed] [Google Scholar]
- 49.Le TQ, Gochin M, Featherstone JDB, Li W, DenBesten PK. Comparative calcium binding of leucine-rich amelogenin peptide and full-length amelogenin. Eur. J. Oral Sci. 2006;114(Suppl. 1):320–326. doi: 10.1111/j.1600-0722.2006.00313.x. [DOI] [PubMed] [Google Scholar]
- 50.Miller S, Janin J, Lesk AM, Chothia C. Interior and surface of monomeric proteins. J. Mol. Biol. 1987;196:641–656. doi: 10.1016/0022-2836(87)90038-6. [DOI] [PubMed] [Google Scholar]
- 51.Kohn WD, Kay CM, Hodges RS. Salt effects on protein stability: two-stranded alpha-helical coiled-coils containing inter- or intrahelical ion pairs. J. Mol. Biol. 1997;267:1039–1052. doi: 10.1006/jmbi.1997.0930. [DOI] [PubMed] [Google Scholar]