Abstract
Genome wide association studies have been increasingly used to study the genetics of complex human diseases. Within the field of cardiac electrophysiology, this technique has been applied to traits such as AF, the QT interval, and several electrocardiographic parameters. While these studies have identified multiple genomic regions associated with each trait, questions remain including the best way to explore the pathophysiology of each association and the potential for clinical utility. This review will summarize recent genome wide association study results within cardiac electrophysiology, and discuss their broader implications in basic science and clinical medicine.
Introduction
In the last three years, genome wide association studies (GWAS) have been used to identify the genetic underpinnings of over 100 traits including electrocardiographic parameters and atrial fibrillation (AF). While increasingly popular, a number of fundamental questions exist. Specifically, what are the implications of these genetic studies? Will the results have utility beyond the basic research setting? Should we as electrophysiologists care about the results? Will these studies add clinical information beyond what is currently available in patient care?
In this review, we seek to describe the methods for performing a GWAS, summarize the latest results of GWAS in cardiac electrophysiology, and explore the implications of these studies in research and patient management.
What is a genome wide association study (GWAS)?
Traditional approaches to understanding the genetic basis of heritable traits or diseases have focused on single gene defects with clear modes of inheritance. These diseases or traits are often referred to as Mendelian, familial, or monogenic. In electrophysiology, a classic example is autosomal dominant long QT syndrome. Using linkage analysis, investigators identify genetic loci and ultimately a single genetic defect that causes the disease. A limitation of this approach is the need for large families with multiple affected individuals, and a definitive diagnosis for affected and unaffected family members. Once a causative gene has been identified in a Mendelian family, investigators will then use this information to guide candidate gene resequencing efforts in other families to identify mutations in related genes. As sequencing technology has advanced and costs declined, candidate gene sequencing has been increasingly employed when family structure does not permit linkage analysis.
In contrast to monogenic traits, which are typically rare, many human diseases and traits are genetically complex. Complex conditions are believed to be caused by the joint effects of many common genetic variants, and potentially, environmental factors. Complex traits may be heritable, and yet have no straightforward mode of inheritance. Diseases such as coronary artery disease and hypertension are complex traits that have proven refractory to linkage analysis. The effect sizes of rare, causal genetic variants in monogenic conditions are large, in contrast to the much smaller effect sizes attributable to common modifying variants in complex traits.
The use of GWAS has led to the identification of hundreds of genetic loci for many complex diseases and traits. These loci serve both as markers of novel pathophysiological signaling pathways and as disease susceptibility loci. Advantages of GWAS over linkage analysis include the ability to use available epidemiologic study populations, and the fact that large families are not necessary. Technological advances permit association testing of a million genetic markers simultaneously. These markers are single nucleotide polymorphisms (SNPs) or commonly occurring variations in single bases throughout the genome. SNPs can be directly associated with disease or may serve as proxies for disease-causing variant in close proximity. Genetic variants that lie in close physical proximity to one another are typically inherited together, and when linked in this way are said to be in linkage disequilibrium. Thus, in order to demonstrate a genetic association, investigators do not have to identify or even test the causative variant, so long as they test a nearby variant that is closely linked. However, identification of the causative genetic variant is a critical step for moving beyond the statistical association and providing a greater understanding of disease mechanism.
In a typical GWAS, investigators might select individuals with or without myocardial infarction (MI) from a population. Each individual’s DNA is characterized at several hundred thousand locations using a SNP chip. Each of these SNPs is then tested for association with MI using after adjusting for differences in ancestry between cases and controls. Variants associated with MI beyond a stringent significance threshold that is adjusted for the large number of independent tests performed (e.g., P<5×10−8) are subsequently tested in an independent replication cohort for association with MI.
Several limitations of GWAS exist. False-positive associations are inevitable with multiple testing, and therefore GWAS must employ very stringent significance thresholds. Because of the stringent significance thresholds and modest effect sizes of each genetic variant, thousands of subjects are generally necessary. An additional disadvantage of GWAS is that racial variations in SNP frequencies and linkage patterns can confound associations unless population ancestry is properly identified. Studies to date have largely been performed in subjects of European descent, so the applicability to other ethnicities may be limited. Additionally, GWAS are expensive, with genotyping costs at ~$400 per person. By design, GWAS will not detect rare variants that are associated with a disease. Finally, a missing feature of almost all GWAS is the effect of environmental factors that influence the trait. It is especially difficult to account for gene-environment interactions where a particular genotype modifies the susceptibility to an environmental exposure.
GWAS in Cardiac Electrophysiology
Within cardiac electrophysiology, GWAS have been applied to several ECG traits as well as specific arrhythmias. Electrocardiographic traits include the QT interval, but also PR and RR intervals. The diseases studied to date include AF and sudden cardiac death (although SCD was not investigated as a GWAS, but in follow-up to the QT studies.) One of the themes that emerges is that the effect sizes of the genetic variants identified are fairly small. Additionally, it is clear that GWAS identify a genetic region, but not the specific gene or SNP that is responsible for the trait.
Atrial fibrillation
The first AF susceptibility region was identified on chromosome 4q25 using a GWAS in 2007.1 The study included 3,580 subjects with AF and 19,256 controls of European ancestry. The most significantly associated SNP at this locus had a risk allele frequency of approximately 20%. The robust association with AF demonstrated in this study has been replicated in subsequent studies,2–5 including an analysis of 333 cases and 2,836 controls of Asian ancestry (P=0.0006). The strongest signal at this locus lies in a large interval that contains no known or putative genes. The closest gene, PITX2 (paired-like homeodomain transcription factor 2), is located approximately 150,000 bases away and is an interesting candidate gene for AF. PITX2 is known to play an important role in embryonic development including asymmetric morphogenesis of the gut and heart.6 One transcript variant of PITX2 is critical for the suppression of a default sinoatrial node in the left atrium and in specifying pulmonary venous myocardium.7
The recognition that large sample sizes are necessary for the identification of common variants associated with disease has prompted investigators from independent cohorts to combine their study populations. Analyses from two independent consortia identified a novel susceptibility locus for AF on chromosome 16q22 in individuals of European ancestry.4,8 The most significantly associated variants from each study had risk allele frequencies of approximately 20%, and fell within the ZFHX3 (zinc finger homeobox 3) gene. Little is known about the role of ZFHX3 in the heart or cardiovascular system, but it is interesting to note that the top variant in one of the two GWAS of AF8 was also strongly associated with Kawasaki disease in a separate GWAS.9
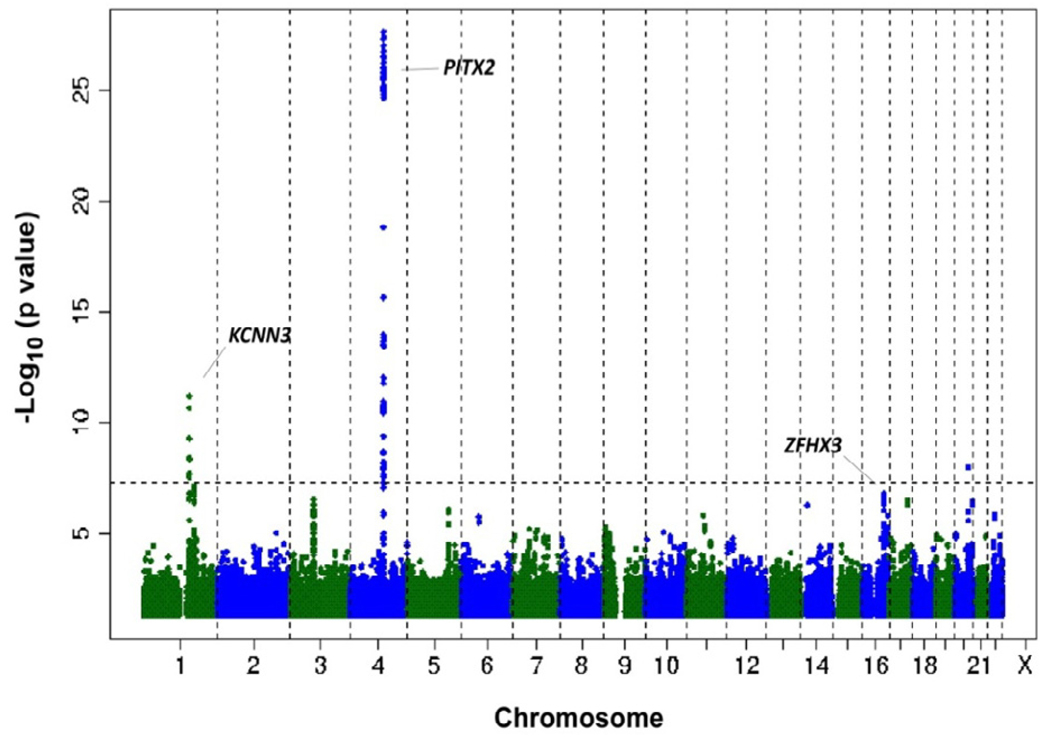
A further collaborative study of lone AF was performed in a meta-analysis of five cohorts using 1,335 cases of lone AF and 12,844 referent subjects. The results of the meta-analysis for lone AF are illustrated in the Manhattan plot where each point on the graph indicates the strength of association between a SNP and AF (Figure 2). The strongest association with lone AF was at the chromosome 4q25 or PITX2 locus. A second locus was identified at chromosome 1q21 near a calcium-activated potassium channel, KCNN3 (KCa2.3 or SK3). The KCNN channels are expressed in the brain, heart and vascular endothelium. KCNN3 has been shown to participate in regulation of blood pressure via the endothelium derived hyperpolarization factor vasodilator pathway.10,11 Although KCNN3 remains a strong candidate gene at this locus, further work will be needed to clarify the relation between KCNN3 and AF.
Figure 2. Genetic loci for AF identified by GWAS.
To date, three genetic loci associated with AF have been identified near KCNN3, PITX2, and ZFHX3. The -log10(p value) is plotted against the physical positions of each SNP on each chromosome. The threshold for genome-wide significance, P <5×10−8, is indicated by the dashed line. Reprinted with permission from Nature Genetics 42, 240–244 (2010).
PR interval and relation to AF
Two GWAS of the PR interval conducted in subjects of European ancestry also tested the top PR-related variants for association with AF.12,13 Variants in six of nine genes associated with the PR interval were also associated with AF, albeit at less stringent thresholds (Table 2).12 The plausibility of the PR interval as an intermediate phenotype for AF is buttressed by observations that PR interval prolongation is associated with AF in the community,14,15 though not all SNPs associated with an increased risk of AF were associated with PR prolongation in these GWAS.12,13
Table 2.
GWAS Results for Atrial Fibrillation
In both studies that tested for associations between PR-related variants and AF, variants in CAV1 were identified.12,13 CAV1 and CAV2 encode caveolin proteins, which line small invaginations in cellular membranes involved in signal transduction.16 CAV1 is expressed in atrial but not ventricular myocytes. Mice deficient in CAV1 develop dilated cardiomyopathy,17 pulmonary hypertension,17 and cardiac hypertrophy.18 In the study by Pfeufer et al.,12 additional variants in SCN5A, SCN10A, NKX2-5, and SOX5 were associated with both the PR interval and AF. In the study by Holm et al.,13 variants in TBX5 were also associated with both phenotypes. SCN5A and SCN10A are two adjacent voltage gated sodium channels. Genetic variants in SCN5A underlie numerous arrhythmias, including the sick sinus syndrome, long QT syndrome, the Brugada syndrome, and AF, as mentioned above.19 SCN10A has not been previously implicated in cardiac arrhythmias. The study by Holm et al.13 did not replicate the association between variants in SNC10A and AF, despite a -high degree of linkage disequilibrium between the variant tested in this study and that by Pfeufer et al.12 Interestingly, three transcription factors necessary for cardiac development were related to PR including NKX2-5, SOX5, and TBX5a.
QT Interval
The first GWAS reported for an electrocardiographic trait was performed in 200 subjects who were found to lie at the extremes of a population-based QT interval distribution of 3,966 subjects.20 This GWAS identified a signal that lies upstream of the gene NOS1AP (CAPON) that was associated with QT interval. NOS1AP is a regulator of neuronal nitric oxide synthase, and was a new candidate gene for modulation of cardiac repolarization.
Subsequently, two larger GWAS of the QT interval were reported in early 2009. In the QTGEN study, three genome-wide association studies involving 13,685 individuals of European ancestry were meta-analyzed for SNPs associated with QT interval duration. QTGEN investigators observed associations with variants in or near NOS1AP, KCNQ1, KCNE1, KCNH2 and SCN5A.21–24 All of these genes were previously known to be involved in myocardial repolarization and/or long-QT syndromes.24,25 Five new loci were also identified on 16q21 near NDRG4 and GINS3, 6q22 near PLN, 1p36 near RNF207, 16p13 near LITAF, and 17q12 near LIG3 and RFFL. In the accompanying paper, genome-wide data from five population-based cohorts with a total of 15,842 individuals of European ancestry confirmed associations between variants in NOS1AP, KCNQ1, KCNH2, SCN5A and KCNJ2 and the QT interval.26 Two other loci included ATP1B1 and PLN, genes with established electrophysiological function, whereas three mapped to RNF207, near LITAF and within NDRG4-GINS3-SETD6-CNOT1, none of which had previously been implicated in cardiac electrophysiology.
There are several noteworthy findings. First, all three studies of QT interval have consistently identified a region near NOS1AP. Second, the two large GWAS of QT interval identified common variants in 5 of the known Long QT syndrome genes. These results can be viewed as a proof of concept since these genes had the highest pretest probability of influencing QT interval in the general population. Finally, even the novel loci in the two large QT GWAS studies have significant overlap, highlighting genomic regions that do not contain known ion channels or genes with established roles in cardiac electrophysiology.
Currently, genetic testing is able to identify mutations in known long QT genes in approximately 75% of probands. The novel loci identified by these GWAS may contain new long QT genes that might be responsible for some of the remaining cases, though in order to demonstrate this, causal genes will need to be identified. Even if these loci do not identify novel monogenic long QT syndrome genes, the causal genes may modify the severity of the Long QT phenotype, as has been demonstrated for NOS1AP in one large South African long QT population. 27 Further work will be required to determine whether this result is applicable outside the single highly inbred family where it was found. These genetic signals may also yield information about susceptibility to drug-induced QT prolongation and torsades de pointes (TdP). It is known that baseline QT prolongation is a risk factor for drug-induced QT prolongation and TdP, but whether any of the genetic associations in baseline QT interval will be major determinants of this risk remains to be seen.
Sudden Cardiac Death
Longer QT intervals are associated with increased risk of sudden cardiac death.28–30 Therefore, a logical next step was to determine if variation at the most strongly associated locus, near NOS1AP, alters the risk of sudden cardiac death. In a study of two populations of more than 19,000 subjects and 498 episodes of SCD, a significant association was observed with two different SNPs at the NOS1AP locus.31 The first SNP was correlated with longer QT interval and increased risk of sudden death, while the second SNP was not correlated with QT interval, but seemed to be associated with decreased risk of sudden death. The effects were only observed in white subjects and the discordance in the associations between blacks and whites may represent the result of lower statistical power (due to inappropriate tagging SNPs and smaller numbers of events) in blacks, or it may represent a real genetic difference.
RR interval
A GWAS in 8,842 samples from Korea reported two genomic loci that were associated with heart rate.32 The strongest signal was found in a region with no likely candidate gene and was responsible for <1 beat per minute change in heart rate per variant allele. The next signal was responsible for approximately 0.5 beats per minute variation. The closest gene has an unknown function, but the connexin gene GJA1 is 400kb away.32 A second GWAS examined ~10,000 individuals of European descent and found a single locus for heart rate at the gene encoding alpha-myosin heavy chain (MYH6).13 The two loci identified in the Korean population failed to replicate in this study, perhaps because of population differences. The MYH6 genetic variant explains about 0.1% of the variability in heart rate. Finally, a third GWAS for heart rate identified a SNP within the GPR133 gene,33 a G-protein coupled receptor that interestingly had been previously reported to be associated with height.34
QRS Duration
Researchers at deCODE genetics recently published four genetic loci associated with QRS duration.13 Two of these loci, near TBX5 and SCN10A were also implicated in PR interval, suggesting potential roles in generalized myocyte impulse propagation. The other two loci did not contain obvious candidate genes. The first linked region contains only a single gene, cyclin dependent kinase inhibitor 1A, and the second region contained several genes, the closest is DKK1, an inhibitor of the Wnt signaling pathway. Together these variants explain only 0.6% of the variance in QRS duration, but they may identify novel signaling, developmental, or regulatory pathways that are important in cardiac electrophysiology.
Clinical implications of these findings
From a clinical perspective, the value of these findings remains to be determined. While, in general, individual magnitudes of effects are too small to be of clinical predictive value, a combination of genetic risk factors may provide meaningful information. Additional studies will be needed to determine whether these variants might influence response to specific therapies or identify subtypes of disease states.
Atrial Fibrillation
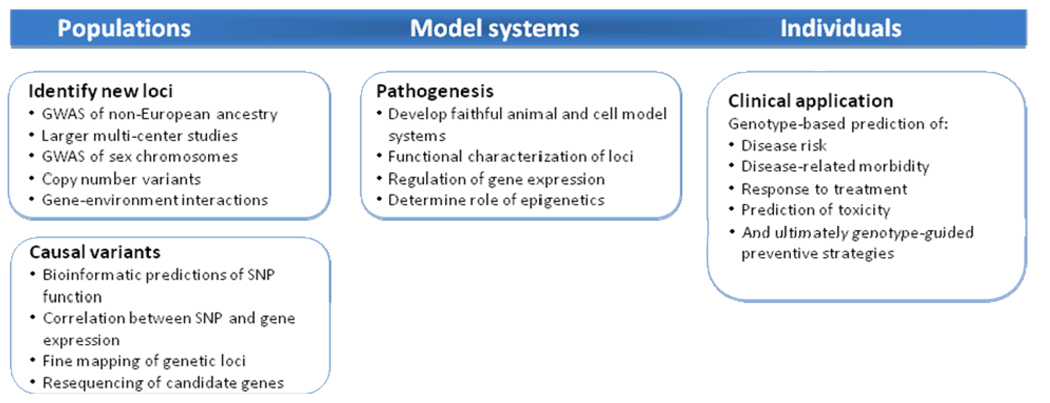
In the case of AF, the effect sizes for the genetic variants identified to date are sufficiently large to be expected to eventually provide clinically useful risk profiling. Although genotyping at the 4q25 locus is commercially available to identify individuals at risk of AF, we do not believe that routine testing is currently warranted. Genetic risk stratification will ultimately incorporate testing at all of the major AF loci and its utility will be dependent on whether such testing alters clinical practice. Potential applications include prediction of response to pharmacological or procedural therapies, progression of disease, or risk prediction of outcomes such as heart failure, stroke, and death (Figure 4).
Figure 4. Future directions in AF genomics.
An outline of potential research questions and clinical applications to arise from studies of AF genetics.
QT Interval and Sudden Cardiac Death
The effect estimates for the common genetic variants associated with the QT interval and SCD are small. Therefore, genotyping NOS1AP to predict SCD risk is unlikely to be useful in clinical testing. However, in the case of QT interval, there are environmental factors, chief among them drug exposure, that pose a significant health hazard to a minority of individuals. It is possible that one or a combination of many SNPs associated with the QT interval may identify individuals at greater risk for drug-induced QT prolongation and TdP, but at present such testing is not warranted.
Future research directions
Although the clinical utility of GWAS data at present remains limited, each genetic locus that is associated with a phenotype identifies a relevant pathway influencing human physiology. To a large extent, enthusiasm within the scientific community for GWAS is based on the fact that, unlike the study of model organisms in which human relevance is always in question, GWAS is an unbiased approach to understanding physiology and disease in humans. Novel loci uncovered by this method will require further investigation using a variety of animal models, cell culture and genetic approaches to understand the fundamental mechanisms responsible for the original genetic signal. Years of work may lie ahead in unraveling such mechanisms, but their relevance to human physiology is already established (Figure 4).
The modest effect sizes expected with common variants35 suggest that larger sample sizes may be necessary to identify additional novel signals. These signals will likely have even smaller effect sizes than the ones already discovered and discussed in this review. However, it is hoped that patterns will emerge that allow identification of entire signaling pathways and networks that regulate a given complex trait. Regions of the genome that have not been extensively analyzed, such as the X and Y chromosomes or mitochondria, have the potential to harbor variants that affect phenotypic diversity. Moreover, the search for variants that interact with environmental variables may reveal loci that have effects only among those individuals exposed to specific triggers or with a particular substrate.
Studying the genetic basis of disease in populations of non-European ancestry has great potential to enhance our understanding of the biological mechanisms underlying disease. Most of the understanding of AF epidemiology, genetic and otherwise, is derived from the study of individuals of European descent.36 As a result of the migration patterns of human populations, linkage disequilibrium patterns differ between different racial and ethnic groups. As such, high density genotyping, or fine mapping, in different ancestral populations may help narrow the boundaries of a common disease susceptibility locus. AF prevalence has significant racial and ethnic differences,37 which may potentially be explained by different frequencies of common susceptibility variants or distinct susceptibility loci.
Genetic variation only represents one aspect of a complementary network of signals that influence phenotypic variation.38 Epigenetic phenomena such as DNA methylation and gene silencing, as well as complex interactions between distinct genetic elements, differentially expressed transcripts, and metabolic and proteomic signals, may also explain a substantial proportion of phenotypic diversity. Bioinformatic techniques, improved phenotyping informed by molecular, electrophysiological, or refined clinical features may enhance the discovery of loci and networks that govern disease pathogenesis.
Conclusions
GWAS have identified novel biological signals involved in human electrophysiology and disease. Findings from GWAS present tremendous opportunities to understand the mechanisms by which these novel pathways relate to human health and disease. Future research will focus on whether consideration of variants identified in GWAS can facilitate the identification of individuals at risk for disease, guide clinical management decisions, and relate to prognosis.
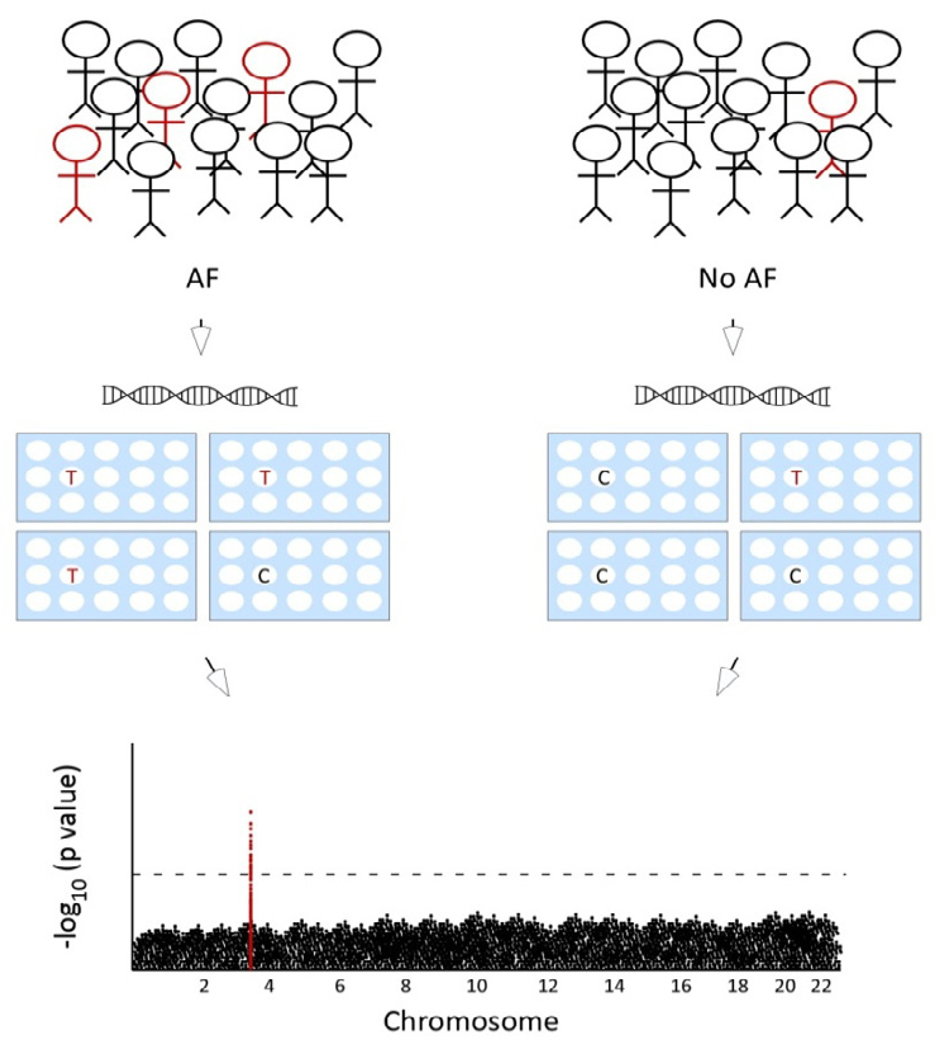
Figure 1. Overview of a GWAS.
To perform a GWAS, cases and control subjects are genotyped at 500,000 to 1 million common SNPs throughout the genome to identify regions associated with disease. On top is a population with carriers of a risk allele indicated in red. In the middle, a sample of DNA is obtained from each subject and genome wide genotyping is performed. On bottom, a comparison is made of the frequency of each SNP throughout the genome in cases versus controls. The genetic interval associated with disease is indicated in red.
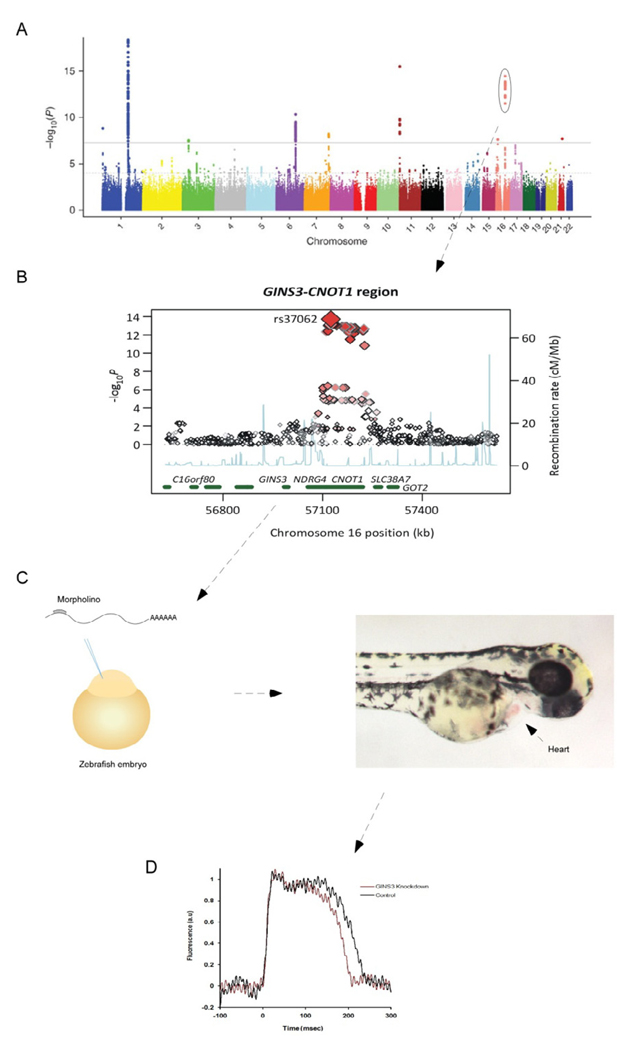
Figure 3. Functional characterization of a genetic locus for the QT interval.
Manhattan plot of SNPs associated with the QT interval with the GINS3 locus encircled (A). A regional plot of the GINS3 locus illustrating the SNPs associated with the QT interval (B). Genes in this region were knocked down in the zebrafish embryo by injecting a morpholino directed against GINS3 (C). Hearts from 48 hour old zebrafish were optically mapped and the action potential duration determined in wild-type and GINS3 morpholino knockout fish (D). Knockdown of GINS3 results in a shortening of the action potential duration. Panels A and B reprinted with permission from Nature Genetics 41, 399–406 (2009).
Table 1.
GWAS Results for Electrocardiographic Intervals.
| ECG traits / Locus | Gene name | References |
|---|---|---|
| PR interval | ||
| MEIS1 | Meis homeobox 1 | 12 |
| SCN5A | Sodium channel, voltage-gated, type V | 12 |
| SCN10A | Sodium channel, voltage-gated, type X | 12 |
| ARHGAP24 | Rho GTPase activating protein 24 | 12 |
| NKX2-5 | NK2 transcription factor related, locus 5 | 12 |
| CAV1/CAV2 | Caveolin 1, 2 | 12 |
| WNT11 | Wingless-type MMTV integration site family, member 11 | 12 |
| SOX5 | Sex determining region Y-box 5 | 12 |
| TBX5/TBX3 | T-box 5, 3 | 12 |
| QT interval | ||
| RNF207 | Ring finger protein 207 | 26,39 |
| NOS1AP | Nitric oxide synthase 1 adaptor protein | 26,39 |
| ATP1B1 | ATPase, Na+/K+ transporting, beta 1 polypeptide | 26 |
| SCN5A-SCN10A | Sodium channel, voltage gated, types V and X | 26,39 |
| PLN | Phospholamban | 26,39,40 |
| KCNH2 | HERG / Potassium voltage-gated channel, subfamily H (eag- related), member 2 |
26,39 |
| KCNQ1 | Potassium voltage-gated channel, KQT-like subfamily, member 1 | 26,39 |
| LITAF | Lipopolysaccharide-induced TNF factor | 26,39 |
| NDRG4 | NDRG family member 4 | 26,39 |
| RFFL | Ring finger and FYVE-like domain containing 1 | 39 |
| KCNJ2 | Potassium inwardly-rectifying channel, subfamily J, member 2 | 26 |
| KCNE1 | Potassium voltage-gated channel, Isk-related family, member 1 | 39 |
| RR interval | ||
| MYH6 | Alpha myosin heavy chain | 13 |
| GJA1 | Gap junction protein, alpha 1, 43kDa | 32 |
| CD34 | CD34 molecule | 32 |
| GPR133 | G-protein Coupled Receptor | 32 |
| QRS duration | ||
| TBX5 | T-box 5 | 13 |
| SCN10A | Sodium channel, voltage-gated, type X | 13 |
| CDKN1A | Cyclin Dependent Kinase Inhibitor 1A | 13 |
| DKK1 | Dickkopf-related protein 1 | 13 |
Acknowledgments
Supported by grants from the NIH (K08HL076361, R21DA026982, HL092577, DA027021, T32HL007575), Fondation Leducq (07-CVD 03), German National Genome Research Network (NGFN, 01GS0838), German AF-Net (01 GI 0204/N), and Exzellenzinitiative at Ludwig-Maximilians University, Munich.
Glossary
- Candidate gene resequencing
A candidate gene is hypothesized to be biologically related to a particular condition, based on preexisting knowledge of physiology, proximity to an association signal, or some other prior information. Candidate gene resequencing is the screening of genes for mutations in a cohort of individuals with a given condition.
- Genome wide significance
When performing multiple tests of association, the chances for a false positive finding are increased. Adjusting the significance threshold for the number of tests performed will mitigate false positives, but results in smaller P value requirements in order to declare true associations. GWAS usually employ a conservative Bonferroni adjustment, dividing the traditional significance threshold of 0.05 by the number of independent tests performed. A commonly accepted threshold is P < 5 × 10−8.
- Haplotype
Alleles on the same chromosome that are inherited together form a haplotype. Haplotypes are bounded by areas where recombination is likely to occur during meiosis.
- Human Genome Project
The large scale effort to sequence every one of the 3 billion nucleotides in the human genome. The completion of this project has provided investigators a roadmap for the genome that is now readily searchable at www.ucsc.genome.org or www.ncbi.org.
- Linkage analysis
A technique to identify a locus or genetic region associated with a disease. In a Mendelian family, the disease gene and the known marker will be linked or shared in common with all affected individuals and not by the unaffected individuals.
- Linkage disequilibrium
The non-random association of two alleles. Alleles in linkage disequilibrium are found together more often than expected by chance alone.
- Manhattan plot
A plot of the –log10 of the p value versus chromosomal location for each SNP. Higher points on the Y axis indicate stronger associations (resembling the Manhattan skyline).
- Single nucleotide polymorphism (SNP)
Common genetic variation at a single base pair of DNA in the genome. Typically SNPs are biallelic or due to the presence of one of two variations at a given base pair. For example: GATTC versus GACTC. SNPs account for an estimated 90% of the variation between individuals.
- Risk allele
The form of a single nucleotide polymorphism that is associated with disease.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gudbjartsson DF, Arnar DO, Helgadottir A, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 2.Kaab S, Darbar D, van Noord C, et al. Large scale replication and meta-analysis of variants on chromosome 4q25 associated with atrial fibrillation. Eur Heart J. 2009;30:813–819. doi: 10.1093/eurheartj/ehn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Body SC, Collard CD, Shernan SK, et al. Variation in the 4q25 chromosomal locus predicts atrial fibrillation after coronary artery bypass graft surgery. Circ Cardiovasc Genet. 2009;2:499–506. doi: 10.1161/CIRCGENETICS.109.849075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benjamin EJ, Rice KM, Arking DE, et al. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat Genet. 2009;41:879–881. doi: 10.1038/ng.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellinor PT, Lunetta KL, N.L G, et al. Common Variants in KCNN3 are Associated with Lone Atrial Fibrillation. Nat Genet. 2010;42:240–244. doi: 10.1038/ng.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Logan M, Pagan-Westphal SM, Smith DM, Paganessi L, Tabin CJ. The transcription factor Pitx2 mediates situs-specific morphogenesis in response to left-right asymmetric signals. Cell. 1998;94:307–317. doi: 10.1016/s0092-8674(00)81474-9. [DOI] [PubMed] [Google Scholar]
- 7.Mommersteeg MT, Brown NA, Prall OW, et al. Pitx2c and Nkx2-5 are required for the formation and identity of the pulmonary myocardium. Circ Res. 2007;101:902–909. doi: 10.1161/CIRCRESAHA.107.161182. [DOI] [PubMed] [Google Scholar]
- 8.Gudbjartsson DF, Holm H, Gretarsdottir S, et al. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet. 2009;41:876–878. doi: 10.1038/ng.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burgner D, Davila S, Breunis WB, et al. A genome-wide association study identifies novel and functionally related susceptibility Loci for Kawasaki disease. PLoS Genet. 2009;5:e1000319. doi: 10.1371/journal.pgen.1000319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brahler S, Kaistha A, Schmidt VJ, et al. Genetic deficit of SK3 and IK1 channels disrupts the endothelium-derived hyperpolarizing factor vasodilator pathway and causes hypertension. Circulation. 2009;119:2323–2332. doi: 10.1161/CIRCULATIONAHA.108.846634. [DOI] [PubMed] [Google Scholar]
- 11.Taylor MS, Bonev AD, Gross TP, et al. Altered expression of small-conductance Ca2+-activated K+ (SK3) channels modulates arterial tone and blood pressure. Circ Res. 2003;93:124–131. doi: 10.1161/01.RES.0000081980.63146.69. [DOI] [PubMed] [Google Scholar]
- 12.Pfeufer A, van Noord C, Marciante KD, et al. Genome-wide association study of PR interval. Nat Genet. 2010;42:153–159. doi: 10.1038/ng.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holm H, Gudbjartsson DF, Arnar DO, et al. Several common variants modulate heart rate, PR interval and QRS duration. Nat Genet. 2010;42:117–122. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- 14.Schnabel RB, Sullivan LM, Levy D, et al. Development of a risk score for atrial fibrillation (Framingham Heart Study): a community-based cohort study. Lancet. 2009;373:739–745. doi: 10.1016/S0140-6736(09)60443-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng S, Keyes MJ, Larson MG, et al. Long-term outcomes in individuals with prolonged PR interval or first-degree atrioventricular block. JAMA. 2009;301:2571–2577. doi: 10.1001/jama.2009.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gratton JP, Bernatchez P, Sessa WC. Caveolae and caveolins in the cardiovascular system. Circ Res. 2004;94:1408–1417. doi: 10.1161/01.RES.0000129178.56294.17. [DOI] [PubMed] [Google Scholar]
- 17.Zhao YY, Liu Y, Stan RV, et al. Defects in caveolin-1 cause dilated cardiomyopathy and pulmonary hypertension in knockout mice. Proc Natl Acad Sci U S A. 2002;99:11375–11380. doi: 10.1073/pnas.172360799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen AW, Park DS, Woodman SE, et al. Caveolin-1 null mice develop cardiac hypertrophy with hyperactivation of p42/44 MAP kinase in cardiac fibroblasts. Am J Physiol Cell Physiol. 2003;284:C457–C474. doi: 10.1152/ajpcell.00380.2002. [DOI] [PubMed] [Google Scholar]
- 19.Ruan Y, Liu N, Priori SG. Sodium channel mutations and arrhythmias. Nat Rev Cardiol. 2009;6:337–348. doi: 10.1038/nrcardio.2009.44. [DOI] [PubMed] [Google Scholar]
- 20.Arking DE, Pfeufer A, Post W, et al. A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nat Genet. 2006;38:644–651. doi: 10.1038/ng1790. [DOI] [PubMed] [Google Scholar]
- 21.Sanguinetti MC, Jiang C, Curran ME, Keating MT. A mechanistic link between an inherited and an acquired cardiac arrhythmia: HERG encodes the IKr potassium channel. Cell. 1995;81:299–307. doi: 10.1016/0092-8674(95)90340-2. [DOI] [PubMed] [Google Scholar]
- 22.Wang Q, Curran ME, Splawski I, et al. Positional cloning of a novel potassium channel gene: KVLQT1 mutations cause cardiac arrhythmias. Nat Genet. 1996;12:17–23. doi: 10.1038/ng0196-17. [DOI] [PubMed] [Google Scholar]
- 23.Splawski I, Timothy KW, Tateyama M, et al. Variant of SCN5A sodium channel implicated in risk of cardiac arrhythmia. Science. 2002;297:1333–1336. doi: 10.1126/science.1073569. [DOI] [PubMed] [Google Scholar]
- 24.Splawski I, Tristani-Firouzi M, Lehmann MH, Sanguinetti MC, Keating MT. Mutations in the hminK gene cause long QT syndrome and suppress IKs function. Nat Genet. 1997;17:338–340. doi: 10.1038/ng1197-338. [DOI] [PubMed] [Google Scholar]
- 25.Splawski I, Timothy KW, Vincent GM, Atkinson DL, Keating MT. Molecular basis of the long-QT syndrome associated with deafness. N Engl J Med. 1997;336:1562–1567. doi: 10.1056/NEJM199705293362204. [DOI] [PubMed] [Google Scholar]
- 26.Pfeufer A, Sanna S, Arking DE, et al. Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nat Genet. 2009;41:407–414. doi: 10.1038/ng.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crotti L, Monti MC, Insolia R, et al. NOS1AP is a genetic modifier of the long-QT syndrome. Circulation. 2009;120:1657–1663. doi: 10.1161/CIRCULATIONAHA.109.879643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Algra A, Tijssen JG, Roelandt JR, Pool J, Lubsen J. QTc prolongation measured by standard 12-lead electrocardiography is an independent risk factor for sudden death due to cardiac arrest. Circulation. 1991;83:1888–1894. doi: 10.1161/01.cir.83.6.1888. [DOI] [PubMed] [Google Scholar]
- 29.Schouten EG, Dekker JM, Meppelink P, et al. QT interval prolongation predicts cardiovascular mortality in an apparently healthy population. Circulation. 1991;84:1516–1523. doi: 10.1161/01.cir.84.4.1516. [DOI] [PubMed] [Google Scholar]
- 30.Straus SM, Kors JA, De Bruin ML, et al. Prolonged QTc interval and risk of sudden cardiac death in a population of older adults. J Am Coll Cardiol. 2006;47:362–367. doi: 10.1016/j.jacc.2005.08.067. [DOI] [PubMed] [Google Scholar]
- 31.Kao WH, Arking DE, Post W, et al. Genetic variations in nitric oxide synthase 1 adaptor protein are associated with sudden cardiac death in US white community-based populations. Circulation. 2009;119:940–951. doi: 10.1161/CIRCULATIONAHA.108.791723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho YS, Go MJ, Kim YJ, et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet. 2009;41:527–534. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- 33.Marroni F, Pfeufer A, Aulchenko YS, et al. A genome-wide association scan of RR and QT interval duration in 3 European genetically isolated populations: the EUROSPAN project. Circ Cardiovasc Genet. 2009;2:322–328. doi: 10.1161/CIRCGENETICS.108.833806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tonjes A, Koriath M, Schleinitz D, et al. Genetic variation in GPR133 is associated with height: genome wide association study in the self-contained population of Sorbs. Hum Mol Genet. 2009;18:4662–4668. doi: 10.1093/hmg/ddp423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benjamin EJ, Chen PS, Bild DE, et al. Prevention of atrial fibrillation: report from a National Heart, Lung, and Blood Institute workshop. Circulation. 2009;119:606–618. doi: 10.1161/CIRCULATIONAHA.108.825380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soliman EZ, Alonso A, Goff DC., Jr Atrial fibrillation and ethnicity: the known, the unknown and the paradox. Future Cardiol. 2009;5:547–556. doi: 10.2217/fca.09.49. [DOI] [PubMed] [Google Scholar]
- 38.Oti M, Huynen MA, Brunner HG. Phenome connections. Trends Genet. 2008;24:103–106. doi: 10.1016/j.tig.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 39.Newton-Cheh C, Eijgelsheim M, Rice KM, et al. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat Genet. 2009;41:399–406. doi: 10.1038/ng.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nolte IM, Wallace C, Newhouse SJ, et al. Common genetic variation near the phospholamban gene is associated with cardiac repolarisation: meta-analysis of three genome-wide association studies. PLoS One. 2009;4:e6138. doi: 10.1371/journal.pone.0006138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaab S, Darbar D, van Noord C, et al. Large scale replication and meta-analysis of variants on chromosome 4q25 associated with atrial fibrillation. Eur Heart J. 2009 doi: 10.1093/eurheartj/ehn578. [DOI] [PMC free article] [PubMed] [Google Scholar]