Abstract
Cytochrome oxidase (COX) activity varies between individuals and low activities associate with Alzheimer’s disease. Whether genetic heterogeneity influences function of this multimeric enzyme is unknown. To explore this we sequenced 3 mitochondrial DNA (mtDNA) and 10 nuclear COX subunit genes from at least 50 individuals. 20% had non-synonymous mtDNA COX gene polymorphisms, 12% had a COX4I1 non-synonymous G to A transition, and other genes rarely contained non-synonymous polymorphisms. Frequent untranslated region (UTR) polymorphisms were seen in COX6A1, COX6B1, COX6C, and COX7A1; heterogeneity in a COX7A1 5′ UTR Sp1 site was extensive. Synonymous polymorphisms were common and less frequent in the more conserved COX1 than the less conserved COX3, suggesting at least in mtDNA synonymous polymorphisms experience selection pressure and are not functionally silent. Compound gene variations occurred within individuals. To test whether variations could have functional consequences we studied the COX4I1 G to A transition and an AGCCCC deletion in the COX7A1 5′ UTR Sp1 site. Cells expressing the COX4I1 polymorphism had reduced COX Vmax activity. In reporter construct-transduced cells where green fluorescent protein expression depended on the COX7A1 Sp1 site, AGCCCC deletion reduced fluorescence. Our findings indicate COX subunit gene heterogeneity is pervasive and may mediate COX functional variation.
Keywords: Alzheimer’s disease, cytochrome oxidase, mitochondria, mitochondrial DNA, polymorphisms
INTRODUCTION
Cytochrome oxidase (COX), the terminal complex of the mitochondrial electron transport chain (ETC), is the major site of cell oxygen consumption [1, 2]. It exists as a 13 subunit holoenzyme. Three subunit proteins are encoded by mitochondrial DNA (mtDNA) genes. The remaining 10 subunits are encoded by nuclear DNA (nDNA) genes, but because tissue-specific isoforms exist the actual number of nDNA COX genes actually exceeds 10 [3–6].
COX Vmax activities are reported from human subjects with and without particular diseases. While it is not possible to compare absolute activities across studies using different tissues or assay methods, several generalizations are possible. First, severe COX dysfunction occurs in certain rare diseases and is associated with devastating health consequences [7]. Second, most persons with Alzheimer’s disease (AD), a common disorder, have systemically low or below normal range COX activities [8, 9]. Third, the “normal” population shows substantial activity variation. In many studies control group standard deviations are 20–45% of their mean [8, 10–15]. For studies providing control subject COX activity scatter plots the highest values may exceed the lowest values by over 100% [8, 14, 15]. While some of this variability likely reflects assay precision factors, the magnitude of the variability strongly suggests COX Vmax activity varies greatly between individuals.
COX gene mutations probably reduce COX activity. COX2 (mt-CO2) is an mtDNA gene, it accumulates somatic mutations, and heteroplasmy is possible. Heteroplasmic COX2 mutation levels are known to inversely correlate with brain COX activity [16]. Heteroplasmic deletion of mtDNA COX genes also occurs, interferes with mtDNA COX gene expression, and likely impacts holoenzyme function [17]. To our knowledge, though, no one has evaluated whether inherited COX gene polymorphisms are functionally relevant to the general population. To evaluate this question we sequenced 3 mtDNA and 10 nuclear DNA COX subunit genes from at least 50 people and determined potential functional consequences of common representative polymorphisms.
MATERIALS AND METHODS
Human Subjects and DNA Preparation
Potassium EDTA tubes were used to collect human subject blood samples. Subject recruitment was mostly accomplished by soliciting volunteers at chapter meetings of civic groups located in the Commonwealth of Virginia; several subjects were patients or spouses of patients evaluated at the University of Virginia Health System. Prior to phlebotomy subjects signed the institutional review board (IRB)-approved consent form for this IRB-approved study. Human participation in this study was in accord with the Helsinki Declaration of 1975. Subjects had to be functionally independent and could not be diagnosed with a neurodegenerative disease. Most were male. Genomic DNA prepared from many of these subjects constituted the “control” group in a previously reported Parkinson’s disease case-control association study, and additional characteristics of these subjects are included in that report [18]. Genomic DNA was prepared using a Qiagen blood DNA purification kit.
DNA Amplification, Sequencing, and Restriction Analysis
DNA sequences for 13 COX subunit genes were obtained from Ensembl (http://www.ensembl.org/index.html). For subunits in which tissue specific isoforms exist, we selected one isoform for sequence analysis. The open reading frames for each gene analyzed were amplified and sequenced using a dideoxynucleotide sequencing approach. In addition to all open reading frame (ORF) regions, complete (COX4I1, COX6A1, COX6B1, COX6C, COX7A1) or partial (COX5A, COX5B, COX7B, COX7C, COX8A) 5′ and 3′ untranslated region (UTR) sequences were also determined. Table 10 lists the genes that were sequenced along with the PCR primers used to amplify them. For each gene analyzed, DNA from at least 50 different individuals was amplified and sequenced. For mtDNA genes, nucleotide numbering is per the reference Cambridge Sequence (rCS)[19].
Table 10.
COX subunit genes studied and PCR primers used to amplify them.
| Gene/Location/Ensembl Gene ID | PCR Primers |
|---|---|
| mt-CO1 (COX1) mtDNA nucleotides 5,905–7,446 ENSG00000198804 |
5-AGCACCCTAATCAACTGGCTTCAA-3 (rCS 5701-57204) 5-CTTCGCAGGCGGCAAAGACTA-3 (rCS 10659–10679) |
| mt-CO2 (COX2) mtDNA nucleotides 7,587–8,270 ENSG00000198712 |
5-AGCACCCTAATCAACTGGCTTCAA-3 5-CTTCGCAGGCGGCAAAGACTA-3 |
| mt-CO3 (COX3) mtDNA nucleotides 9,208–9,988 ENSG00000198938 |
5-AGCACCCTAATCAACTGGCTTCAA-3 5-CTTCGCAGGCGGCAAAGACTA-3 |
| COX4 Isoform 1 (COX4I1) 16q24.1 (Ensembl); 16q22-qter (Entrez) Ensembl nucleotides 85,833,196-85,840,608 Entrez nuceotides 84,390,697-84,398,109 ENSG00000131143 |
Exon 1/2U: 5-CTCCTGGAAAAGCGACTCG-3 Exon 1/2L: 5-CTTTATACACAGCAGGAGCAAA-3 Exon 3/4/5U: 5-TCCAGGGTTTCAAGGCGT-3 Exon 3/4/5L: 5-CAGGTTTCCAGTAAATAGGCA-3 |
| COX5A 15q24.1 (Ensembl); 15q24.1 (Entrez) Ensembl nucleotides 75,212,619-75,230,495 Entrez nucleotides 72,999,672-73,017,438 ENSG00000178741 |
Exon 1U: 5-TCACCTGACCAGAGACAAG-3 Exon 1L: 5-TTTCAGGTCCTCCACTACTC-3 Exon 2U:5-TGCTGCCACAACATATATAGTCAAC-3 Exon 2L: 5-GTGCCTGCCTTAAAATCCTG-3 Exon 3U: 5-CCCAGACAGATAAGATCATACATCA-3 Exon 3L: 5-ATGCTGCCAAAGTAGCCTCT-3 Exon 4U: 5-TCTGTCCTACCTGCCTCTGC-3 Exon 4L: 5-GCTCACGGCCATTACCTCTA-3 Exon 5U: 5- CAGTGATTTCCCTGGTTGTAGCAC-3 Exon 5L: 5-TGTAAGAGGGCAGCAAAACCA-3 |
| COX5B 2q11.2 (Ensembl); 2cen-q13 (Entrez) Ensembl nucleotides 98,262,521-98,264,654 Entrez nucleotides 97,628,953-97,631,089 ENSG00000135940 |
Exon 1U: 5-TCCCAGCGTTATTAAAGG-3 Exon 1L: 5-AGAATTACAGGGAGATGC-3 Exon 2/3U: 5-GGTACTTGGTGGTTCTTAGG-3 Exon 2/3L: 5-ATAATGAGGCTTGAGAGTGTC-3 Exon 4U: 5-GCCTCAATTTCTTCATCTG-3 Exon 4L: 5-ATCTAAGCACTATACACTGG-3 |
| COX6A1 12q24.31 (Ensembl); 12q24.2 (Entrez) Ensembl nucleotides 120,875,904-120,878,532 Entrez nucleotides 119,360,287-119,362,915 ENSG00000111775 |
Exon 1/2U: 5-CGCACGAAGGAAACGGTAAAGC-3 Exon 1/2L: 5-GCACTAAGGCACACATAACGAAAAGAG-3 Exon 3U: 5-CCATCACAGTGTCTCCCGATACTACCC-3 Exon 3L: 5-TGAGCCACCGCACCTGACCAAG-3 |
| COX6B1 19q13.12 (Ensemble); 19q13.1 (Entrez) Ensemble nucleotides 36,139,155-36,149,683 Entrez nucleotides 40,830,995-40,841,524 ENSG00000126267 |
Exon 1U: 5-CCCAATAGAAAGTCGTAG-3 Exon 1L: 5-AACCGTATCATTGTTAGC-3 Exon 2U: 5-AACAGGCTCAGAGATACC-3 Exon 2L: 5-GGCTATACAGAGACTTGC-3 Exon 3U: 5-GCCCTCTTCCATTGATCC-3 Exon 3L: 5-TGAACAGCAGGTGATGAG-3 Exon 4U: 5-CCCAGGAGGAGGTAGAGG-3 Exon 4L: 5-AGAGGCATCAGCAGAAGG-3 Exon 5U: 5-GTCCATCCGTTCAGTTTCC-3 Exon 5L: 5-CAGAGATCGTGCCATTGC-3 |
| COX6C 8q22.2 (Ensembl); 8q22-q23 (Entrez) Ensembl nucleotides 100,890,372-100,905,895 Entrez nucleotides 100,959,548-100,975,071 ENSG00000164919 |
Exon 1U: 5-TGAACTCCTTCGGCTACCG-3 Exon 2L: 5-ACACAGTCACGACTAAATCC-3 Exon 2U: 5-GCTCCAATCAATGCTTCCAG-3 Exon 2L: 5-AGTCTGTGGTTCTTTGTTACG-3 Exon 3U: 5-ATCTGTCACCACCTCCACC-3 Exon 3L: 5-GTATCACTTCCACACCATCG-3 Exon 4U: 5-AATAGACCTCAGTTGATCCTC-3 Exon 4L: 5-TATGCTAGTAAGTGCCCTGC-3 |
| COX7A1 19q13.12 (Ensembl); 19q13.1 (Entrez) Ensemble nucleotides 36,641,824-36,643,771 Entrez nucleotides 41,333,664-41,335,611 ENSG00000161281 |
Exon 1U: 5-TGCTGCACCCCGTTGACTG-3 Exon 1L: 5-GCGTATTAGGGCGGGGTTTTCT-3 Exon 2/3U: 5-ATCCAAGCTATTACCCTCCTC-3 Exon 2/3L: 5-GAAACCAGCCACCTCTTACTC-3 Exon 4U: 5-GGCTTTACAATAGGAGTCC-3 Exon 4L: 5-TGAGAGGTAGCAACATCC-3 |
| COX7B Xq21.1 (Ensembl); Xq21.1 (Entrez) Ensembl nucleotides 77,154,971-77,160,881 Entrez nucleotides 77,041,617-77,047,537 ENSG00000131174 |
Exon 1U: 5-ACCACCCACCATACGTCATT-3 Exon 1L: 5-AAAGGAAAGCACACGAGAGG-3 Exon 2U: 5-TTCCTTGGCTTTCCTGATTG-3 Exon 2L: 5-CACAATCTCTCAAGTCCCTCCT-3 Exon 3U: 5-AGGACAGCAATCAGGTTTGG-3 Exon 3L: 5-GGGACAAAGTGAGACTTCGT-3 |
| COX7C 5q14.3 (Ensembl); 5q14 (Entrez) Ensembl nucleotides 85,913,784-85,916,581 Entrez nucleotides 85,949,540-85,952,339 ENSG00000127184 |
Exon 1U: 5-GGTCTGAACTACAATTCC-3 Exon 1L: 5-TTTCTGGCTATCATCTCC-3 Exon 2U: 5-TTAAGCAGATGATTTGAGATTC-3 Exon 2L: 5-GCCACATCTACACATAACC-3 Exon 3/4U: 5-TATAGAACACTAGGATGATTGG-3 Exon 3/4L: 5-AATAATAAAGTAGGCAAATGGC-3 |
| COX8A 11q13.1 (Ensemble); 11q12-q13 (Entrez) Ensembl nucleotides 63,742,079-63,774,014 Entrez nucleotides 63,498,655-63,500,591 ENSG00000176340 |
Exon 1U: 5-TGTCAATTGGCTGTTTCGAG-3 Exon 1L: 5-AGGCATTCTCAGGACCACCT-3 Exon 2U: 5-AGGGTTAGCGTAGCTTTGGACCTGCT-3 Exon 2L: 5-GCAGAAGAGGTGACTGGAAT-3 |
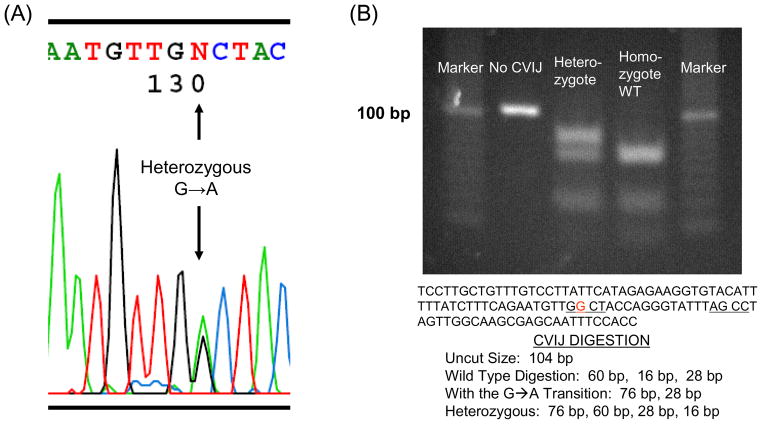
A COX4I1 polymorphism (rs11557187) was also assessed using a restriction fragment length polymorphism (RFLP) assay. For this assay, a 104 base pair amplicon was amplified from genomic DNA using the following primers: upper primer 5-TCCTTGCTGTTTGTCCTTAT-3, lower primer 5-GGTGGAAATTGCTCGCTT-3. The amplicon was digested with CviJ I (Chimerx, Milwaukee, WI) according to the manufacturer’s instructions. The digestion products were electrophoresed through a 4% agarose gel.
Lentivirus Constructs
Self-inactivating, third generation lentiviral vector constructs were prepared as described below. All final constructs were verified by sequencing.
A human COX4I1 cDNA was amplified from a pSPORT vector containing the COX4I1 cDNA (Open Biosystems catalog number IHS1380-97430887; Thermo Fisher Scientific). PCR amplification was accomplished using the following primers: upper primer 5′-gaggGAATTCgcggcgggcagtggcgg -3′, lower primer 5′-aggcgGCGGCCGCcatctctcgcttcttcc-3′; EcoRI (upper primer) and NOTI (lower primer) restriction sites were incorporated to facilitate ligation of the PCR product into a pCDH-EF1-MCS-T2A-copGFP expression lentivector (System Biosciences, catalogue number CD521A-1). This vector contains an elongation factor 1 (EF1) promoter as well as a green fluorescent protein (GFP) reporter gene expression cassette. To create a second vector a QuickChange II XL Site-Directed Mutagenesis Kit (Stratagene) was used to convert the rs11557187 COX4I1 single nucleotide polymorphism (SNP) from guanine to adenine prior to insertion into the lentivirus vector.
Lentivirus vector constructs containing short hairpin sequences that produce small inhibitor RNA (siRNA) molecules directed against the COX4I1 5′UTR or 3′UTR were also prepared. This was accomplished by inserting the following sequences into the pPS-H1-LCS vector (System Biosciences, catalogue number LF 523A-1): 5′-gagtcttcctcgatcccgtggtgct-3′ (directed against the 5′UTR) or 5′-cgccatgcaactccatgcctattt-3′ (directed against the 3′UTR).
Lentivirus constructs designed to assess COX7A1 5′UTR Sp1 site transcription activity were generated using double stranded oligonucleotides containing the COX7A1 5′UTR Sp1 binding region and its flanking sequence. The oligonucleotide corresponded to ENSG00000161281 nucleotides 305–395. One oligonucleotide was wild type sequence, and the other contained the rs72107438 AGCCCC COX7A1 exon 1 deletion. To allow for the creation of EcoRI and SpeI overhangs, an AATC nucleotide stretch was added to the 5′ end of the oligonucleotide and an additional adenine was added to the 3′ end. To create the modified vector we digested the pTRF1-NFkB-dscGFP vector (System Biosciences, catalogue number TR103PA-1) with EcoRI and SpeI, which allowed us to replace this vector’s NFkB transcription recognition element with our COX7A1 5′UTR Sp1 site-containing oligonucleotides.
Transfection, Transduction, and Identification of Stably Transduced Cells
Lentivector constructs were packaged using HEK 293T cells (which contain the simian virus 40 T antigen) co-transfected with the packaging plasmid psPAX2 and the envelope plasmid pMD2G (kindly provided by Dr. D. Trono, School of Life Sciences, Swiss Institute of Technology, Lausanne). ExGen 500 transfection reagent (Fermentas) was used to transfect the 293T cells, which were maintained in medium consisting of DMEM supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin. The complete lentiviral particles were harvested, concentrated using PEG-IT (System Biosciences), and the viral particles were used to transduce NT2 teratocarcinoma cells. For some transductions NT2 cells were exposed to only one species of viral particle, and for others they were exposed to multiple species of viral particles. Following transduction NT2 cells were cultured in DMEM with 10% FBS and penicillin-streptomycin. Approximately one week later, a FACSAria IIu fluorescence activated cell sorter (FACS) running DiVa6 software (BD Biosciences) was used to identify and isolate stably transduced NT2 cells, as the pCDH-EF1-MCS-T2A-copGFP, pPS-H1-LCS, and pTRF1-NFkB-dscGFP vectors express a gene for GFP.
Immunochemistry
Approximately 2 weeks after FACS, cells treated with different lentivirus particles were harvested and lysed using M-PER lysis buffer (Pierce). Lysate protein concentrations were measured using a DC Protein Assay Kit (Biorad). 50 mg of protein lysate were loaded into wells of 4–15% polyacrylamide gradient gels, electrophoresed at 200V for approximately 30 minutes, and transferred to nitrocellulose membranes at 100V for 1 hour. The membranes were stained with a primary antibody to COX4I1 (Invitrogen; catalogue number A21348) at a 0.5 microgram/ml final concentration, and then with a horseradish peroxidase-conjugated anti-mouse secondary antibody (Santa Cruz) at a 1:2000 dilution. To ensure equivalent loading between wells the membranes were also stained with Ponceau S.
Enzyme Assays
COX and citrate synthase (CS) Vmax activities were determined as previously described [20]. Vmax activities were measured in the native NT2 cell line, GFP-expressing NT2 cells exposed to the two siRNA vectors as well as the wild type COX4I1 vector, and NT2 cells exposed to the two siRNA vectors as well as the mutagenized COX4I1 vector. COX Vmax activities were first normalized for inter-sample differences in mitochondrial mass by dividing the COX Vmax activity of a given sample by the citrate synthase Vmax activity for that sample. Second, day-to-day inter-assay variation for the transfected cell groups was reduced by dividing the COX/CS activity from a particular transfected cell population on a particular day by the COX/CS activity of concurrently assayed native NT2 cells. Five independent assays were performed for each group.
Sp1 Reporter Assay
Approximately 1 week after exposure to lentivirus particles containing the wild type COX7A1 5′UTR or the AGCCCC-deleted version, transduced NT2 cells were identified and isolated by FACS. At this time the fluorescence intensity of the GFP-expressing cells was also determined. For each group of successfully transduced cells the mean fluorescence intensity was calculated. Three independent experiments were performed for each condition.
Statistical Analyses
For continuous variable data, mean ± SEM values from independent experiments were calculated. To compare group means we used a two-tailed Student’s t-test. P values less than 0.05 were considered significant. For categorical data, frequencies were calculated and compared using Fisher’s Exact test, with p values less than 0.05 considered significant.
RESULTS
Open Reading Frame Variations
Synonymous mtDNA COX gene polymorphisms were extremely common (Table 1). Over half (27/50) of our subjects had at least one synonymous polymorphism in COX1, COX2, or COX3. 13 subjects had multiple synonymous mtDNA COX gene polymorphisms. All polymorphisms were single nucleotide polymorphism (SNP) substitutions. 34 of the 36 unique substitutions we detected are listed by Mitomap as known coding region sequence polymorphisms. The remaining two, a T7142C SNP in COX1 and a T7861C SNP in COX2, are listed in Mitomap as “unpublished polymorphisms”. The most common individual synonymous mtDNA SNPs were T9656C and T9716 SNPs in COX3. Both were present in 8% of the subjects. The frequency of synonymous SNPs per nucleotides sequenced was greatest for CO3 (22 synonymous SNPs per 39,050 sequenced nucleotides) and least for CO1 (17 synonymous SNPs per 77,050 sequenced nucleotides). These frequencies were significantly different (p<0.05) (Table 2).
Table 1.
Synonymous mtDNA COX Gene Polymorphisms.
| Gene | # of Subjects | # of Unique SNPs | Total SNPs |
|---|---|---|---|
| mt-CO1 | 11/50 | 16 | 17 |
| mt-CO2 | 9/50 | 10 | 11 |
| mt-CO3 | 18/50 | 10 | 22 |
| Mt-CO1, CO2, or CO3 | 27/50* | 36 | 40 |
7 subjects had SNPs in two different genes, and 2 had SNPs in all 3 genes.
Table 2.
Distribution of mtDNA synonymous SNPs.
| Gene | Nucleotide Length | Subjects Sequenced | Nucleotides Sequenced | Synonymous SNPs | SNPs per Nucleotides Sequenced |
|---|---|---|---|---|---|
| Mt-CO1 | 1542 | 50 | 77,100 | 17 | 1/4535 |
| Mt-CO2 | 684 | 50 | 34,200 | 11 | 1/3109 |
| Mt-CO3 | 781 | 50 | 39,050 | 22 | 1/1775 |
Non-synonymous mtDNA COX gene polymorphisms were found in 20% (Table 3). There were seven unique SNPs that changed amino acids. 50% of these SNPs were in COX3, 40% in COX1, and 10% in COX2. Three SNPs were present in more than one person, but no SNP was found in more than 4%. Compound non-synonymous mtDNA COX gene SNPs were not observed for the 50 subjects analyzed. All non-synonymous mtDNA COX gene SNPs are listed in Mitomap as known variations.
Table 3.
Non-synonymous mtDNA COX Gene Polymorphisms.
| Gene | SNP | Amino Acid Change | Frequency |
|---|---|---|---|
| mt-CO1 | T6253C | Met→Thr | 1/50 |
| G6261A | Ala→Thr | 2/50 | |
| G6366A | Val→Ile | 1/50 | |
| mt-CO2 | G7859A | Asp→Asn | 1/50 |
| mt-CO3 | G9477A | Val→Ile | 2/50 |
| A9667G | Asn→Ser | 1/50 | |
| G9966A | Val→Ile | 2/50 | |
| Mt-CO1, CO2, or CO3 | - | - | 10/50 |
Nine distinct variations were found within nuclear COX gene open reading frames (ORFs). Six were synonymous (Table 4) and three were non-synonymous (Table 5). Five of the six synonymous variations were previously reported polymorphisms. The fifth, a previously unreported C→T substitution in COX5B, was present in 4%. The most common synonymous ORF substitution was a COX6C T→C transition. It was found in 30%, and all carriers were heterozygous. The Hardy-Weinberg calculation predicted 5 homozygotes would have been expected. The second most common synonymous ORF substitution was a COX6B1 C→T transition. It was found in 16%, one subject was homozygous, and allele distributions reflected Hardy-Weinberg equilibrium.
Table 4.
Synonymous nuclear COX Gene open reading frame polymorphisms.
| Gene/Exon | Nucleotide Change/Codon Change | Nucleotide Number/Amino Acid | Previously Reported? | Homozygous/Heterozygous |
|---|---|---|---|---|
| COX4I1 Exon3 |
C→T CTG→TTG |
254 Leu |
rs2599091 | 1/50 0/50 |
| COX4I1 Exon 3 |
T→C GAT→GAC |
298 Asp |
rs4885 | 1/50 0/50 |
| COX5B Exon 1 |
C→T CGC→CGT |
68 Arg |
No | 2/50 0/50 |
| COX6B Exon 3 |
C→T ACC→ACT |
137 Thr |
rs7991 | 7/50 1/50 |
| COX6C Exon 3 |
T→C TAC→TAT |
228 Tyr |
rs1130569 | 15/50 0/50 |
| COX8A Exon 1 |
C→A ATC→ATA |
185 Ile |
rs61759492 | 3/54 0/54 |
The nucleotide number refers to the nucleotide number of the cDNA (COX4I1=ENST00000253452; COX5b=ENST00000258424; COX6b=ENST00000392201; COX6C= ENST00000297564; COX8a=ENST00000314133).
Table 5.
Non-synonymous nuclear COX gene open reading frame polymorphisms.
| Gene/Exon | Nucleotide Change/Amino Acid Change | Nucleotide Number*/Amino Acid Number | Previously Reported? | Homozygous/Heterozygous |
|---|---|---|---|---|
| COX4I1 Exon 2 |
G→A Ala→Thr |
77 3 |
rs11557187 | 12/100 0/100 |
| COX5A Exon 3 |
C→T Thr→Ile |
381 76 |
No | 1/50 0/50 |
| COX5B Exon 4 |
A→G Gln→Arg |
421 125 |
rs71429371 | 1/50 0/50 |
The nucleotide number refers to the nucleotide number of the cDNA (COX4I1=ENST0000025342;COX5A=ENST00000322347;COX5B=ENST00000258424).
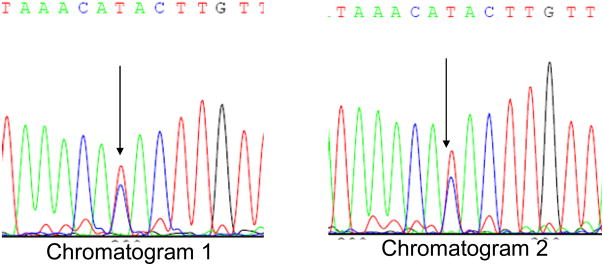
Only one common non-synonymous nuclear COX gene variation was found. This was a G→A substitution in COX4I1 exon 2 that changes the third amino acid in the protein from alanine to threonine (Figure 1). This is a previously recognized SNP. It was found in 12% of our subjects, making it the single most frequently encountered non-synonymous ORF COX gene substitution. All carriers were heterozygous. The other two non-synonymous SNPs were found only in single subjects. One of these was a previously reported COX5B SNP. The other, a COX5B exon 3 C→T substitution that changes threonine to isoleucine at amino acid 76, is not reported in dbSNP (Figure 2).
Figure 1.
A common non-synonymous COX4I1 substitution. (A) The chromatogram from a heterozygous subject. (B) The substitution was confirmed by RFLP.
Figure 2.
A novel non-synonymous COX5A substitution. The DNA sample was amplified and sequenced two independent times. Both chromatograms showed a heterozygous C→T substitution.
Untranslated Region Variations
Four different nuclear COX genes had UTR polymorphisms (Table 6). The most common was a C→T substitution in the COX6A1 3′UTR that was found in 49%. Both heterozygous and homozygous carriers were seen, and the heterozygous-homozygous distribution reflected Hardy-Weinberg equilibrium. There was a G→A substitution in the COX6B1 5′UTR that was present in 22%. One of the 11 carriers was homozygous. 28% carried an A→G substitution in the COX6C 5′UTR; all were heterozygous. The COX6A1, COX6B1, and COX6C UTR SNPS were previously reported.
Table 6.
Nuclear COX Gene untranslated region polymorphisms.
| Gene/Exon | Nucleotide Change/5′ or 3′ UTR | Nucleotide Number/Transcript Number | Previously Reported? | Homozygous/Heterozygous |
|---|---|---|---|---|
| COX6A1 Exon3 |
C→T 3′UTR |
546 ENST00000229379 |
rs8903 | 21/51 4/51 |
| COX6B Exon 1 |
G→A 5′UTR |
18 ENST00000246554 |
rs10420252 | 10/50 1/50 |
| COX6C Exon 2 |
A→G 5′UTR |
33 ENST00000297564 |
rs1130474 | 14/50 0/50 |
| COX7A1 Exon 1 |
A→C 5′UTR |
77 ENST00000292907 |
rs753420 | 17/51 4/51 |
| COX7A1 Exon 1 |
AGCCCC Deletion 5′UTR |
334–339 ENST00000292907 |
rs72107438 | 10/51 2/51 |
| COX7A1 Exon 1 |
AGCCCC Insertion 5′UTR |
340–345 ENST00000292907 |
rs72107438 | 2/51 0/51 |
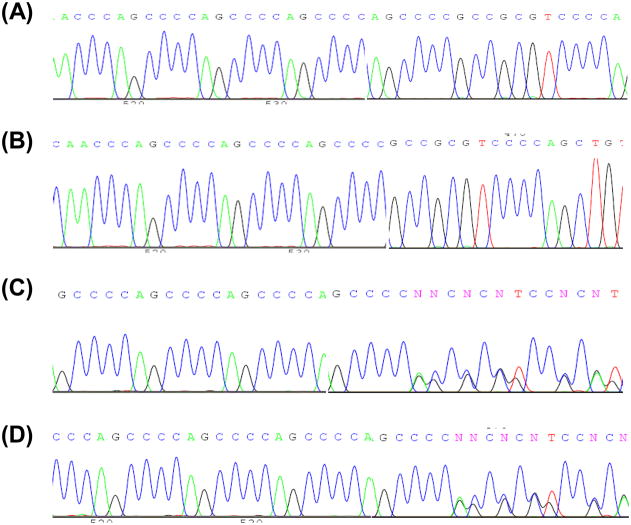
Three variants were found in the COX7A1 5′UTR (Table 6). The first, an A→C SNP at nucleotide 77 of the cDNA, was present in 41%. There were both heterozygous and homozygous carriers and the distribution reflected Hardy-Weinberg equilibrium. This SNP was previously reported. Further downstream variation was seen in a region containing AGCCCC hexanucleotide repeats (Figure 3). The wild type sequence contains four repeats, but an AGCCCC insertion-deletion variant is noted in dbSNP. We found 24% had an AGCCCC deletion; 4% of all subjects were homozygous. There was also an AGCCCC insertion in 4%. AGCCCC repeat number variation only occurred when the upstream SNP at nucleotide 77 was an adenine. Using these common COX7A1 exon 1 5′UTR variations it is possible to define four alleles (Table 7).
Figure 3.
A hexanucleotide insertion-deletion variation in the COX7A1 5′UTR. (A) The wild type sequence has four AGCCCC repeats. (B) Homozygous AGCCCC deletion. (C) Heterozygous AGCCCC deletion. (D) Heterozygous AGCCCC insertion.
Table 7.
Exon 1 5′UTR-defined COX7A1 alleles.
| Allele | Distribution Across 102 Alleles | Frequency |
|---|---|---|
| 77A, 4 repeat AGCCCC | 61/102 | 60% |
| 77A, 3 repeat AGCCCC | 14/102 | 14% |
| 77A, 5 repeat AGCCCC | 2/102 | 2% |
| 77C, 4 repeat AGCCCC | 25/102 | 25% |
Compound Variation and Overall Gene Variability
We characterized the non-synonymous SNP distribution pattern and determined if non-synonymous genotype variations occurred in combination. All but two amino acid-changing polymorphisms were in CO1, CO2, CO3, or COX4I1; we obtained complete sequence data for these four genes from 50 individual subjects. 28% of these subjects had at least one non-synonymous polymorphism and 6% had compound non-synonymous polymorphisms (Table 8). For these 50 subjects the non-synonymous polymorphisms defined 11 distinct genomes.
Table 8.
Non-synonymous SNP distribution.
| CO1 | CO2 | CO3 | COX4I1 | COX5A | COX5B | |
|---|---|---|---|---|---|---|
| 1 | G9477A | |||||
| 2 | C381T | A421G | ||||
| 3 | G6366A | |||||
| 4 | G9966A | |||||
| 5 | G9477A | |||||
| 6 | G6261A | |||||
| 7 | G9966A | G77A | ||||
| 8 | G77A | |||||
| 9 | T6253C | |||||
| 10 | G77A | |||||
| 11 | G7859A | |||||
| 12 | A9667G | |||||
| 13 | G6261A | G77A | ||||
| 14 | G77A |
Complete sequences for all 13 analyzed COX genes were obtained from 27 subjects (Table 9). When both ORF and UTR polymorphisms were considered, 26 of these subjects had at least one COX gene variation. Only one subject had no variations. COX6C polymorphisms in exon 2 and exon 3 formed a haplotype. No two subjects had exactly the same overall COX genotype.
Table 9.
Genotype variation among subjects sequenced completely for all 13 COX genes.
| CO1 | CO2 | CO3 | COX4I1 | COX5A | COX5B | COX6A | COX6B | COX6C | COX7A1 | COX7B | COX7C | COX8A | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | T9698T | T298C | C546T | ||||||||||
| 2 | C546T | A77C | |||||||||||
| 3 | T6827C | T9698C | C68T | ||||||||||
| 4 | A7385G | A7768G | G9477A | C546T | A77C | ||||||||
| 5 | A77C | ||||||||||||
| 6 | C381T | A421G | C546T | G18A | |||||||||
| 7 | T9656C | A33G C228T |
|||||||||||
| 8 | G7013A | T9716C | C546T | A33G C228T |
334-9 del | ||||||||
| 9 | |||||||||||||
| 10 | G6366A G6962A |
G9380A | C546T | C137T | A77C | ||||||||
| 11 | G18A | A33G C228T |
A77C | ||||||||||
| 12 | C546T | A33G C228T |
A77C | ||||||||||
| 13 | G18A | A33G C228T |
340-5 ins | ||||||||||
| 14 | A9254G T9386C |
A33G C228T |
A77C | ||||||||||
| 15 | G7912A | A77C | |||||||||||
| 16 | C546T | G18A | A33G C228T |
||||||||||
| 17 | T7142C | A77C | |||||||||||
| 18 | C546T | C137T | |||||||||||
| 19 | T6221C C6371T |
334-9 del | |||||||||||
| 20 | A7768G | T9540C | C546T | C137T | |||||||||
| 21 | T6216C G6260A |
C546T | G18A | A77C | C185A | ||||||||
| 22 | C546T | A33G C228T |
|||||||||||
| 23 | G7859A | T9899C | C546T | ||||||||||
| 24 | A9667G | A33G C228T |
|||||||||||
| 25 | A9254G G9380A |
334-9 del | |||||||||||
| 26 | G6261A | G77A | C546T | C137T | 334-9 del | ||||||||
| 27 | T7861C | T9656C | G18A | C228T | A77C |
Functional Consequences of Common Variants
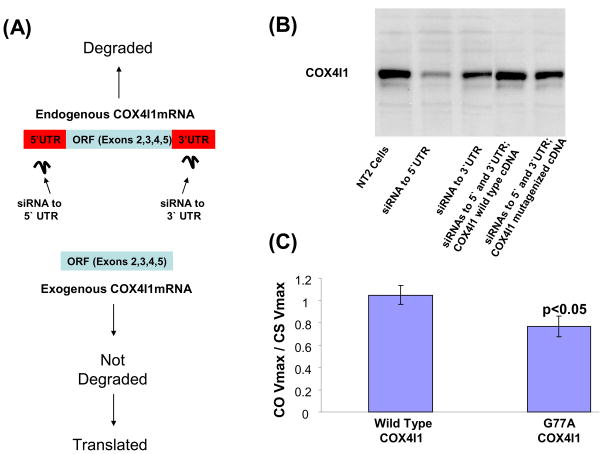
We further evaluated whether the most common non-synonymous polymorphism, the COX4I1 G77A SNP that changes an alanine to a threonine, could have functional consequences. This was accomplished using a lentiviral transduction strategy (Figure 4a). First, we prepared four different lentiviral vectors. The vectors contained one of the following inserts: a hairpin siRNA that targets the COX4I1 5′UTR, a hairpin siRNA that targets the COX4I1 3′UTR, a wild type COX4I1 cDNA sequence, and a COX4I1 cDNA in which the sequence was mutagenized from guanine to adenine at cDNA nucleotide position 77. In addition to our inserts, these vectors also contain a gene for GFP. This allowed us to sort for stably transduced cells following lentivirus exposures. Next, we showed stable transduction of NT2 teratocarcinoma cells with either the 5′UTR or 3′UTR siRNA vector reduced levels of endogenously-encoded COX4I1 protein (Figure 4b). We also showed that when NT2 cells were exposed to a mixture of three viral vectors (the 3′UTR siRNA vector plus the 5′UTR siRNA vector plus either the wild type COX4I1 vector or the mutagenized COX4I1 vector), COX4I1 protein levels were higher than they were when either of the siRNA plasmids was used in isolation (Figure 4b). Since the COX4I1 cDNAs lack 5′ and 3′ UTRs and are not targeted for degradation by the siRNAs, persistence of normal COX4I1 protein levels in cells also exposed to the siRNA vectors indicates expression of COX4I1 from the transduced COX4I1 cDNAs compensates for knock down of endogenous COX4I1 expression. Finally, we measured COX Vmax activities in cells expanded from GFP-producing NT2 cells originally exposed to both siRNA vectors and the wild type COX4I1 vector, and from GFP-producing NT2 cells originally exposed to both siRNA vectors and the mutagenized COX4I1 vector. To account for potential differences in mitochondrial mass that might exist between the different cell populations, COX activities were normalized to citrate synthase (CS) activity, and to minimize variability between independent experiments COX/CS values from the transfected cells were also normalized to COX/CS values from concurrently assayed non-transduced NT2 cells. For cells in which the wild type COX4I1 vector was used the mean relative COX/CS activity (1.16 ± 0.13) was significantly higher than it was for cells in which the mutagenized COX4I1 vector was used (0.78 ± 0.07) (Figure 4c).
Figure 4.
Evaluation of the COX4I1 G77A substitution. (A) The strategy for evaluating this polymorphism’s functional consequences took advantage of a commercially available COX4I1 cDNA’s lack of 5′ and 3′ UTRs. NT2 cells were co-exposed to three lentiviral constructs. One contained a hairpin siRNA to the COX4I1 5′UTR, one contained a hairpin siRNA to the COX4I1 3′UTR, and one contained either a wild type or G77A mutagenized COX4I1 cDNA. The siRNAs were designed to knock down expression of the endogenous COX4I1 protein, and the exogenous COX4I1 cDNA without these siRNA target sites was intended to restore COX4I1 protein. (B) Western blot using a COX4I1 antibody shows robust reduction of COX4I1 protein with expression of the 5′UTR siRNA and modest reduction with expression of the 3′UTR siRNA. To further ensure good endogenous COX4I1 knock down cells were treated with both siRNAs; co-treatment with either wild type or mutagenized COX4I1 cDNA in the siRNA-treated cells allowed COX4I1 protein levels to persist at a normal level. (C) COX Vmax activity was lower in NT2 cells expressing a COX4I1 cDNA with the G77A SNP than it was in NT2 cells expressing a COX4I1 cDNA with the wild type sequence. COX activities were normalized to citrate synthase activities to account for potential differences in mitochondrial mass, and to minimize variability between independent experiments COX/CS values for the transduced cells was also normalized to COX/CS values from concurrently assayed non-transduced NT2 cells.
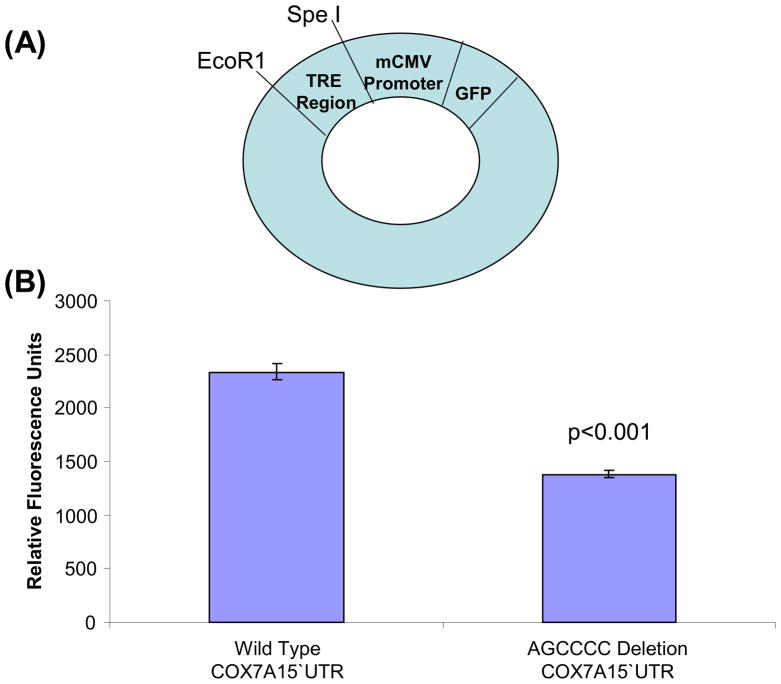
Although UTR substitutions do not change amino acid sequences we considered the possibility that UTR polymorphisms could still have functional consequences. Because the COX7A1 AGCCCC deletion falls within a COX7A1 Sp1 promoter site, we specifically evaluated if this deletion might affect gene expression. Two oligonucleotides containing the COX7A1 5′UTR Sp1 binding site and its flanking sequence were prepared, one with the wild type COX7A1 sequence and one containing an AGCCCC deletion. These oligonucleotides were ligated into lentiviral vectors in which a GFP gene is located downstream of a minimum CMV (mCMV) promoter. This vector is transcriptionally incompetent unless a functional transcriptional response element (TRE) is inserted upstream of the mCMV promoter region. Inserting either the wild type or deletion variant COX7A1 5′UTR oligonucleotide upstream of the mCMV promoter rendered it functional, most likely due to the presence of the COX7A1 5′UTR Sp1 binding site. NT2 cells transduced with this construct expressed GFP. GFP-expressing cells were identified using fluorescence activated cell sorting (FACS), isolated, and the fluorescence intensity of the GFP-expressing cells was determined. GFP fluorescence was greater in NT2 cells transduced with the vector that contained the wild type COX7A1 oligonucleotide sequence (2338 ± 78 relative fluorescence units) than it was in NT2 cells transduced with the construct that contained the AGCCCC-deleted COX7A1 oligonucleotide sequence (1379 ± 35 relative fluorescence units) (Figure 5).
Figure 5.
The wild type COX7A1 5′UTR Sp1 region is a more effective transcriptional response element (TRE) than the AGCCCC-deleted COX7A1 5′UTR region. (A) The different 5′UTR regions were inserted upstream of a minimum CMV (mCMV) promoter in a GFP expression plasmid, and served as a TRE. Successfully transduced NT2 cells were identified by fluorescence activated cell sorting. (B) When the TRE contained the wild type 5′UTR region fluorescence was increased relative to when the AGCCCC-deleted 5′UTR region served as the TRE.
DISCUSSION
We find unrelated individuals are unlikely to have identical cytochrome oxidase gene sequences. We provide evidence that suggests common cytochrome oxidase gene variations have functional consequences.
ORF regions from 13 cytochrome oxidase genes, complete UTRs from five cytochrome oxidase genes, and partial UTRs from five cytochrome oxidase genes were sequenced from 27 individuals. No two subjects had identical sequences. Sequencing the remaining UTRs, promoter regions, intron-exon borders, and unevaluated isoforms would likely reveal additional variation.
The importance of cytochrome oxidase gene variation to human health depends on how much it influences holoenzyme function. In general, the more a variant adversely affects function the more selection pressure it encounters [21, 22]. Non-synonymous SNPs were less common than synonymous SNPs and UTR variations, and are expected to incur the greatest degree of functional consequence. The number of persons with non-synonymous SNPs, though, was still relatively large. For the 50 individuals we sequenced COX1, COX2, COX3, and COX4I1 (the four genes containing most of the non-synonymous SNPs), 28% had at least one non-synonymous SNP.
Non-synonymous SNPs were more common in mtDNA genes than in nuclear DNA genes. This could reflect random occurrence, less overall conservation among the mtDNA genes, or the fact that mtDNA accumulates mutations more rapidly than nuclear DNA [23]. We suspect the latter possibility is most likely.
The single most common non-synonymous SNP was in the nuclear COX4I1 gene. To test the effects of the G77A transition we expressed a COX4I1 cDNA minor allele in a neuronal cell line while concurrently knocking down expression of the major allele. This required transducing cells with up to three vectors. It is known that simultaneously exposing cells to multiple lentiviral vectors creates cells that express the different vectors [24]. Despite this, we suspect heterogeneous populations of single, double, and triple-transduced cells were biochemically analyzed. It is also important to note the exogenous cDNA did not contain UTRs and that we do not know the ratio of minor to major allele expression in our experimental system. To minimize the impact of these variables we generated a control cell population that underwent the same exact transduction and selection procedure; the only experimental variable was whether nucleotide 77 was guanine or adenine. The most reasonable interpretation of our experiment is that having threonine instead of alanine at the COX4I1 third amino acid position reduces the cytochrome oxidase Vmax activity. Because of the factors discussed above, though, we believe it is best to assume that although activity is reduced by threonine substitution the magnitude of the reduction is not established. Genotyping additional subjects could provide further insight into this SNP’s biological relevance. All minor allele carriers we found were heterozygous. Failure to find homozygous minor allele carriers would suggest the G77A transition is disadvantageous.
Polymorphisms concentrated within UTRs. At least four of the 10 nuclear COX genes had common UTR variants. While not a cause of amino acid substitutions, UTR variations can and do influence gene expression. For example, microRNAs bind mRNA 3′UTRs; the presence or absence of microRNA binding to mRNAs can profoundly affect expression [25]. UTRs affect mRNA stability, target mRNAs to particular ribosome populations, and influence translation efficiency [26–29]. UTRs can also contain promoter binding regions [30]. We found AGCCCC deletion in a COX7A1 5′UTR Sp1 site was quite common. We used a transcription reporter system to experimentally test the effects of the AGCCCC deletion on gene expression and found it reduced expression of an adjacent GFP ORF. It is important to emphasize we did not critically evaluate the deletion’s affect on cytochrome oxidase holoenzyme function. Our experimental results, though, are consistent with the possibility that it could affect holoenzyme function by reducing levels of COX7A1 protein. This could be directly tested by measuring COX7A1 levels and cytochrome oxidase activity in AGCCCC deletion carriers.
Synonymous SNPs were not evenly distributed throughout the mtDNA COX genes; the synonymous SNP frequency was higher in COX3 than in COX1. The reasons for this are unclear but several explanations are possible. Primary sequence characteristics, DNA coiling, or protein associations might render COX3 more susceptible to mutation than COX1. Alternatively, COX1 may not tolerate variation as well as COX3. Increased phylogenetic conservation of COX1 is more consistent with the latter explanation [31–33]. Increased phylogenetic conservation indicates selection pressure against functionally consequential but not functionally inconsequential mutations should be greater in COX1. If correct, the relative reduction in COX1 synonymous SNPs would indicate functional relevance. Experimental data from eukaryotes [34] and prokaryotes [35, 36] also suggest synonymous SNPs are functionally important. In bacteria, replacing a specific codon with a synonymous codon alters translation efficiency. Changes in translation efficiency affect protein levels, protein folding, and ultimately protein function [35, 36]. These findings from bacteria are especially pertinent since mitochondria evolved from bacteria, retain prokaryotic characteristics, and have a protein-translation apparatus that resembles that of prokaryotes [37–40].
Our study revealed several other unique cytochrome oxidase subunit gene features which, to our knowledge, constitute novel observations. At least one gene, COX6C, shows haplotype variation. The COX7A1 5′UTR AGCCCC polymorphisms arose on the ancestral allele after the G77C SNP variant was established. Compound polymorphisms can define the cytochrome oxidase holoenzyme; it would be interesting to know how certain combinations interact to ultimately affect function. We also report one novel synonymous and one novel non-synonymous SNP, which raises a cautionary point. Some diseases have sporadic and Mendelian variants [41]. In some cases mutations in genes that cause the Mendelian variants are known. As sequencing technologies advance more and more persons with sporadic diseases are being tested for mutations in genes that cause Mendelian diseases. Our study suggests if enough sporadic patients are sequenced novel “mutations” in Mendelian disease-causing genes will be found. We predict in coming years these novel mutations will increasingly cause clinical and scientific angst.
It is worth considering how genes encoding such an important enzyme accumulated so much variation. One possibility is experimental design limitations or data misinterpretation led us to wrongly conclude at least some common cytochrome oxidase gene variations have functional consequences. If the variations we detected are functionally silent they should not experience selection pressure and could easily expand in human populations. Selection pressure is minimized, though, if a gene variant’s adverse effects primarily manifest after reproductive senescence occurs. The most common cytochrome oxidase-associated disease, AD, is a late-life disorder [42]. The common late-onset form is a pseudo-sporadic disease since Mendelian inheritance is not obvious in most but a positive family history does imply increased risk [43]. Persons with an affected father or mother have an increased lifetime AD risk, but that risk is greater when the affected parent is the mother [44]. Multiple studies using different approaches have recently concluded a maternally inherited genetic factor or factors contribute to presymptomatic AD carrier states or AD endophenotypes [45–47]. Our study does not address the role of cytochrome oxidase function or cytochrome oxidase genes in AD or AD risk, but cytochrome oxidase genetics could explain why AD subjects have systemically low cytochrome oxidase activity. Cytochrome oxidase genetics are also compatible with a pseudo-sporadic epidemiology in which gene contributions from both parents influence risk but maternal inheritance contributes more than paternal inheritance [48, 49].
Acknowledgments
This work was supported by the National Institutes of Health (AG022407 to R.H.S.) and the Parkinson’s Foundation of the Heartland (local grant to R.H.S.). The University of Kansas School of Medicine’s flow cytometry core lab is funded by the National Institutes of Health COBRE program of the National Center for Research Resources (P20 RR016443). The core is directed by Joyce Slusser, PhD.
References
- 1.Capaldi RA. Structure and assembly of cytochrome c oxidase. Arch Biochem Biophys. 1990;280:252–262. doi: 10.1016/0003-9861(90)90327-u. [DOI] [PubMed] [Google Scholar]
- 2.Capaldi RA. Structure and function of cytochrome c oxidase. Annu Rev Biochem. 1990;59 :569–596. doi: 10.1146/annurev.bi.59.070190.003033. [DOI] [PubMed] [Google Scholar]
- 3.Lenka N, Vijayasarathy C, Mullick J, Avadhani NG. Structural organization and transcription regulation of nuclear genes encoding the mammalian cytochrome c oxidase complex. Prog Nucleic Acid Res Mol Biol. 1998;61:309–344. doi: 10.1016/s0079-6603(08)60830-2. [DOI] [PubMed] [Google Scholar]
- 4.Taanman JW. Human cytochrome c oxidase: structure, function, and deficiency. J Bioenerg Biomembr. 1997;29:151–163. doi: 10.1023/a:1022638013825. [DOI] [PubMed] [Google Scholar]
- 5.Huttemann M, Kadenbach B, Grossman LI. Mammalian subunit IV isoforms of cytochrome c oxidase. Gene. 2001;267 :111–123. doi: 10.1016/s0378-1119(01)00385-7. [DOI] [PubMed] [Google Scholar]
- 6.Huttemann M, Muhlenbein N, Schmidt TR, Grossman LI, Kadenbach B. Isolation and sequence of the human cytochrome c oxidase subunit VIIaL gene. Biochim Biophys Acta. 2000;1492:252–258. doi: 10.1016/s0167-4781(00)00087-7. [DOI] [PubMed] [Google Scholar]
- 7.Shoubridge EA. Cytochrome c oxidase deficiency. Am J Med Genet. 2001;106 :46–52. doi: 10.1002/ajmg.1378. [DOI] [PubMed] [Google Scholar]
- 8.Parker WD, Jr, Filley CM, Parks JK. Cytochrome oxidase deficiency in Alzheimer’s disease. Neurology. 1990;40 :1302–1303. doi: 10.1212/wnl.40.8.1302. [DOI] [PubMed] [Google Scholar]
- 9.Swerdlow RH, Kish SJ. Mitochondria in Alzheimer’s disease. Int Rev Neurobiol. 2002;53 :341–385. doi: 10.1016/s0074-7742(02)53013-0. [DOI] [PubMed] [Google Scholar]
- 10.Bosetti F, Brizzi F, Barogi S, Mancuso M, Siciliano G, Tendi EA, Murri L, Rapoport SI, Solaini G. Cytochrome c oxidase and mitochondrial F1F0-ATPase (ATP synthase) activities in platelets and brain from patients with Alzheimer’s disease. Neurobiol Aging. 2002;23 :371–376. doi: 10.1016/s0197-4580(01)00314-1. [DOI] [PubMed] [Google Scholar]
- 11.Mancuso M, Filosto M, Bosetti F, Ceravolo R, Rocchi A, Tognoni G, Manca ML, Solaini G, Siciliano G, Murri L. Decreased platelet cytochrome c oxidase activity is accompanied by increased blood lactate concentration during exercise in patients with Alzheimer disease. Exp Neurol. 2003;182 :421–426. doi: 10.1016/s0014-4886(03)00092-x. [DOI] [PubMed] [Google Scholar]
- 12.Mutisya EM, Bowling AC, Beal MF. Cortical cytochrome oxidase activity is reduced in Alzheimer’s disease. J Neurochem. 1994;63 :2179–2184. doi: 10.1046/j.1471-4159.1994.63062179.x. [DOI] [PubMed] [Google Scholar]
- 13.Reichmann H, Florke S, Hebenstreit G, Schrubar H, Riederer P. Analyses of energy metabolism and mitochondrial genome in post-mortem brain from patients with Alzheimer’s disease. J Neurol. 1993;240 :377–380. doi: 10.1007/BF00839971. [DOI] [PubMed] [Google Scholar]
- 14.Valla J, Schneider L, Niedzielko T, Coon KD, Caselli R, Sabbagh MN, Ahern GL, Baxter L, Alexander G, Walker DG, Reiman EM. Impaired platelet mitochondrial activity in Alzheimer’s disease and mild cognitive impairment. Mitochondrion. 2006;6 :323–330. doi: 10.1016/j.mito.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Zuylen AJ, Bosman GJ, Ruitenbeek W, Van Kalmthout PJ, De Grip WJ. No evidence for reduced thrombocyte cytochrome oxidase activity in Alzheimer’s disease. Neurology. 1992;42 :1246–1247. doi: 10.1212/wnl.42.6.1246. [DOI] [PubMed] [Google Scholar]
- 16.Lin MT, Simon DK, Ahn CH, Kim LM, Beal MF. High aggregate burden of somatic mtDNA point mutations in aging and Alzheimer’s disease brain. Hum Mol Genet. 2002;11 :133–145. doi: 10.1093/hmg/11.2.133. [DOI] [PubMed] [Google Scholar]
- 17.Hsieh RH, Hou JH, Hsu HS, Wei YH. Age-dependent respiratory function decline and DNA deletions in human muscle mitochondria. Biochem Mol Biol Int. 1994;32 :1009–1022. [PubMed] [Google Scholar]
- 18.Swerdlow RH, Weaver B, Grawey A, Wenger C, Freed E, Worrall BB. Complex I polymorphisms, bigenomic heterogeneity, and family history in Virginians with Parkinson’s disease. J Neurol Sci. 2006;247:224–230. doi: 10.1016/j.jns.2006.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 20.Ghosh S, Patel N, Rahn D, McAllister J, Sadeghi S, Horwitz G, Berry D, Wang KX, Swerdlow RH. The thiazolidinedione pioglitazone alters mitochondrial function in human neuron-like cells. Mol Pharmacol. 2007;71 :1695–1702. doi: 10.1124/mol.106.033845. [DOI] [PubMed] [Google Scholar]
- 21.Schmidt S, Gerasimova A, Kondrashov FA, Adzhubei IA, Kondrashov AS, Sunyaev S. Hypermutable non-synonymous sites are under stronger negative selection. PLoS Genet. 2008;4 :e1000281. doi: 10.1371/journal.pgen.1000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kryukov GV, Schmidt S, Sunyaev S. Small fitness effect of mutations in highly conserved non-coding regions. Hum Mol Genet. 2005;14 :2221–2229. doi: 10.1093/hmg/ddi226. [DOI] [PubMed] [Google Scholar]
- 23.Brown WM, George M, Jr, Wilson AC. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci U S A. 1979;76 :1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albagli-Curiel O, Lecluse Y, Pognonec P, Boulukos KE, Martin P. A new generation of pPRIG-based retroviral vectors. BMC Biotechnol. 2007;7 :85. doi: 10.1186/1472-6750-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136 :215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andreassi C, Riccio A. To localize or not to localize: mRNA fate is in 3′UTR ends. Trends Cell Biol. 2009;19:465–474. doi: 10.1016/j.tcb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Chabanon H, Mickleburgh I, Hesketh J. Zipcodes and postage stamps: mRNA localisation signals and their trans-acting binding proteins. Brief Funct Genomic Proteomic. 2004;3 :240–256. doi: 10.1093/bfgp/3.3.240. [DOI] [PubMed] [Google Scholar]
- 28.Hughes TA. Regulation of gene expression by alternative untranslated regions. Trends Genet. 2006;22 :119–122. doi: 10.1016/j.tig.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 29.Mendell JT, Dietz HC. When the message goes awry: disease-producing mutations that influence mRNA content and performance. Cell. 2001;107 :411–414. doi: 10.1016/s0092-8674(01)00583-9. [DOI] [PubMed] [Google Scholar]
- 30.Yu M, Jaradat SA, Grossman LI. Genomic organization and promoter regulation of human cytochrome c oxidase subunit VII heart/muscle isoform (COX7AH) Biochim Biophys Acta. 2002;1574 :345–353. doi: 10.1016/s0167-4781(02)00228-2. [DOI] [PubMed] [Google Scholar]
- 31.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3 :294–299. [PubMed] [Google Scholar]
- 32.Garcia-Horsman JA, Barquera B, Rumbley J, Ma J, Gennis RB. The superfamily of heme-copper respiratory oxidases. J Bacteriol. 1994;176 :5587–5600. doi: 10.1128/jb.176.18.5587-5600.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown WM. The mitochondrial genome of animals. In: MacIntyre RJ, editor. Molecular Evolutionary Genetics. New York: Plenum Press; 1985. pp. 95–130. [Google Scholar]
- 34.Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315 :525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 35.Komar AA, Guillemet E, Reiss C, Cullin C. Enhanced expression of the yeast Ure2 protein in Escherichia coli: the effect of synonymous codon substitutions at a selected place in the gene. Biol Chem. 1998;379 :1295–1300. [PubMed] [Google Scholar]
- 36.Komar AA, Lesnik T, Reiss C. Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett. 1999;462 :387–391. doi: 10.1016/s0014-5793(99)01566-5. [DOI] [PubMed] [Google Scholar]
- 37.Emelyanov VV. Rickettsiaceae, rickettsia-like endosymbionts, and the origin of mitochondria. Biosci Rep. 2001;21 :1–17. doi: 10.1023/a:1010409415723. [DOI] [PubMed] [Google Scholar]
- 38.Punj V, Chakrabarty AM. Redox proteins in mammalian cell death: an evolutionarily conserved function in mitochondria and prokaryotes. Cell Microbiol. 2003;5 :225–231. doi: 10.1046/j.1462-5822.2003.00269.x. [DOI] [PubMed] [Google Scholar]
- 39.Spremulli LL, Coursey A, Navratil T, Hunter SE. Initiation and elongation factors in mammalian mitochondrial protein biosynthesis. Prog Nucleic Acid Res Mol Biol. 2004;77:211–261. doi: 10.1016/S0079-6603(04)77006-3. [DOI] [PubMed] [Google Scholar]
- 40.Smeitink JA, Elpeleg O, Antonicka H, Diepstra H, Saada A, Smits P, Sasarman F, Vriend G, Jacob-Hirsch J, Shaag A, Rechavi G, Welling B, Horst J, Rodenburg RJ, van den Heuvel B, Shoubridge EA. Distinct clinical phenotypes associated with a mutation in the mitochondrial translation elongation factor EFTs. Am J Hum Genet. 2006;79 :869–877. doi: 10.1086/508434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swerdlow RH. The Neurodegenerative Mitochondriopathies. J Alzheimers Dis. 2009 doi: 10.3233/JAD-2009-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swerdlow RH. Is aging part of Alzheimer’s disease, or is Alzheimer’s disease part of aging? Neurobiol Aging. 2007;28:1465–1480. doi: 10.1016/j.neurobiolaging.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 43.Swerdlow RH. Pathogenesis of Alzheimer’s disease. Clin Interv Aging. 2007;2 :347–359. [PMC free article] [PubMed] [Google Scholar]
- 44.Edland SD, Silverman JM, Peskind ER, Tsuang D, Wijsman E, Morris JC. Increased risk of dementia in mothers of Alzheimer’s disease cases: evidence for maternal inheritance. Neurology. 1996;47 :254–256. doi: 10.1212/wnl.47.1.254. [DOI] [PubMed] [Google Scholar]
- 45.Mosconi L, Brys M, Switalski R, Mistur R, Glodzik L, Pirraglia E, Tsui W, De Santi S, de Leon MJ. Maternal family history of Alzheimer’s disease predisposes to reduced brain glucose metabolism. Proc Natl Acad Sci U S A. 2007;104 :19067–19072. doi: 10.1073/pnas.0705036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Debette S, Wolf PA, Beiser A, Au R, Himali JJ, Pikula A, Auerbach S, Decarli C, Seshadri S. Association of parental dementia with cognitive and brain MRI measures in middle-aged adults. Neurology. 2009;73 :2071–2078. doi: 10.1212/WNL.0b013e3181c67833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Honea R, Swerdlow R, Vidoni E, Goodwin J, Burns J. Reduced gray matter volume in normal adults with a maternal family history of Alzheimer disease. Neurology. doi: 10.1212/WNL.0b013e3181c918cb. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swerdlow RH, Khan SM. A “mitochondrial cascade hypothesis” for sporadic Alzheimer’s disease. Med Hypotheses. 2004;63 :8–20. doi: 10.1016/j.mehy.2003.12.045. [DOI] [PubMed] [Google Scholar]
- 49.Swerdlow RH, Khan SM. The Alzheimer’s disease mitochondrial cascade hypothesis: an update. Exp Neurol. 2009;218 :308–315. doi: 10.1016/j.expneurol.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]