Abstract
MiR-107 is a microRNA (miRNA) that has been shown to have decreased expression in the temporal cortical gray matter of humans early in the progression of Alzheimer’s disease (AD). Here we study a new group of well-characterized brain samples (N=19). MiR-107 expression was assessed, normalized to other miRNAs. Quantitative AD histopathology was performed on adjacent tissue. Statistical regression analyses were performed comparing neuritic plaque (NP) and neurofibrillary tangle (NFT) counts. Correlation was observed between decreased miR-107 expression and NP counts (P<0.05), and NFT counts (P<0.02). Among these samples adjusted miR-107 and BACE1 mRNA levels tend to be correlated negatively as detected with qPCR (trend with regression P<0.07). In sum, miR-107 expression tended to be lower relative to other miRNAs as AD progresses.
Introduction
MicroRNAs (miRNAs) are short (~22 nucleotides) non-coding regulatory RNAs that serve vital functions in the mammalian brain. For example, miRNAs are important players in neurodevelopment and synapse function [1, 2]. Moreover, miRNAs also participate in (thus far mostly unknown) neuroprotective functions because after miRNA depletion neurons tend to degenerate relatively quickly [3–6].
Aside from their relevance to basic neurobiological processes, miRNAs may also contribute to specific neurodegenerative diseases. Aberrant miRNA biology has been implicated in Alzheimer’s disease (AD) [7–13]. However, this field is still in its infancy and the initial findings need to be refined, replicated, and better understood.
We previously described that the expression of a particular miRNA (miR-107) becomes downregulated in parallel with the progression of AD in human cerebral cortical gray matter [11]. This led to a specific hypothesis about how miRNAs can contribute to AD pathogenesis because miR-107 targets the beta-site amyloid precursor protein catalyzing enzyme 1 (BACE1) which is upregulated in AD brains [11].
Although miRNAs have been studied in AD brains, there has been limited validation of prior results. Here we sought to analyze samples from the University of Kentucky Alzheimer’s Disease Center (UK ADC) autopsy series to either validate or refute the earlier finding that miR-107 is altered in the course of AD. AD is defined neuropathologically by the presence of neurofibrillary tangles (NFTs) and neuritic plaques (NPs) [14]. Thus we assessed the level of miR-107 in the brain in correlation with NFT and NP counts using biochemical methods and histopathology in adjacent tissue sections.
Materials and Methods
RNA isolation from a human cerebral cortex
All analyses were performed blind with respect to patient information. Cases were selected on the bases of representing the full spectrum of clinical/pathological progression of AD, and of their being available more recently than our prior study [15] on miRNAs in AD brain. RNA was extracted from snap-frozen brain tissue in the superior and middle temporal gyri (SMT; Brodmann Areas 21/22) from the UK ADC under a University of Kentucky IRB protocol. Premortem clinical evaluations and pathological assessments were as described previously [15–17]. The inclusion criteria that were applied: post-mortem interval (PMI) <4hrs; no argyrophilic grains; no cortical Lewy bodies (LB); no evidence of frontotemporal dementia; no cancer in the brain parenchyma; and no large infarctions in the brain, or microinfarcts found within 3cm of the brain tissue samples. These neuropathological confounds were assessed using standard neuropathological procedures as described in detail elsewhere [16, 18]. NFTs and NPs were counted in the SMT as described previously in detail [16]. Included patient characteristics, along with pathological lesion counts in the same cases, are shown on Table 1.
Table 1.
Demographic and pathologic information along with RNA data on patient cohort.
| Case | Braak stage (0-VI) |
Age | Sex | PMI (hrs) |
Brain Weight (gms) |
SMT NP Counts |
SMT NFT Counts |
ApoE Alleles |
miR-107 (normalized arbitrary units) |
BACE1 mRNA (normalized arbitrary units) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | I | 75 | F | 3.50 | 1330 | 0 | 0 | 3/3 | 0.59 | 0.71 |
| 2 | I | 100 | F | 2.25 | 1320 | 0 | 0.2 | 2/3 | 0.61 | 0.71 |
| 3 | I | 84 | F | 3.00 | 900 | 0 | 0 | 3/3 | 0.86 | 0.47 |
| 4 | I | 84 | F | 2.50 | 1100 | 0 | 0 | 3/3 | 0.92 | 0.63 |
| 5 | IV | 96 | F | 2.50 | 970 | 0 | 1 | 3/3 | 0.68 | 0.42 |
| 6 | II | 92 | F | 3.00 | 1010 | 0 | 0 | 3/3 | 1.03 | 0.40 |
| 7 | IV | 78 | M | 1.20 | 1130 | 0 | 0 | 3/3 | 0.85 | 0.76 |
| 8 | IV | 91 | M | 2.00 | 1210 | 4.4 | 2.4 | 3/4 | 0.79 | 0.68 |
| 9 | IV | 84 | M | 3.50 | 1350 | 5.6 | 1.2 | 3/4 | 0.65 | 0.64 |
| 10 | III | 86 | F | 3.25 | 1130 | 5.8 | 0 | NA | 0.75 | 0.49 |
| 11 | III | 92 | F | 3.25 | 1220 | 8.2 | 1.2 | 2/3 | 0.71 | 0.72 |
| 12 | 0 | 92 | M | 3.33 | 1220 | 10 | 0 | 3/3 | 0.81 | 0.79 |
| 13 | V | 94 | F | 3.00 | 1070 | 10.8 | 15.8 | 3/3 | 0.82 | 1.20 |
| 14 | V | 94 | M | 2.00 | 1110 | 13.6 | 12.6 | 3/3 | 0.72 | 0.63 |
| 15 | III | 88 | M | 3.00 | 1130 | 16.4 | 0 | 3/3 | 0.69 | 1.27 |
| 16 | VI | 86 | M | 3.25 | 1150 | 16.6 | 41.4 | 3/4 | 0.52 | 0.94 |
| 17 | IV | 77 | M | 2.75 | 1340 | 16.6 | 5.6 | 3/4 | 0.80 | 0.46 |
| 18 | VI | 99 | F | 2.10 | 1150 | 20 | 23.6 | 3/4 | 0.55 | 0.94 |
| 19 | VI | 73 | M | 2.00 | 1130 | 35 | 42.6 | 3/3 | 0.59 | 1.31 |
19 cases were used in the study, arranged from lowest to highest in terms of neuritic plaque density. RNAs are quantified with qPCR--miR-107 is normalized to miR-124 and let-7a. BACE1 mRNA levels were normalized to beta-Actin mRNA. PMI=post-mortem interval. Brain weight is in grams. SMT=superior and mid-temporal cortical gyri. NP = Neuritic plaque. NFT = Neurofibrillary tangle. “Counts” refer to lesion densities (average of five highest 20× fields). N/A=Not available.
RNA was isolated as previously described in detail [11, 15]. Briefly, Prior to RNA extraction, gray matter was dissected away from white matter and only gray matter was used for these studies. RNA was isolated from brain tissue cut from superior and middle temporal cortex. Adjacent tissue was evaluated neuropathologically for all samples. Tissue (1–3 gms) that had been snap-frozen in liquid nitrogen and then transferred to a −80°C freezer was thawed in isotonic lysis buffer with RNAsin (Promega, Madison, WI; 250 U/ml) and Complete protease inhibitor pills (Roche, Basel Switzerland). Trizol LS (Invitrogen, Carlsbad, CA) was used according to manufacturer’s instructions, except for an additional overnight −20°C precipitation step during isopropanol precipitation. RNA quality was confirmed using A260/A280 readings.
RTqPCR and expression analyses
As with the RNA isolation, all steps prior to final data analyses were performed blindly in regard to clinical and pathological information. For qPCR analysis of miRNAs, total RNAs were subjected to RT-qPCR using miRNA detection kits (Ambion-Applied Biosystems, Foster City, CA). qPCR was performed using ABI Prism 7000 Sequence Detection System (Applied Biosystems, Foster City, CA) with separate standard curves for each miRNA. Three separate qPCR runs (with three replicates each) were performed on all the data and the data represent averaged values of the qPCR readouts. For qPCR analysis of BACE1 and β-Actin, RNAs were reverse-transcribed with qScript cDNA SuperMix (Quanta Biosciences, Gaithersburg, MD), amplified using TaqMan 2X PCR Master Mix (Applied Biosystems, Branchburg, NJ) and SYBR as detector. BACE1 levels were normalized to β-Actin levels.
MiR-107 expression, as indicated by qPCR, was normalized to the expressions of miR-124 and let-7a in the same cases. These control miRNAs were selected because we evaluated them previously in AD and nondemented brains and they were not altered in AD brain [11]. No other miRNA levels were evaluated.
Results
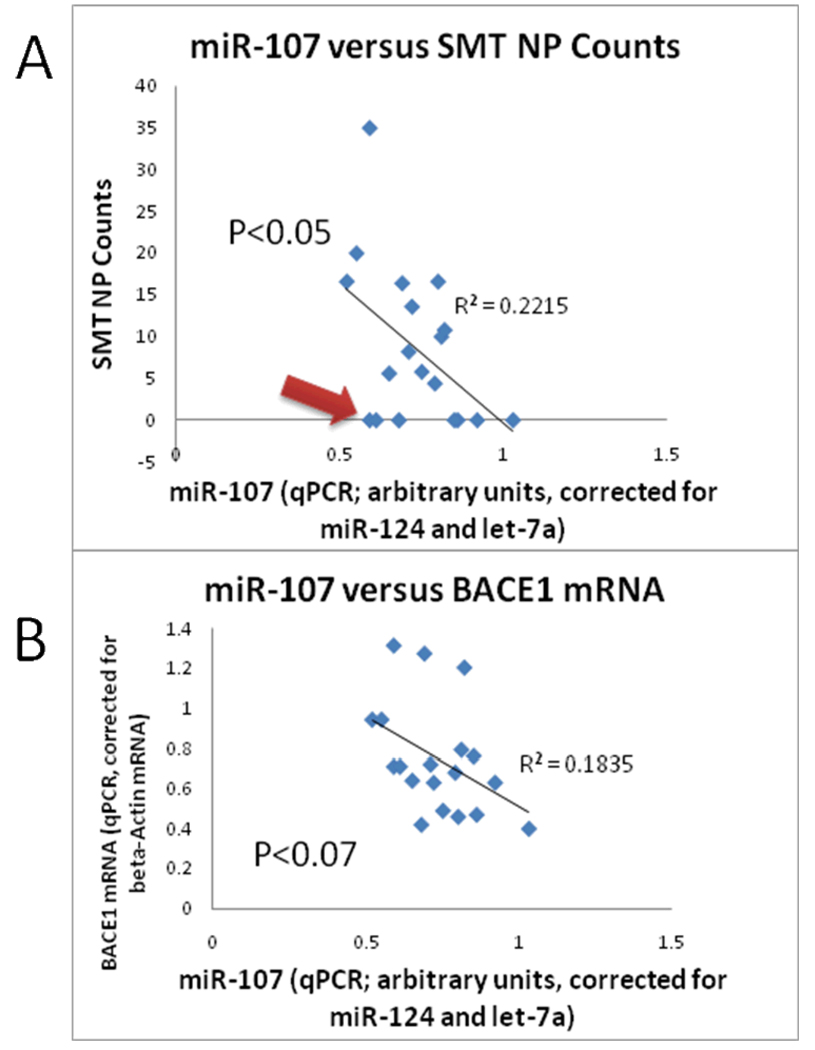
Levels of miR-107 (normalized to miR-124 and let-7a) were assessed and correlated with the amount of AD-type pathology in adjacent tissue sections. This approach was utilized rather than correlating solely with Braak Staging [19] of NFTs or Consortium to Establish a Registry for Alzheimer’s Disease scores of NP densities because the latter pathological rating scales provide ordinal rather than continuous variables for regression analyses. Linear regression data correlating the amount of AD-type pathology with the normalized miR-107 expression (Figure 1) indicated that miR-107 expression tended to correlate in a negative fashion with NPs (P<0.07) and NFTs (P<0.04). By contrast, miR-107 expression did not correlate in any way with age of death or PMI. When assessing the correlation between miR-107 expression and NP density in these human neocortical samples, the R-squared correlation coefficient was 0.19 (Figure 1). Note that there are several cases with relative low miR-107 levels despite also having low amount of AD-type pathology in adjacent tissue (Arrow in Figure 1).
Figure 1.
Charts shows the results of 19 cases evaluated, correlating the normalized expression of miR-107 with neuritic plaque lesion density counts in adjacent tissue sections from superior-mid temporal cortical (SMT) gyri (A) and BACE1 mRNA levels (B). There is a trend for decreased miR-107 in cases with more neuritic plaques (regression P<0.05). Note, however, that there were cases (red arrow) with low miR-107 expression despite lacking any SMT neuritic plaques. The negative correlation between miR-107 and BACE1 mRNA levels does not reach statistical significance (P<0.07).
Discussion
Brain samples from the UK ADC autopsy series were used as a validation cohort to test how miR-107 expression correlates with AD pathological markers. Gray matter tissue from the SMT was used and RNA isolated as described previously. MiRNAs were quantified using qPCR. This study supports the prior observation miR-107 expression decreases relative to other miRNAs as AD progresses[11].
There are limitations to the current study. The results are circumscribed – thus providing only an incremental gain of information – and pertain to the measurement of a few miRNAs in human brain. It has been shown previously that miRNA levels can be artifactually (and unpredictably) changed in the human brain postmortem [20]. In this study we used short PMI cases and did not see an effect in miRNAs that could be correlated with PMI. Nor was PMI different in high-pathology versus low-pathology cases. However, there is a possibility that unanticipated biases related to differential miRNA stability could affect our results.
Although we find in this validation cohort that miR-107 levels in brain tend to decrease in correlation to increased AD pathology, we also find (as in the prior study) that there is considerable overlap in miR-107 levels across the gamut of cases. For example, as shown in Figure 1, miR-107 levels are still relatively low in two brains that lacked appreciable AD pathology. In any case it is unknown how miR-107 levels can be determined in cerebrospinal fluid for in vivo analyses. These considerations are important because there have been suggestions of using miRNA levels as biomarkers for neurodegenerative diseases including AD [7]. MiR-107 expression in this portion of the brain does not appear to be a good candidate biomarker to differentiate the diseased from non-diseased brain.
If there is a trend for miR-107 to decrease in AD brain, what does it mean? We have previously identified a specific target of miR-107 (BACE1) that could provide links to AD pathogenesis [11]. This raises questions about biological factors upstream to miR-107 gene expression regulation. Evidence points to the roles of miR-107 in cellular metabolism [21, 22]. Levels of miR-107 expression in cultured cells are relatively high in high-glucose or low folate culture media conditions [21, 23]. In all known vertebrates, the genes for miR-107 and miR-103 reside within introns of pantothenate kinase genes. Pantothenate kinase polypeptides catalyze the enzymatic rate-determining step in Acetyl-CoA formation [22]. It is tempting to speculate that the miR-107 story may help link the known metabolic dysfunction seen very early in AD brains [24, 25] with the development of pathology in the disease course.
Acknowledgement
We are profoundly grateful to patients from the University of Kentucky Alzheimer’s Disease Center clinic. We thank Ms Willa Huang for technical and collegial assistance in the project, and Dr Zissimos Mourelatos for advice and insights. This study was supported by grants R01 NS061933 and K08 NS050110 from the National Institutes of Health, Bethesda, MD, and a New Investigator Research Grant from the Alzheimer Association.
References
- 1.Smalheiser NR, Lugli G. microRNA Regulation of Synaptic Plasticity. Neuromolecular Med. 2009 doi: 10.1007/s12017-009-8065-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kosik KS, Krichevsky AM. The Elegance of the MicroRNAs: A Neuronal Perspective. Neuron. 2005;47:779–782. doi: 10.1016/j.neuron.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 3.Cuellar TL, Davis TH, Nelson PT, Loeb GB, Harfe BD, Ullian E, McManus MT. Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in the absence of neurodegeneration. Proc Natl Acad Sci U S A. 2008;105:5614–5619. doi: 10.1073/pnas.0801689105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schaefer A, O'Carroll D, Tan CL, Hillman D, Sugimori M, Llinas R, Greengard P. Cerebellar neurodegeneration in the absence of microRNAs. J Exp Med. 2007;204:1553–1558. doi: 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 6.Bilen J, Liu N, Burnett BG, Pittman RN, Bonini NM. MicroRNA pathways modulate polyglutamine-induced neurodegeneration. Mol Cell. 2006;24:157–163. doi: 10.1016/j.molcel.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 7.Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, Kelnar K, Kemppainen J, Brown D, Chen C, Prinjha RK, Richardson JC, Saunders AM, Roses AD, Richards CA. Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 8.Hebert SS, Horre K, Nicolai L, Bergmans B, Papadopoulou AS, Delacourte A, De Strooper B. MicroRNA regulation of Alzheimer's Amyloid precursor protein expression. Neurobiol Dis. 2009;33:422–428. doi: 10.1016/j.nbd.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 9.Hebert SS, Horre K, Nicolai L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, Kauppinen S, Delacourte A, De Strooper B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer's disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci U S A. 2008;105:6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel N, Hoang D, Miller N, Ansaloni S, Huang Q, Rogers JT, Lee JC, Saunders AJ. MicroRNAs can regulate human APP levels. Mol Neurodegener. 2008;3:10. doi: 10.1186/1750-1326-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, Rigoutsos I, Nelson PT. The expression of microRNA miR-107 decreases early in Alzheimer's disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lukiw WJ, Zhao Y, Cui JG. An NF-kappaB-sensitive micro RNA-146a-mediated inflammatory circuit in Alzheimer disease and in stressed human brain cells. J Biol Chem. 2008;283:31315–31322. doi: 10.1074/jbc.M805371200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson PT, Wang WX, Rajeev BW. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol. 2008;18:130–138. doi: 10.1111/j.1750-3639.2007.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Consensus recommendations for the postmortem diagnosis of Alzheimer's disease. The National Institute on Aging, and Reagan Institute Working Group on Diagnostic Criteria for the Neuropathological Assessment of Alzheimer's Disease. Neurobiol Aging. 1997;18:S1–S2. [PubMed] [Google Scholar]
- 15.Wang WX, Wilfred BR, Baldwin DA, Isett RB, Ren N, Stromberg A, Nelson PT. Focus on RNA isolation: obtaining RNA for microRNA (miRNA) expression profiling analyses of neural tissue. Biochim Biophys Acta. 2008;1779:749–757. doi: 10.1016/j.bbagrm.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson PT, Jicha GA, Schmitt FA, Liu H, Davis DG, Mendiondo MS, Abner EL, Markesbery WR. Clinicopathologic correlations in a large Alzheimer disease center autopsy cohort: neuritic plaques and neurofibrillary tangles "do count" when staging disease severity. J Neuropathol Exp Neurol. 2007;66:1136–1146. doi: 10.1097/nen.0b013e31815c5efb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmitt FA, Davis DG, Wekstein DR, Smith CD, Ashford JW, Markesbery WR. "Preclinical" AD revisited: neuropathology of cognitively normal older adults. Neurology. 2000;55:370–376. doi: 10.1212/wnl.55.3.370. [DOI] [PubMed] [Google Scholar]
- 18.Nelson P, Jicha GA, Schmitt FA, Liu HL, Davis DG, Mendiondo MS, Abner EL, Markesbery WR. Clinicopathological Correlations in a Large Alzheimer’s Disease Center Autopsy Cohort: Neuritic Plaques and Neurofibrillary Tangles ‘Do Count’ When Staging Disease Sseverity. J Neuropathol Exp Neurol. 2007 doi: 10.1097/nen.0b013e31815c5efb. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 20.Sethi P, Lukiw WJ. Micro-RNA abundance and stability in human brain: Specific alterations in Alzheimer's disease temporal lobe neocortex. Neurosci Lett. 2009;459:100–104. doi: 10.1016/j.neulet.2009.04.052. [DOI] [PubMed] [Google Scholar]
- 21.Tang X, Muniappan L, Tang G, Ozcan S. Identification of glucose-regulated miRNAs from pancreatic {beta} cells reveals a role for miR-30d in insulin transcription. RNA. 2009;15:287–293. doi: 10.1261/rna.1211209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilfred BR, Wang WX, Nelson PT. Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol Genet Metab. 2007;91:209–217. doi: 10.1016/j.ymgme.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marsit CJ, Eddy K, Kelsey KT. MicroRNA responses to cellular stress. Cancer Res. 2006;66:10843–10848. doi: 10.1158/0008-5472.CAN-06-1894. [DOI] [PubMed] [Google Scholar]
- 24.Scarmeas N, Habeck CG, Hilton J, Anderson KE, Flynn J, Park A, Stern Y. APOE related alterations in cerebral activation even at college age. J Neurol Neurosurg Psychiatry. 2005;76:1440–1444. doi: 10.1136/jnnp.2004.053645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reiman EM, Caselli RJ, Yun LS, Chen K, Bandy D, Minoshima S, Thibodeau SN, Osborne D. Preclinical evidence of Alzheimer's disease in persons homozygous for the epsilon 4 allele for apolipoprotein E. N Engl J Med. 1996;334:752–758. doi: 10.1056/NEJM199603213341202. [DOI] [PubMed] [Google Scholar]