Abstract
Synthetic three-dimensional (3D) scaffolds for cell and tissue engineering routinely utilize peptide ligands to provide sites for cell adhesion and to promote cellular activity. Given the fact that recent studies have dedicated great attention to the mechanisms by which cell behavior is influenced by various ligands and scaffold material properties, it is surprising that little work to date has been carried out to investigate the influence of covalently-bound ligands on hydrogel material properties. Herein we report the influence of 3 common ligands utilized in tissue engineering, namely RGD, YIGSR, and IKVAV on the mechanical properties of cross-linked poly(ethylene glycol) (PEG) hydrogels. The effect of the ligands on hydrogel storage modulus, swelling ratio, mesh size, and also on the diffusivity of bovine serum albumin (BSA) through the hydrogel were investigated in detail. We identified conditions at which these ligands strikingly influence the properties of the material: the extent of influence and whether the ligand increases or decreases a specific property is linked to ligand type and concentration. Further, we pinpoint mechanisms by which the ligands interact with the PEG network. This work thus provides specific evidence for interactions between peptide ligands and cross-linked PEG hydrogels that significantly impact hydrogel material and transport properties. As a result, this work may have important implications for interpreting cell experiments carried out with ligand-modified hydrogels because the addition of ligand may affect not only the scaffold’s biological properties, but also key physical properties of the system.
Keywords: ligand, PEG hydrogel, swelling ratio, mesh size, storage modulus
1. Introduction
The adhesive ligand sequence RGD (arg-gly-asp) from fibronectin was first discovered in 1984 by Pierschbacher and Ruoslahti et al. [1]. It was recognized as the minimal active amino acid sequence necessary to promote cell adhesion. Soon after this report, it was confirmed that the RGD sequence is the cell attachment site of many other adhesive proteins such as collagen type I and laminin [2–6]. Since then, RGD, as well as other adhesive ligands such as the laminin derived YIGSR (tyr-ile-gly-ser-arg) and the fibrinogen derived IKVAV (ile-lys-val-ala-val), have been used in hundreds of applications to induce specific cell behaviors [7–10]. These applications include bone biology [11], cardioprogenitor differentiation [12], fibroblast cell migration [13, 14], vascular healing [15], axon guidance [16], support of differentiating marrow stromal osteoblasts [17], and stem cell applications [18].
Despite the fact that ligands retain only 10% to 30% of their biological activity as compared to the whole protein [19, 20], chemical conjugation of polymers with short peptides has several advantages: 1) higher stability against conformational change; 2) control of ligand density; 3) control of ligand orientation to possibly provide more favorable ligand-receptor interactions and cell adhesion; 4) benefits in terms of minimizing immune response and infection; 5) higher stability during sterilization; 6) easier storage and characterization; and 7) cost effectiveness [21].
While adhesive ligands have also been incorporated into natural materials (e.g., collagen [22, 23], fibrin [24], and laminin [25]) to enhance cell adhesion and proliferation, they have become a crucial component of synthetic materials for tissue engineering. Even though natural materials are inherently biocompatible and bioactive, synthetic materials offer a wider range of properties and lend themselves to easier processing and modification [26]. Furthermore, synthetic materials are an invaluable resource when deciphering the intricate relationship between the matrix components and their possible individual influence on cell behavior. For example, synthetic materials have well defined and controllable mechanical properties so they can serve as the “blank slate” or inert structural backbone of a scaffolding matrix. However, since the scaffolds lack bioactivity, whole proteins or short ligands must be incorporated to afford biological properties to the system. This approach allows for delineation of the effects of mechanical properties from the biological properties of the materials, a recognized issue of growing importance for the field of tissue engineering. Scaffold mechanical properties, such as stiffness, which is well represented by the storage modulus of the material, have been shown to have a tremendous effect on cell behavior and to have important implications for development, differentiation, disease progression and regeneration [27]. Specifically, material stiffness has been linked to promoting malignant behavior in tumors [28], to directing stem cell lineage [29], to guiding fibroblast cells movement [30], and to the spreading and organization of aortic smooth muscle cells [31].
To date, it has been generally assumed that an addition of a small amount of ligand to a matrix does not interfere significantly with the mechanical properties of the material and that these two properties can be tuned independently. While a tremendous amount of studies have been carried out on the influence of ligands on cell behavior, few studies have been undertaken to investigate the possible influence of ligands on the mechanical properties of the material [31].
This project was aimed at investigating the influence of the 3 most commonly used ligands in tissue engineering, namely RGDS, YIGSR, and IKVAV on the mechanical properties of a cross-linked poly(ethylene glycol) (PEG) hydrogel. Ligands covalently bound to the PEG hydrogel were compared to a negative control of bound monofunctional PEG-thiol (PEG-SH) of similar molecular weight. We tested the effect of the ligands on hydrogel stiffness, swelling ratio, mesh size, and also on the diffusivity of a model protein, bovine serum albumin (BSA), through the hydrogel. We have identified conditions under which the ligands profoundly influence hydrogel properties and performed additional tests in order to pinpoint the possible causes for these effects.
2. Materials and methods
All reagents were acquired from Fisher Scientific or Sigma Aldrich unless otherwise noted.
2.1. Synthesis of 4-arm poly(ethylene glycol)-vinyl sulfone (4-arm PEG-VS)
The synthesis of 4-arm PEG-VS was adapted from a previous protocol [32, 33] where 4-arm PEG-OH (10 kDa; Nektar, Huntsville, LA) was modified in the presence of excess divinyl sulfone. Briefly, PEG was dried by azeotropic distillation in toluene using a Dean-Stark trap and then dissolved in dry dichloromethane. Sodium hydride was added under Ar at a 5-fold molar excess over OH groups. After hydrogen evolution divinyl sulfone was added immediately at a 50-fold molar excess to OH groups. The solution was allowed to react for 3 d at room temperature under Ar and with constant stirring. The solution was then filtered and reduced in volume (~30 ml) by rotary evaporation. The polymer was recovered in ice-cold diethyl ether and dried under vacuum. The dry polymer was then dissolved in deionized water containing sodium chloride and extracted three times in dichloromethane. After drying with sodium carbonate, the polymer product was reduced in volume, precipitated, and dried as described above. The product was stored under Ar at −20°C until use. Derivatization was confirmed by 1H NMR (CDCL3): 3.6 ppm (982H, PEG backbone), 6.1 ppm (4H, 1H, =CH2), 6.4 ppm (4H, 1H, =CH2), 6.8 ppm (4H, 1H, −SO2CH=). The typical yield from this procedure was 80–90% and the degree of end group conversion, as shown by NMR, was 93–98%.
2.2. Adhesive ligands
The adhesive ligands GRCD-YIGSR-PD, GRCD-IKVAV-PD, GRCD-RGDS-PD and GRCD-YIGSR were purchased from CPC Scientific Inc., San Jose, CA (>90% purity; Table 1). The italic font designates the bioactive peptide sequence. The ligands were terminated on one end with the sequence GRCD which contains a cysteine residue to allow for covalent attachment to 4-arm PEG-VS [34]. The PD flanking group was chosen based on work by Lutolf et. al. [35].
Table 1.
Properties of adhesive ligands and PEG-SH control.
| Peptide Sequence | Net Charge | pI | Hydrophobicity | Molecular Weight, Da | Abbreviation |
|---|---|---|---|---|---|
| GRCD-YIGSR | +1 | 8.9 | 0.5 | 1026 | YIGSR |
| GRCD-RGDS-PD | −1 | 4.2 | 1.6 | 1077 | RGDS |
| GRCD-YIGSR-PD | 0 | 6.2 | 0.7 | 1238 | YIGSRPD |
| GRCD- IKVAV-PD | 0 | 6.2 | 0.5 | 1172 | IKVAV |
| PEG-SH | 0 | N/A | N/A | 1000 | PEG-SH |
To minimize weighing error, the ligand was aliquoted in advance in 5% acetic acid solution. The acidic solution kept the thiol groups of the cysteine residue protonated and thus prevented disulfide bonding between the peptides prior to reaction with PEG-VS. Each aliquot was used for up to 2 wks before discarding. The ligands were chosen such as to represent a large span of biochemical properties (Table 1) and thus a range of possible effects on hydrogel structure and function. Additionally, the sequences RGDS, IKVAV, YIGSR are the most commonly used in tissue engineering, including a system of our particular interest, neural tissue engineering [36, 37].
2.3. Formation of PEG-based hydrogels
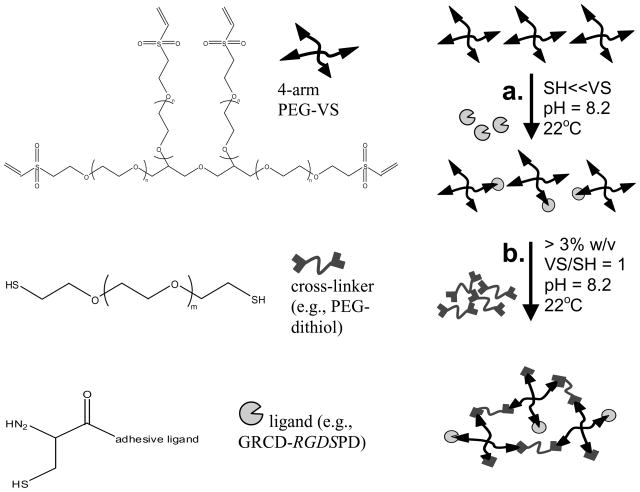
PEG hydrogels were made with a PEG-dithiol cross-linker (Laysan Bio Inc., Arab AL) and functionalized with adhesive ligands (Figure 1) with negative controls consisting of hydrogels without ligand and hydrogels made with mono-functional PEG-thiol (PEG-SH, 1000 Da) which is the same molecular weight as the adhesive peptide ligands but has the properties of the hydrogel backbone polymer.
Figure 1.
Schematic of the Michael-type addition reaction to form PEG hydrogels with covalently incorporated ligands. In the first step (a), 4-arm PEG-VS is functionalized with adhesive ligands via an unpaired cysteine residue. The ligand is added at a large stoichiometric deficit to VS groups. In a second cross-linking step (b), a PEG-dithiol cross-linker is added such that total VS/SH = 1:1 to form a 3D hydrogel network.
Briefly, adhesive ligand was added to 4-arm PEG-vinyl sulfone (PEG-VS) dissolved in 0.3 M triethanolamine (TEA) solution of pH 8.2 at a large stoichiometric deficit and left to react for 30 min. The cross-linker, PEG-dithiol was then added to give a final ratio of VS:SH of 1:1 for all hydrogel densities and types. TEA of pH 8.2 was used to dilute reagents to the desired final PEG concentration and hydrogel volume. The solution was vortexed for 30 sec and transferred to the center of a glass slide pre-treated with RainX (Sopus Products, Houston, TX) to provide a hydrophobic surface. Silicone spacers (1- or 2.5-mm thick cut from CoverWell perfusion chambers, Grace Bio-Labs, Bend, OR) were placed at the ends of the glass slide and a second hydrophobic slide was placed on top. The two slides were clamped together over the spacers with binder clips. The slides were then transferred to a humidified incubator to allow the hydrogel to cross-link at 37°C. Gelation occurred in several minutes but the hydrogels were left in the incubator for 1–2 h to achieve maximum cross-linking.
2.4. Efficiency of ligand incorporation in the PEG hydrogel
The fluorescently labeled ligand 5FAM-GRCD-RGDS-PD was used to estimate the efficiency of ligand incorporation in the PEG hydrogel, where 5FAM stands for 5-carboxyfluorescein (λex = 490 nm, λem = 520 nm). The ligand was added to the PEG-VS as described above and allowed to react for 30 min. The solution was then diluted with PBS and the unbound ligand was removed via a centrifugal filter device (Millipore, Bedford, MA; 5 kDa molecular weight cut-off). The amount of unbound ligand was quantified via measurement of the filtrate fluorescence with a Cary Eclipse spectrophotometer (Varian Inc., Walnut Creek, CA) and comparison to a calibration curve with lower ligand sensitivity limit of 0.1 μM.
In order to determine the extent to which ligand diffused out of the hydrogel prior to further characterization of the hydrogel properties (at 24 h), the hydrogels were prepared as described above. The hydrogels were then positioned in a cuvette filled with 10 mM PBS, pH 7.4 and were kept at room temperature and not moved until the end of the experiment. Over 24–72 h the fluorescence of the hydrogel and the supernatant PBS solution was measured. The PBS was transferred to a separate cuvette via a syringe and its fluorescence as well as that of the hydrogel itself were determined separately. After the measurement, the PBS was returned to the original cuvette containing the hydrogel. In cases where only the fluorescence of the hydrogel was measured, the supernatant PBS was replaced with fresh PBS prior to each measurement. To improve the accuracy of the measurements and avoid evaporation of the PBS, the solution was kept capped at all times. The experiment was performed at room temperature. The solution containing cuvette was covered with foil at all times (except during measurements) to prevent fluorophore photobleaching.
2.5. Hydrogel characterizations
Rheological measurements were performed with an AR 2000ex rheometer (TA Instruments) in parallel plate geometry with a 20-mm diameter acrylic upper plate, at 22°C, a frequency of 1–10 rad/s, and a constant 2% strain [38]. The hydrogel samples were prepared to yield discs of 20-mm diameter and 1-mm thick (thickness < diameter/4 per TA Instruments guide) following swelling in 10 mM phosphate buffer saline (PBS), pH 7.4. The excess water from the hydrogel surface was carefully blotted before measurement. Storage modulus (G′) at 1 rad/s was reported for each sample.
To estimate swelling ratio, hydrogels (50 μL) were soaked in 10 mM PBS, pH 7.4. Hydrogel samples were collected at regular intervals and their mass after swelling (MS) was measured. The hydrogels were then dried in an oven at 80°C for 24 h and their dry mass (MD) was measured. The swelling ratio based on hydrogel mass (QM) was calculated using Equation 1 [39]:
| (1) |
Flory-Rehner calculations were used to determine hydrogel mesh size (ξ). First the molecular weight between cross-links (Mc) was calculated by Equation 2 [40]:
| (2) |
where is the number-average molecular weight of the un-cross-linked hydrogel, V1 is the molar volume of the solvent (18 cm3/mol for water), ν2 is the polymer volume fraction in the equilibrium swollen hydrogel, v̄ is the specific volume of the polymer (ρs/ρp), and χ1 is the polymer-solvent interaction parameter (0.426 for PEG-water [39, 40]).
Mesh size was then determined as described by Canal and Peppas [41]. The root-mean-square end to end distance of the polymer chain in the unperturbed state ( ) was calculated using Equation 3:
| (3) |
where l is the average bond length (0.146 nm [42, 43]), Cn is the characteristic ratio of the polymer (typically 4.0 for PEG [43, 44]) and n is the number of bonds in the crosslink [13]:
| (4) |
where Mr is the molecular weight of the repeat unit (44 for PEG). Mesh size was then calculated by
| (5) |
The diffusion coefficient was estimated from bulk diffusion experiments. Briefly, the hydrogels were cross-linked in the presence of the solute BSA to achieve a final solute concentration in the hydrogel of 2% w/v. The hydrogels were then placed in 15-ml tubes filled with 10 mM PBS of pH 7.4 and mixed end-over-end at 22°C. At specified sample collection times, 1 ml of solution was transferred to a microfuge tube and was replaced in the 15-ml tube with fresh PBS. The solute content of each sample was analyzed with the Bio-Rad protein assay using the manufacturer’s microassay procedure.
Diffusion coefficient was calculated via a modified form of the Fick’s law for short release times [45]:
| (6) |
From the Equation 6, it follows that Mi/Minf is directly proportional to t1/2 and therefore a plot of Mi/Minf versus t1/2 can be used to find De. A mass balance was performed to calculate Mi:
| (7) |
where Ci is the concentration of solute in the release solution at time i, V is the total volume of the release solution (15 ml) and Vs is the sample volume (1 ml).
2.6. Statistical analysis
The results of all experiments are the mean values (± SD ) of triplicate samples performed in minimum three independent experiments. Comparisons between multiple samples were performed with single factor analysis of variance (ANOVA). Comparisons between two samples were performed with two-tailed Student’s t-test. Differences between data sets were considered significant when p < 0.05.
3. Results and Discussion
3.1. Incorporation of ligand into the PEG hydrogel
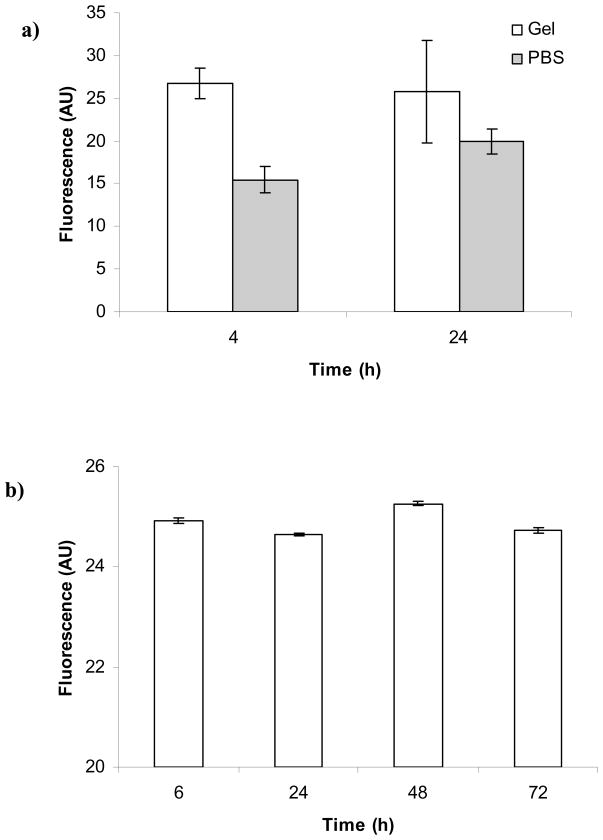
The fluorescent ligand 5FAM-GRCD-RGDS-PD was incorporated into the PEG hydrogel and its fluorescence was measured over time to estimate whether any unbound peptide was released from the hydrogel. Figure 2a shows that the fluorescence of the ligand in the hydrogel or in the supernatant did not change appreciably over the course of the experiment (4 and 24 h). The average fluorescence of the PBS solution was lower than the average fluorescence of the hydrogel indicating that some ligand was not bound covalently to the hydrogel and therefore was free to diffuse out of the hydrogel during the soaking in PBS. Figure 2b shows that the fluorescence of the hydrogel did not change over 72 h which also indicated that all of the ligand was released initially (in < 4 h) and the ligand remaining in the hydrogel was stably incorporated; therefore, characterization of hydrogel properties (e.g., G′) was not affected by unbound ligand. Additionally, fluorescent ligand was reacted with PEG-VS and after the reaction was complete, the solution was filtered to recover the unbound ligand. The unbound ligand from this experiment was estimated to be 36.1 ± 0.3%.
Figure 2.
PEG hydrogel with covalently incorporated fluorescent ligand was used to determine that unbound ligand: a) was released in the supernatant PBS initially prior to 4 h, but b) was not released from the hydrogel over time (i.e., differences between sample fluorescence at time points up to 72 h are not statistically distinct).
There are several possible explanations for the apparent incomplete incorporation of the ligand. First, it is possible that a small amount of free dye is present in solution. The excess dye was cleaned with a one-step high-performance liquid chromatography (HPLC) purification (CPC Scientific) and the purification efficiency was not tested subsequently. Second, it is possible that despite the preventative measures, a fraction of the ligands had reacted with each other forming dimers by disulfide bonding thus taking up thiol groups that otherwise would be covalently bound to the VS group of the PEG polymer. Disulfide formation could have occurred in the aliquot or after the ligand was added to the PEG-VS solution. The competitive disulfide formation after the peptide was added to the PEG-VS solution would be closely related to the initial concentration of ligand: more ligand would lead to the formation of more disulfides [34, 46]. Lastly, the reaction efficiency of the cysteine thiol binding to the VS group of the PEG by Michael-type addition strongly depends on the surrounding amino acids and their charges: Lutolf et al. [34] demonstrated that positively charged amino acids positioned near the thiol resulted in a faster addition reaction rate. Specifically, the binding efficiency constant for the sequence GRCD (used in our project) to PEG-diacrylate (PEGDA) was 58.3 L/mol.min compared to 124.0 L/mol.min for GRCR. Therefore, it is possible that choosing a peptide with a sequence such as GRCR-RGDS-PD, would improve the binding efficiency.
3.2. Effect of ligand type on material properties: general findings
QM is defined as the mass of the swollen hydrogel divided by the mass of the dry hydrogel and is a measure of the hydrophilicity of the polymer. When the polymer is cross-linked and forms a 3D structure, QM is also a measure of how much water is incorporated into the structure (i.e., the water to polymer ratio). Given a mesh network (a good approximation of the PEG hydrogel) as shown in Figure 3a: more cross-link points will result in tighter mesh; consequently, less water will be incorporated in the network. On the other hand, if some of the cross-links are disrupted, local “pockets” with larger mesh would be created that are able to incorporate more water. Hence, by adding ligand and consequently inhibiting the complete cross-linking of the hydrogel, we should see increase in QM (up to a certain maximum peptide concentration that would occupy sufficient cross-linking sites to prevent polymer-polymer bonds and result in a soluble macropolymer). If this hypothesis holds true, it would indicate that the properties of the ligand and the PEG polymer are comparable and there is no ligand-polymer interaction.
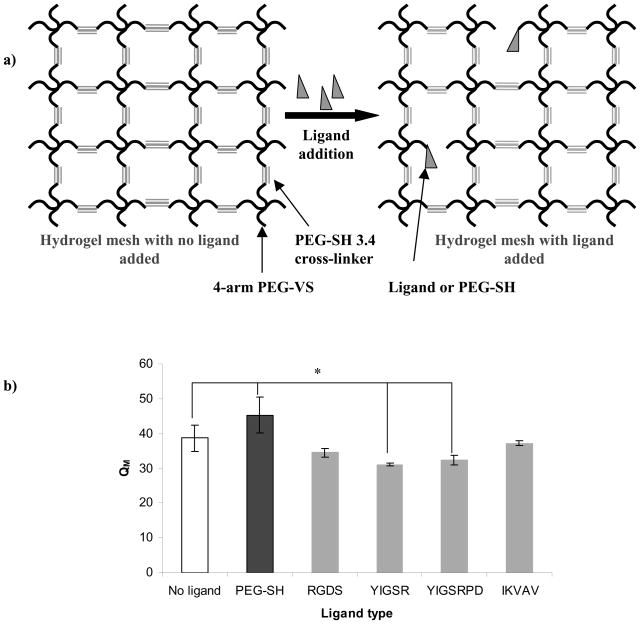
Figure 3.
a) Schematic representation of hydrogel mesh disruption upon covalent binding of ligand or PEG-SH to the 4-arm PEG-VS; b) Influence of ligand type on PEG hydrogel swelling ratio (QM). All hydrogels were prepared as 10% w/v polymer with 100 μM ligand. Asterisks designate significant differences.
It is also likely that QM would not be affected by the incorporation of ligand. Note that at 100 μM ligand or PEG-SH, only up to 0.7% of the VS groups would be occupied. The rest of the VS groups would be free for gelation with the PEG-dithiol cross-linker. This decrease in available cross-linkable VS sites may have a negligible effect; thus the net homogeneity of the resultant hydrogel network would not be affected and QM would not be altered.
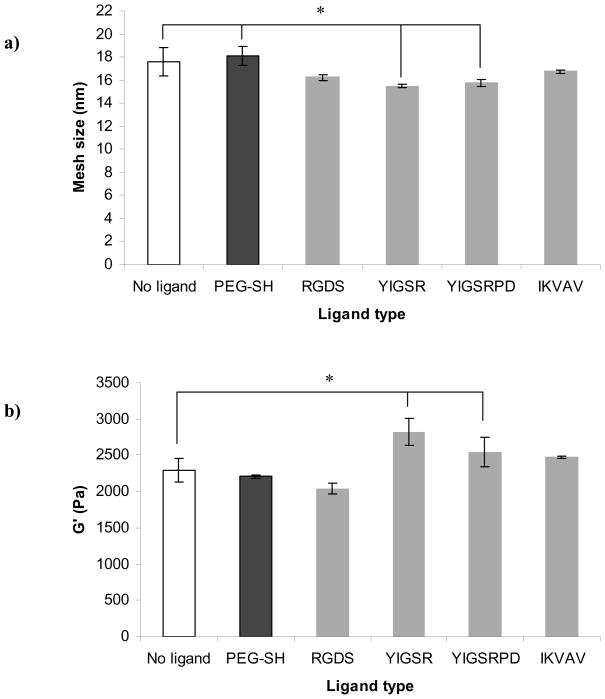
Figure 3b shows the influence of ligand type on QM of the PEG hydrogel. Compared to hydrogels made without ligand or PEG-SH, QM was ~14% higher when PEG-SH was incorporated into the hydrogel, which was expected because the PEG-SH occupied cross-link sites but was not expected to interact with the surrounding PEG network. For the peptide ligands, the addition of ligand either had no effect (as for RGDS and IKVAV), or decreased QM (as for YIGSR and YIGSRPD).
For all general purposes, especially cell attachment and protein adhesion, PEG is considered an inert polymer [47], an observation which would not explain the above findings. However, Kokufuta et. al. [48] and Xia et. al. [49] have shown that PEG was able to form a complex with pepsin under the appropriate conditions. The complex formation had been attributed to hydrogen bonding of the carboxyl and phenolic OH groups in pepsin. However, the hydrogen bonding would only be possible when these acidic side chains are protonated, i.e., this effect would depend on the pH of the surrounding solution. The pH of the PEG solution was 8.2 during gelation and 7.4 after gelation when the fully cross-linked 3D hydrogel was soaked in PBS. Considering the full amino acid sequence of all ligands, one may note that the only amino acid present that contains a carboxylic side chain is D (aspartic acid). It has a pKa of 3.9 [50] and thus would be protonated at pH ~3 or below. Therefore, under the selected experimental conditions, the carboxylic acid of D would be unprotonated and could not participate in hydrogen bonding with the ether oxygen of the PEG polymer. Furthermore, D is present in all ligands (in the flanking sequences GRCD and PD) and thus could not explain the differences in their influence over the material properties.
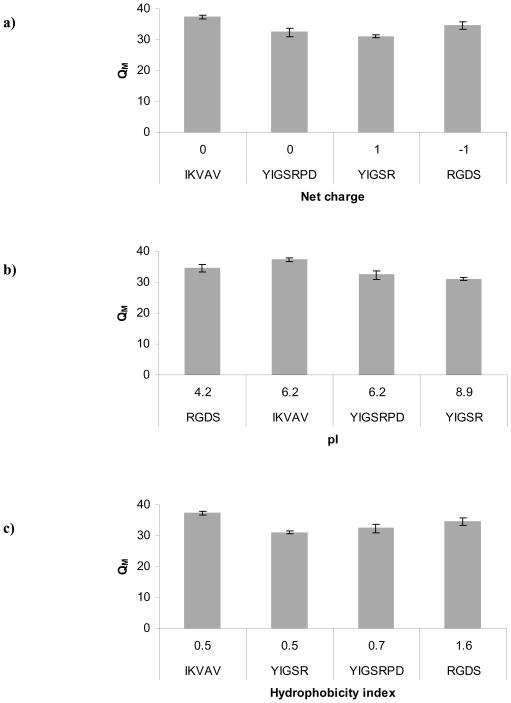
All ligands as well as the control PEG-SH were of similar molecular weight (~1000 Da), thus it is not likely that ligand size alone could explain the observed results. Therefore, the effect of ligand incorporation could not be generalized and specific ligand properties must be considered to explain their various effects. We first examined the individual properties of the ligand peptide sequences used in this work (Figure 4). Because we observed a completely different influence associated with addition of PEG-SH to the hydrogel as compared to addition of peptide ligands, we examined the major properties that clearly distinguish the two. First, we considered the net charge of the ligands (Figure 4a). PEG itself (including PEG-SH) is uncharged, while each of the ligands contained charged polar amino acids. However, considering net charge alone, we did not observe obvious trends in the data.
Figure 4.
Influence of ligand type on PEG hydrogel swelling ratio (QM). Each plot highlights a different ligand property: a) net charge; b) hydrophobicity index; c) pI. All hydrogels were prepared as 10% w/v polymer with 100 μM ligand.
Next, we considered the possibility that negatively charged or polar uncharged amino acids were forming ionic or weaker Van der Waals bonds with the partially positively charged ether oxygen of the PEG. An interaction of this type could potentially cause the ligand to align parallel to the PEG polymer chain or even create a weak bond (temporary or permanent) with an opposing PEG chain and thus influence the mesh structure. Such bond is more likely to be created at solution pH close to that of the ligand pI. Therefore, we examined whether any trends existed between QM and ligand pI (Figure 4b). Again, we did not observe a clear trend in the data but in general the ligands with the highest pI, YIGSR and YIGSRPD, were the ones that had the greatest effect on QM versus gels without ligand (see also Figure 3b). The YIGSR ligand with the largest influence (20% lower QM than hydrogels with no ligand) had the highest pI of 8.9, which is the closest to the pH of the hydrogel solution during cross-linking (pH 8.2). Therefore, it is possible that some ligand types (in our case YIGSR and YIGSRPD) bonded via weaker ionic or Van der Waals interactions with the 4-arm PEG-VS after they had been covalently incorporated into the PEG network (via the unpaired cysteine residue). It is also possible that these interactions resembled the manner of a cross-linker, connecting (permanently or temporarily) opposing PEG chains or creating folds within a single PEG chain. Such an interaction would not only result in a disrupted non-homogeneous mesh but would also lead to a smaller mesh since the molecular weight of the ligand (1000 Da) is 3-fold smaller than that of the cross-linker used (3400 Da).
Because QM is a measure of the hydrophilicity of the network environment, it is possible that the addition of ligand introduced local areas of hydrophobicity to the otherwise fully hydrophilic polymer. To explore this hypothesis, we specifically examined the hydrophobicity index of each ligand type (Figure 4c). We did not observe a clear trend in the data. In fact, it appeared that the ligand with highest hydrophobicity index, RGDS, did not exhibit a significant influence on QM versus hydrogels without ligand. Therefore, change in the hydrophobicity of the hydrogel environment alone could not explain the observed results.
To explore other possible effects of ligand type on hydrogel properties, we also determined hydrogel ξ and G′. The concept of QM is closely related to ξ and these two hydrogel properties are proportional. Figure 5a shows the effect of ligand type on ξ of the hydrogel, and as expected, the data followed the same trends observed in Figure 3b. As discussed earlier, addition of PEG-SH to the hydrogel occupied sites that otherwise would be available for cross-linking thus leading to an increase in ξ. On the other hand, ligands also occupied sites available for cross-linking but interactions with the PEG chains either counter-balanced that effect or lead to a decrease in the overall ξ of the hydrogel.
Figure 5.
a) Influence of ligand type on PEG hydrogel mesh size; b) Influence of ligand type on PEG hydrogel storage modulus (G′). All hydrogels were prepared as 10% w/v polymer with 100 μM ligand. Asterisks designate significant differences.
Another important scaffold property that could be influenced by the addition of ligand is G′. Figure 5b shows the influence of the ligand type on PEG hydrogel G′. Based on the data presented in Figure 3b and 5a we expected that the addition of the control PEG-SH would lead to an increase in G′ but we did not see a significant difference in G′ between the hydrogels made without ligand and hydrogels made with PEG-SH. This result could be explained with the fact that even though PEG-SH occupied cross-linking sites, the overall polymer density was not altered. As with QM and ξ, addition of RGDS and IKVAV also did not have a significant effect on G′ but the influence of YIGSRPD and YIGSR ligands in this case was most pronounced and resulted in 10% and 19% increases in G′, respectively. This outcome confirmed our initial observation that some inherent property of the YIGSR and YIGSRPD ligands may be responsible for altering PEG hydrogel properties.
3.3. Effect of ligand type and pH on material properties: focus on YIGSR
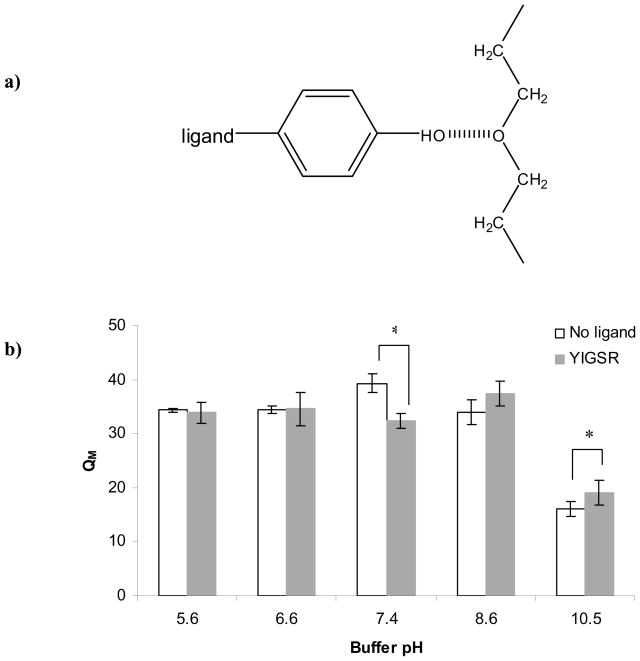
To further explore the concept that the difference in material properties observed upon addition of ligand is inherently linked to ligand type, we examined the two ligands that had the most effect on QM, namely YIGSR and YIGSRPD. The unique amino acids that they have in common but do not share with the other ligands were Y (tyrosine; polar, uncharged and contains a benzene ring) and I (isoleucine; non-polar and hydrophobic). Y is also unique in that it contains a phenolic OH group [50]. The pKa of the Y side chain is 10.1 [50]. Under the current experimental conditions, the side chain would be protonated and therefore able to form a hydrogen bond with the ether oxygen of the PEG polymer (Figure 6a).
Figure 6.
a) Schematic representation of hydrogen bonding between the phenolic OH group of the tyrosine (Y) amino acid of the YIGSR and YIGSRPD ligands and the ether oxygen of the PEG polymer; b) Influence of buffer pH on the swelling ratios of PEG hydrogels containing no ligand or 100 μM YIGSR. All hydrogels were 10% w/v. Asterisks designate significant differences.
The impact of a bond between Y and the PEG network could have several outcomes. For example, this hydrogen bond could reduce polymer hydrophilicity by altering the nature of the ether oxygen of the backbone chain or more importantly reducing the mesh size of the network by connecting opposing PEG chains or creating entanglements in a single PEG chain. A hydrogen bond between the YIGSR or YIGSRPD and the PEG polymer chain could therefore explain the decrease in swelling ratio and mesh size and increase in G′ upon addition of these ligands to a PEG-based hydrogel.
A pH experiment was specifically designed to test whether the interaction between the YIGSR ligand and the PEG polymer was due to hydrogen bonding between the phenolic OH group of the Y amino acid and the ether oxygen of the PEG repeat unit (Figure 6a). Figure 6b shows the effect of pH on PEG hydrogels made with or without YIGSR ligand. In the pH range of 5.6–10.5 we noted three distinct effects of YIGSR incorporation. First, in the acidic pH range (5.6 and 6.6) there was no difference in the QM of the hydrogels made with and without YIGSR. This finding supported our hypothesis that an acidic environment provides an abundance of protons that would compete with the phenolic OH group of the Y amino acid, occupy hydrogen bonding sites and thus decrease or completely eliminate the influence of the ligand on the bulk PEG properties. As noted previously (Figure 3a), at pH 7.4 the presence of ligand lead to a decrease in hydrogel QM. However, in a basic environment (pH 8.6 and 10.5), we observed a shift in the hydrogel QM: hydrogels made with YIGSR had higher QM than hydrogels without ligand. The difference between the two types of hydrogels was significant at pH 10.5, which is above the pKa of the Y side chain. This shift could be explained by the fact that the phenolic OH groups became deprotonated at basic pH and were no longer available for hydrogen bonding with the PEG polymer. With deprotonated phenolic OH groups, the YIGSR ligand behaved more like the control PEG-SH (i.e., did not interact with the PEG polymer but simply occupied cross-linking sites).
3.4. Effect of ligand concentration on material properties
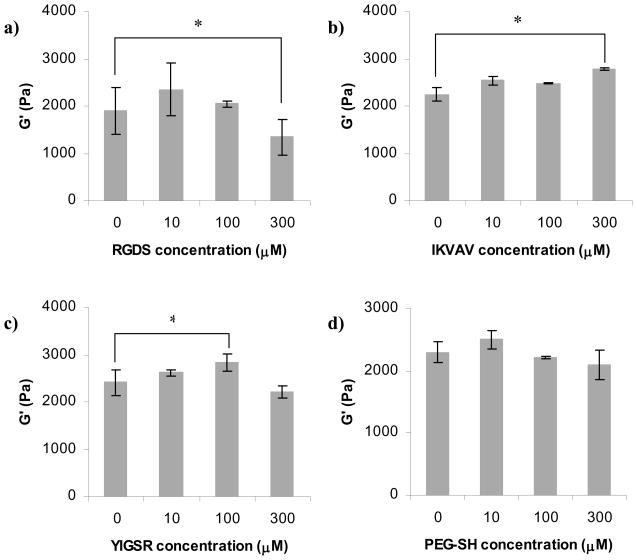
All of the above data was collected at a single ligand concentration. However, in building tissue engineering scaffolds, ligands are used at a range of concentrations. For example, comparing between RGDS, YIGSR and IKVAV ligands, Gunn et. al. [37] has shown that the concentration of ligand required for optimal neurite extension on 2D PEGDA hydrogels depended on which specific ligand was incorporated. In this work, the authors had also linked the extent of neurite outgrowth to the change in mechanical properties of the hydrogels without attempting to establish a connection between the ligand concentration and the resulting hydrogel mechanical properties. In general, adhesive ligand concentration which influences neurite outgrowth independent of scaffold mechanical properties, has not been consistently controlled, nor quantitatively measured [51]. Thus, we have chosen an array of RGDS, IKVAV and YIGSR concentrations widely used in building tissue engineering scaffolds (10, 100, 300 μM) [35] and tested the influence of ligand concentration on hydrogel materials properties. (Between the two similar sequences, YIGSR and YIGSRPD, we chose YIGSR for further testing as it showed the greatest impact on material properties.)
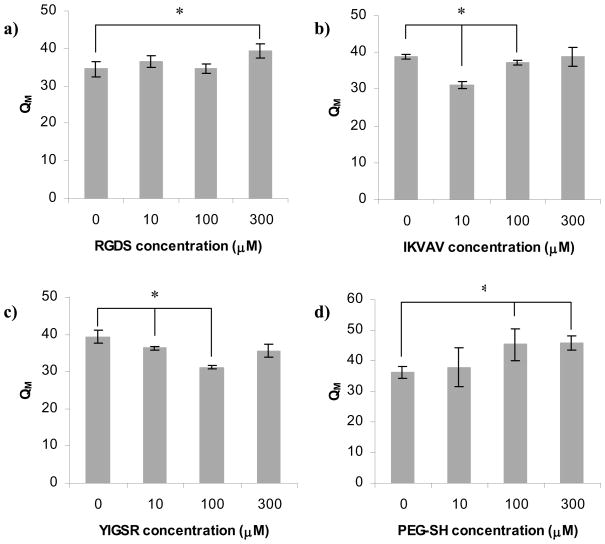
Figure 7a shows that the only RGDS concentration to have a significant influence on QM was 300 μM, which led to a 12% increase in QM compared to hydrogels synthesized without ligand. On the other hand, Figures 7b and 7c indicate that addition of IKVAV and YIGSR ligand either had no effect, or in fact resulted in reduced values of QM. The greatest influence of IKVAV was noted at 10 μM ligand (20% decrease in QM versus hydrogels without ligand), whereas the greatest impact of YIGSR was observed at 100 μM (21% decrease). Hence, in contrast to the results presented in Figure 3b (which showed that the effect of YIGSR on QM was larger than that of IKVAV), we saw that both IKVAV and YIGSR had the same overall influence on the hydrogel QM but at different concentrations. Figure 7d shows that addition of 10 μM of the control PEG-SH did not affect the hydrogel QM while addition of 100 and 300 μM both lead to a 20% increase in QM, which is consistent with Figure 3b.
Figure 7.
Influence of ligand concentration on swelling ratio (QM) of PEG hydrogels: a) RGDS ligand; b) IKVAV ligand; c) YIGSR ligand; d) PEG-SH control. All hydrogels were prepared as 10% w/v polymer. Asterisks designate significant differences.
Thus, we observed that both IKVAV and YIGSR exhibited an “optimal” concentration at which their influence on the material QM was at its maximum, but RGDS ligand did not exhibit the same behavior within the range of ligand concentrations explored in this study. In fact, at the highest concentration (300 μM) the RGDS influence on the material properties resembled that of the PEG-SH. Based on this observation, it is possible that RGDS did not react in any significant way with the PEG hydrogel and did not change the hydrogel network environment.
Note that RGDS has the highest hydrophobicity index. Since the addition of a high quantity of the ligand resulted in an increase in QM rather than a decrease, we can conclude that the decrease in the overall hydrophilicity of the PEG network upon addition of ligand under these conditions was negligible and not responsible for the overall change in hydrogel properties. Additionally, the pI of the RGDS ligand (4.2, the lowest for all studied ligand types), however, was far below the pH of the hydrogel solution upon cross-linking (8.2) and after cross-linking (7.4). Therefore, it is unlikely that the RGDS ligand would interact with the PEG polymer (apart from the covalent addition of the cysteine thiol to the PEG’s VS reactive group).
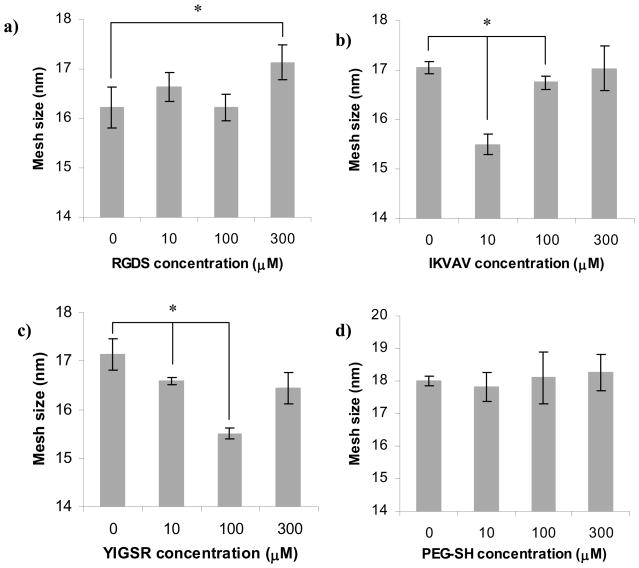
Figure 8 shows the influence of ligand concentration on hydrogel ξ. These data follow the same trend as measurements of QM (Figure 7). The addition of RGDS did not significantly influence ξ at concentrations below 300 μM (Figure 8a). Both IKVAV (at 10 μM) and YIGSR (at 100 μM) lead to ~3 nm decrease in ξ (Figure 8b and 8c, respectively). Addition of the control PEG-SH did not significantly alter in the studied concentration range (Figure 8d).
Figure 8.
Influence of ligand concentration on mesh size of PEG hydrogels: a) RGDS ligand; b) IKVAV ligand; c) YIGSR ligand; d) PEG-SH control. All hydrogels were prepared as 10% w/v polymer. Asterisks designate significant differences.
Figure 9 shows the influence of ligand concentration on G′. The addition of RGDS did not significantly alter G′ at concentrations below 300 μM (Figure 9a). Both IKVAV (at 300 μM) and YIGSR (at 100 μM) lead to ~15% increase in G′ (Figure 9b and 9c, respectively). Addition of the control PEG-SH did not change G′ significantly in the studied concentration range (Figure 9d).
Figure 9.
Influence of ligand concentration on storage modulus (G′) of PEG hydrogels: a) RGDS ligand; b) IKVAV ligand; c) YIGSR ligand; d) PEG-SH control. All hydrogels were prepared as 10% w/v polymer. Asterisks designate significant differences.
Figures 7–9 indicate that IKVAV influenced the hydrogel properties in a similar fashion as YIGSR, albeit at different concentrations. Upon considering the amino acid sequence for IKVAV, we note that the amino acids that are not shared with the other ligands are lysine (K), valine (V) and aspartic acid (A). A and V are non-polar hydrophobic amino acids, but as mentioned above, we hypothesize that the hydrophobicity of the tested ligands does not significantly alter hydrogel material properties. K is a basic amino acid with a pKa of the side chain of 10.79. Therefore, it was possible that at the pH of the hydrogel microenvironment, these fully protonated basic groups form hydrogen bonds with PEG, a phenomenon that had been observed by Azegami et. al. [52] when mixing human serum albumin with PEG at pH 8.
The other basic amino acid found in all tested ligands was R (arginine) where it was present in the flanking sequence GRCD and was also part of the active sequence for YIGSR, YIGSRPD and RGDS. However, based on its proximity to cysteine (C), the site of covalent bonding to 4-arm PEG-VS, we would expect that only the R of YIGSR and YIGSRPD would be available for hydrogen bonding. (In the case of GRCD sequence, R was an α substituent and in the case of RGDS (GRCDRGDSPD) it was a β substituent and potentially physically restricted, i.e., not available for hydrogen bonding).
In summary, the results of the ligand types and concentrations presented in Figures 7–9 lead to the conclusion that ligand concentration must be considered when discussing the influence of ligand type on the material properties of hydrogels because a single concentration may not result in similar effects or trends in all cases.
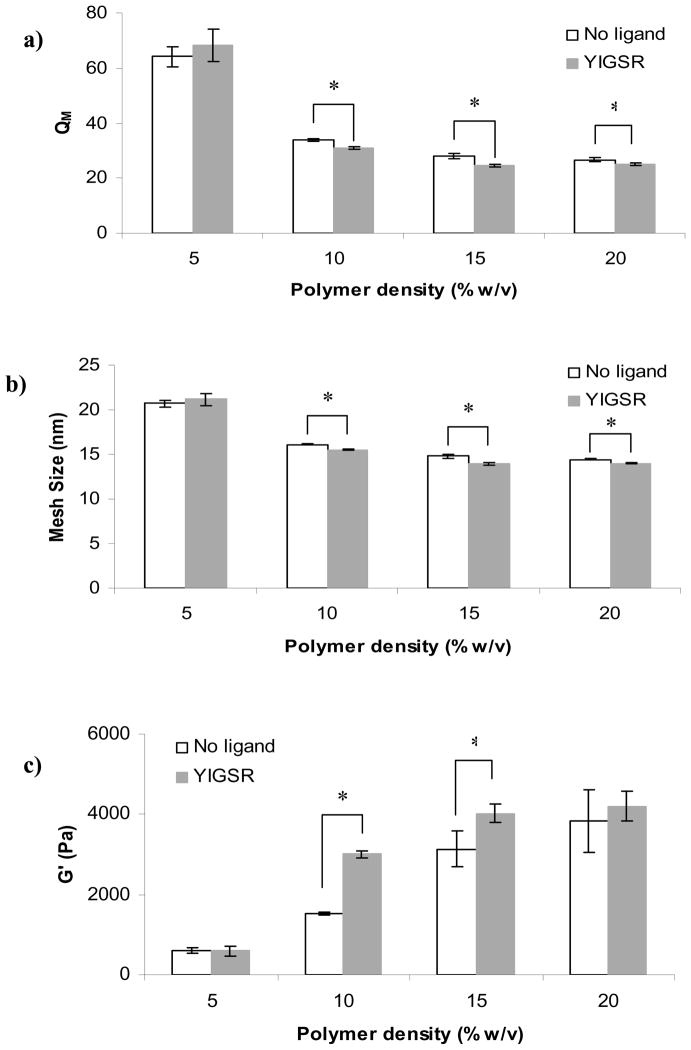
3.5. Effect of polymer density on material properties
Figure 10 depicts the influence of YIGSR ligand on material properties examined in hydrogels of 5–20% w/v polymer density. At the lowest polymer density, the addition of YIGSR did not have an effect on QM (Figure 10a), ξ (Figure 10b), or G′ (Figure 10c). For higher polymer concentrations (10, 15, 20% w/v), addition of ligand resulted in decreased QM and ξ and increased G′ (though the difference in G′ at 20% was not significant). Since ligand concentration was kept constant for all polymer densities, and in the range of 10–20% w/v polymer, no other trends were noted, these results ruled out the possibility that the ratio of ligand to polymer concentration had a significant influence. Moreover, if the influence of the ligand on hydrogel properties was due to consumption of cross-linking sites, the extent of this effect would have been inversely proportional to polymer density; this result was generally not observed. The lack of influence of ligand on the properties of the 5% w/v hydrogels could be explained by the very large QM of the hydrogel at this polymer density. With high swelling, it is possible that weak bonds (such as hydrogen bonds) were insufficiently strong and temporary in effect; thus, the overall contribution of entanglements resulting from such bonds could be negligible.
Figure 10.
Comparison between PEG hydrogels with varying polymer density prepared with 100 μM YIGSR ligand or no ligand and the effects on: a) swelling ratio, b) mesh size, c) and storage modulus. Asterisks designate significant differences.
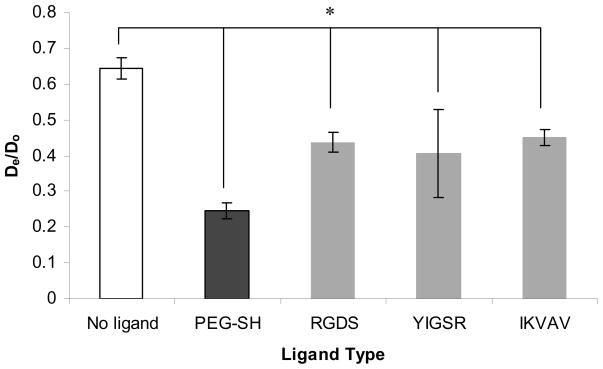
3.6. Effect of ligand on bulk diffusion in the PEG hydrogels
Figure 11 shows the effect of ligand on BSA diffusivity in the PEG hydrogels. All ligand types resulted in decreased diffusivity by ~30%, whereas addition of the control PEG-SH decreased diffusivity of BSA by ~60%. The charge and hydrophobicity of the ligand amino acids likely interact with BSA and obstruct its diffusivity in the otherwise inert hydrogel. However, the discussion above may suggest another ligand-specific mechanism that could restrict the diffusion of soluble molecules in the PEG hydrogels. The ~2-fold decrease in BSA diffusivity in hydrogels made with PEG-SH as compared to hydrogels made with peptide ligands could be explained by the fact that covalently-bound PEG-SH did not interact with the PEG polymer chain. Instead, it may act as an additional free chain rotating in the mesh of the hydrogel, thus obstructing the diffusion of BSA inside the hydrogel. The peptide ligands on the other hand, may interact with PEG polymer chains as described above. In this way, the net effect of ligands in the hydrogel was a reduced obstacle to the diffusion of BSA inside the mesh.
Figure 11.
Effect of ligand type on BSA diffusivity (De) in PEG hydrogels. All hydrogels were prepared as 10% w/v polymer with 100 μM ligand. Asterisks designate significant differences. The BSA diffusivity in the hydrogel is normalized by that of BSA diffusivity in water (Do = 0.941×10−5 mm2/s).
4. Conclusions
The peptide ligands studied here, namely RGDS, IKVAV, YIGSR and YIGSRPD are recognized as adhesive sequences derived from larger proteins that promote integrin-mediated cell adhesion and therefore are routinely used for adding biological activity to inert synthetic materials. In general, their influence on the cell behavior is typically assessed without note as to their influence on the material properties when the ligands are added to hydrogel scaffolds.
This work determined the influence of ligands covalently bound to PEG hydrogels on the materials’ physical, mechanical and transport properties. We demonstrated that the ligands affect the QM, ξ, and G′ of the PEG hydrogel, as well as BSA diffusivity through the hydrogel and the extent of this effect was specific to the ligand type and concentration. The YIGSR and the YIGSRPD ligands had the most pronounced effect on the material properties due to hydrogen bonding between the phenolic OH group of the Y and the ether oxygen of the PEG polymer as confirmed by additional testing of hydrogel properties at various pH. Generally, the addition of YIGSR, YIGSRPD and IKVAV ligands lead to a decrease in QM and ξ as well as an increase in G′, while addition of a control monofunctional PEG-SH and RGDS ligand had the opposite effect on the material properties. The addition of all ligands as well as the PEG-SH control led to a decrease in BSA diffusivity. The concentrations of RGDS, IKVAV, YIGSR and PEG-SH at which the greatest change in properties was observed were 300 μM, 10 μM, 100 μM and >100 μM, respectively; this finding emphasizes the specificity of the ligand/polymer interaction. Polymer densities wherein incorporation of ligands affected hydrogel material properties occurred at 10 and 20% w/v, but not at 5% w/v possibly due to large QM at this lowest polymer density.
This work provides specific evidence for interactions between peptide ligands and cross-linked PEG hydrogels that significantly impact hydrogel material and transport properties. Therefore, this work may have important implications for interpreting the results of cell experiments carried out with ligand-modified hydrogels because the addition of ligand may affect not only the scaffold’s biological properties, but also key physical properties of the system.
Supplementary Material
Acknowledgments
We thank Theresa Good for valuable technical discussions. This work was supported by NIH-NINDS (R01NS065205), the Henry Luce Foundation and UMBC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pierschbacher MD, Ruoslahti E. Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature. 1984;309:30–3. doi: 10.1038/309030a0. [DOI] [PubMed] [Google Scholar]
- 2.Pierschbacher MD, Ruoslahti E. Variants of the cell recognition site of fibronectin that retain attachment-promoting activity. Proc Natl Acad Sci U S A. 1984;81:5985–8. doi: 10.1073/pnas.81.19.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki S, Oldberg A, Hayman EG, Pierschbacher MD, Ruoslahti E. Complete amino acid sequence of human vitronectin deduced from cDNA. Similarity of cell attachment sites in vitronectin and fibronectin. Embo J. 1985;4:2519–24. doi: 10.1002/j.1460-2075.1985.tb03965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruoslahti E, Pierschbacher MD. New perspectives in cell adhesion: RGD and integrins. Science. 1987;238:491–7. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- 5.Davis GE. Affinity of integrins for damaged extracellular matrix: alpha v beta 3 binds to denatured collagen type I through RGD sites. Biochem Biophys Res Commun. 1992;182:1025–31. doi: 10.1016/0006-291x(92)91834-d. [DOI] [PubMed] [Google Scholar]
- 6.Grant DS, Tashiro K, Segui-Real B, Yamada Y, Martin GR, Kleinman HK. Two different laminin domains mediate the differentiation of human endothelial cells into capillary-like structures in vitro. Cell. 1989;58:933–43. doi: 10.1016/0092-8674(89)90945-8. [DOI] [PubMed] [Google Scholar]
- 7.Hern DL, Hubbell JA. Incorporation of adhesion peptides into nonadhesive hydrogels useful for tissue resurfacing. J Biomed Mater Res. 1998;39:266–76. doi: 10.1002/(sici)1097-4636(199802)39:2<266::aid-jbm14>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 8.Shin H, Jo S, Mikos AG. Biomimetic materials for tissue engineering. Biomaterials. 2003;24:4353–64. doi: 10.1016/s0142-9612(03)00339-9. [DOI] [PubMed] [Google Scholar]
- 9.Fittkau MH, Zilla P, Bezuidenhout D, Lutolf MP, Human P, Hubbell JA, Davies N. The selective modulation of endothelial cell mobility on RGD peptide containing surfaces by YIGSR peptides. Biomaterials. 2005;26:167–74. doi: 10.1016/j.biomaterials.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 10.VandeVondele S, Voros J, Hubbell JA. RGD-grafted poly-L-lysine-graft-(polyethylene glycol) copolymers block non-specific protein adsorption while promoting cell adhesion. Biotechnol Bioeng. 2003;82:784–90. doi: 10.1002/bit.10625. [DOI] [PubMed] [Google Scholar]
- 11.Schaffner P, Dard MM. Structure and function of RGD peptides involved in bone biology. Cell Mol Life Sci. 2003;60:119–32. doi: 10.1007/s000180300008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kraehenbuehl TPZP, Van der Vlies AJ, Schoenmakers RG, Lutolf MP, Jaconi ME, Hubbell JA. Three-dimensional extracellular matrix-directed cardioprogenitor differentiation: systematic modulation of a synthetic cell-responsive PEG-hydrogel. Biomaterials. 2008;29:2757–66. doi: 10.1016/j.biomaterials.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 13.Raeber GP, Lutolf MP, Hubbell JA. Molecularly engineered PEG hydrogels: a novel model system for proteolytically mediated cell migration. Biophys J. 2005;89:1374–88. doi: 10.1529/biophysj.104.050682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raeber GP, Lutolf MP, Hubbell JA. Mechanisms of 3-D migration and matrix remodeling of fibroblasts within artificial ECMs. Acta Biomaterialia. 2007;3:615–29. doi: 10.1016/j.actbio.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 15.Seliktar D, Zisch AH, Lutolf MP, Wrana JL, Hubbell JA. MMP-2 sensitive, VEGF-bearing bioactive hydrogels for promotion of vascular healing. Journal of Biomedical Materials Research Part A. 2004;68A:704–16. doi: 10.1002/jbm.a.20091. [DOI] [PubMed] [Google Scholar]
- 16.Luo Y, Shoichet MS. A photolabile hydrogel for guided three-dimensional cell growth and migration. Nature Materials. 2004;3:249–53. doi: 10.1038/nmat1092. [DOI] [PubMed] [Google Scholar]
- 17.Behravesh E, Mikos AG. Three-dimensional culture of differentiating marrow stromal osteoblasts in biomimetic poly(propylene fumarate-co-ethylene glycol)-based macroporous hydrogels. J Biomed Mater Res A. 2003;66:698–706. doi: 10.1002/jbm.a.10003. [DOI] [PubMed] [Google Scholar]
- 18.Santiago LY, Nowak RW, Peter Rubin J, Marra KG. Peptide-surface modification of poly(caprolactone) with laminin-derived sequences for adipose-derived stem cell applications. Biomaterials. 2006;27:2962–9. doi: 10.1016/j.biomaterials.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 19.Hautanen A, Gailit J, Mann DM, Ruoslahti E. Effects of modifications of the RGD sequence and its context on recognition by the fibronectin receptor. J Biol Chem. 1989;264:1437–42. [PubMed] [Google Scholar]
- 20.Ruoslahti E. RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 21.Hersel U, Dahmen C, Kessler H. RGD modified polymers: biomaterials for stimulated cell adhesion and beyond. Biomaterials. 2003;24:4385–415. doi: 10.1016/s0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]
- 22.Itoh S, Takakuda K, Samejima H, Ohta T, Shinomiya K, Ichinose S. Synthetic collagen fibers coated with a synthetic peptide containing the YIGSR sequence of laminin to promote peripheral nerve regeneration in vivo. J Mater Sci Mater Med. 1999;10:129–34. doi: 10.1023/a:1008977221827. [DOI] [PubMed] [Google Scholar]
- 23.Duan X, McLaughlin C, Griffith M, Sheardown H. Biofunctionalization of collagen for improved biological response: scaffolds for corneal tissue engineering. Biomaterials. 2007;28:78–88. doi: 10.1016/j.biomaterials.2006.08.034. [DOI] [PubMed] [Google Scholar]
- 24.Schense JC, Bloch J, Aebischer P, Hubbell JA. Enzymatic incorporation of bioactive peptides into fibrin matrices enhances neurite extension. Nat Biotechnol. 2000;18:415–9. doi: 10.1038/74473. [DOI] [PubMed] [Google Scholar]
- 25.Streuli CH, Schmidhauser C, Bailey N, Yurchenco P, Skubitz AP, Roskelley C, Bissell MJ. Laminin mediates tissue-specific gene expression in mammary epithelia. J Cell Biol. 1995;129:591–603. doi: 10.1083/jcb.129.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peter SJ, Miller MJ, Yasko AW, Yaszemski MJ, Mikos AG. Polymer concepts in tissue engineering. J Biomed Mater Res. 1998;43:422–7. doi: 10.1002/(sici)1097-4636(199824)43:4<422::aid-jbm9>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 27.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–43. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 28.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Tensional Homeostasis and the Malignant Phenotype. Cancer Cell. 2005;8:241–54. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 29.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 30.Lo C-M, Wang H-B, Dembo M, Wang Y-L. Cell Movement is Guided by the Rigidity of the Substrate. Biophysical Journal. 2000;79:144–52. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Engler AJ, Bacakova L, Newman C, Hategan A, Griffin M, Discher DE. Substrate compliance versus ligand density in cell on gel responses. Biophysical Journal. 2004;86:617–28. doi: 10.1016/S0006-3495(04)74140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lutolf MP, Hubbell JA. Synthesis and pysiochemical characterization of end-linked poly(ethylene glycol)-co-peptide hydrogels formed by Michael-type addition. Biomacromolecules. 2003;4:713–22. doi: 10.1021/bm025744e. [DOI] [PubMed] [Google Scholar]
- 33.Zustiak SP, Leach JB. Hydrolytically degradable poly(ethylene glycol) hydrogel scaffolds with tunable degradation and mechanical properties. Biomacromolecules. 2010 doi: 10.1021/bm100137q. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lutolf MP, Tirelli N, Cerritelli S, Cavalli L, Hubbell JA. Systematic modulation of Michael-type reactivity of thiols through the use of charged amino acids. Bioconjug Chem. 2001;12:1051–6. doi: 10.1021/bc015519e. [DOI] [PubMed] [Google Scholar]
- 35.Lutolf MP, Lauer-Fields JL, Schmoekel HG, Metters AT, Weber FE, Fields GB, Hubbell JA. Synthetic matrix metalloproteinase-sensitive hydrogels for the conduction of tissue regeneration: engineering cell-invasion characteristics. PNAS. 2003;100:5413–8. doi: 10.1073/pnas.0737381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rao SS, Winter JO. Adhesion molecule-modified biomaterials for neural tissue engineering. Front Neuroengineering. 2009;2:6. doi: 10.3389/neuro.16.006.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gunn JW, Turner SD, Mann BK. Adhesive and mechanical properties of hydrogels influence neurite extension. J Biomed Mater Res A. 2005;72:91–7. doi: 10.1002/jbm.a.30203. [DOI] [PubMed] [Google Scholar]
- 38.Mann BK, Gobin AS, Tsai AT, Schmedlen RH, West JL. Smooth muscle cell growth in photopolymerized hydrogels with cell adhesive and proteolytically degradable domains: synthetic ECM analogs for tissue engineering. Biomaterials. 2001;22:3045–51. doi: 10.1016/s0142-9612(01)00051-5. [DOI] [PubMed] [Google Scholar]
- 39.Leach JB, KAB, CWPJ, Schmidt CE. Photocrosslinked Hyaluronic Acid Hydrogels: Natural, Biodegradable Tissue Engineering Scaffolds. Biotechnology and Bioengineering. 2003;82:578–89. doi: 10.1002/bit.10605. [DOI] [PubMed] [Google Scholar]
- 40.Lu S, Anseth KS. Release Behavior of High Molecular Weight Solutes from Poly(ethylene-glycol)-Based Degradable Networks. Macromolecules. 2000;33:2509–15. [Google Scholar]
- 41.Canal T, Peppas NA. Correlation Between Mesh Size and Equilibrium Degree of Swelling of Polymeric Networks. Journal of Biomedical Materials Research. 1989;23:1183–93. doi: 10.1002/jbm.820231007. [DOI] [PubMed] [Google Scholar]
- 42.Cruise GM, Scharp DS, Hubbell JA. Characterization of permeability and network structure of interfacially photopolymerized poly(ethylene glycol) diacrylate hydrogels. Biomaterials. 1998;19:1287–94. doi: 10.1016/s0142-9612(98)00025-8. [DOI] [PubMed] [Google Scholar]
- 43.Mellott MB, Searcy K, Pishko MV. Release of protein from highly cross-linked hydrogels of poly(ethylene glycol) diacrylate fabricated by UV polymerization. Biomaterials. 2001;22:929–41. doi: 10.1016/s0142-9612(00)00258-1. [DOI] [PubMed] [Google Scholar]
- 44.Merrill EW, Dennison KA, Sung C. Partitioning and diffusion of solutes in hydrogels of poly(ethylene oxide) Biomaterials. 1993;14:1117–26. doi: 10.1016/0142-9612(93)90154-t. [DOI] [PubMed] [Google Scholar]
- 45.Ritger PL, Peppas NA. A simple equation for description of solute release: I. Fickian and non-Fickian release from non-swellable devices in the form of slabs, spheres, cylinders or disks. Journal of Controlled Release. 1987;5:23–36. [PubMed] [Google Scholar]
- 46.Salinas CN, Cole BB, Kasko AM, Anseth KS. Chondrogenic differentiation potential of human mesenchymal stem cells photoencapsulated within poly(ethylene glycol)-arginine-glycine-aspartic acid-serine thiol-methacrylate mixed-mode networks. Tissue Eng. 2007;13:1025–34. doi: 10.1089/ten.2006.0126. [DOI] [PubMed] [Google Scholar]
- 47.Krsko P, Libera M. Biointeractive hydrogels. Materials Today. 2005;8:38–44. [Google Scholar]
- 48.Kokufuta E, Nishimura H. Complexation of pepsin poly(ethylene glycol) Polymer Bulletin. 1991;26:277–82. [Google Scholar]
- 49.Xia J, Dubin PL, Kokufuta E. Dynamic and electrophoretic light scattering of a water-soluble complex formed between pepsin and polyethylene glycol. Macromolecules. 1993;26:6688–90. [Google Scholar]
- 50.Garrett RH, Grisham CM. Biochemistry. 3. Thompson Brooks/Cole; Belmont, CA: 2005. [Google Scholar]
- 51.Buettner HM, Pittman RN. Quantitative effects of laminin concentration on neurite outgrowth in vitro. Dev Biol. 1991;145:266–76. doi: 10.1016/0012-1606(91)90125-m. [DOI] [PubMed] [Google Scholar]
- 52.Azegami S, Tsuboi A, Izumi T, Hirata M, Dubin PL, Wang B, Kokufuta E. Formation of an intrapolymer complex from human serum albumin and poly(ethylene glycol) Langmuir. 1999;15:940–7. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.