Summary
Werner syndrome (WS) is an autosomal recessive disorder, the hallmarks of which are premature aging and early onset of neoplastic diseases (Orren 2006; Bohr 2008). The gene, whose mutation underlies the WS phenotype, is called WRN. The protein encoded by the WRN gene, WRNp, has DNA helicase activity (Gray et al. 1997; Orren 2006; Bohr 2008; Opresko 2008). Extensive evidence suggests that WRNp plays a role in DNA replication and DNA repair (Chen et al. 2003; Hickson 2003; Orren 2006; Turaga et al. 2007; Bohr 2008). However, WRNp function is not yet fully understood. In this study, we show that WRNp is involved in de novo DNA methylation of the promoter of the Oct4 gene, which encodes a crucial stem cell transcription factor. We demonstrate that WRNp localizes to the Oct4 promoter during retinoic acid-induced differentiation of human pluripotent cells, and associates with the de novo methyltransferase Dnmt3b in the chromatin of differentiating pluripotent cells. Depletion of WRNp does not affect demethylation of lysine 4 of the histone H3 at the Oct4 promoter, nor methylation of lysine 9 of H3, but it blocks recruitment of Dnmt3b to the promoter and results in reduced methylation of CpG sites within the Oct4 promoter. The lack of DNA methylation was associated with continued, albeit greatly reduced, Oct4 expression in WRN-deficient, retinoic acid-treated cells, which resulted in attenuated differentiation. The presented results reveal a novel function of WRNp, and demonstrate that WRNp controls a key step in pluripotent stem cell differentiation.
Keywords: Werner syndrome, Oct4, Dnmt3b, de novo methylation, aging, stem cells
Introduction
Werner syndrome (WS) is an autosomal recessive disorder, the hallmarks of which are premature aging and the early onset of degenerative and neoplastic diseases (Orren 2006). Gene expression in WS closely resembles that of normal aging and supports the use of WS as a model of aging (Orren 2006). The gene, whose mutation underlies the WS phenotype, is called WRN. Mutations in WRN result in the instability of WRN mRNA, as well as truncation of the protein with loss of the nuclear localization signal (NLS) and all or some enzymatic domains of the protein (Orren 2006; Bohr 2008). The protein encoded by the WRN gene, WRNp, has DNA helicase activity (Gray et al. 1997). WRNp is a member of the RecQ DNA helicase family, which in humans includes four other members [RecQ1, Bloom Syndrome Protein (BLM), RecQ4 and RecQ5 (Hickson 2003)]. WS cells exhibit high sensitivity to the topoisomerase I poison camptothecin (Lebel & Leder 1998). These and other data suggest that WRNp plays a role in DNA replication, recombination and repair (Orren 2006; Bohr 2008). In addition, it has been shown that WRNp is involved in telomere maintenance (Opresko et al. 2004). Finally, the similarities in transcriptional profiles of aged and WS cells suggest that WRNp might be involved in transcriptional regulation (Kyng et al. 2003). However, WRNp function is not yet fully understood, nor it is known if WRNp plays a role in cellular processes that are unique to certain cell types and that are at the same time crucial for the well-being of the whole organism. An example of such cell type-specific process is stem cell differentiation.
Stem cells are undifferentiated cells that are capable of self-renewal and differentiation. Most human tissues are composed of a majority of differentiated cells with a limited life span. These cells die and the tissue shrinks, unless replenished by new cells. These new cells originate from tissue stem cells, which compose only a small minority of the tissue cells.
A subspecies of stem cells are embryonic stem (ES) cells. These are pluripotent cells, which can be obtained from early stage embryos (blastocyst) and can differentiate into all three primary germ layers (Okita & Yamanaka 2006).
ES cells are characterized by the expression of stem cell transcriptional factors (ESTF), which include Oct4, Nanog and Sox2 (Boiani & Scholer 2005; Okita & Yamanaka 2006; Sun et al. 2006; Loh et al. 2008; Hu et al. 2009). Animal and other studies have suggested that ESTFs are crucial for self-renewal of ES cells and pluripotency (Okita & Yamanaka 2006). This hypothesis was recently confirmed by reprogramming adult somatic cells into induced pluripotent stem (iPS) cells, which posses ES cell properties (Wernig et al. 2007; McDevitt & Palecek 2008). The reprogramming was achieved by re-introduction of ESTFs into the somatic cells and it has been very recently shown that Oct4 alone is sufficient to achieve reprogramming (Kim et al. 2009). However, recent studies demonstrated that Oct4 is not present solely in ES cells (or iPS cells), but that an Oct4-expressing subpopulation of stem cells also exists in adults (Jiang et al. 2002; D'Ippolito et al. 2004; Kucia et al. 2006; Pallante et al. 2007; Ratajczak et al. 2007). These Oct4-expressing cells might be critical to the adult regenerative capacity (Edelberg & Ballard 2008).
During stem cell differentiation, the Oct4 gene is rapidly, and irreversibly repressed (Feldman et al. 2006). This process of Oct4 inactivation involves a cascade of histone and DNA modifications. First to occur is deacetylation of lysines 9 and 14 of histone H3. This is accompanied by demethylation of the lysine 4 of H3 (H3K4). Later, trimethylation of lysine 9 of histone H3 (H3K9) can be observed. The histone methyltransferase responsible for this event is G9a (also known as EHMT2). Trimethylated H3K9 (H3K9me3) serves as a binding site for the heterochromatin protein 1 beta (HP1β). Finally, Oct4 promoter DNA is methylated by the de novo DNA methyltransferase Dnmt3a/3b, the recruitment of which depends on G9a (Feldman et al. 2006; Li et al. 2007; Yeo et al. 2007; Epsztejn-Litman et al. 2008). DNA methylation makes Oct4 inactivation irreversible, and thus prevents Oct4 re-expression. Although the process of Oct4 promoter suppression was thus described in some detail, fundamental questions remain. One of these is the identity of putative cellular co-factors that may cooperate with Dnmt3a/3b. In this study, we tested the hypothesis that the Werner syndrome protein, WRNp, plays such a role and is required to ensure efficient DNA methylation at the Oct4 promoter.
Results
As a cell model to study the potential role of WRNp in stem cell differentiation, we have chosen NCCIT cells. NCCIT are a developmentally pluripotent cell line that can differentiate into derivatives of all three embryonic germ layers and extraembryonic cell lineages (Damjanov et al. 1993; Taranger et al. 2005). NCCIT cells express stem cell markers at high levels, and extracts of NCCIT cells were shown to reprogram somatic cells into pluripotent stem cells (Taranger et al. 2005). Following retinoic acid (RA) stimulation to differentiate, NCCIT cells readily suppress, in an ES cell-like manner, Oct4 expression (Cheng et al. 2007; Dahl & Collas 2007).
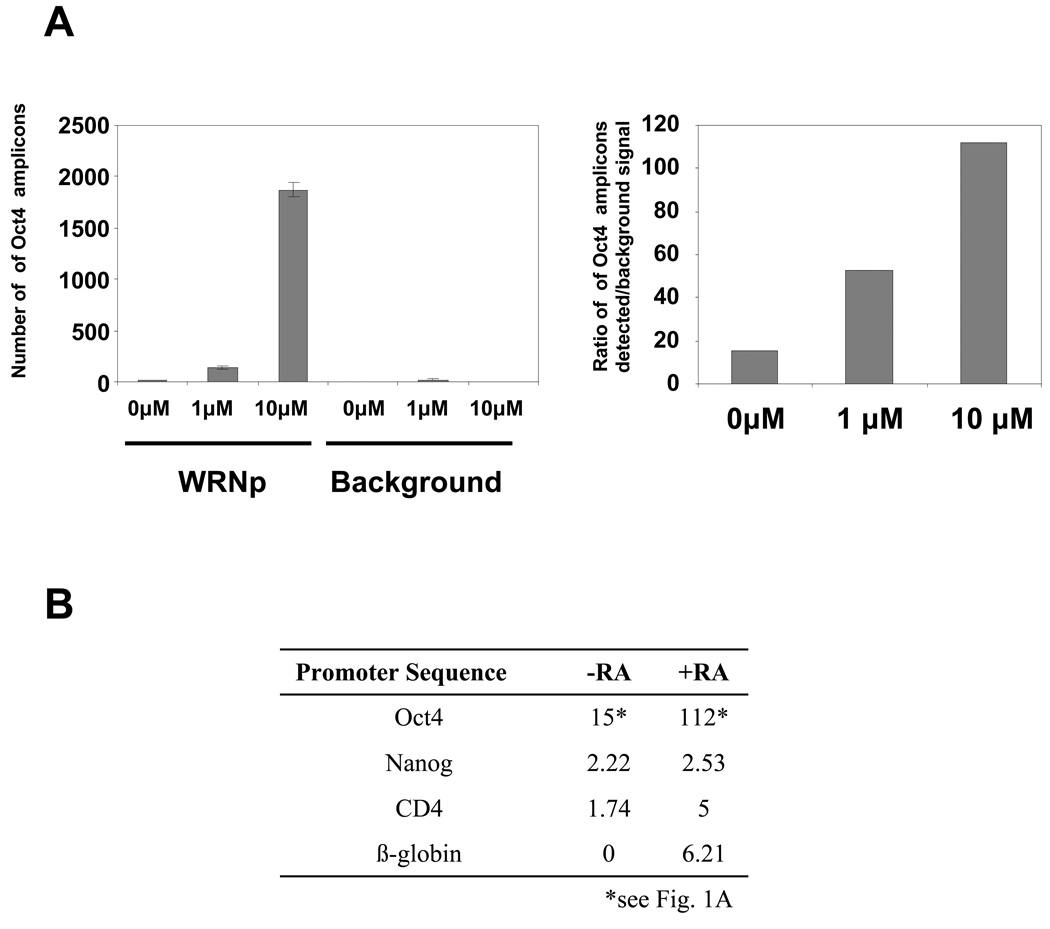
It has been noted that the RecQ helicases, including WRNp, engage in dynamic subnuclear relocalizations in response to different stimuli, which include DNA damage and aberrations in chromatin structure (Turaga et al. 2007; Bohr 2008). We first examined WRNp association with the Oct4 promoter in undifferentiated cells and cells stimulated to differentiate with varying amounts of RA using quantitative chromatin immunoprecipitation (Q-ChIP). We observed a high degree of enrichment of WRNp at the Oct4 promoter of undifferentiated cells (15 fold over background, Fig. 1). To determine if WRNp associates with the Oct4 promoter during differentiation, we have examined WRNp accumulation at the Oct4 promoter of RA-induced cells. We observed an accumulation of WRNp at three days after RA addition (53 fold over background in cells stimulated with 1 µM RA and 112 fold over background in cells stimulated with 10 µM RA, Fig. 1A). We conclude that WRNp associates with the Oct4 promoter in pluripotent cells, and accumulates at the promoter during RA-induced differentiation.
Fig. 1. Enrichment of WRNp at the Oct4 promoter.
(A) Q-ChIP analysis of WRNp levels at the Oct4 promoter in undifferentiated pluripotent cells and cells stimulated to differentiate with either 1 or 10 µM RA for three days. WRNp was immunoprecipitated from NCCIT lysates and the amount of associated Oct4 promoter DNA was measured by real-time PCR using the Oct4 B probe and primers (See Supplementary Table 2). Left – raw data, Oct4 promoter amplicons detected in samples with 0, 1, or 10 µM RA, immunoprecipitated with a WRN antibody (WRNp), or without antibody to determine extent of background signal (Background). Error bars indicate standard deviation. Right – ratio of Oct4 promoter amplicons detected in lysates immunoprecipitated with the antibody over the signal detected in no antibody control samples. (B) WRNp enrichment at the Nanog, CD4 and β-globin promoters in the presence and absence of RA (fold enrichment over background). Cells were treated as in A.
To determine if the observed association of WRNp with the Oct4 promoter could be due to an overall WRNp association with genomic DNA, we have investigated the presence of WRN on the Nanog, CD4 promoters and β-globin promoters. The Nanog promoter is RA-responsive, whereas the CD4 and β-globin promoters do not appear likely to respond to RA (Feldman et al. 2006). We have found only a very weak association of WRNp with these promoters (Fig. 1B). We conclude that the observed WRNp association with a promoter is highly specific for Oct4.
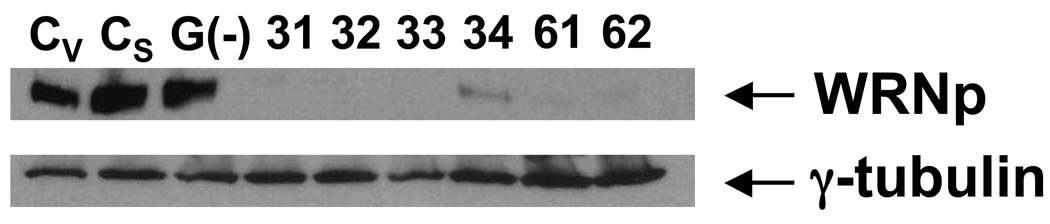
To determine if WRNp plays a role in Oct4 regulation, we have transfected NCCIT cells with shRNAs targeting WRN. Following selection in puromycin, we have obtained six clones that expressed very low or undetectable levels of WRNp (Fig. 2). Four of these clones were transfected with shRNA targeting nucleotides 1362 – 1390 of WRN (clones 31–34, see Supplementary Table 3) and two clones (61 and 62) were transfected with shRNA targeting nucleotides 3743 – 3771 of WRN. We have also developed a clone, in which G9a was knocked down by shRNA. G9a knockdown did not affect intracellular levels of WRNp (Fig. 2). These and control clones that were transfected with control plasmids (either the empty vector or the vector encoding a non-effective scrambled shRNA cassette) were then stimulated with RA to differentiate.
Fig. 2. shRNA-mediated knockdown of WRNp in NCCIT cells.
NCCIT cells were transfected with plasmids encoding one of two types of shRNA against WRN, or shRNA against G9a, or one of two control plasmids (CV – empty vector, CS – vector control expressing a scrambled, non-targeting sequence, G(−) – G9a-deficient clone, 31–34, 61, and 62 – WRNp deficient clones, see Supplementary Table 3). Transfected cells were selected in puromycin and individual clones were isolated and subjected to western blotting analysis with a WRNp antibody or a γ-tubulin control.
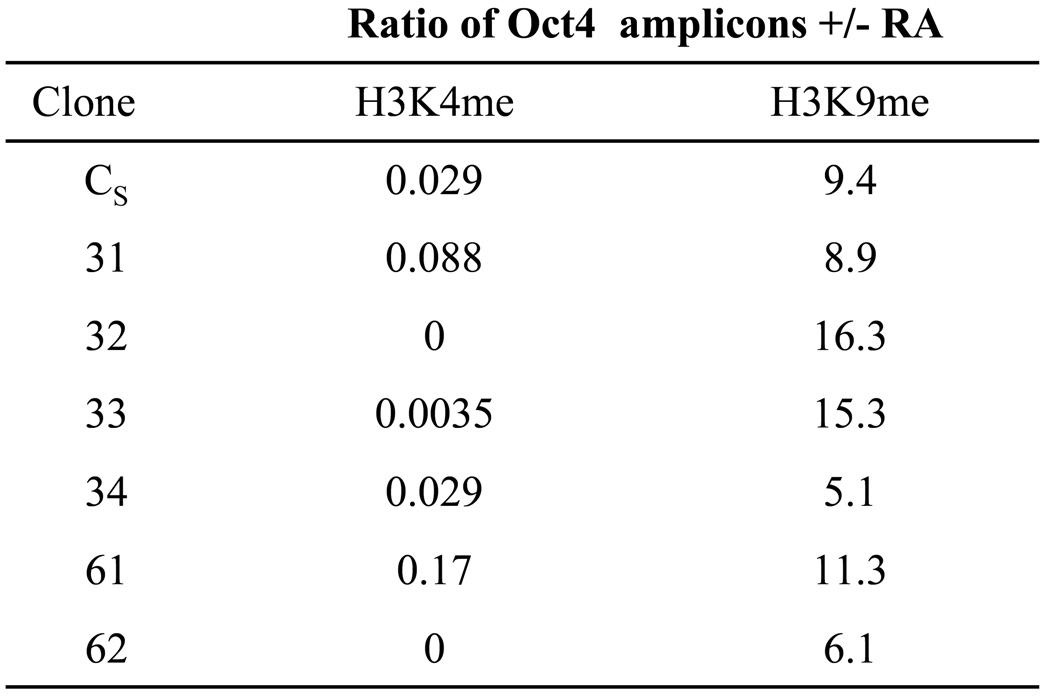
First we investigated whether WRNp associates with the Oct4 promoter in WRNp-deficient cells. As expected, we did not find enrichment of WRNp at the promoter (Supplementary Fig. 1). We then studied the effect of WRNp knockdown on H3K4 demethylation, which is an early histone modification during RA-induced differentiation. Three days after RA addition, control, WRNp-expressing cells exhibited 97% decrease in the H3K4me2 at the Oct4 promoter, when compared to nontreated cells (Fig. 3). RA treatment decreased H3K4me2 at the Oct4 promoters of all WRNp-deficient clones, with the decrease ranging from 83% to no detectable H3K4me2 after RA treatment (Fig. 3). We conclude that H3K4 demethylation appears to proceed normally in all WRNp-deficient clones. We then examined H3K9 methylation. RA was reported to induce a dramatic accumulation of trimethylated H3K9 (H3K9me3) at the Oct4 promoter (Feldman et al. 2006). We observed that at three days after RA addition, control cells showed 9.4 fold upregulation of H3K9me3 when compared to untreated cells. WRNp-deficient, RA-stimulated clones exhibited similar increases of H3K9me3 at the Oct4 promoter (5.1 to 16.3 fold, Fig. 3). We conclude that WRNp knockdown does not reduce H3K4 demethylation, nor does it reduce H3K9 methylation at the Oct4 promoter.
Fig. 3. WRNp knockdown does not affect changes in histone methylation during pluripotent cell differentiation.
Q-ChIP analysis of Oct4 promoter DNA associated with H3K4me2 (dimethylated lysine 4 of H3) and H3K9me3 (trimethylated lysine 9 of H3) in undifferentiated and RA treated cells. Results are presented as a ratio of amplicons (Oct4 B primer/probe set, see Supplementary Table 2) detected in differentiated over non-differentiated samples. Cells were treated with 10 µM RA for 72 hrs, when histone methylation changes were shown to peak (Feldman et al. 2006).
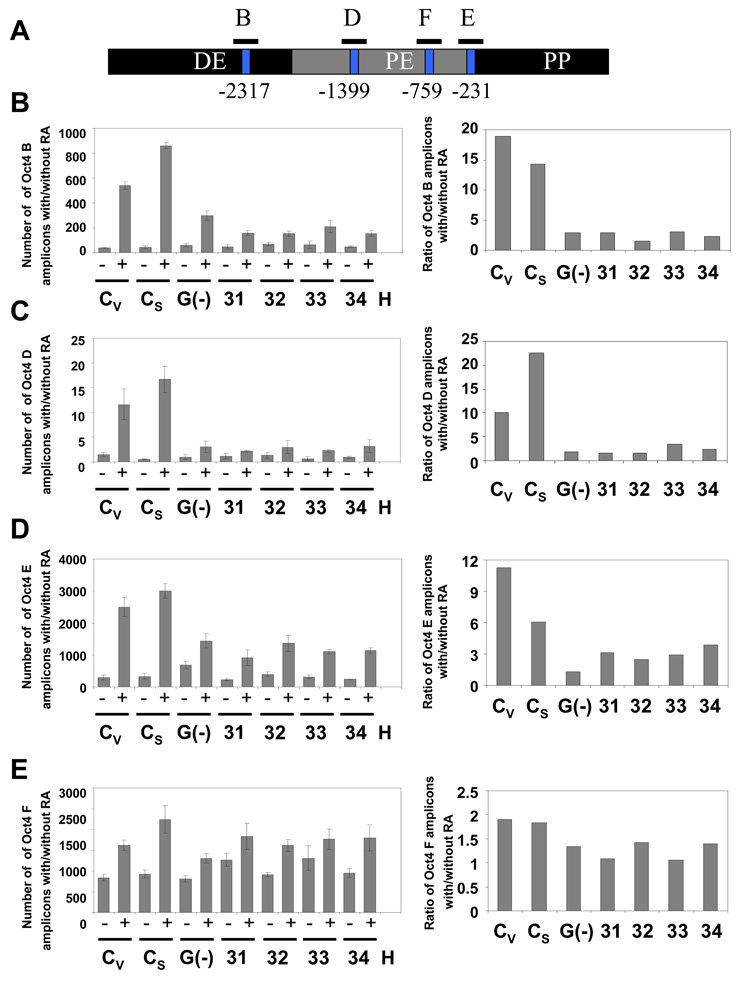
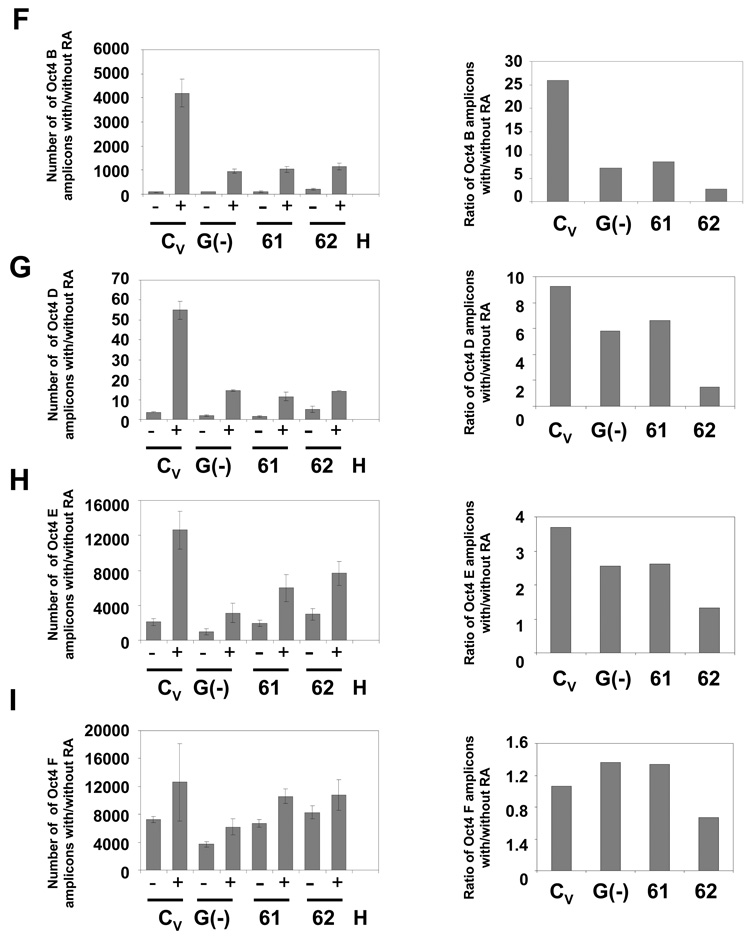
The last step of Oct4 repression is DNA methylation of its promoter, which results in its irreversible inactivation (Feldman et al. 2006; Epsztejn-Litman et al. 2008). To measure the WRNp effect on this process, we have developed an HpaII and HhaI-based assay. These two enzymes only cut unmethylated CpG sites. We have identified two HpaII sites – one in the distal enhancer and another in the proximal enhancer – of the Oct4 promoter (GenBank No. AJ297527, positions −2317 and −1399, Fig. 4A), and two HhaI sites in the proximal enhancer (−759 and −231, Fig. 4A). We have digested DNA from undifferentiated and RA treated cells with these enzymes and measured methylation of CpG sites in the Oct4 promoter using real-time PCR instead of the more traditional Southern blotting assay. The real-time PCR assay allows us an accurate quantitation of DNA methylation in the cultured cell population. CpG methylation blocks HpaII or HhaI digestion and thus increases the PCR signal. WRNp-deficient and control cells were treated with RA for seven days. As shown in Fig. 4 (and Supplementary Fig. 2), RA treatment induces methylation at CpG sites in the distal enhancer (DE) and proximal enhancer (PE) regions of the Oct4 promoter of control cells. We then analyzed DNA methylation at Oct4 promoters of the WRNp-deficient clones 31–34 (see Fig. 2). We observed a dramatic reduction in DNA methylation in the DE region of the promoter: 78–92% reduction in the B segment of DE (spaning the −2317 site, Fig. 4B), and 66%-93% reduction in DNA methylation in the D segment (Fig. 4C). 37%-78% reduction was also found in the E segment of the PE region (Fig. 4D). No significant reduction was observed in the F segment (Fig. 4E). Similarly, WRNp deficient clones 61 and 62 showed a 67%-90% reduction in DNA methylation in the DE region (B segment, Fig. 4F, Supplementary Fig. 3) and 29%-84% and 30%-65% reduction at the sites in the PE region (D and E segment, Fig. 4G and 4H). No significant RA-induced differences in DNA methylation were observed at one CpG site in the PE region (F segment, Fig. 4I). As expected, G9a-deficient cells showed a similar degree of reduction in DNA methylation (Fig. 4). To confirm that the observed reduction in DNA methylation is due to WRNp knockdown, we have transduced NCCIT cells with lentiviral particles that contain three expression constructs encoding shRNA targeting WRN. The three sequences target nucleotides 3927 – 3945, 3974 – 3992, and 4312 – 4330. A polyclonal population of puromycin-resistant cells expressed reduced levels of WRNp, when compared to control cells (Supplementary Fig. 4A). An analysis of DNA methylation at the Oct4 promoter of these cells demonstrated a 69% and 51% drop in the rate of methylation in the B and D segments respectively of the WRNp-deficient population when compared to the control population (Supplementary Fig. 4B). Taken together, the presented evidence indicates that WRNp plays a role in DNA methylation at the Oct4 promoter.
Fig. 4. Deficient DNA methylation of the Oct4 promoter in WRNp-deficient clones.
During the differentiation of NCCIT cells, Oct4 promoter DNA is progressively methylated and thus becomes resistant to the methylation-sensitive enzymes HpaII and HhaI. Cells were stimulated with 10 µM RA for 7 days then harvested, DNA extracted and digested with the methylation-sensitive enzymes HpaII or HhaI. The digested DNA was then subjected to real-time PCR with primers targeting segments of the Oct4 promoter that flank the restriction sites. Error bars indicate standard deviation. (A) Map of the Oct4 promoter with the location of the HpaII and HhaI restriction sites and DNA methylation sites (GeneBank No. AJ297527) and amplified Oct4 segments (B, D, E, and F). (B) CpG methylation status in the Oct4 B region of undifferentiated and RA treated WRNp-deficient, G9a-deficient and control cells (see Fig. 2 for terminology). The results are shown as the number of amplicons obtained with DNA from undifferentiated cells (−) as well as cells that were stimulated to differentiate with RA (+) (left panels). Right – a ratio of the number of amplicons detected in stimulated vs. non-stimulated samples (after adjustment to DNA input according to qPCR signal of undigested samples). (C) CpG methylation status in the Oct4 D region. (D) CpG methylation status in the Oct4 E region. (E) CpG methylation status in the Oct4 F region. (F) CpG methylation status in the Oct4 B region of undifferentiated and RA treated WRNp-deficient clones 61 and 62 and control cells. Cells were treated as in Fig. 4A-E. (G) CpG methylation status in the Oct4 D region of 61 and 62 clones. (H) CpG methylation status in the Oct4 E region. (I) CpG methylation status in the Oct4 F region of 61 and 62 clones. See Supplementary Figs. 2 and 3 for additional information.
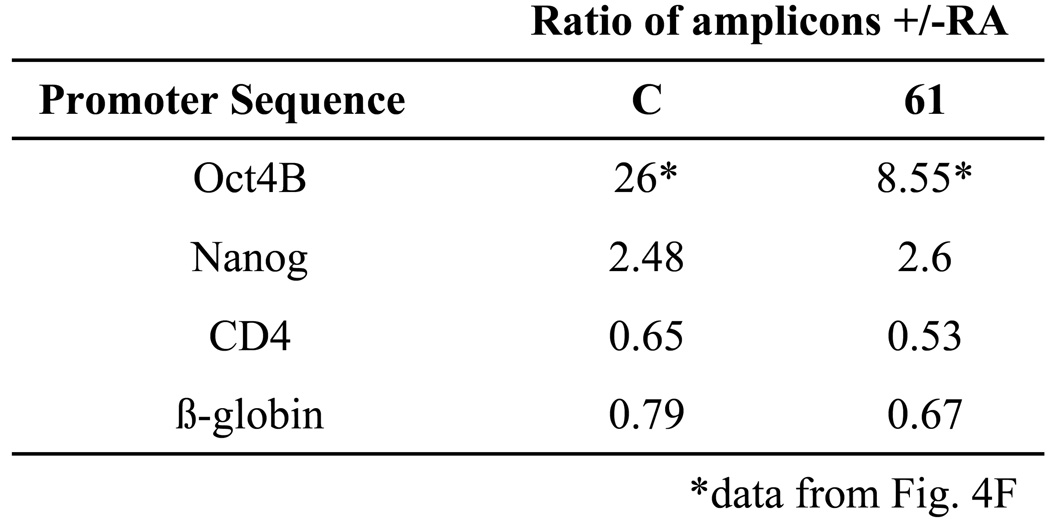
To determine if WRNp may play this role on different promoters, we have identified potential methylation sites in the Nanog, CD4 and β-globin promoters. We note that the Nanog promoter may be predominantly controlled by RA-induced histone modifications, rather than DNA methylation, although some DNA methylation has been reported to occur (Feldman et al. 2006; Dahl & Collas 2007; Barrand & Collas 2010). The CD4 promoter was not reported to be methylated in response to RA; however, it does contain a potential methylation site. We did not identify a potential methylation site in the β-globin promoter. As shown in Fig 5, the Nanog promoter is somewhat methylated in response to RA treatment, but to a much lower degree than the Oct4 promoter. Nanog promoter methylation occurs even in WRNp-deficient cells and is thus not dependent on WRNp. The CD4 promoter is not methylated in response to RA (Fig. 5). We conclude that the WRNp role in methylation of promoter DNA appears to be specific for the Oct4 promoter, and correlates with the presence of WRNp on the promoter (see Fig. 1).
Fig. 5. Methylation on Nanog, CD4 and β-globin promoters.
Cells were treated with RA as described in Fig. 4. DNA was extracted and digested with HpaII. The digested DNA was then subjected to real-time PCR with primers targeting segments of the Nanog and CD4 promoter that flank the HpaII restriction sites in these promoters. No such restriction site was found in the β-globin promoter. Control cells – CV (see Fig. 4), WRNp-deficient cells – clone 61 (see Fig. 4).
Finally, we have analyzed the potential WRNp role in the maintenance of Oct4 promoter methylation in somatic cells. We have treated normal primary fibroblasts with WRNp siRNA, or knocked down WRNp using an shRNA approach. We have observed that the Oct4 promoter is methylated in these cells, and the levels of promoter methylation do not depend on WRNp (Supplementary Fig. 5). This result may suggest that WRNp does not control the maintenance of Oct4 promoter methylation, and its effect may be limited to de novo methylation, as demonstrated above (Fig. 4). However, we cannot exclude the possibility that other differences between NCCIT cells and primary fibroblasts may underlie different effects of WRNp knockdown in the two settings.
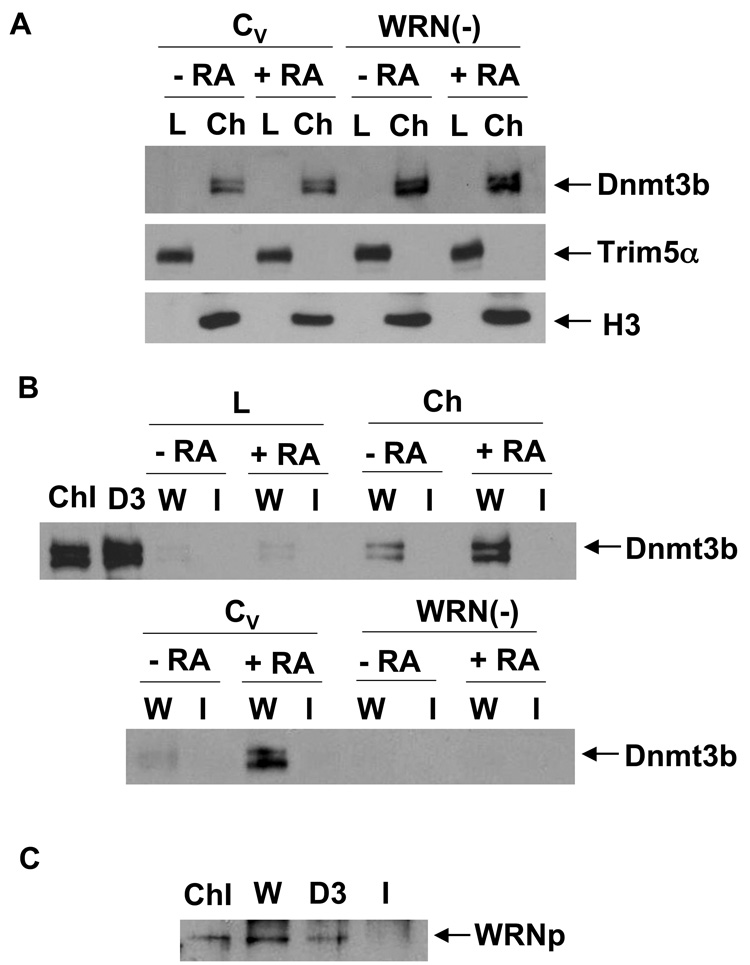
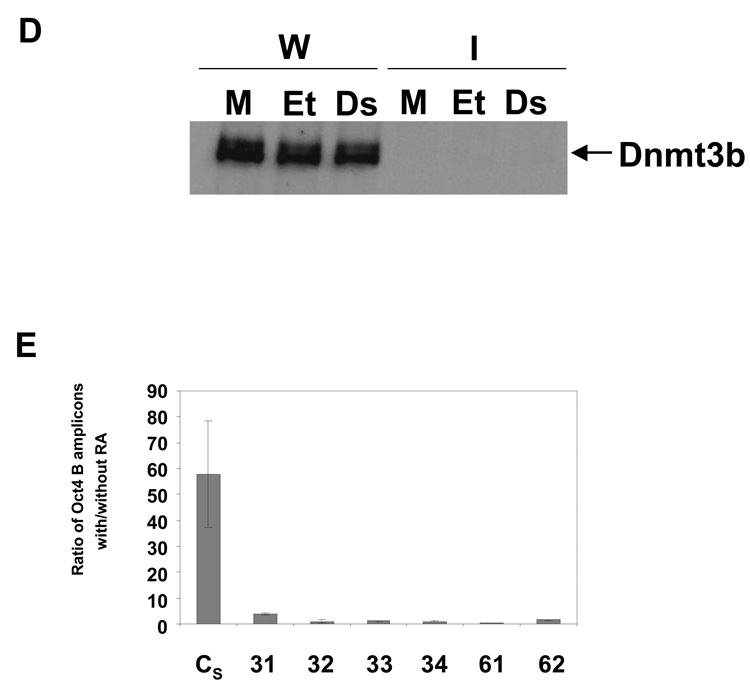
Knockout experiments demonstrated that DNA methylation at the Oct4 promoter depends on Dnmt3b (Feldman et al. 2006). Since WRNp appears to control this methylation, we examined the putative interaction between Dnmt3b and WRNp. A western blotting analysis demonstrated that Dnmt3b is present in the chromatin fraction of both control and WRNp-deficient cells (Fig. 6A). We note that Dnmt3b migrates as two bands on SDS-PAGE, which likely reflect different Dnmt3b isoforms [Fig. 6A, (Okano et al. 1998; Xie et al. 1999)]. An immunoprecipitation analysis revealed that Dnmt3b is present in the chromatin fraction of WRNp immunoprecipitates (Fig. 6B, top). RA treatment upregulates the amount of Dnmt3b which immunoprecipitates with WRNp (Fig. 6B). To test the possibility that Dnmt3b is recognized by the WRNp antibody, we performed the immunoprecipitation analysis on WRNp-deficient cells (Fig. 6B, bottom). We did not detect the presence of Dnmt3b in WRNp immunoprecipitates in these cells. In addition, we performed reverse co-immunoprecipitation and investigated the presence of WRNp in Dnmt3b immunoprecipitates from RA treated cells. Fig. 6C shows that WRNp co-immunoprecipitates with Dnmt3b, when the latter is immunoprecipitated with the Dnmt3b antibody. We conclude that Dnmt3b is present in a protein complex that contains WRNp, and the accumulation of this complex increases upon the induction of differentiation. A densitometry analysis showed that less than 10% of chromatin bound Dnmt3b associates with WRNp in RA-treated cells (Fig. 6B, data not shown). On the other hand, approximately 20% of chromatin-bound WRNp immunoprecipitates with Dnmt3b in RA-treated cells (Fig. 6C, data not shown). Finally, we investigated the possibility that the WRNp and Dnmt3b association is mediated by DNA. We used a dual approach. In some samples, we treated the cell lysates with DNAse, which has been employed to this purpose previously (Otterlei et al. 2006). Other samples were treated with ethidium bromide (EtBr) in immunoprecipitation, because EtBr was shown to reduce association of proteins with DNA (Lai & Herr 1992; Monahan et al. 1998). However, neither of these treatments reduced the association of Dnmt3b and WRNp (Fig. 6D). We conclude that the observed association of WRNp and Dnmt3b is due to a protein-protein interaction.
Fig. 6. WRNp is in a complex with and is required for Dnmt3b recruitment to the Oct4 promoter.
(A) Dnmt3b is localized in the chromatin fraction of control and WRNp-deficient cells. CV and WRNp-deficient cells (clone 61) were treated with 10 µM RA for 3 days. Chromatin fraction was then separated and both chromatin-lacking lysates (L) and the chromatin fraction (Ch) were analyzed by western blotting. H3 – histone H3, loading control for the chromatin fraction, Trim5α – cytoplasmic fraction loading control. (B) Dnmt3b is present in WRNp immunoprecipitates. Top: WRNp-proficient control cells (CV, see Fig. 2) were stimulated with 10 µM RA for 3 days. WRNp then was immunoprecipitated from the chromatin-lacking lysates and from the chromatin fraction of undifferentiated and RA treated cells. WRNp immunoprecipitates were resolved on SDS-PAGE and Dnmt3b was detected by western blotting analysis. ChI – whole chromatin input lysate of RA-treated cells (1/10 of the immunoprecipitation input), D3 – chromatin input lysate immunoprecipitated with the Dntm3b antibody, W – lysates immunoprecipitated with a WRNp antibody, I – lysates immunoprecipitated with normal rabbit IgG. Bottom: Control cells and WRNp-deficient cells (clone 61) were stimulated with 10 µM RA for 3 days. WRNp then was immunoprecipitated from the chromatin fraction of undifferentiated and RA treated cells. (C) WRNp is present in Dnmt3b immunoprecipitates. WRNp-proficient control cells (CV, see Fig. 2) were stimulated with 10 µM RA for 3 days. WRNp and Dnmt3b then were immunoprecipitated from the chromatin fraction. Immunoprecipitates were resolved on SDS-PAGE and WRNp was detected by western blotting analysis. ChI – whole chromatin input lysate of RA-treated cells (1/10 of the immunoprecipitation input), W – lysates immunoprecipitated with a WRNp antibody, D3 - lysates immunoprecipitated with a Dnmt3b antibody, I – lysates immunoprecipitated with normal rabbit IgG. (D) Effect of EtBr and DNAse on Dnmt3b and WRNp co-immunoprecipitation. WRNp-proficient control cells (CV, see Fig. 2) were stimulated with 10 µM RA for 3 days. WRNp was immunoprecipitated from the chromatin fraction as in Fig. 6B and C, except cell lysates were treated with EtBr (100 µg/ml) or DNAse prior to immunoprecipitation (see Methods). Immunoprecipitates were resolved on SDS-PAGE and Dnmt3b was detected by western blotting analysis. Et – EtBr-treated lysates, Ds – DNAse-treated lysates, M – mock-treated lysates, W – lysates immunoprecipitated with a WRNp antibody, I – lysates immunoprecipitated with normal rabbit IgG. (E) Q-ChIP analysis of Dnmt3b levels at the Oct4 promoter regions of control and WRNp-deficient cells. Cells were treated with RA for three days as described above and then subjected to Q-ChIP analysis using a Dnmt3b antibody.
The case for a Dnmt3b role in DNA methylation at the Oct4 promoter is based on the finding that the Oct4 promoter remains unmethylated in Dnmt3b-deficient cells (Feldman et al. 2006). This result suggests that Dnmt3b should accumulate at the Oct4 promoter in RA-treated NCCIT cells. To test this hypothesis, we performed the Q-ChIP assay and determined that Dnmt3b is highly enriched at the promoter of RA-treated control cells. The number of Oct4 amplicons associated with Dnmt3b in RA-treated control cells was about 58 fold higher than the number amplicons detected by Q-ChIP analysis of untreated cells (Fig. 6E). On the other hand, WRNp-deficient cells displayed minimal enrichment of Dnmt3b at the Oct4 promoter in RA-treated WRNp-deficient clones when compared to untreated WRNp-deficient cells (0.88–3.68 fold enrichment, Fig. 6E). We conclude that WRNp facilitates Dnmt3b recruitment to the Oct4 promoter. The co-immunoprecipitation analysis suggests that WRNp may physically interact with Dnmt3b in this process.
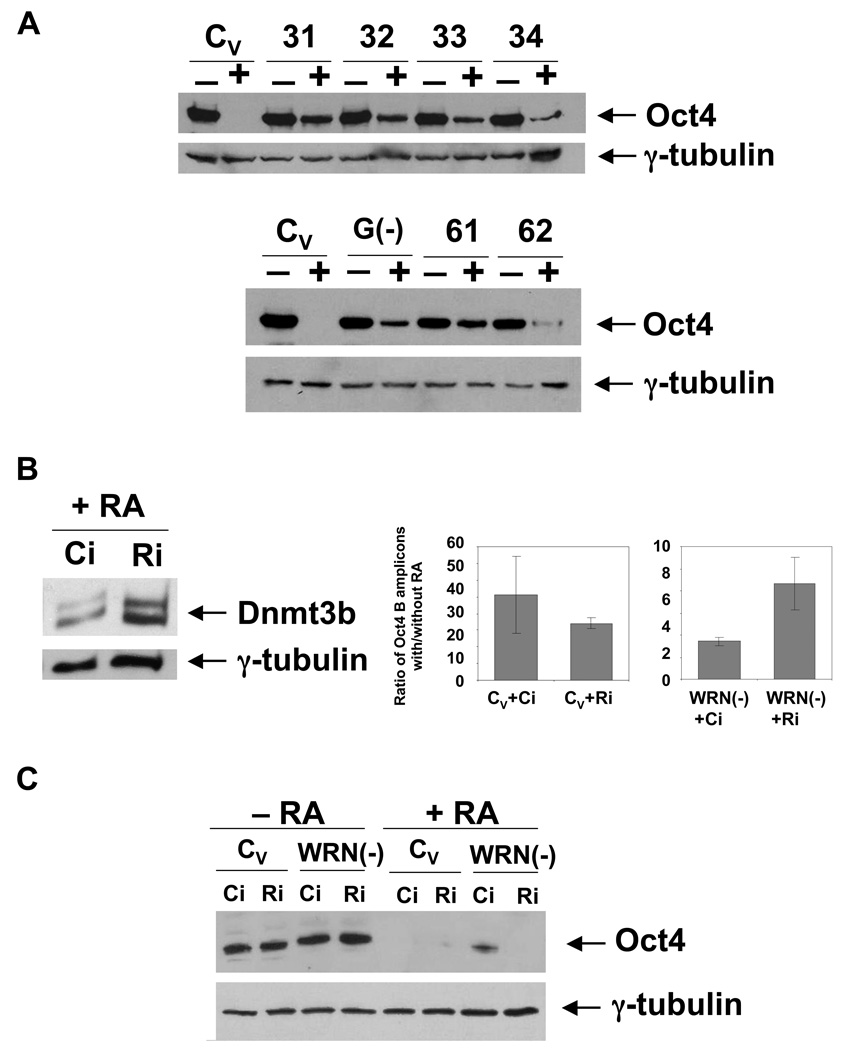
RA treatment of ES cells results in a complete inactivation of the Oct4 promoter and disappearance of the Oct4 protein. We demonstrated in this study that the Oct4 inactivation process in NCCIT cells mirrors that of ES cells. We then tested if the Oct4 protein is absent in RA-treated NCCIT cells. Seven days of RA treatment results in complete repression of Oct4 expression in WRNp-expressing NCCIT cells (Fig. 7A). As expected, G9a knockdown resulted in continued, but weaker Oct4 expression in RA-treated cells. Interestingly, WRNp-deficient NCCIT cells also continue to express the Oct4 protein after RA treatment, albeit at highly reduced levels (Fig. 7A). This was an unexpected finding, since DNA methylation of the Oct4 promoter is secondary in ES cells to chromatin changes and not necessary to achieve full suppression of Oct4 (Feldman et al. 2006). To determine if the observed “leaky” expression of Oct4 can be attributed to the deficient DNA methylation at the Oct4 promoter, we have asked if increased Dnmt3b expression can revert this phenotype and induce full suppression of Oct4. Dnmt3b expression is controlled by retinoblastoma like protein 2 [Rbl2, (Benetti et al. 2008; Sinkkonen et al. 2008)]. We have knocked down Rbl2 expression in RA-treated cells by siRNA treatment and measured Dnmt3b expression in these cells. We observed that Dnmt3b is strongly upregulated in RA-treated, Rbl2-deficient cells (Fig. 7B, left). We then asked if this treatment results in increased methylation of the Oct4 promoter. As shown in Fig. 7B (right), Rbl2 knockdown reverts the hypomethylation of the Oct4 promoter in WRNp-deficient cells and increases the methylation by about 3 fold. Finally, we assayed for Oct4 expression. The Oct4 protein was completely absent in RA-treated Rbl2- and WRNp-deficient cells, but not in RA-treated WRNp-deficient cells that were treated with control siRNA (Fig. 7C).
Fig. 7. Oct4 expression and Oct4 promoter methylation status in RA-treated WRNp-deficient cells.
(A) RA-treated WRNp-deficient cells continue to express Oct4. WRNp-deficient (31–62), G9a-deficient [G(−)] and control (CV) cells were treated with 10 µM RA for 7 days, after which cells were harvested and Oct4 expression analyzed by western blotting (+, cells treated with RA; −, untreated, control samples). (B) Rbl2 knockdown stimulates Dnmt3b expression and reverses the Oct4 promoter methylation defect of WRNp-deficient cells. The day following RA addition, cells were treated with siRNA targeting Rbl2. Left – Dnmt3b expression in WRNp-deficient cells (61) treated with control (Ci) or Rbl2 siRNA (Ri) at 5 days after RA addition. Right – DNA methylation in the Oct4 promoter B region. (C) Oct4 expression in differentiated (+RA) and undifferentiated (−RA) control and WRNp-deficient cells treated with siRNA. Cells were treated as in B. Oct4 expression was analyzed by western blotting 7 days after RA addition.
Finally, we have performed a Dnmt3b overexpression experiment. We transfected an expression vector encoding Dnmt3b into control and WRNp-deficient cells, both in the presence and absence of RA. Similarly to Rbl2 knockdown, Dnmt3b overexpression extinguished the residual Oct4 expression in RA-treated WRNp-deficient cells (Supplementary Fig. 6A). Analysis of methylation at the Oct4 promoter in these cells demonstrated increased methylation of promoter DNA, similar to that obtained by Rbl2 knockdown (Supplementary Fig. 6B). We conclude that at least in a fraction of cells, DNA methylation is necessary for a complete suppression of Oct4 expression, and increased Dnmt3b expression can compensate for the reduced Oct4 promoter DNA methylation in WRNp-deficient cells.
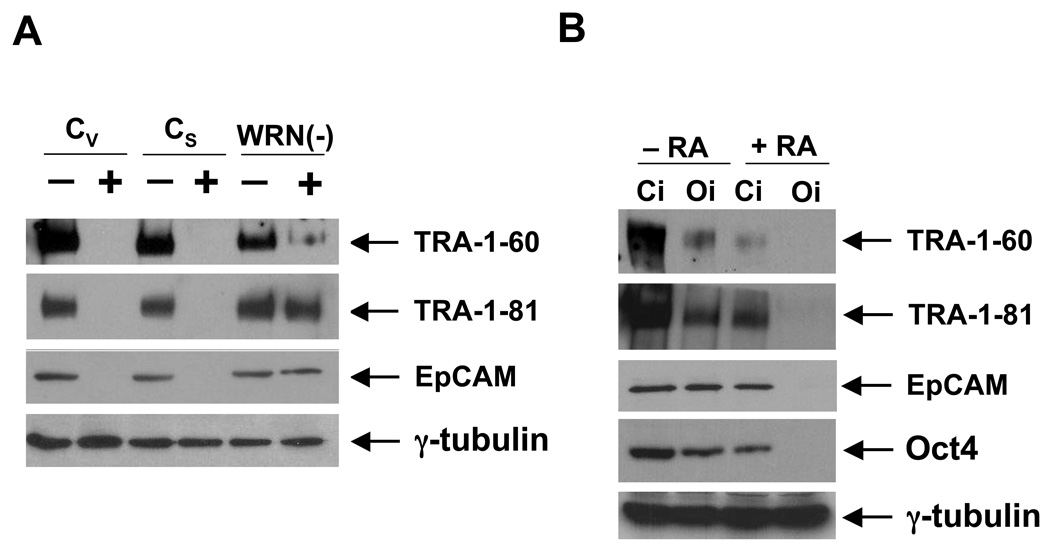
The primary function of the Oct4 protein is to maintain pluripotency (Boiani & Scholer 2005). The residual Oct4 expression in RA-treated cells might thus suggest a potential defect in differentiation. To investigate this possibility, we have assayed the expression of stem cell markers in RA-treated control and WRNp-deficient cells. We have analyzed the expression of TRA-1-60 and TRA-1-81 markers, as well as the presence of EpCAM, which was recently identified as a marker of undifferentiated human ES cells (Damjanov et al. 1993; Lu et al. 2010; Ng et al. 2010). RA treatment highly reduced or extinguished expression of these markers in control cells (Fig. 8A). However, WRNp-deficient cells still expressed significant amounts of these proteins one week after they were stimulated to differentiate (Fig. 8A). To investigate the relationship between the residual Oct4 expression and the expression of the above mentioned stem cell markers, we knocked down Oct4 expression using siRNA treatment. Using this approach, Oct4 expression was completely extinguished in RA-treated WRNp-deficient cells (Fig. 8B). We then again assayed the expression of stem cell markers. As shown in Fig. 8B, Oct4 knockdown resulted in suppression of all examined markers. We conclude that the residual Oct4 expression in RA-treated WRNp-deficient cells leads to the maintenance of stem cell markers in cultures. These data suggest that WRNp deficiency attenuates stem cell differentiation.
Fig. 8. Expression of stem cell markers in WRNp-deficient cells.
(A) Control (CV, CS) and WRNp-deficient cells (31) were treated with RA for 7 days. Cells were then harvested and the presence of stem cell markers was analyzed by western blotting (+, cells treated with RA; −, untreated control samples). TRA-1-60, TRA-1-81 and EpCAM bands are indicated. γ-tubulin served as a loading control. (B) WRNp-deficient cells (31) were treated with RA for 7 days. Cells were then transfected with Oct4 siRNA (Oi) or control siRNA (Ci). Three days after transfection, cells were harvested and the expression of stem cell markers analyzed as described above (−RA – untreated cells, +RA − RA treated cells).
Discussion
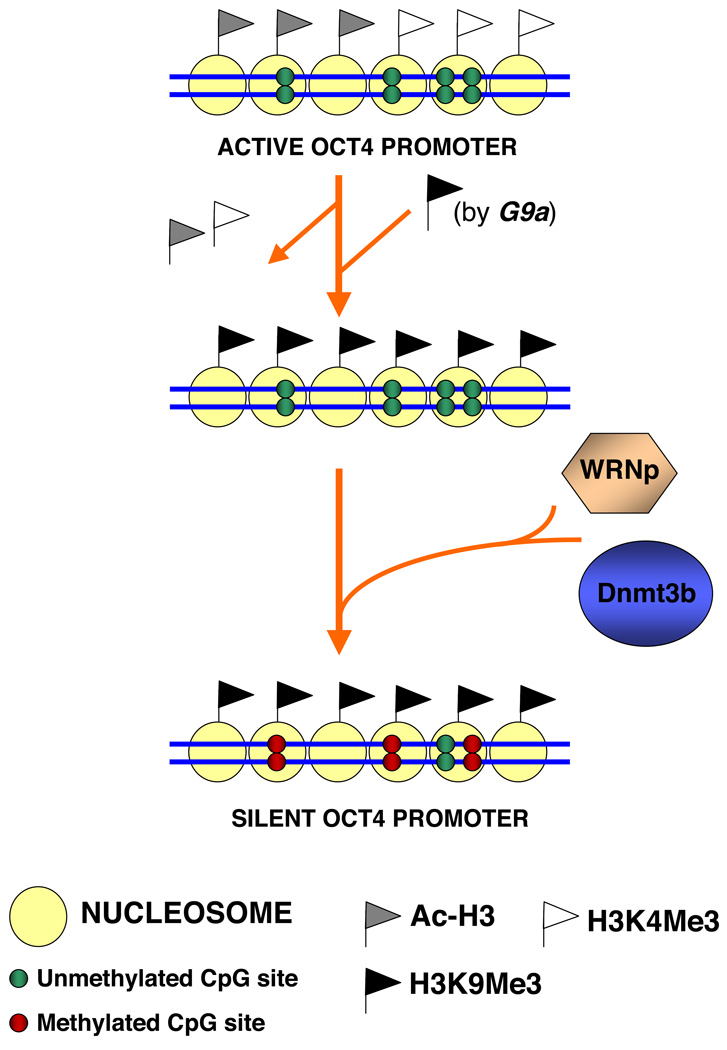
In this study, we identify a previously unknown function of the Werner syndrome protein. We demonstrate that WRNp translocates to the promoter of a stem cell transcription factor during retinoic acid-induced differentiation, and regulates recruitment of a de novo DNA methyltransferase to this promoter (Fig. 9). Our data show for the first time that WRNp is involved in de novo DNA methylation, and a key step of stem cell differentiation, which is silencing of the Oct4 gene. Studies in plants, and recently mice, suggest that DNA helicases of the SNF2 family are involved in DNA methylation (Bourc'his & Bestor 2002; Xi et al. 2009). In contrast, the human WRN gene belongs to the RecQ family of DNA helicases, which was shown to play a role in replication-associated DNA repair (Hickson 2003; Bohr 2008). The presented results thus open up the possibility that the RecQ family plays a wider role in cellular processes than previously suspected.
Fig. 9. Model of the WRNp role in the regulation of Oct4 inactivation in differentiating pluripotent cells.
For details see text.
We have observed that WRNp accumulates at the Oct4 promoter in RA-induced pluripotent cells. To study the potential WRNp role in regulation of the Oct4 promoter, we have developed WRNp-deficient cells. We have used an shRNA approach, since inactivation of Oct4 in differentiating cells is a process that takes up to one week and thus long-term suppression of WRNp is needed. To minimize any potential effects that could be due to either clonal selection or to some shRNA effect that is not due to WRNp suppression, we have used a three-pronged approach. We have developed clones, where WRNp was suppressed by one of two different shRNAs, and we have also employed a pool of clones, where WRNp was suppressed by a mixture of lentivirus-delivered shRNAs (Fig. 2 and Supplementary Fig. 3). All of the clones showed defects in de novo methylation at the Oct4 promoter. These results indicate that the observed phenotype is indeed due to knockdown of WRNp.
In contrast, WRNp knockdown in differentiated cells (primary fibroblasts) did not affect the maintenance of Oct4 methylation in these cells. This finding again supports the role of WRNp in de novo, but not maintenance methylation.
Inactivation of Oct4 is a multistep process, in which a key role is played by the G9a histone methyltransferase. G9a methylates the H3K9 residue, and is required for the recruitment of Dnmt3b (Feldman et al. 2006). We observed that WRNp-deficient clones show a lack of DNA methylation similar to that of G9a-deficient cells (c). These data led us to examine the G9a function in RA-induced WRNp-deficient cells. However, we observed that G9a-dependent H3K9 methylation does not appear to be affected in WRNp-deficient cells. This result places WRNp downstream of G9a in the process of Oct4 inactivation. On the other hand, co-immunoprecipitation results indicate that WRNp is in a protein complex with Dnmt3b. The formation of this complex is inducible by RA, and the complex is present almost exclusively in the chromatin of cell lysates. In the absence of WRNp, Dnmt3b recruitment to the Oct4 promoter is dramatically reduced. Consistently, WRNp-deficient cells show profound differences in Dnmt3b-dependent DNA methylation. These data indicate that WRNp acts exclusively at the last step of Oct4 inactivation.
What is the mechanism involved in Dnmt3b recruitment? One possibility is a direct interaction between the WRNp and Dnmt3b proteins, which are apparently present in the same protein complex. Dnmt3b was reported to directly bind to the ankyrin domain of G9a, and this interaction was shown to be necessary for Dnmt3b recruitment to the Oct4 promoter (Epsztejn-Litman et al. 2008). One possible explanation for the WRNp effect on Dnmt3b recruitment is that WRNp may enhance the association of Dnmt3b with G9a. However, WRNp could also facilitate Dnmt3b binding to Oct4 promoter DNA, possibly through its own binding to DNA. We cannot also exclude a possibility that the WRNp helicase activity may play a role, and changes in DNA structure due to this activity may contribute to Dnmt3b recruitment. A deletion analysis, included in our future experiments, will determine if the helicase function, or even possibly the N-terminus of WRNp, which has a nuclease function, are involved.
It has been proposed very recently that one of the main causes of aging might be a reduced ability of stem cells to differentiate (Edelberg & Ballard 2008). We have observed that the residual expression of Oct4 in RA-treated WRNp-deficient cells leads to persistence of stem cell makers in the culture and thus appears to attenuate differentiation. The presented results are consistent with the hypothesis that inefficient stem cell differentiation may contribute to aging, due to a reduced number of some cell types in developed organs. We also note that Oct4 expression was recently reported, in addition to human ES cells, in adult stem cells (Edelberg & Ballard 2008). It will be intriguing to investigate whether the regulation of Oct4 expression in these cells changes with age, due to either epigenetic inactivation or mutations in WRNp, or other proteins that are involved in Oct4 regulation. However, we note that given the fact that both WRNp-deficient individuals and mice develop relatively normally (Lebel & Leder 1998; Lombard et al. 2000), alternative, WRNp-independent mechanisms that control de novo methylation must exist, even if they may not be as efficient as normal WRNp. To test this hypothesis, we have investigated methylation of the Oct4 promoter in differentiated fibroblasts of two WS patients. We found in one case, hypomethylation at the D segment of the promoter. However, in the other case, the Oct4 promoter was fully methylated (data not shown). It thus appears that other, alternative pathways may, to some extent, replace the WRNp function. It remains to be seen if these pathways involve DNA helicases and/or other proteins that may repress Oct4 expression, such as histone deacetylases, and methyltransferases.
Finally, the role of WRNp in Oct4 inactivation may suggest a new link between tumor formation and WRNp. WRN is a tumor suppressor gene and its mutations or epigenetic inactivation have been associated with tumorigenesis (Hickson 2003; Esteller 2007; Bohr 2008). The exact mechanism of how a loss of WRNp contributes to tumor formation is not yet known, although the relationship between DNA repair deficiencies and cancer is well established. One can thus assume that there is a causal link between the role of this protein in DNA repair and cancer development in its absence. However, it has also been repeatedly shown that aberrant DNA methylation is associated with cancer (Laird & Jaenisch 1996; Linhart et al. 2007; Meissner et al. 2008). A loss of WRNp may affect de novo methylation by Dnmt3b and contribute to cancer development in this way. In addition, several types of cancer stem cells were recently shown to express Oct4, and ectopic expression of Oct4 promotes tumorigenesis in mice (Hochedlinger et al. 2005; Ben-Porath et al. 2008; Tang et al. 2008; Du et al. 2009; Suva et al. 2009). A lack of DNA methylation of the Oct4 promoter due to a loss of WRNp function may lead to Oct4 re-expression, and thus contribute to tumor development.
Methods
shRNA and siRNA reagents
Plasmids encoding shRNA were transfected with Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. shRNA against WRNp was purchased from Origene [TI333413 (clones 31–34) and TI333416 (clones 61 and 62), (Rockville, MD, USA)]. An empty control plasmid (TI20003, Origene) and control non-targeting shRNA (TI30003, Origene) were utilized. A plasmid encoding shRNA against G9a was also stably transduced into NCCIT cells (TI1319244, Origene). siRNA targeting Rbl2, Oct4, and control siRNA were obtained from Dharmacon (M-003299-03, M-019591-03, and D-001206-13, and D-001210-02-05, Lafayette, CO, USA). siRNA targeting WRN was obtained from Qiagen (SI02663759) and the control siRNA for this experiment was purchased from Dharmacon (D-001210-02-05). Lentiviral particles encoding 3 shRNAs targeting WRN (sc-36843-v) or scrambled control shRNA (sc-108080) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). See Supplementary Table 3.
Cell culture and transfections
NCCIT cells (ATCC, Manassas, VA, USA) were propagated in RPMI 1640 medium with 10% fetal bovine serum, non-essential amino acids, and antibiotics. The primary fibroblasts (GM00200) were obtained from the Coriell Institute for Medical Research (Camden, NJ) and propagated in the above described medium. NCCIT were stimulated with 10 µM retinoic acid (Sigma, St. Louis, MO, USA) to differentiate for the indicated time periods. To knock down WRNp, cells were transfected with plasmids encoding shRNA (Supplementary Table 3) and a puromycin resistance marker using Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA, USA). Two days post-transfection, cells were selected for puromycin resistance by addition of the drug [2 µg/ml (MP Biomedicals, Solon, OH, USA)]. Individual puromycin-resistant clones were then analyzed by western blotting to determine WRNp levels. siRNA against Rbl2, Oct4 or WRN were transfected with Lipofectamine™ RNAiMAX (Invitrogen) as described in the manufacturers instructions. The Dnmt3b-encoding plasmid was obtained from Origene (RC223206). It and a control noncoding plasmid (pcDNA3.1) were transfected into cells using Lipofectamine™ 2000 (see above).
Western blot analyses and co-immunoprecipitations
Cells were lysed in a lysis buffer containing 1% Triton X-100 and the chromatin fraction was separated from the rest of the lysate as described (Smith et al. 2008). For western blotting analyses, lysates were run on SDS-PAGE. Blots were probed with antibodies specific for WRN, Oct4, Dnmt3b, and histone H3, as listed in Supplementary Table 1. Bands were detected using chemiluminiscence (see Supplementary Table 1 for antibodies used). For co-immunoprecipitations, lysates of 1×106 cells per sample were used. Chromatin fraction was resuspended in 250 µl of the lysis buffer, briefly sonicated to break DNA and cleared by centrifugation prior to addition of the antibody (WRN or IgG control antibody). For DNAse treatment, lysates were treated with 300 U/ml DNAse for 30 min at room temperature prior to immunoprecipitation. Antibody-protein complexes were then isolated by binding to protein A/protein G+ agarose (Calbiochem, Gibbstown, NJ, USA) and analyzed on 7.5% SDS-PAGE.
Chromatin immunoprecipitation
Q-ChIP experiments were performed essentially as described (Dahl & Collas 2007). NCCIT cells were treated with RA (Sigma, St. Louis, MO, USA) for indicated periods of time and equal amounts of cells (1–3×106 per sample) were then harvested, treated with crosslinking agents and processed for Q-ChIP. Immunoprecipitations were performed with following antibodies: anti-WRNp, H3K4me2, H3K9me3 and Dnmt3b. For real-time PCR, 2 µl of the 50 µl DNA extract was used, with Oct4 primers and probes indicated in Supplementary Table 2.
DNA methylation assay
Cells were stimulated with 10 µM RA (Sigma, St. Louis, MO, USA) for 7 days, then harvested, DNA extracted and digested (600ng) with the methylation-sensitive enzymes HpaII or HhaI [R0171 and R0139 respectively (New England Biolabs, Ipswich, MA, USA)], see Fig. 4 for location of restriction sites]. The digested DNA was then subjected to real-time PCR with primers targeting regions of the Oct4 promoter that flank the restriction sites.
Real-time PCR
Primer/probe sets were designed using the IDT RealTime PCR SciTool. FAM™/Iowa Black® FQ dual-labeled probes were utilized [PrimeTime qPCR Assays, (Integrated DNA Technologies, Coralville, IA, USA)]. Real-time PCR was performed using a LightCycler 1.5 with software 3.5.3 (Roche) or a Stratagene Mx3000P with the MxPro software. Reaction mixtures contained QuantiFast Probe 2x mix (Qiagen, Valencia, CA, USA), 100nM probe, and 200nM primers. The standard cycling conditions were 95° for 3 min followed by 50 cycles at 95 ° for 3 s and 60 ° for 30 s. Samples were run in triplicate, except Supplementary Fig. 4, where they were run in duplicate.
Supplementary Material
Supplementary Fig. 1. Q-ChIP analysis of WRNp levels at the Oct4 promoter in WRNp-deficient cells. WRNp-deficient cells (clone 31) were treated with 10 µM RA for three days. Q-ChIP analysis was performed as described in Fig. 1.
Supplementary Fig. 2. Deficient DNA methylation of the Oct4 promoter in WRNp-deficient clones 31–34. Result adjusted according to input DNA detected in undigested samples. During differentiation of NCCIT cells, Oct4 promoter DNA is progressively methylated and thus becomes resistant to the methylation-sensitive enzymes HpaII and HhaI. Control, G9a-deficient and WRNp-deficient cells were stimulated with 10 µM RA for 7 days. Cells were then harvested, DNA extracted and digested with the methylation-sensitive enzymes HpaII or HhaI. The digested DNA was then subjected to real-time PCR with primers targeting segments of the Oct4 promoter that flank the restriction sites. The results are shown as a number of amplicons (DNA copies) obtained with DNA from cells that were not treated with RA (−) and DNA from cells that were stimulated with RA (+). (A) CpG methylation status in the Oct4 B segment of undifferentiated and RA treated WRNp-deficient, G9a-deficient and control cells. (B) CpG methylation status in the Oct4 D segment. (C) CpG methylation status in the Oct4 E segment. (D) CpG methylation status in the Oct4 E segment. (E) Undigested DNA, Oct4 D segment, used to normalize HpaII digestions. Error bars indicate standard deviation. The Oct4 E segment was quantified to normalize HhaI digestions, data not shown.
Supplementary Fig. 3. Deficient DNA methylation of the Oct4 promoter in WRNp-deficient clones 61 and 62. Result adjusted according to input DNA detected in undigested samples. Raw data are presented in Fig. 4 F-I. Cells were stimulated with 10 µM RA and processed as outlined in Fig. 4. Results are shown as a number of amplicons (DNA copies) obtained with DNA from cells that were not treated with RA (−) and DNA from cells that were stimulated to differentiate with RA (+). (A) CpG methylation status in the Oct4 B segment of undifferentiated and RA treated WRNp-deficient and control cells. (B) CpG methylation status in the Oct4 D segment. (C) CpG methylation status in the Oct4 E segment. (D) CpG methylation status in the Oct4 F segment. (E) Undigested DNA, Oct4 B segment. Error bars indicate standard deviation.
Supplementary Fig. 4. Deficient DNA methylation of the Oct4 promoter in a polyclonal population of NCCIT cells that were transduced with a lentiviral vector carrying WRN shRNA. (A) Lentivirus-delivered shRNA-mediated knockdown of WRNp in NCCIT cells. Cells that were infected with a lentiviral vector carrying WRN or control shRNA. Infected cells were selected in puromycin and the polyclonal population was subjected to western blotting analysis with a WRNp antibody. γ-tubulin served as a loading control. (B) Control and WRNp-deficient cells were stimulated with 10 µM RA and processed as outlined in Fig. 4. Real-time PCR assays were performed as described above. We tested Oct4 B and D segments, which showed the strongest differences between control and WRNp-deficient clones as shown in Fig. 4 and 5. Results are shown as a ratio of the number of amplicons obtained with DNA from cells that were stimulated with RA vs. DNA from cells that were not treated with RA. Results were adjusted according to the input DNA amount of undigested samples. Left – CpG methylation status in the Oct4 B segment of undifferentiated vs. RA treated control and WRNp-deficient cells. Right – CpG methylation status in the Oct4 D segment.
Supplementary Fig. 5. Oct4 promoter methylation in WRNp-deficient primary fibroblasts. Normal primary fibroblasts were treated with siRNA against WRN (Wi) or control siRNA (Ci) for 3 days. Alternatively, cells were stably transduced with lentiviral particles encoding WRN shRNA (see the bottom part of the Figure) and selected with puromycin, as described in Supplementary Fig. 4. DNA was then extracted, digested with HpaII and real-time PCR performed as described above. Results are plotted as a ratio of Oct4B amplicons in digested vs. undigested DNA. We note that the number of Oct4B amplicons was even higher in digested samples than undigested samples. This is probably due to a higher accessibility of DNA that was fragmented by enzymatic digestion to the PCR machinery, providing the digestion occurs outside of the PCR-amplified region.
Supplementary Fig. 6. Dnmt3b overexpression suppresses residual Oct4 expression in RA treated WRNp-deficient cells. (A) WRNp-deficient (clone 31) and control cells (CV) were treated with 10 µM RA. Three days after the addition of RA, cells were transfected with a Dnmt3b-encoding plasmid (DN) or a control empty plasmid (P). Seven days after addition of RA, cells were harvested and Oct4 expression was analyzed by western blotting (+RA – cells stimulated to differentiate with RA, −RA – unstimulated cells). (B) Dnmt3b overexpression reverses the Oct4 promoter methylation defect of WRNp-deficient cells. DNA was extracted from transfected and RA-treated WRNp-deficient cells [treated and transfected as described in (A)]. Oct4 promoter methylation in the B region was analyzed as described in Fig. 4.
Acknowledgements
This work has been supported by NIH grants CA125272, CA135214 and a W.W. Smith Foundation AIDS Research Award to R.D. We thank Dr. Chen’s laboratory (TJU, Center for Translational Medicine) for technical help with real-time PCR experiments.
Footnotes
Authors’ Contributions
J.A.S. developed WRNp-deficient and control clones, performed DNA methylation assays, real-time PCR assays, and siRNA knockdown experiments. A.M.N. performed Q-ChIP assays in Fig. 1, Fig. 3 and Fig. 6. K.G. performed the Q-Chip assay with H3K9me3 antibody (Fig. 3). O.I. provided support for analysis of stem cell differentiation and M.L. contributed to data analysis. R.D. wrote the manuscript and performed co-immunoprecipitation assays and western blotting analyses.
Competing interests statement: The authors declare that they have no competing financial interests.
Contributor Information
Johanna A. Smith, Email: Johanna.Smith@jefferson.edu.
Abibatou M. N. Ndoye, Email: abindoye@gmail.com.
Kyla Geary, Email: Kyla.Geary@jefferson.edu.
Michael P. Lisanti, Email: michael.lisanti@kimmelcancercenter.org.
Olga Igoucheva, Email: Olga.Igoucheva@jefferson.edu.
References
- Barrand S, Collas P. Chromatin states of core pluripotency-associated genes in pluripotent, multipotent and differentiated cells. Biochem Biophys Res Commun. 2010;391:762–767. doi: 10.1016/j.bbrc.2009.11.134. [DOI] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetti R, Gonzalo S, Jaco I, Munoz P, Gonzalez S, Schoeftner S, Murchison E, Andl T, Chen T, Klatt P, Li E, Serrano M, Millar S, Hannon G, Blasco MA. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol. 2008;15:998. doi: 10.1038/nsmb0908-998b. [DOI] [PubMed] [Google Scholar]
- Bohr VA. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem Sci. 2008;33:609–620. doi: 10.1016/j.tibs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiani M, Scholer HR. Regulatory networks in embryo-derived pluripotent stem cells. Nat Rev Mol Cell Biol. 2005;6:872–884. doi: 10.1038/nrm1744. [DOI] [PubMed] [Google Scholar]
- Bourc'his D, Bestor TH. Helicase homologues maintain cytosine methylation in plants and mammals. Bioessays. 2002;24:297–299. doi: 10.1002/bies.10078. [DOI] [PubMed] [Google Scholar]
- Chen L, Huang S, Lee L, Davalos A, Schiestl RH, Campisi J, Oshima J. WRN, the protein deficient in Werner syndrome, plays a critical structural role in optimizing DNA repair. Aging Cell. 2003;2:191–199. doi: 10.1046/j.1474-9728.2003.00052.x. [DOI] [PubMed] [Google Scholar]
- Cheng L, Sung MT, Cossu-Rocca P, Jones TD, MacLennan GT, De Jong J, Lopez-Beltran A, Montironi R, Looijenga LH. OCT4: biological functions and clinical applications as a marker of germ cell neoplasia. J Pathol. 2007;211:1–9. doi: 10.1002/path.2105. [DOI] [PubMed] [Google Scholar]
- D'Ippolito G, Diabira S, Howard GA, Menei P, Roos BA, Schiller PC. Marrow-isolated adult multilineage inducible (MIAMI) cells, a unique population of postnatal young and old human cells with extensive expansion and differentiation potential. J Cell Sci. 2004;117:2971–2981. doi: 10.1242/jcs.01103. [DOI] [PubMed] [Google Scholar]
- Dahl JA, Collas P. Q2ChIP, a quick and quantitative chromatin immunoprecipitation assay, unravels epigenetic dynamics of developmentally regulated genes in human carcinoma cells. Stem Cells. 2007;25:1037–1046. doi: 10.1634/stemcells.2006-0430. [DOI] [PubMed] [Google Scholar]
- Damjanov I, Horvat B, Gibas Z. Retinoic acid-induced differentiation of the developmentally pluripotent human germ cell tumor-derived cell line, NCCIT. Lab Invest. 1993;68:220–232. [PubMed] [Google Scholar]
- Du Z, Jia D, Liu S, Wang F, Li G, Zhang Y, Cao X, Ling EA, Hao A. Oct4 is expressed in human gliomas and promotes colony formation in glioma cells. Glia. 2009;57:724–733. doi: 10.1002/glia.20800. [DOI] [PubMed] [Google Scholar]
- Edelberg JM, Ballard VL. Stem cell review series: regulating highly potent stem cells in aging: environmental influences on plasticity. Aging Cell. 2008;7:599–604. doi: 10.1111/j.1474-9726.2008.00404.x. [DOI] [PubMed] [Google Scholar]
- Epsztejn-Litman S, Feldman N, Abu-Remaileh M, Shufaro Y, Gerson A, Ueda J, Deplus R, Fuks F, Shinkai Y, Cedar H, Bergman Y. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol. 2008;15:1176–1183. doi: 10.1038/nsmb.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum Mol Genet. 2007;16(Spec No 1):R50–R59. doi: 10.1093/hmg/ddm018. [DOI] [PubMed] [Google Scholar]
- Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, Cedar H, Bergman Y. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol. 2006;8:188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- Gray MD, Shen JC, Kamath-Loeb AS, Blank A, Sopher BL, Martin GM, Oshima J, Loeb LA. The Werner syndrome protein is a DNA helicase. Nat Genet. 1997;17:100–103. doi: 10.1038/ng0997-100. [DOI] [PubMed] [Google Scholar]
- Hickson ID. RecQ helicases: caretakers of the genome. Nat Rev Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Jahagirdar BN, Reinhardt RL, Schwartz RE, Keene CD, Ortiz-Gonzalez XR, Reyes M, Lenvik T, Lund T, Blackstad M, Du J, Aldrich S, Lisberg A, Low WC, Largaespada DA, Verfaillie CM. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature. 2002;418:41–49. doi: 10.1038/nature00870. [DOI] [PubMed] [Google Scholar]
- Kim JB, Zaehres H, Arauzo-Bravo MJ, Scholer HR. Generation of induced pluripotent stem cells from neural stem cells. Nat Protoc. 2009;4:1464–1470. doi: 10.1038/nprot.2009.173. [DOI] [PubMed] [Google Scholar]
- Kucia M, Reca R, Campbell FR, Zuba-Surma E, Majka M, Ratajczak J, Ratajczak MZ. A population of very small embryonic-like (VSEL) CXCR4(+)SSEA-1(+)Oct-4+ stem cells identified in adult bone marrow. Leukemia. 2006;20:857–869. doi: 10.1038/sj.leu.2404171. [DOI] [PubMed] [Google Scholar]
- Kyng KJ, May A, Kolvraa S, Bohr VA. Gene expression profiling in Werner syndrome closely resembles that of normal aging. Proc Natl Acad Sci U S A. 2003;100:12259–12264. doi: 10.1073/pnas.2130723100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai JS, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci U S A. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird PW, Jaenisch R. The role of DNA methylation in cancer genetic and epigenetics. Annu Rev Genet. 1996;30:441–464. doi: 10.1146/annurev.genet.30.1.441. [DOI] [PubMed] [Google Scholar]
- Lebel M, Leder P. A deletion within the murine Werner syndrome helicase induces sensitivity to inhibitors of topoisomerase and loss of cellular proliferative capacity. Proc Natl Acad Sci U S A. 1998;95:13097–13102. doi: 10.1073/pnas.95.22.13097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JY, Pu MT, Hirasawa R, Li BZ, Huang YN, Zeng R, Jing NH, Chen T, Li E, Sasaki H, Xu GL. Synergistic function of DNA methyltransferases Dnmt3a and Dnmt3b in the methylation of Oct4 and Nanog. Mol Cell Biol. 2007;27:8748–8759. doi: 10.1128/MCB.01380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhart HG, Lin H, Yamada Y, Moran E, Steine EJ, Gokhale S, Lo G, Cantu E, Ehrich M, He T, Meissner A, Jaenisch R. Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing. Genes Dev. 2007;21:3110–3122. doi: 10.1101/gad.1594007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Ng JH, Ng HH. Molecular framework underlying pluripotency. Cell Cycle. 2008;7:885–891. doi: 10.4161/cc.7.7.5636. [DOI] [PubMed] [Google Scholar]
- Lombard DB, Beard C, Johnson B, Marciniak RA, Dausman J, Bronson R, Buhlmann JE, Lipman R, Curry R, Sharpe A, Jaenisch R, Guarente L. Mutations in the WRN gene in mice accelerate mortality in a p53-null background. Mol Cell Biol. 2000;20:3286–3291. doi: 10.1128/mcb.20.9.3286-3291.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu TY, Lu RM, Liao MY, Yu J, Chung CH, Kao CF, Wu HC. Epithelial cell adhesion molecule regulation is associated with the maintenance of the undifferentiated phenotype of human embryonic stem cells. J Biol Chem. 2010 doi: 10.1074/jbc.M109.077081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDevitt TC, Palecek SP. Innovation in the culture and derivation of pluripotent human stem cells. Curr Opin Biotechnol. 2008;19:527–533. doi: 10.1016/j.copbio.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan SJ, Grinstead LA, Olivieri W, Parris DS. Interaction between the herpes simplex virus type 1 origin-binding and DNA polymerase accessory proteins. Virology. 1998;241:122–130. doi: 10.1006/viro.1997.8953. [DOI] [PubMed] [Google Scholar]
- Ng VY, Ang SN, Chan JX, Choo AB. Characterization of epithelial cell adhesion molecule as a surface marker on undifferentiated human embryonic stem cells. Stem Cells. 2010;28:29–35. doi: 10.1002/stem.221. [DOI] [PubMed] [Google Scholar]
- Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- Okita K, Yamanaka S. Intracellular signaling pathways regulating pluripotency of embryonic stem cells. Curr Stem Cell Res Ther. 2006;1:103–111. doi: 10.2174/157488806775269061. [DOI] [PubMed] [Google Scholar]
- Opresko PL. Telomere ResQue and preservation--roles for the Werner syndrome protein and other RecQ helicases. Mech Ageing Dev. 2008;129:79–90. doi: 10.1016/j.mad.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Opresko PL, Otterlei M, Graakjaer J, Bruheim P, Dawut L, Kolvraa S, May A, Seidman MM, Bohr VA. The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol Cell. 2004;14:763–774. doi: 10.1016/j.molcel.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Orren DK. Werner syndrome: molecular insights into the relationships between defective DNA metabolism, genomic instability, cancer and aging. Front Biosci. 2006;11:2657–2671. doi: 10.2741/1999. [DOI] [PubMed] [Google Scholar]
- Otterlei M, Bruheim P, Ahn B, Bussen W, Karmakar P, Baynton K, Bohr VA. Werner syndrome protein participates in a complex with RAD51, RAD54, RAD54B and ATR in response to ICL-induced replication arrest. J Cell Sci. 2006;119:5137–5146. doi: 10.1242/jcs.03291. [DOI] [PubMed] [Google Scholar]
- Pallante BA, Duignan I, Okin D, Chin A, Bressan MC, Mikawa T, Edelberg JM. Bone marrow Oct3/4+ cells differentiate into cardiac myocytes via age-dependent paracrine mechanisms. Circ Res. 2007;100:e1–e11. doi: 10.1161/01.RES.0000253487.02398.85. [DOI] [PubMed] [Google Scholar]
- Ratajczak MZ, Machalinski B, Wojakowski W, Ratajczak J, Kucia M. A hypothesis for an embryonic origin of pluripotent Oct-4(+) stem cells in adult bone marrow and other tissues. Leukemia. 2007;21:860–867. doi: 10.1038/sj.leu.2404630. [DOI] [PubMed] [Google Scholar]
- Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG, Zavolan M, Svoboda P, Filipowicz W. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol. 2008;15:259–267. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- Smith JA, Wang FX, Zhang H, Wu KJ, Williams KJ, Daniel R. Evidence that the Nijmegen breakage syndrome protein, an early sensor of double-strand DNA breaks (DSB), is involved in HIV-1 post-integration repair by recruiting the ataxia telangiectasia-mutated kinase in a process similar to, but distinct from, cellular DSB repair. Virol J. 2008;5:11. doi: 10.1186/1743-422X-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Li H, Yang H, Rao MS, Zhan M. Mechanisms controlling embryonic stem cell self-renewal and differentiation. Crit Rev Eukaryot Gene Expr. 2006;16:211–231. doi: 10.1615/critreveukargeneexpr.v16.i3.20. [DOI] [PubMed] [Google Scholar]
- Suva ML, Riggi N, Stehle JC, Baumer K, Tercier S, Joseph JM, Suva D, Clement V, Provero P, Cironi L, Osterheld MC, Guillou L, Stamenkovic I. Identification of cancer stem cells in Ewing's sarcoma. Cancer Res. 2009;69:1776–1781. doi: 10.1158/0008-5472.CAN-08-2242. [DOI] [PubMed] [Google Scholar]
- Tang Y, Kitisin K, Jogunoori W, Li C, Deng CX, Mueller SC, Ressom HW, Rashid A, He AR, Mendelson JS, Jessup JM, Shetty K, Zasloff M, Mishra B, Reddy EP, Johnson L, Mishra L. Progenitor/stem cells give rise to liver cancer due to aberrant TGF-beta and IL-6 signaling. Proc Natl Acad Sci U S A. 2008;105:2445–2450. doi: 10.1073/pnas.0705395105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taranger CK, Noer A, Sorensen AL, Hakelien AM, Boquest AC, Collas P. Induction of dedifferentiation, genomewide transcriptional programming, and epigenetic reprogramming by extracts of carcinoma and embryonic stem cells. Mol Biol Cell. 2005;16:5719–5735. doi: 10.1091/mbc.E05-06-0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turaga RV, Massip L, Chavez A, Johnson FB, Lebel M. Werner syndrome protein prevents DNA breaks upon chromatin structure alteration. Aging Cell. 2007;6:471–481. doi: 10.1111/j.1474-9726.2007.00301.x. [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Xi S, Geiman TM, Briones V, Tao YG, Xu H, Muegge K. Lsh Participates in DNA Methylation and Silencing of Stem Cell Genes. Stem Cells. 2009 doi: 10.1002/stem.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S, Wang Z, Okano M, Nogami M, Li Y, He WW, Okumura K, Li E. Cloning, expression and chromosome locations of the human DNMT3 gene family. Gene. 1999;236:87–95. doi: 10.1016/s0378-1119(99)00252-8. [DOI] [PubMed] [Google Scholar]
- Yeo S, Jeong S, Kim J, Han JS, Han YM, Kang YK. Characterization of DNA methylation change in stem cell marker genes during differentiation of human embryonic stem cells. Biochem Biophys Res Commun. 2007;359:536–542. doi: 10.1016/j.bbrc.2007.05.120. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. Q-ChIP analysis of WRNp levels at the Oct4 promoter in WRNp-deficient cells. WRNp-deficient cells (clone 31) were treated with 10 µM RA for three days. Q-ChIP analysis was performed as described in Fig. 1.
Supplementary Fig. 2. Deficient DNA methylation of the Oct4 promoter in WRNp-deficient clones 31–34. Result adjusted according to input DNA detected in undigested samples. During differentiation of NCCIT cells, Oct4 promoter DNA is progressively methylated and thus becomes resistant to the methylation-sensitive enzymes HpaII and HhaI. Control, G9a-deficient and WRNp-deficient cells were stimulated with 10 µM RA for 7 days. Cells were then harvested, DNA extracted and digested with the methylation-sensitive enzymes HpaII or HhaI. The digested DNA was then subjected to real-time PCR with primers targeting segments of the Oct4 promoter that flank the restriction sites. The results are shown as a number of amplicons (DNA copies) obtained with DNA from cells that were not treated with RA (−) and DNA from cells that were stimulated with RA (+). (A) CpG methylation status in the Oct4 B segment of undifferentiated and RA treated WRNp-deficient, G9a-deficient and control cells. (B) CpG methylation status in the Oct4 D segment. (C) CpG methylation status in the Oct4 E segment. (D) CpG methylation status in the Oct4 E segment. (E) Undigested DNA, Oct4 D segment, used to normalize HpaII digestions. Error bars indicate standard deviation. The Oct4 E segment was quantified to normalize HhaI digestions, data not shown.
Supplementary Fig. 3. Deficient DNA methylation of the Oct4 promoter in WRNp-deficient clones 61 and 62. Result adjusted according to input DNA detected in undigested samples. Raw data are presented in Fig. 4 F-I. Cells were stimulated with 10 µM RA and processed as outlined in Fig. 4. Results are shown as a number of amplicons (DNA copies) obtained with DNA from cells that were not treated with RA (−) and DNA from cells that were stimulated to differentiate with RA (+). (A) CpG methylation status in the Oct4 B segment of undifferentiated and RA treated WRNp-deficient and control cells. (B) CpG methylation status in the Oct4 D segment. (C) CpG methylation status in the Oct4 E segment. (D) CpG methylation status in the Oct4 F segment. (E) Undigested DNA, Oct4 B segment. Error bars indicate standard deviation.
Supplementary Fig. 4. Deficient DNA methylation of the Oct4 promoter in a polyclonal population of NCCIT cells that were transduced with a lentiviral vector carrying WRN shRNA. (A) Lentivirus-delivered shRNA-mediated knockdown of WRNp in NCCIT cells. Cells that were infected with a lentiviral vector carrying WRN or control shRNA. Infected cells were selected in puromycin and the polyclonal population was subjected to western blotting analysis with a WRNp antibody. γ-tubulin served as a loading control. (B) Control and WRNp-deficient cells were stimulated with 10 µM RA and processed as outlined in Fig. 4. Real-time PCR assays were performed as described above. We tested Oct4 B and D segments, which showed the strongest differences between control and WRNp-deficient clones as shown in Fig. 4 and 5. Results are shown as a ratio of the number of amplicons obtained with DNA from cells that were stimulated with RA vs. DNA from cells that were not treated with RA. Results were adjusted according to the input DNA amount of undigested samples. Left – CpG methylation status in the Oct4 B segment of undifferentiated vs. RA treated control and WRNp-deficient cells. Right – CpG methylation status in the Oct4 D segment.
Supplementary Fig. 5. Oct4 promoter methylation in WRNp-deficient primary fibroblasts. Normal primary fibroblasts were treated with siRNA against WRN (Wi) or control siRNA (Ci) for 3 days. Alternatively, cells were stably transduced with lentiviral particles encoding WRN shRNA (see the bottom part of the Figure) and selected with puromycin, as described in Supplementary Fig. 4. DNA was then extracted, digested with HpaII and real-time PCR performed as described above. Results are plotted as a ratio of Oct4B amplicons in digested vs. undigested DNA. We note that the number of Oct4B amplicons was even higher in digested samples than undigested samples. This is probably due to a higher accessibility of DNA that was fragmented by enzymatic digestion to the PCR machinery, providing the digestion occurs outside of the PCR-amplified region.
Supplementary Fig. 6. Dnmt3b overexpression suppresses residual Oct4 expression in RA treated WRNp-deficient cells. (A) WRNp-deficient (clone 31) and control cells (CV) were treated with 10 µM RA. Three days after the addition of RA, cells were transfected with a Dnmt3b-encoding plasmid (DN) or a control empty plasmid (P). Seven days after addition of RA, cells were harvested and Oct4 expression was analyzed by western blotting (+RA – cells stimulated to differentiate with RA, −RA – unstimulated cells). (B) Dnmt3b overexpression reverses the Oct4 promoter methylation defect of WRNp-deficient cells. DNA was extracted from transfected and RA-treated WRNp-deficient cells [treated and transfected as described in (A)]. Oct4 promoter methylation in the B region was analyzed as described in Fig. 4.