Abstract
Advances in molecular genetics of hypertrophic cardiomyopathy (HCM) have led to identification of mutations in 11 genes coding for sarcomeric proteins. In addition, mutations in gene coding for the γ subunit of AMP-activated protein kinase and triplet-repeat syndromes, as well as in mitochondrial DNA have been identified in patients with HCM. Mutations in genes coding for the β-myosin heavy chain, myosin binding protein-C, and cardiac troponin T account for approximately 2/3 of all HCM cases. Accordingly, HCM is considered a disease of contractile sarcomeric proteins. Genotype-phenotype correlation studies show mutations and the genetic background affect the phenotypic expression of HCM. The final phenotype is the result of interactions between the causal genes, genetic background (modifier genes), and probably the environmental factors. The molecular pathogenesis of HCM is not completely understood. The initial defects caused by the mutant proteins are diverse. However, despite their diversity, they converge into common final pathway of impaired cardiac myocyte function. The latter leads to an increased myocyte stress and subsequent activation of stress-responsive signaling kinases and trophic factors, which activate the transcriptional machinery inducing cardiac hypertrophy, interstitial fibrosis and myocyte disarray, the pathological characteristics of HCM. Studies in transgenic animal models show that cardiac hypertrophy, interstitial fibrosis, and myocyte disarray are potentially reversible. These findings raise the possibility of reversal of evolving phenotype or prevention of phenotypes in human patients with HCM. Elucidation of the molecular genetic basis and the pathogenesis of HCM could provide the opportunity for genetic based diagnosis, risk stratification, and implementation of preventive and therapeutic measures in those who have inherited the causal mutations for HCM.
Keywords: Cardiomyopathy, hypertrophic, genetics - Genes - Mutation - Death, sudden, cardiac
Hypertrophic cardiomyopathy (HCM) is a primary disease of the myocardium caused by mutations in contractile sarcomeric proteins. It is clinically diagnosed by the presence of left ventricular hypertrophy (LVH) in the absence of an increased external load (unexplained LVH). Myocyte hypertrophy, disarray, and interstitial fibrosis are the commonly pathological features and myocyte disarray is considered the pathological hallmark.1, 2 LV cavity is small and and LV global systolic function, as indicated by the ejection fraction, is preserved, but diastolic function is impaired. The latter commonly leads to an increased left ventricular end diastolic pressure and symptoms of heart failure.
LVH is often asymmetric and interventricular septum is the predominant site of involvement. In apical HCM, which is more common in Japan, LVH is restricted to apex. Extent of LVH is variable and is, in part, determined by the underlying mutations, genetic background, age, gender, and possibly the environmental factors.3 LVH frequently accelerates during adolescence and puberty.4, 5 LVH, interstitial fibrosis, and myocyte disarray are the major determinants of mortality and morbidity in HCM.6–9 Myocyte hypertrophy and disarray are more prominent in the interventricular septum, but scattered myocyte disarray is often present throughout the myocardium.2 Myocyte disarray comprises <5% of the myocardium in the normal hearts and >20 to 30% of the myocardium in HCM.
The true prevalence of HCM is unknown. Its prevalence, detected by echocardiography, in 25–35 years old individuals is ~ 1:500.10 It is expected to be higher in older subjects because the penetrance is age-dependent and many affected individuals do not exhibit the phenotype until later in life.4, 11
The clinical manifestations of HCM are also variable. The majority of patients with HCM are asymptomatic or mildly symptomatic. The main symptoms are dyspnea, chest pain, palpitations, and infrequently syncope. Cardiac arrhythmias, in particular atrial fibrillation and non-sustained ventricular tachycardia are relatively common and Wolff-Parkinson-White syndrome is present in approximately 5% of patients. Symptoms of heart failure are predominantly due to left ventricular diastolic dysfunction. Sudden cardiac death (SCD) is often the first manifestation of HCM in the young. Indeed, HCM is the most common cause of SCD in the competitive athletes.12
Physical findings are reflective of LVH and LV outflow tract obstruction. Physical examination may be completely normal or detect subtle abnormalities in subjects without significant LVH or outflow tract obstruction. The typical arterial pulse has two components of percussion and tidal waves. The jugular vein may show a prominent a wave reflective of poor right ventricular compliance. Apical impulse is strong and commonly bifid. A harsh, crescendo-decrescendo mid-systolic murmur in left sternal border is the most common and often the only finding on physical examination. The murmur rarely radiates to carotid arteries. Maneuvers that diminish left ventricular volume such as inhalation of amyl nitrate, or increases left ventricular contractility, such as infusion of inotropic agents, accentuate the murmur and those that increase ventricular volume, such as squatting diminish with intensity of the murmur. A loud S4 is commonly present.
HCM, the most common cause of SCD in the young athletes,12 is a relatively benign disease in adult population. The estimated annual mortality rate of HCM is <0.7%.13 Several potential risk factors for SCD have been identified and those with certain causal mutations, modifier genes, early age of onset, history of syncope, family history of SCD, exercise-induced hypotension, or malignant arrhythmias are considered at high risk for SCD (Table I).
Table I.
Potential risk factors for sudden cardiac death in hypertrophic cardiomyopathy.
| History of SCD |
| Family history of premature death |
| Causal mutations |
| Modifier genes |
| History of syncope |
| Magnitude of left ventricular hypertrophy |
| Extent of myocyte disarray |
| Extent of interstitial fibrosis |
| Early onset of the disease |
| Myocardial ischemia on perfusion tomography |
| Abnormal blood pressure response to exercise |
| Presence of non-sustained VT on Holter |
Genetic basis of HCM
Clinical genetics
HCM is a genetic disease with an autosomal dominant mode of inheritance with the exception of those caused by mutations in the mitochondrial genome, which show a matrilinear transmission. Approximately 2/3 of patients have familial HCM and the remainder is sporadic, caused by de novo mutations. Sporadic cases transmit the mutation and thus the disease to their offspring. No founder effect is uncommon and the majority of mutations occur independently.14–16
Molecular genetics of HCM
Dr. Seidman and her group mapped the first locus to chromosome 14q1 and identified the R403Q missense mutation in the β-MyHC as the first causal mutation for HCM.17, 18 Subsequently, mutations in cardiac troponin T (cTnT) and a-tropomyosin and other components of thin and thick filaments of sarcomere were identified, leading to the notion that HCM is a disease of contractile sarcomeric proteins.19 Thus far, more than 100 different mutations in 11 genes encoding contractile sarcomeric proteins have bben identified. The list of the causal genes for HCM, shown in Table II, includes those encoding b-MyHC, cTnT, myosin binding protein-C (MyBP-C), cardiac troponin I (cTnI), cardiac troponin C (cTnC), titin, cardiac α-actin, and essential and regulatory light chains (ELC1 and ECL2). Mutations in non-sarcomeric genes are also known to cause a phenotype similar to HCM (Table III). Mutations in a gene coding for the AMP-activated γ2 non-catalytic subunit of protein kinase A, a regulator of cell bioenergetics, have been identified in families with Wolff-Parkinson-White syndrome and HCM.20,21 Furthermore, mutations in mitochondrial genome have been associated with HCM and multi-organ disorders.22 HCM is also often observed in patients with the triplet repeat syndromes.23 Collectively, these data suggest that HCM, defined as hypertrophy in the absence of an increased external load, occurs primarily as a result of mutations in genes encoding the contractile sarcomeric proteins. However, HCM, often in conjunction with other cardiac and non-cardiac phenotypes, also occurs because of mutations in non-sarcomeric proteins, such as mitochondrial DNA.
Table II.
Causal genes for HCM: genes coding for sarcomeric proteins.
| Gene | Symbol | Locus | Frequency | Mutations |
|---|---|---|---|---|
| β-myosin heavy chain | MYH7 | 14q12 | ~35% | >70, predominantly missense mutations |
| Myosin binding protein-C | MYBPC3 | 11p11.2 | ~20% | >40, Predominantly splice junction and insertion/deletion mutations |
| Cardiac troponin T | TNNT2 | 1q32 | ~20% | > 15, Mostly missense |
| α-tropomyosin | TPM1 | 15q22.1 | ~5% | > 5 missense mutations |
| Cardiac troponin I | TNNI3 | 19p13.2 | ~5% | 3 missense and 1 deletion mutations |
| Essential myosin light chain | MYL3 | 3p21.3 | <5% | 2 missense mutations |
| Regulatory myosin light chain | MYL2 | 12q23-24.3 | <5% | 7 missense and 1 truncation mutations |
| Cardiac α-actin | ACTC | 15q11 | <5% | 2 missense mutations |
| Titin | TTN | 2q24.1 | <5% | 1 missense mutation |
| α-myosin heavy chain | MYH6 | 14q1 | Rare | 1 missense and 1 rearrangement mutations |
| Cardiac troponin C | TNNC1 | 3p21.3-3p14.3 | Rare | 1 missense mutations in a patient with HCM |
Table III.
Causal genes for HCM: genes coding for non-sarcomeric proteins.
| Gene | Symbol | Locus | Frequency | Mutations |
|---|---|---|---|---|
| AMP-activated protein kinase, γ2 regulatory subunit | PRKAG2 | 7q35-q36 | <5% | 3 point and 1 insertion mutations |
| Mitochondrial DNA | MTTI | Mitochondrial | Rare | tRNA Isoleucine and tRNA glycine |
| Frataxin (Friedreich’ ataxia) | FRDA | 9q13 | >200 GAA in intron 1 | |
| Myotonin protein kinase (Myotonic dystrophy) | DMPK | 19q13 | Uncommon | >50 CTG in 3′-UTR |
Mutations in sarcomeric proteins
MYH7, MYBPC3, and TNNT2 are the three most common causal genes for HCM and collectively account for approximately three-fourths of all HCM cases. The β-MyHC gene (MYH7) is the most common gene responsible for HCM, accounting for approximately 35–50% of all HCM cases.24 MYH7 is comprised of 40 exons and codes for a 6 kb mRNA and a 220 kD protein.25 Over 60 different mutations in the β-MyHC have been identified and the majority are missense mutations. Codons 403 and 719 are considered hot spots for mutations.26, 27 The majority of the mutations are located in the globular head of the myosin molecule.22 Missense and deletion mutations28–30 and an insertion/deletion mutation changing amino acids 395-40431 in the rod and tail regions also have been described. The frequency of each particular MYH7 mutation is relatively low.
The second most common causal gene for human HCM is the MYBPC3 gene on chromosome.11, 32 Mutations in MYBPC3 account for an approximately 20% to 25% of all HCM cases.3, 4, 33 The MYBPC3 gene has a complex structure comprised of 35 exons spanning approximately 23 kb of DNA.32 MyBP-C has a cardiac specific motif comprised of 9 amino acids translated from exon.8 Over 40 different mutations in the MYBPC3 gene have been identified and the frequency of each mutation is low.4, 32–35 Unlike mutations in the MYH7 gene, which are mostly missense mutations, the majority of mutations in the MYBPC3 are deletion/insertion or splice junction mutations.33 Deletion/insertion mutations are expected to result in frame-shift or truncation of the MyBP-C proteins either leading to severe structural and functional defects in the protein or immediate degradation of the expressed protein.
More than 20 mutations in cardiac troponin T gene (TNNT2), located on chromosome 1q3, have been identified. Mutations in TNNT2, probably the third most common causal gene for HCM, account for approximately 20% of all HCM cases.36 The majority of the mutations are missense mutations and codon 92 is considered a hot spot for mutations.19, 37 Deletion mutations that involve the splice donor sites and lead to truncated proteins 19 also have been reported.
Mutations in two other components of the troponin-tropomyosin complex, namely troponin I,38, 39 α-tropomyosin,16, 19, 40, 41 troponin C,42 have been identified in patients with HCM. Other relatively uncommon causal genes for HCM are ACTC, encoding cardiac α-actin;43, 44 TTN, encoding titin;45 MYL3, encoding MLC1,46 and MYL2, coding for MLC2.46, 47 Overall, mutations in the above components of thin and thick filaments are uncommon causes of HCM in humans.
HCM as a consequence of mutations in non-sarcomeric proteins
Cardiac hypertrophy is the common response of the heart to all forms of stress. Therefore, it is not surprisingly that HCM phenotype, i.e. hypertrophy in the absence of an increased external load, is also observed in a variety of conditions including triplet repeat syndromes, mitochondrial disorders, and metabolic disorders (Table III). Recently, mutations in PRKAG2 gene, encoding the γ2 regulatory subunit of AMP-activated protein kinase (AMPK), in families with HCM and Wolff-Parkinson-White syndrome were identified.20, 21 The described phenotype varies from that of pre-excitation and conduction defects and minimal hypertrophy to that of severe and early onset hypertrophy with minority of patients showing pre-excitation.
HCM also occurs in myotonic muscular dystrophy and Friedreich’ ataxia, two triplet repeat syndromes due to expansion of trinucleotide repeat sequences in their respective genes (Table III). Myotonic muscular dystrophy, is an autosomal dominant disorder due to expansion of GCT trinucleotide repeats in in the 3′ untranslated region of dystrophia myotonica protein kinase (DMPK) gene from less than 40 in normal individuals to more than several hundreds and thousands.48 Expansion of the repeats results in unstable DNA and decreased levels of mRNA and protein. It is the most common form of muscular dystrophy in adults and commonly manifests as progressive degeneration of muscles and myotonia, cardiomyopathy, male pattern baldness, infertility, premature cataracts, mental and endocrine abnormalities.48 Cardiac involvement is common and often includes conduction defects, dilated cardiomyopathy and less often HCM.49 The severity of the disease correlates with the number of the GCT repeats. Friedreich’ ataxia is an autosomal recessive neuro-degenerative disease also caused by expansion of trinucleotide repeat sequences.50 The mutation is expansion of the GAA trinucleotides located within intron 1 of FRDA, which codes for frataxin, a soluble mitochondrial protein with 210 amino acids.50 Friedreich’ ataxia involves both central and peripheral nervous system and cardiac involvement includes dilated and hypertrophic cardiomyopathy. The severity of clinical manifestations of Friedreich’ ataxia also correlates with the size of the repeats.51
Mutations in mitochondrial DNA also have been found in patients with HCM.52 Mutations in mitochondrial DNA often give rise to a complex phenotype involving multiple organs including the heart.52 Mitochondrial DNA is a circular double-stranded genome of approximately 16.5 kilobases, which codes for 13 polypeptides of the respiratory chain complexes I, III, IV, and V subunits, 28 ribosomal RNAs, and 22 tRNAs. Each mitochondrion has multiple copies of its own DNA and each cells contains thousands of mitochondrial DNA. Therefore, mutations result in a significant degree of heteroplasmy (combination of wild type and mutant mitochondrial DNA), which makes establishing the causality of mitochondrial DNA mutations in HCM difficult. It is estimated >80 to 90% of mitochondrial DNA need to mutate before it causes a significant clinical phenotype.53 An example of a disease caused by mitochondrial DNA mutation is the Kearns-Sayre syndrome (KSS), which is characterized by a triad of progressive external ophthalmoplegia, pigmentary retinopathy, and cardiac conduction defects.54 Almost all patients with KSS exhibit sporadically occurring mutations in mitochondrial DNA.54 While the classic cardiac abnormality in KSS is conduction defects, dilated and hypertrophic cardiomyopathies are also often observed but a lower frequency.
Thus, HCM, a genetic model of cardiac hypertrophy, is caused by a diverse array of mutations in a variety of genes, with the pure form (no other cardiac or non-cardiac phenotype) resulting from mutations in contractile sarcomeric proteins.
Genotype-phenotype correlation studies
Elucidation of the molecular genetic basis of HCM has led to a significant enthusiasm in identifying the genetic determinants of cardiac phenotypes in HCM, in particular the role of mutations in MYH7, TNNT2, and MYBPC3 in predicting the risk of SCD in HCM.28, 33, 55–59 Unfortunately, the results of genotype-phenotype correlation studies are subjects to a large number of confounding factors. Potential confounders are the small size of the families, small number of families with identical mutations due to low frequency of each mutation, variability in the phenotypic expression in affected individuals within the same family or amongst families with identical mutations, influence of modifier genes, influence of non-genetic factors, and rarely homozygosity for causal mutations and compound mutations.60, 61 Collective data indicate that mutations exhibit highly variable clinical, electrocardiographic, and echocardiographic manifestations and no particular phenotype is mutation-specific.3 Keeping the limitations of the existing genotype-phenotype correlation studies, the data suggest mutations affect the phenotypic expression of HCM, in particular the magnitude of cardiac hypertrophy and the risk of SCD. In general, mutations in the β-MyHC are associated with an early onset of disease, more extensive hypertrophy, and a higher incidence of SCD than others.4, 62 Inversely, mutations in the MyBP-C are associated with a relatively mild hypertrophy and late onset of clinical manifestations.4, 33, 62 Mutations in cTnT are usually associated with a mild degree of LVH but a high incidence of SCD and more extensive disarray.9, 58
Mutations in MYBPC3 gene are often associated with a low penetrance, mild hypertrophy, and a low incidence of SCD.4 The phenotype often develops late, which may coincide with the concomitant presence of hypertension. It has been suggested that hypertensive hypertrophic cardiomyopathy of the elderly may be a form of HCM caused by mutations in the MyBP-C and the concomitant hypertension.4 Despite the overall benign nature of mutations in the MyBP-C, significant variability in the phenotypic expression of HCM also exists and the so-called “malignant” mutations also have been reported in the MYBPC3 genes.33
Mutations in α-tropomyosin are generally associated with a benign phenotype and mild left ventricular hypertrophy. Despite mild degree of LVH, a high incidence of SCD also has been described.41 Mutations in essential and regulatory myosin light chains have been associated with mid-cavity obstruction in HCM and skeletal myopathy in some 46 but not in others.47 Mutations in titin45 and α-actin43, 44 are uncommon and have been observed in a small number of families. With regard to HCM caused by mutations in PRKAG2, the phenotype is variable. In some families the predominant phenotype is pre-excitation and conduction abnormalities20 and cardiac hypertrophy is present in the minority of the patients.20 In others, early cardiac hypertrophy predominates and pre-excitation is present in a fraction of the cases.21
Mutations, regardless of the causal gene or mutation, exhibit an age-dependent penetrance. Therefore, a normal physical examination and clinical testing at an early age do not effectively exclude the possibility of developing HCM later in life.11 This is particularly the case for HCM caused by mutations in MyBP-C, since the phenotype often develops in the fifth or sixth decades of life.11 In general, mutations associated with milder hypertrophy, late onset of HCM, and a low penetrance are associated with a relatively benign prognosis.63 In contrast, those associated with more extensive hypertrophy, high penetrance, and an early age of onset of HCM carry a higher risk of SCD.
Modifier genes
The phenotype of single gene disorders, particularly autosomal dominant disorders, is also affected by genetic factors other than the causal mutation. Genetic background, often referred to as the modifier genes is known to affect phenotypic expression of single-gene disorders, such as HCM. It should be noted that the modifier genes do not cause the disease but simply affect the severity of its phenotypic expression. Despite the significance of modifier genes in HCM, they remain unknown but several candidates have emerged.64–66 Association studies suggest that functional variants of angiotensin-1 converting enzyme-1 (ACE-1) gene, which are associated with an increased risk of SCD64 and the magnitude of left ventricular hypertrophy,65 endothelin-1, and tumor necrosis factor-α,67, 68 are potential modifier genes for HCM. The results of association studies should be considered preliminary and large-scale studies are needed to identify the potential modifier genes.
Functional studies of mutant sarcomeric proteins
Elucidation of the molecular genetic basis of HCM has led to a variety of in vitro and in vivo structure-function studies that collectively provide significant insight into the pathogenesis of HCM. The results are summarized here.
Effects of mutations on sarcomere and myofibril formation
Mutant sarcomeric proteins, when expressed in vitro, incorporate into myofilaments and sarcomeres. However, when expressed at high levels, they also could induce sarcomere dysgenesis69 and myofibrillar disarray.70 The efficiency of incorporation of mutant protein into sarcomere also is reduced for mutations such as a truncated MyBP-C protein in transgenic mice.70 Overall, the majority of the mutant sarcomeric proteins assemble into myofilaments and sarcomere and do not cause immediate sarcomere dysgenesis or myofibrillar degeneration.71, 72
Effects of mutations on interaction of actin and myosin
Mutations impact the interaction of actin filaments with myosin and the net outcome varies for different mutations and experimental conditions, such as Ca+2 concentration.73–80 The majority of the studies show mutant MyHC proteins reduce the ability of the myosin molecules to dislocate the thin actin filaments. Reduced velocity of actin dislocation is partly because of the reduced affinity of the mutant myosin for the actin filaments as evidenced by an increased dissociation rate. Similarly, the cross-bridging kinetics between the thin and thick filaments and the excitation-contraction coupling of myocytes carrying the mutant MyHC are also impaired.81, 82 The degree of impairment of acto-myosin interaction is associated with the prognostic significance of the causal mutations. Those associated with a poor prognosis exert a more pronounced effect than those associated with a benign prognosis.73–75
Mutant cTnT and α-tropomyosin also impair acto-myosin interactions and the results vary for different mutations and experimental conditions. CTnT mutations could increase the rate of actin displacement, without affecting the affinity of the troponin complex for tropomyosin, or that of troponin/tropomyosin complex for actin, or the affinity of the thin filaments for myosin.78, 80 Mutations in α-tropomyosin show a Ca+2-dependent effect on sliding velocity of actin filaments by myosin77 and increase the displacement rate under activating conditions (pCa5).
Effects of mutations on myosin ATPase activity
Mutations in the β-MyHC could affect the ATP binding site in the globular head of β-MyHC and thus, the rate of ATP hydrolysis.76, 83, 84 Reduced affinity of the mutant myosin for thin actin filaments could reduce actin-activated ATPase activity, but not the intrinsic ATPase activity of the MyHC molecule.76 In contrast to the previous findings, a recent study showed single myosin molecules isolated from the heart of α-MyHC-403 homozygous mice had a 2.3 fold increase in the actin-activated ATPase activity, while the unitary forces and displacement were unchanged.84
Effects of mutations on Ca+2 sensitivity of myofibrils and myocytes
Ca+2 modulates the cyclic interaction of the Myosin and actin. Ca+2 also activates a variety of Ca+2-sensitive signaling molecules. A variety of studies have explored the effects of mutations on Ca+2 sensitivity of the acto-myosin interaction 85–91. Overall, the effects of mutations on Ca+2 sensitivity of the myofibrils and myocytes vary for different mutations and according to the experimental conditions. The effect is mutation-specific and the majority enhance Ca+2 sensitivity of contractile apparatus in muscle cells.61, 86, 91, 92
Effects of mutations on contraction of cardiac and non-cardiac myocytes
Muscle fibers isolated from the slow skeletal muscles, expressing mutant β-MyHC, show reduced mechanical performance, which correlates with the severity of the cardiac phenotype.93, 94 Similarly, cardiac myocytes isolated from α-MyHC-403 heterozygote mice exhibit impaired contraction and relaxation.95 In contrast, studies of single myosin molecules isolated from mice homozygous for the α-MyHC-mutation show a 1.6-fold faster actin filament sliding in in vitro motility assay and a 2.2-fold greater average force generation.84 The dichotomy may reflect the potential limitations of single myosin molecules not duplicating the in vivo heterozygous state in HCM.
Expression of mutant cTnT and α-tropomyosin proteins in cardiac myocytes72, 96, 97 and in myotubes98, 99 led to impairment of mechanical performance. The net effect varies according to the loading conditions and Ca+2 concentration. Collectively, these data suggest power output of cardiac myocytes expressing the mutant sarcomeric proteins is reduced.
Genetically engineered animal models of HCM
To elucidate the pathogenesis of human HCM, several transgenic and knock out/knock-in mouse models, a rat model, and a rabbit model have been generated (Table V). The genetically engineered animal models provide the opportunity to delineate the pathogenesis of HCM and explore new therapeutic targets for treatment and reversal of cardiac phenotypes in HCM. The α-MyHC-Q403+/− and MyBPC−/+ mouse models mimic the genotype of human HCM, while transgenic models offer the opportunity for dose-titration studies (effects of various levels of expression of the mutant proteins on cardiac structure and function). The α-MyHC-Q403+/− and MyBPC−/+, cTnT-Q-92,100, 101 α-tropomyosin,102 and truncated MyBP-C70 mouse models exhibit a variety of cardiac phenotypes, including myocyte disarray, interstitial fibrosis, and systolic and diastolic dysfunction. However, the existing mouse models do not show significant cardiac hypertrophy, the phenotypic hallmark of HCM in humans, or show only mild hypertrophy that develops late and varies according to the genetic background. In contrast, cardiac hypertrophy develops in mice homozygous for the mutant sarcomeric proteins,103, 104 a transgenic rat model expressing a truncated cTnT and only after exercise,105 and in a transgenic rabbit model expressing mutant β-MyHC-Q403.106 Premature death is also relatively uncommon in the transgenic animal models. Global systolic function in adult life is often impaired in transgenic mouse models, but preserved or increased function in the very young mice (5-week-old).103, 107 Transgenic rabbits expressing β-MyHC-Q403 show reduced tissue Doppler systolic and diastolic velocities, cardiac hypertrophy, disarray, interstitial fibrosis.106 A variety of cellular and biochemical abnormalities, such as reduced crossbridge kinetics 81, altered calcium sensitivity of myocytes and myofibrils,70, 108, 109 reduced myocyte contractility,95 myocyte atrophy,108 altered energetics,110 impaired excitation-contraction coupling82 and electrophysiological abnormalities111, 112 in transgenic mouse models and reduced tissue Doppler velocities in a transgenic rabbit model113 also have been described. Altogether, data in transgenic animal models point to the diversity of the molecular and cellular mechanisms involved in the pathogenesis of final phenotypes of HCM, which are summarized in Table V.
Table V.
Genetically engineered animal models of HCM
| Phenotype | |
|---|---|
| Knock out/in models | |
| α–MyHC-Q403 mice | Myocyte disarray, interstitial fibrosis, hypertrophy mild and late, enlarged left atrium, premature death, neonatal dilated cardiomyopathy in homozygous mice 131, systolic and diastolic dysfunction,95, 115 increased contractile performance in very early life,107 heterogeneous ventricular conduction, inducible ventricular tachycardia 111, 112, 132, reduced crossbridge kinetics, increased force generation of single myosin molecules and [Ca+2]I sensitivity,81, 82, 84, 116 reduced [PCr], and increased [Pi] 110, increased actin-activated ATPase activity. |
| α–MyHC knock out mice | Embryonic lethality in −/−, +/− show fibrosis, sarcomere disarray, impaired contractility and relaxation120 |
| α–Tropomyosin knock-out | Embryonic lethality in homozygotes, no phenotype in heteozygotes, normal cardiac function121, 122 |
| Transgenic mice | |
| α–MyHC-Q403/ΔAA468-527 | Myocyte disarray, interstitial fibrosis, ventricular hypertrophy in female and dilatation in male mice, Increased ANF133, 134 |
| α–MyHC-Δ LCBD | Myocyte disarray, hypertrophy (only in homozygote), valvular thickening, decreased Ca+2 sensitivity and diastolic dysfunction103 |
| cTnT-ΔC-terminus | Myocyte disarray, interstitial fibrosis, myocyte atrophy and drop out, cardiac atrophy, premature death, systolic and diastolic dysfunction108 |
| cTnT-Q92 | Myocyte disarray, interstitial fibrosis, myocyte drop out, cardiac atrophy, systolic and diastolic dysfunction100, 101, 108 |
| cTnT-N179 | Normal, normal survival, no hypertrophy, increased Ca+2 sensitivity of ATPase activity and force generation, increased rate of contraction and relaxation, lower maximum force/cross section area and ATPase135 |
| MyBP-C-ΔC-terminus | Myocyte disarray, sarcomere dysgenesis, interstitial fibrosis, no cardiac hypertrophy109 |
| Truncated MyBP-C | Neonatal dilated cardiomyopathy in homozygous mice expressing <10% of the truncated protein, disarray, minimal or mild hypertrophy, decreased contractility and diastolic dysfunction136 |
| ELC-V149 (human gene) | Papillary muscle hypertrophy, altered stretch-activation response104 |
| ELC-V149 (mouse cDNA) | Normal, no hypertrophy, increased Ca+2 sensitivity and impaired relaxation137 |
| α–Tropomyosin-N175 | Myocyte disarray and hypertrophy, impaired contractility and relaxation, Increased Ca+2 sensitivity and decreased relaxation102, 135 |
| cTnI-G146 | Myocyte disarray, interstitial fibrosis, premature death. Increased Ca+2 sensitivity, hypercontractility, and diastolic dysfunction138 |
| Transgenic rat | |
| cTnT-ΔExon 16 | Normal, no cardiac hypertrophy, systolic and diastolic dysfunction. After 6 months of exercise hypertrophy, myofibrillar disarray105 |
| Transgenic rabbit | |
| β-MyHC-Q403 | Cardiac hypertrophy, myocyte disarray, interstitial fibrosis, increased mortality and SCD, systolic and diastolic dysfunction, preserved global systolic function, reduced myocardial contraction and relaxation velocities106, 118 |
[Ca+2]I: Intracellular Ca+2 concentration; [PCr]: phosphocreatinine; ↑[Pi]: Inorganic phosphate; −/−: null (homozygous for the deletion); +/−: heterozygous, LCBD: Light chain binding domain, MyHC: myosin heavy chain; cTnT: Cardiac troponin T: cTnI: Cardiac troponin I; MyBP-C: Myosin binding protein C, ELC: Essential light chain.
Limitations and utility of genetically engineered animal models
A concern with the mouse models of human HCM, is the presence of major differences in the composition of sarcomeric proteins between humans and mice, which could limit the utility of the transgenic mouse models in deciphering the pathogenesis of human HCM. In contrast to mouse hearts, in which α-MyHC predominates, the β-MyHC is the predominant MyHC isoform in human and rabbit hearts.114 It comprises >90% of the total myosin pool in the human heart.114 Therefore, the concern regarding the utility of mouse model to decipher the pathogenesis of HCM is particularly relevant to HCM caused by mutations in the β-MyHC, since β-MyHC is expressed only at a very low level in the adult mouse heart and α-MyHC predominates.114 The presence of major differences in the rate of Mg-ATPase activity and acto-myosin kinetics between α (fast) and the β (slow) MyHC isoforms could affect the phenotypic response of the heart to mutant sarcomeric proteins. The lack of significant or mild cardiac hypertrophy, the clinical hallmark of HCM, in transgenic mouse models100, 101, 109, 115 and in some cases the presence of cardiac and myocyte atrophy,101 further underscores this concern. Differences in calcium handling of myofibrils, implicated in the pathogenesis of HCM,116 between rabbits and mice also favor the use of transgenic rabbits to elucidate the pathogenesis of human HCM.117
The β-MyHC-Q403 transgenic rabbit model for human HCM fully recapitulates the phenotype of human HCM and exhibit cardiac hypertrophy (50% increase in wall thickness), interstitial fibrosis (2–3-fold increase), myocyte disarray (10–20% of the myocardium), premature death particularly during the first 3 months of life, and diastolic dysfunction.106 Myocardial contraction and relaxation velocities are reduced in all mutant transgenic rabbits, including those without detectable hypertrophy.118 Thus, β-MyHC-Q403 transgenic rabbits fully recapitulate the phenotype of HCM in human patients caused by the β-MyHC-Q403 mutation.
Pathogenesis of HCM
The results of in vitro and in vivo studies suggest that mutations cause a diverse array of initial defects in the structure and function of sarcomeric proteins. Type of the mutations (missense, frame shift, deletion, etc.), their topography, and the function of the affected protein account for the diversity of the initial defects. However, despite the diversity of the initial defects, the final phenotype is cardiac hypertrophy, fibrosis, and disarray. The intermediary pathways that connect the initial defects to the final phenotype of hypertrophy, disarray, and fibrosis at the present time are largely unknown. Overall, it appears that the initial phenotypes are mostly functional, followed by the molecular phenotype, which are likely the intermediary phenotypes, and subsequent structural phenotypes.
The majority of mutation in HCM are missense mutations and do not appear to interfere with initial assembly and the proper alignment of myofilaments and sarcomeres. Therefore, proteins with missense mutations are likely to function as “poison-peptides” exerting a dominant-negative effect on myocyte function following incorporation into myofibrils.17 Deletion or truncation mutations that abolish the stop codon or the polyadenylation site or encode truncated proteins that are likely to degrade immediately after translation,30, 119 could alter stoichiometry of the sarcomeric proteins. Such mutation could function as a “null-allele” and exert an effect through “haplo-insufficiency”. Gene targeting experiment in mice suggest that “haplo-insufficiency” may be gene-specific, since ablation of one copy of the murine α-MyHC gene120 led to alteration in sarcomeric structure and myocardial dysfunction, in contrast, ablation of one copy of α-tropomyosin did not induce a detectable morphological or functional abnormalities.121, 122
Functional defects precede cardiac hypertrophy
Experimental and clinical data suggest cardiac hypertrophy, the clinical hallmark of HCM, is a compensatory phenotype. LVH often occurs late and is absent in a significant number of patients who have inherited the causal mutation for HCM. Structure-function studies in adult cardiac myocytes and in intact hearts suggest that functional abnormalities precede cardiac hypertrophy in human subjects with HCM.96–98, 113 A recent study showed that tissue Doppler velocities of myocardial contraction and relaxation were reduced in human subjects with the mutations even in the absence of detectable cardiac hypertrophy.123 Similarly, myocardial contraction and relaxation velocities are reduced in the β-MyHC-Q403 transgenic rabbits prior to development of cardiac hypertrophy or interstitial fibrosis.113 Furthermore, gene-transfer studies in adult cardiac myocytes show that their function is impaired prior to the development of discernible sarcomere or myofibrillar disarray.96, 97 Moreover, skeletal myotubes and muscle fibers, isolated from the skeletal muscles of patients with HCM, show reduced force generation in the absence of structural abnormality.93, 94 Finally, myocytes isolated from the hearts of transgenic mice expressing a mutant α-MyHC protein95 show impaired mechanical performance. Collectively these results suggest the functional impairment precedes the structural changes in HCM.
Evidence for compensatory nature of LVH
Cardiac hypertrophy, the ubiquitous response of the myocardium to all forms of stress, commonly develops late in humans with HCM. Several lines of evidence suggest that LVH is a compensatory process due to yet to be defined impetus provided by the mutant sarcomeric proteins (Table VI). As discussed earlier, the primary defect induced by the mutant proteins are diverse and the link between the primary defects and subsequent evolution of LVH remain unknown.
Table VI.
Evidence in support of compensatory hypertrophy in HCM.
|
Current hypothesis
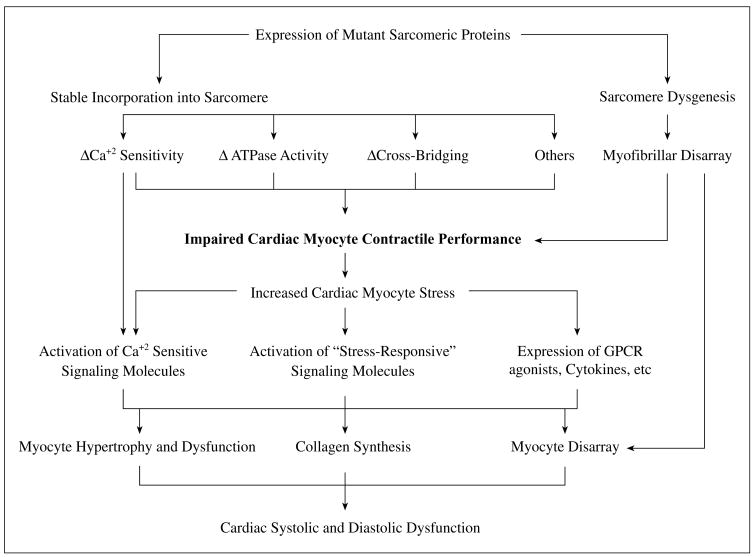
We and others have proposed that mutant sarcomeric proteins cause a diverse array of initial defects that converge into a common phenotype of impaired cardiac myocyte function and subsequent development of compensatory hypertrophy, fibrosis, and disarray (Fig. 1). Accordingly, the common defect in HCM, regardless of the diversity of the causal mutations and the initial defects, is impaired cardiac myocyte mechanical function,124 which increases myocyte stress and leads to activation of stress-responsive intracellular signaling kinases and trophic factors. Collectively stress-responsive signaling kinases and trophic factors activate the transcription machinery leading to cardiac hypertrophy, interstitial fibrosis and other histological and clinical phenotypes of HCM.124 Stimuli are also provided by altered Ca+2 sensitivity of myofibrils, reduced ATPase activity, and sarcomere dysgenesis. Accordingly, myocyte hypertrophy and disarray, interstitial fibrosis, and thickening of the media of intra-mural coronary arteries are “secondary” phenotypes and potentially reversible. Other cellular mechanisms, such as cardiac myocyte atrophy and myocyte “drop-out” possibly due to apoptosis also have been implicated in animal models. However, their significance in human HCM remains to be established.
Fig. 1.
Pathogenesis of HCM.
Pharmacological interventions in genetically engineered animal models could provide new therapeutic opportunities for human patients with HCM
Despite recognition of HCM as a major cause of mortality and morbidity, current pharmacological interventions for treatment of human patients with HCM are empiric and not proven to regress cardiac hypertrophy, fibrosis or disarray. Genetically engineered animal models of HCM provide the opportunity to test the effects of pharmacological interventions that are targeted to specific pathways involved in the pathogenesis of HCM. Such studies provide the opportunity to determine reversibility of the evolving phenotypes or the ability to prevent their development.
Reversal of interstitial fibrosis in the cTnT-Q92 transgenic mice
Interstitial fibrosis is considered as a major risk factor for SCD in human patients with HCM and ventricular arrhythmias.6, 125 In a recent randomized study we tested the effects of blockade of angiotensin II receptor 1 on cardiac phenotype in the cTnT-Q92 transgenic mouse model.126 It is noted that despite the well-established role of blockade of renin-angiotensin-aldosterone system in reversal of cardiac fibrosis in a variety of cardiovascular pathology, they are not currently used in treatment of human patients with HCM. The concern arises from the possible worsening of outflow gradient because of afterload reduction with these agents. In the cTnT-Q92 mice, treatment with losartan reduced interstitial collagen volume by 49% and expression of collagen α1 (I) mRNA and TGF-β1, a known mediator of profibrotic effects of angiotensin II by ~50%. Losartan had no significant effect on the extent of myocyte disarray. Because the cTnT-Q92 mice, unlike the β-MyHC-Q403 rabbits, do not exhibit cardiac hypertrophy,100 the effects of losartan on cardiac hypertrophy could not be assessed.
Reversal of cardiac hypertrophy, interstitial fibrosis and improvement in cardiac function in the β-MyHC-Q403 rabbits: Recent data raises the possibility that HMG-CoA reductase inhibitors could be used to inhibit signaling kinases involved in the pathogenesis of cardiac hypertrophy and thus could potentially attenuate cardiac hypertrophy and fibrosis in pathological state.127–129 We performed a randomized study and determined the effects of simvastatin, a pleiotropic HMG-CoA reductase inhibitor, on cardiac structure and function in the β-MyHC-Q403 transgenic rabbits. Treatment with simvastatin reduced mean left ventricular mass was reduced by 37%, septal and posterior wall thickness by approximately 20%, and collagen volume fraction by ~50%. Indices of left ventricular filling pressure were improved significantly. At mechanistic level, expression of active ERK1/2 was reduced in the treatment group, but levels of other active signaling kinases were unchanged. Thus, simvastatin and probably other HMG-CoA reductase inhibitors are potential candidates for reversal of cardiac hypertrophy and fibrosis, major predictors of mortality and SCD, in human patients with HCM.6, 7
Worsening of cardiac phenotype in α-MyHC-Q403+/− mice following treatment with calcineurin inhibitors: There has been significant controversy regarding the utility of calcineurin inhibitors in treatment and prevention of cardiac hypertrophy in a variety of conditions.130 With regard to HCM, a recent study showed that treatment of α-MyHC-Q403+/− mice with FK506 or cyclosporin A increased cardiac hypertrophy, worsened myocyte hypertrophy, disarray and interstitial fibrosis, increased mortality, and impaired Ca+2 flux 116. Pre-treatment with diltiazem, an L-type Ca+2 channel blocker, prevented the exaggerated cardiac hypertrophic response to inhibitors of calcineurin.
Table IV.
Initial defects conferred by mutant contractile sarcomeric proteins.
| Mechanical defect |
| Impaired acto-myosin interaction |
| Impaired cardiac myocyte, skeletal myoblasts and myofibril contractile performance |
| Biochemical defects |
| Impaired Ca+2 affinity, force and myofibril sensitivity |
| Bioenergetics |
| Impaired ATPase activity |
| Structural defects |
| Impaired sarcomere assembly |
| Impaired subcellular localization of sarcomeric proteins |
| Altered stoichiometry |
Acknowledgments
This work is supported in part by a grant from the National Heart, Lung, and Blood Institute, Specialized Centers of Research (P50-HL42267-01).
Refrences
- 1.Maron BJ, Roberts WC. Quantitative analysis of cardiac muscle cell disorganization in the ventricular septum of patients with hypertrophic cardiomyopathy. Circulation. 1979;59:689–706. doi: 10.1161/01.cir.59.4.689. [DOI] [PubMed] [Google Scholar]
- 2.Maron BJ, Anan TJ, Roberts WC. Quantitative analysis of the distribution of cardiac muscle cell disorganization in the left ventricular wall of patients with hypertrophic cardiomyopathy. Circulation. 1981;63:882–94. doi: 10.1161/01.cir.63.4.882. [DOI] [PubMed] [Google Scholar]
- 3.Marian AJ. On genetic and phenotypic variability of hypertrophic cardiomyopathy: nature versus nurture. J Am Coll Cardiol. 2001;38:331–4. doi: 10.1016/s0735-1097(01)01389-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niimura H, Bachinski LL, Sangwatanaroj S, Watkins H, Chudley AE, McKenna W, et al. Mutations in the gene for cardiac myosin-binding protein C and late-onset familial hypertrophic cardiomyopathy. N Engl J Med. 1998;338:1248–57. doi: 10.1056/NEJM199804303381802. [DOI] [PubMed] [Google Scholar]
- 5.Maron BJ, Spirito P, Wesley Y, Arce J. Development and progression of left ventricular hypertrophy in children with hypertrophic cardiomyopathy. N Engl J Med. 1986;315:610–4. doi: 10.1056/NEJM198609043151003. [DOI] [PubMed] [Google Scholar]
- 6.Shirani J, Pick R, Roberts WC, Maron BJ. Morphology and significance of the left ventricular collagen network in young patients with hypertrophic cardiomyopathy and sudden cardiac death. J Am Coll Cardiol. 2000;35:36–44. doi: 10.1016/s0735-1097(99)00492-1. [DOI] [PubMed] [Google Scholar]
- 7.Spirito P, Bellone P, Harris KM, Bernabo P, Bruzzi P, Maron BJ. Magnitude of left ventricular hypertrophy and risk of sudden death in hypertrophic cardiomyopathy. N Engl J Med. 2000;342:1778–85. doi: 10.1056/NEJM200006153422403. [DOI] [PubMed] [Google Scholar]
- 8.Varnava AM, Elliott PM, Baboonian C, Davison F, Davies MJ, McKenna WJ. Hypertrophic cardiomyopathy: histopathological features of sudden death in cardiac troponin t disease. Circulation. 2001;104:1380–4. doi: 10.1161/hc3701.095952. [DOI] [PubMed] [Google Scholar]
- 9.Varnava AM, Elliott PM, Mahon N, Davies MJ, McKenna WJ. Relation between myocyte disarray and outcome in hypertrophic cardiomyopathy. Am J Cardiol. 2001;88:275–9. doi: 10.1016/s0002-9149(01)01640-x. [DOI] [PubMed] [Google Scholar]
- 10.Maron BJ, Gardin JM, Flack JM, Gidding SS, Kurosaki TT, Bild DE. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA Study. Coronary Artery Risk Development in (Young) Adults. Circulation. 1995;92:785–9. doi: 10.1161/01.cir.92.4.785. [DOI] [PubMed] [Google Scholar]
- 11.Maron BJ, Niimura H, Casey SA, Soper MJ, Wright GB, Seidman JG, et al. Development of left ventricular hypertrophy in adults with hypertrophic cardiomyopathy caused by cardiac myocin-binding protein C gene mutations. J Am Coll Cardiol. 2001;38:315–21. doi: 10.1016/s0735-1097(01)01386-9. [DOI] [PubMed] [Google Scholar]
- 12.Maron BJ, Shirani J, Poliac LC, Mathenge R, Roberts WC, Mueller FO. Sudden death in young competitive athletes. Clinical, demographic, and pathological profiles. JAMA. 1996;276:199–204. [PubMed] [Google Scholar]
- 13.Cannan CR, Reeder GS, Bailey KR, Melton LJ, III, Gersh BJ. Natural history of hypertrophic cardiomyopathy. A population-based study, 1976 through 1990. Circulation. 1995;92:2488–95. doi: 10.1161/01.cir.92.9.2488. [DOI] [PubMed] [Google Scholar]
- 14.Watkins H, Thierfelder L, Hwang DS, McKenna W, Seidman JG, Seidman CE. Sporadic hypertrophic cardiomyopathy due to de novo myosin mutations. J Clin Invest. 1992;90:1666–71. doi: 10.1172/JCI116038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watkins H, Thierfelder L, Anan R, Jarcho J, Matsumori A, McKenna W, et al. Independent origin of identical beta cardiac myosin heavy-chain mutations in hypertrophic cardiomyopathy. Am J Hum Genet. 1993;53:1180–5. [PMC free article] [PubMed] [Google Scholar]
- 16.Watkins H, Anan R, Coviello DA, Spirito P, Seidman JG, Seidman CE. A de novo mutation in alpha-tropomyosin that causes hypertrophic cardiomyopathy. Circulation. 1995;91:2302–5. doi: 10.1161/01.cir.91.9.2302. [DOI] [PubMed] [Google Scholar]
- 17.Geisterfer-Lowrance AA, Kass S, Tanigawa G, Vosberg HP, McKenna W, Seidman CE, et al. A molecular basis for familial hypertrophic cardiomyopathy: a beta cardiac myosin heavy chain gene missense mutation. Cell. 1990;62:999–1006. doi: 10.1016/0092-8674(90)90274-i. [DOI] [PubMed] [Google Scholar]
- 18.Jarcho JA, McKenna W, Pare JA, Solomon SD, Holcombe RF, Dickie S, et al. Mapping a gene for familial hypertrophic cardiomyopathy to chromosome 14q1. N Engl J Med. 1989;321:1372–8. doi: 10.1056/NEJM198911163212005. [DOI] [PubMed] [Google Scholar]
- 19.Thierfelder L, Watkins H, MacRae C, Lamas R, McKenna W, Vosberg HP, et al. Alpha-tropomyosin and cardiac troponin T mutations cause familial hypertrophic cardiomyopathy: a disease of the sarcomere. Cell. 1994;77:701–12. doi: 10.1016/0092-8674(94)90054-x. [DOI] [PubMed] [Google Scholar]
- 20.Gollob MH, Green MS, Tang AS, Gollob T, Karibe A, Hassan AS, et al. Identification of a gene responsible for familial Wolff-Parkinson-White syndrome. N Engl J Med. 2001;344:1823–31. doi: 10.1056/NEJM200106143442403. [DOI] [PubMed] [Google Scholar]
- 21.Blair E, Redwood C, Ashrafian H, Oliveira M, Broxholme J, Kerr B, et al. Mutations in the gamma(2) subunit of AMP-activated protein kinase cause familial hypertrophic cardiomyopathy: evidence for the central role of energy compromise in disease pathogenesis. Hum Mol Genet. 2001;10:1215–20. doi: 10.1093/hmg/10.11.1215. [DOI] [PubMed] [Google Scholar]
- 22.www.uwcm.ac.uk/search/mg/allgenes. 2001.
- 23.Marian AJ. Genetics for Cardiologists. 1. London: REMEDICA Publishing; 2000. pp. 1–53. [Google Scholar]
- 24.Seidman CE. Hypertrophic Cardiomyopathy: from man to mouse. J Clin Invest. 2000;106:S9–13. doi: 10.1172/JCI11093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jaenicke T, Diederich KW, Haas W, Schleich J, Lichter P, Pfordt M, et al. The complete sequence of the human beta-myosin heavy chain gene and a comparative analysis of its product. Genomics. 1990;8:194–206. doi: 10.1016/0888-7543(90)90272-v. [DOI] [PubMed] [Google Scholar]
- 26.Anan R, Greve G, Thierfelder L, Watkins H, McKenna WJ, Solomon S, et al. Prognostic implications of novel beta cardiac myosin heavy chain gene mutations that cause familial hypertrophic cardiomyopathy. J Clin Invest. 1994;93:280–5. doi: 10.1172/JCI116957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dausse E, Komajda M, Fetler L, Dubourg O, Dufour C, Carrier L, et al. Familial hypertrophic cardiomyopathy. Microsatellite haplotyping and identification of a hot spot for mutations in the beta-myosin heavy chain gene. J Clin Invest. 1993;92:2807–13. doi: 10.1172/JCI116900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tesson F, Richard P, Charron P, Mathieu B, Cruaud C, Carrier L, et al. Genotype-phenotype analysis in four families with mutations in beta-myosin heavy chain gene responsible for familial hypertrophic cardiomyopathy. Hum Mutat. 1998;12:385–92. doi: 10.1002/(SICI)1098-1004(1998)12:6<385::AID-HUMU4>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 29.Nakajima-Taniguchi C, Matsui H, Eguchi N, Nagata S, Kishimoto T, Yamauchi-Takihara K. A novel deletion mutation in the beta-myosin heavy chain gene found in Japanese patients with hypertrophic cardiomyopathy. J Mol Cell Cardiol. 1995;27:2607–12. doi: 10.1006/jmcc.1995.0047. [DOI] [PubMed] [Google Scholar]
- 30.Marian AJ, Yu QT, Mares A, Jr, Hill R, Roberts R, Perryman MB. Detection of a new mutation in the beta-myosin heavy chain gene in an individual with hypertrophic cardiomyopathy. J Clin Invest. 1992;90:2156–65. doi: 10.1172/JCI116101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cuda G, Perrotti N, Perticone F, Mattioli PL. A previously undescribed de novo insertion-deletion mutation in the beta myosin heavy chain gene in a kindred with familial hypertrophic cardiomyopathy. Heart. 1996;76:451–2. doi: 10.1136/hrt.76.5.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carrier L, Bonne G, Bahrend E, Yu B, Richard P, Niel F, et al. Organization and sequence of human cardiac myosin binding protein C gene (MYBPC3) and identification of mutations predicted to produce truncated proteins in familial hypertrophic cardiomyopathy. Circ Res. 1997;80:427–34. [PubMed] [Google Scholar]
- 33.Erdmann J, Raible J, Maki-Abadi J, Hummel M, Hammann J, Wollnik B, et al. Spectrum of clinical phenotypes and gene variants in cardiac myosin-binding protein C mutation carriers with hypertrophic cardiomyopathy. J Am Coll Cardiol. 2001;38:322–30. doi: 10.1016/s0735-1097(01)01387-0. [DOI] [PubMed] [Google Scholar]
- 34.Bonne G, Carrier L, Bercovici J, Cruaud C, Richard P, Hainque B, et al. Cardiac myosin binding protein-C gene splice acceptor site mutation is associated with familial hypertrophic cardiomyopathy. Nat Genet. 1995;11:438–40. doi: 10.1038/ng1295-438. [DOI] [PubMed] [Google Scholar]
- 35.Watkins H, Conner D, Thierfelder L, Jarcho JA, MacRae C, McKenna WJ, et al. Mutations in the cardiac myosin binding protein-C gene on chromosome 11 cause familial hypertrophic cardiomyopathy. Nat Genet. 1995;11:434–7. doi: 10.1038/ng1295-434. [DOI] [PubMed] [Google Scholar]
- 36.Seidman CE, Seidman JG. Molecular genetic studies of familial hypertrophic cardiomyopathy. Basic Res Cardiol. 1998;93(Suppl 3):13–6. doi: 10.1007/s003950050196. [DOI] [PubMed] [Google Scholar]
- 37.Forissier JF, Carrier L, Farza H, Bonne G, Bercovici J, Richard P, et al. Codon 102 of the cardiac troponin T gene is a putative hot spot for mutations in familial hypertrophic cardiomyopathy. Circulation. 1996;94:3069–73. doi: 10.1161/01.cir.94.12.3069. [DOI] [PubMed] [Google Scholar]
- 38.Kimura A, Harada H, Park JE, Nishi H, Satoh M, Takahashi M, et al. Mutations in the cardiac troponin I gene associated with hypertrophic cardiomyopathy. Nat Genet. 1997;16:379–82. doi: 10.1038/ng0897-379. [DOI] [PubMed] [Google Scholar]
- 39.Kokado H, Shimizu M, Yoshio H, Ino H, Okeie K, Emoto Y, et al. Clinical features of hypertrophic cardiomyopathy caused by a Lys183 deletion mutation in the cardiac troponin I gene. Circulation. 2000;102:663–9. doi: 10.1161/01.cir.102.6.663. [DOI] [PubMed] [Google Scholar]
- 40.Coviello DA, Maron BJ, Spirito P, Watkins H, Vosberg HP, Thierfelder L, et al. Clinical features of hypertrophic cardiomyopathy caused by mutation of a “hot spot” in the alpha-tropomyosin gene. J Am Coll Cardiol. 1997;29:635–40. doi: 10.1016/s0735-1097(96)00538-4. [DOI] [PubMed] [Google Scholar]
- 41.Karibe A, Tobacman LS, Strand J, Butters C, Back N, Bachinski LL, et al. Hypertrophic cardiomyopathy caused by a novel alpha-tropomyosin mutation (V95A) is associated with mild cardiac phenotype, abnormal calcium binding to troponin, abnormal myosin cycling, and poor prognosis. Circulation. 2001;103:65–71. doi: 10.1161/01.cir.103.1.65. [DOI] [PubMed] [Google Scholar]
- 42.Hoffmann B, Schmidt-Traub H, Perrot A, Osterziel KJ, Gessner R. First mutation in cardiac troponin C, L29Q, in a patient with hypertrophic cardiomyopathy. Hum Mutat. 2001;17:524. doi: 10.1002/humu.1143. [DOI] [PubMed] [Google Scholar]
- 43.Mogensen J, Klausen IC, Pedersen AK, Egeblad H, Bross P, Kruse TA, et al. Alpha-cardiac actin is a novel disease gene in familial hypertrophic cardiomyopathy. J Clin Invest. 1999;103:R39–43. doi: 10.1172/JCI6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olson TM, Doan TP, Kishimoto NY, Whitby FG, Ackerman MJ, Fananapazir L. Inherited and de novo mutations in the cardiac actin gene cause hypertrophic cardiomyopathy. J Mol Cell Cardiol. 2000;32:1687–94. doi: 10.1006/jmcc.2000.1204. [DOI] [PubMed] [Google Scholar]
- 45.Satoh M, Takahashi M, Sakamoto T, Hiroe M, Marumo F, Kimura A. Structural analysis of the titin gene in hypertrophic cardiomyopathy: identification of a novel disease gene. Biochem Biophys Res Commun. 1999;262:411–7. doi: 10.1006/bbrc.1999.1221. [DOI] [PubMed] [Google Scholar]
- 46.Poetter K, Jiang H, Hassanzadeh S, Master SR, Chang A, Dalakas MC, et al. Mutations in either the essential or regulatory light chains of myosin are associated with a rare myopathy in human heart and skeletal muscle. Nat Genet. 1996;13:63–9. doi: 10.1038/ng0596-63. [DOI] [PubMed] [Google Scholar]
- 47.Flavigny J, Richard P, Isnard R, Carrier L, Charron P, Bonne G, et al. Identification of two novel mutations in the ventricular regulatory myosin light chain gene (MYL2) associated with familial and classical forms of hypertrophic cardiomyopathy. J Mol Med. 1998;76:208–14. doi: 10.1007/s001090050210. [DOI] [PubMed] [Google Scholar]
- 48.Korade-Mirnics Z, Babitzke P, Hoffman E. Myotonic dystrophy: molecular windows on a complex etiology. Nucleic Acids Res. 1998;26:1363–8. doi: 10.1093/nar/26.6.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Phillips MF, Harper PS. Cardiac disease in myotonic dystrophy. Cardiovasc Res. 1997;33:13–22. doi: 10.1016/s0008-6363(96)00163-0. [DOI] [PubMed] [Google Scholar]
- 50.Palau F. Friedreich’s ataxia and frataxin: molecular genetics, evolution and pathogenesis. Int J Mol Med. 2001;7:581–9. doi: 10.3892/ijmm.7.6.581. [DOI] [PubMed] [Google Scholar]
- 51.Bit-Avragim N, Perrot A, Schols L, Hardt C, Kreuz FR, Zuhlke C, et al. The GAA repeat expansion in intron 1 of the frataxin gene is related to the severity of cardiac manifestation in patients with Friedreich’s ataxia. J Mol Med. 2001;78:626–32. doi: 10.1007/s001090000162. [DOI] [PubMed] [Google Scholar]
- 52.Simon DK, Johns DR. Mitochondrial disorders: clinical and genetic features. Ann Rev Med. 1999;50:111–27. doi: 10.1146/annurev.med.50.1.111. [DOI] [PubMed] [Google Scholar]
- 53.Williams RS. Canaries in the coal mine: mitochondrial DNA and vascular injury from reactive oxygen species. Circ Res. 2000;86:915–6. doi: 10.1161/01.res.86.9.915. [DOI] [PubMed] [Google Scholar]
- 54.Ashizawa T, Subramony SH. What is Kearns-Sayre syndrome after all? Arch Neurol. 2001;58:1053–4. doi: 10.1001/archneur.58.7.1053. [DOI] [PubMed] [Google Scholar]
- 55.Epstein ND, Cohn GM, Cyran F, Fananapazir L. Differences in clinical expression of hypertrophic cardiomyopathy associated with two distinct mutations in the beta-myosin heavy chain gene. A 908Leu----Val mutation and a 403Arg----Gln mutation [see comments] Circulation. 1992;86:345–52. doi: 10.1161/01.cir.86.2.345. [DOI] [PubMed] [Google Scholar]
- 56.Marian AJ, Mares A, Jr, Kelly DP, Yu QT, Abchee AB, Hill R, Roberts R. Sudden cardiac death in hypertrophic cardiomyopathy. Variability in phenotypic expression of beta-myosin heavy chain mutations. Eur Heart J. 1995;16:368–76. doi: 10.1093/oxfordjournals.eurheartj.a060920. [DOI] [PubMed] [Google Scholar]
- 57.Watkins H, Rosenzweig A, Hwang DS, Levi T, McKenna W, Seidman CE, Seidman JG. Characteristics and prognostic implications of myosin missense mutations in familial hypertrophic cardiomyopathy. N Engl J Med. 1992;326:1108–14. doi: 10.1056/NEJM199204233261703. [DOI] [PubMed] [Google Scholar]
- 58.Watkins H, McKenna WJ, Thierfelder L, Suk HJ, Anan R, O’Donoghue A, et al. Mutations in the genes for cardiac troponin T and alpha-tropomyosin in hypertrophic cardiomyopathy. N Engl J Med. 1995;332:1058–64. doi: 10.1056/NEJM199504203321603. [DOI] [PubMed] [Google Scholar]
- 59.Charron P, Dubourg O, Desnos M, Bennaceur M, Carrier L, Camproux AC, et al. Clinical features and prognostic implications of familial hypertrophic cardiomyopathy related to the cardiac myosin-binding protein C gene. Circulation. 1998;97:2230–6. doi: 10.1161/01.cir.97.22.2230. [DOI] [PubMed] [Google Scholar]
- 60.Ho CY, Lever HM, DeSanctis R, Farver CF, Seidman JG, Seidman CE. Homozygous mutation in cardiac troponin T: implications for hypertrophic cardiomyopathy [In Process Citation] Circulation. 2000;102:1950–5. doi: 10.1161/01.cir.102.16.1950. [DOI] [PubMed] [Google Scholar]
- 61.Jeschke B, Uhl K, Weist B, Schroder D, Meitinger T, Dohlemann C, Vosberg HP. A high risk phenotype of hypertrophic cardiomyopathy associated with a compound genotype of two mutated beta-myosin heavy chain genes. Hum Genet. 1998;102:299–304. doi: 10.1007/s004390050695. [DOI] [PubMed] [Google Scholar]
- 62.Charron P, Dubourg O, Desnos M, Isnard R, Hagege A, Bonne G, et al. Genotype-phenotype correlations in familial hypertrophic cardiomyopathy. A comparison between mutations in the cardiac protein-C and the beta-myosin heavy chain genes. Eur Heart J. 1998;19:139–45. doi: 10.1053/euhj.1997.0575. [DOI] [PubMed] [Google Scholar]
- 63.Abchee A, Marian AJ. Prognostic significance of beta-myosin heavy chain mutations is reflective of their hypertrophic expressivity in patients with hypertrophic cardiomyopathy. J Investig Med. 1997;45:191–6. [PubMed] [Google Scholar]
- 64.Marian AJ, Yu QT, Workman R, Greve G, Roberts R. Angiotensin-converting enzyme polymorphism in hypertrophic cardiomyopathy and sudden cardiac death. Lancet. 1993;342:1085–6. doi: 10.1016/0140-6736(93)92064-z. [DOI] [PubMed] [Google Scholar]
- 65.Lechin M, Quinones MA, Omran A, Hill R, Yu QT, Rakowski H, et al. Angiotensin-I converting enzyme genotypes and left ventricular hypertrophy in patients with hypertrophic cardiomyopathy. Circulation. 1995;92:1808–12. doi: 10.1161/01.cir.92.7.1808. [DOI] [PubMed] [Google Scholar]
- 66.Tesson F, Dufour C, Moolman JC, Carrier L, al Mahdawi S, Chojnowska L, et al. The influence of the angiotensin I converting enzyme genotype in familial hypertrophic cardiomyopathy varies with the disease gene mutation. J Mol Cell Cardiol. 1997;29:831–8. doi: 10.1006/jmcc.1996.0332. [DOI] [PubMed] [Google Scholar]
- 67.Brugada R, Kelsey W, Lechin M, Zhao G, Yu QT, Zoghbi W, et al. Role of candidate modifier genes on the phenotypic expression of hypertrophy in patients with hypertrophic cardiomyopathy. J Investig Med. 1997;45:542–51. [PubMed] [Google Scholar]
- 68.Patel R, Lim DS, Reddy D, Nagueh SF, Lutucuta S, Sole MJ, et al. Variants of trophic factors and expression of cardiac hypertrophy in patients with hypertrophic cardiomyopathy. J Mol Cell Cardiol. 2000;32:2369–77. doi: 10.1006/jmcc.2000.1267. [DOI] [PubMed] [Google Scholar]
- 69.Marian AJ, Yu QT, Mann DL, Graham FL, Roberts R. Expression of a mutation causing hypertrophic cardiomyopathy disrupts sarcomere assembly in adult feline cardiac myocytes. Circ Res. 1995;77:98–106. doi: 10.1161/01.res.77.1.98. [DOI] [PubMed] [Google Scholar]
- 70.Yang Q, Sanbe A, Osinska H, Hewett TE, Klevitsky R, Robbins J. A mouse model of myosin binding protein C human familial hypertrophic cardiomyopathy. J Clin Invest. 1998;102:1292–300. doi: 10.1172/JCI3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Becker KD, Gottshall KR, Hickey R, Perriard JC, Chien KR. Point mutations in human beta cardiac myosin heavy chain have differential effects on sarcomeric structure and assembly: an ATP binding site change disrupts both thick and thin filaments, whereas hypertrophic cardiomyopathy mutations display normal assembly. J Cell Biol. 1997;137:131–40. doi: 10.1083/jcb.137.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Michele DE, Albayya FP, Metzger JM. Direct, convergent hypersensitivity of calcium-activated force generation produced by hypertrophic cardiomyopathy mutant alpha-tropomyosins in adult cardiac myocytes. Nat Med. 1999;5:1413–7. doi: 10.1038/70990. [DOI] [PubMed] [Google Scholar]
- 73.Cuda G, Fananapazir L, Epstein ND, Sellers JR. The in vitro motility activity of beta-cardiac myosin depends on the nature of the beta-myosin heavy chain gene mutation in hypertrophic cardiomyopathy. J Muscle Res Cell Motil. 1997;18:275–83. doi: 10.1023/a:1018613907574. [DOI] [PubMed] [Google Scholar]
- 74.Fujita H, Sugiura S, Momomura S, Omata M, Sugi H, Sutoh K. Characterization of mutant myosins of Dictyostelium discoideum equivalent to human familial hypertrophic cardiomyopathy mutants. Molecular force level of mutant myosins may have a prognostic implication. J Clin Invest. 1997;99:1010–5. doi: 10.1172/JCI119228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sata M, Ikebe M. Functional analysis of the mutations in the human cardiac beta-myosin that are responsible for familial hypertrophic cardiomyopathy. Implication for the clinical outcome. J Clin Invest. 1996;98:2866–73. doi: 10.1172/JCI119115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sweeney HL, Straceski AJ, Leinwand LA, Tikunov BA, Faust L. Heterologous expression of a cardiomyopathic myosin that is defective in its actin interaction. J Biol Chem. 1994;269:1603–5. [PubMed] [Google Scholar]
- 77.Bing W, Redwood CS, Purcell IF, Esposito G, Watkins H, Marston SB. Effects of two hypertrophic cardiomyopathy mutations in alpha-tropomyosin, Asp175Asn and Glu180Gly, on Ca2+ regulation of thin filament motility. Biochem Biophys Res Commun. 1997;236:760–4. doi: 10.1006/bbrc.1997.7045. [DOI] [PubMed] [Google Scholar]
- 78.Lin D, Bobkova A, Homsher E, Tobacman LS. Altered cardiac troponin T in vitro function in the presence of a mutation implicated in familial hypertrophic cardiomyopathy. J Clin Invest. 1996;97:2842–8. doi: 10.1172/JCI118740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Palmiter KA, Tyska MJ, Haeberle JR, Alpert NR, Fananapazir L, Warshaw DM. R403Q and L908V mutant beta-cardiac myosin from patients with familial hypertrophic cardiomyopathy exhibit enhanced mechanical performance at the single molecule level. J Muscle Res Cell Motil. 2000;21:609–20. doi: 10.1023/a:1005678905119. [DOI] [PubMed] [Google Scholar]
- 80.Redwood C, Lohmann K, Bing W, Esposito GM, Elliott K, Abdulrazzak H, et al. Investigation of a truncated cardiac troponin T that causes familial hypertrophic cardiomyopathy: Ca(2+) regulatory properties of reconstituted thin filaments depend on the ratio of mutant to wild-type protein. Circ Res. 2000;86:1146–52. doi: 10.1161/01.res.86.11.1146. [DOI] [PubMed] [Google Scholar]
- 81.Blanchard E, Seidman C, Seidman JG, LeWinter M, Maughan D. Altered crossbridge kinetics in the alphaMHC403/+ mouse model of familial hypertrophic cardiomyopathy. Circ Res. 1999;84:475–83. doi: 10.1161/01.res.84.4.475. [DOI] [PubMed] [Google Scholar]
- 82.Gao WD, Perez NG, Seidman CE, Seidman JG, Marban E. Altered cardiac excitation-contraction coupling in mutant mice with familial hypertrophic cardiomyopathy. J Clin Invest. 1999;103:661–6. doi: 10.1172/JCI5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Roopnarine O, Leinwand LA. Functional analysis of myosin mutations that cause familial hypertrophic cardiomyopathy. Biophys J. 1998;75:3023–30. doi: 10.1016/S0006-3495(98)77743-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tyska MJ, Hayes E, Giewat M, Seidman CE, Seidman JG, Warshaw DM. Single-molecule mechanics of R403Q cardiac myosin isolated from the mouse model of familial hypertrophic cardiomyopathy. Circ Res. 2000;86:737–44. doi: 10.1161/01.res.86.7.737. [DOI] [PubMed] [Google Scholar]
- 85.Harada K, Takahashi-Yanaga F, Minakami R, Morimoto S, Ohtsuki I. Functional consequences of the deletion mutation deltaGlu160 in human cardiac troponin T. J Biochem (Tokyo) 2000;127:263–8. doi: 10.1093/oxfordjournals.jbchem.a022603. [DOI] [PubMed] [Google Scholar]
- 86.Morimoto S, Yanaga F, Minakami R, Ohtsuki I. Ca2+-sensitizing effects of the mutations at Ile-79 and Arg-92 of troponin T in hypertrophic cardiomyopathy. Am J Physiol. 1998;275:C200–7. doi: 10.1152/ajpcell.1998.275.1.C200. [DOI] [PubMed] [Google Scholar]
- 87.Nakaura H, Yanaga F, Ohtsuki I, Morimoto S. Effects of missense mutations Phe110Ile and Glu244Asp in human cardiac troponin T on force generation in skinned cardiac muscle fibers. J Biochem (Tokyo) 1999;126:457–60. doi: 10.1093/oxfordjournals.jbchem.a022473. [DOI] [PubMed] [Google Scholar]
- 88.Szczesna D, Zhang R, Zhao J, Jones M, Guzman G, Potter JD. Altered regulation of cardiac muscle contraction by troponin T mutations that cause familial hypertrophic cardiomyopathy. J Biol Chem. 2000;275:624–30. doi: 10.1074/jbc.275.1.624. [DOI] [PubMed] [Google Scholar]
- 89.Tobacman LS, Lin D, Butters C, Landis C, Back N, Pavlov D, et al. Functional consequences of troponin T mutations found in hypertrophic cardiomyopathy. J Biol Chem. 1999;274:28363–70. doi: 10.1074/jbc.274.40.28363. [DOI] [PubMed] [Google Scholar]
- 90.Elliott K, Watkins H, Redwood CS. Altered regulatory properties of human cardiac troponin I mutants that cause hypertrophic cardiomyopathy. J Biol Chem. 2000;275:22069–74. doi: 10.1074/jbc.M002502200. [DOI] [PubMed] [Google Scholar]
- 91.Bottinelli R, Coviello DA, Redwood CS, Pellegrino MA, Maron BJ, Spirito P, et al. A mutant tropomyosin that causes hypertrophic cardiomyopathy is expressed in vivo and associated with an increased calcium sensitivity. Circ Res. 1998;82:106–15. doi: 10.1161/01.res.82.1.106. [DOI] [PubMed] [Google Scholar]
- 92.Yanaga F, Morimoto S, Ohtsuki I. Ca2+ sensitization and potentiation of the maximum level of myofibrillar ATPase activity caused by mutations of troponin T found in familial hypertrophic cardiomyopathy. J Biol Chem. 1999;274:8806–12. doi: 10.1074/jbc.274.13.8806. [DOI] [PubMed] [Google Scholar]
- 93.Lankford EB, Epstein ND, Fananapazir L, Sweeney HL. Abnormal contractile properties of muscle fibers expressing beta-myosin heavy chain gene mutations in patients with hypertrophic cardiomyopathy. J Clin Invest. 1995;95:1409–14. doi: 10.1172/JCI117795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Malinchik S, Cuda G, Podolsky RJ, Horowits R. Isometric tension and mutant myosin heavy chain content in single skeletal myofibers from hypertrophic cardiomyopathy patients. J Mol Cell Cardiol. 1997;29:667–76. doi: 10.1006/jmcc.1996.0309. [DOI] [PubMed] [Google Scholar]
- 95.Kim SJ, Iizuka K, Kelly RA, Geng YJ, Bishop SP, Yang G, et al. An alpha-cardiac myosin heavy chain gene mutation impairs contraction and relaxation function of cardiac myocytes. Am J Physiol. 1999;276:H1780–7. doi: 10.1152/ajpheart.1999.276.5.H1780. [DOI] [PubMed] [Google Scholar]
- 96.Marian AJ, Zhao G, Seta Y, Roberts R, Yu QT. Expression of a mutant (Arg92Gln) human cardiac troponin T, known to cause hypertrophic cardiomyopathy, impairs adult cardiac myocyte contractility. Circ Res. 1997;81:76–85. doi: 10.1161/01.res.81.1.76. [DOI] [PubMed] [Google Scholar]
- 97.Rust EM, Albayya FP, Metzger JM. Identification of a contractile deficit in adult cardiac myocytes expressing hypertrophic cardiomyopathy-associated mutant troponin T proteins. J Clin Invest. 1999;103:1459–67. doi: 10.1172/JCI6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Watkins H, Seidman CE, Seidman JG, Feng HS, Sweeney HL. Expression and functional assessment of a truncated cardiac troponin T that causes hypertrophic cardiomyopathy. Evidence for a dominant negative action [see comments] J Clin Invest. 1996;98:2456–61. doi: 10.1172/JCI119063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sweeney HL, Feng HS, Yang Z, Watkins H. Functional analyses of troponin T mutations that cause hypertrophic cardiomyopathy: insights into disease pathogenesis and troponin function. Proc Natl Acad Sci USA. 1998;95:14406–10. doi: 10.1073/pnas.95.24.14406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Oberst L, Zhao G, Park JT, Brugada R, Michael LH, Entman ML, et al. Dominant-negative effect of a mutant cardiac troponin T on cardiac structure and function in transgenic mice. J Clin Invest. 1998;102:1498–505. doi: 10.1172/JCI4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tardiff JC, Hewett TE, Palmer BM, Olsson C, Factor SM, Moore RL, et al. Cardiac troponin T mutations result in allele–specific phenotypes in a mouse model for hypertrophic cardiomyopathy. J Clin Invest. 1999;104:469–81. doi: 10.1172/JCI6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Muthuchamy M, Pieples K, Rethinasamy P, Hoit B, Grupp IL, Boivin GP, et al. Mouse model of a familial hypertrophic cardiomyopathy mutation in alpha-tropomyosin manifests cardiac dysfunction. Circ Res. 1999;85:47–56. doi: 10.1161/01.res.85.1.47. [DOI] [PubMed] [Google Scholar]
- 103.Welikson RE, Buck SH, Patel JR, Moss RL, Vikstrom KL, Factor SM, et al. Cardiac myosin heavy chains lacking the light chain binding domain cause hypertrophic cardiomyopathy in mice. Am J Physiol. 1999;276:H2148–58. doi: 10.1152/ajpheart.1999.276.6.H2148. [DOI] [PubMed] [Google Scholar]
- 104.Vemuri R, Lankford EB, Poetter K, Hassanzadeh S, Takeda K, Yu ZX, et al. The stretch-activation response may be critical to the proper functioning of the mammalian heart. Proc Natl Acad Sci USA. 1999;96:1048–53. doi: 10.1073/pnas.96.3.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Frey N, Franz WM, Gloeckner K, Degenhardt M, Muller M, Muller O, et al. Transgenic rat hearts expressing a human cardiac troponin T deletion reveal diastolic dysfunction and ventricular arrhythmias. Cardiovasc Res. 2000;47:254–64. doi: 10.1016/s0008-6363(00)00114-0. [DOI] [PubMed] [Google Scholar]
- 106.Marian AJ, Wu Y, Lim DS, McCluggage M, Youker K, Yu QT, et al. A transgenic rabbit model for human hypertrophic cardiomyopathy. J Clin Invest. 1999;104:1683–92. doi: 10.1172/JCI7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Georgakopoulos D, Christe ME, Giewat M, Seidman CM, Seidman JG, Kass DA. The pathogenesis of familial hypertrophic cardiomyopathy: early and evolving effects from an alpha-cardiac myosin heavy chain missense mutation. Nat Med. 1999;5:327–30. doi: 10.1038/6549. [DOI] [PubMed] [Google Scholar]
- 108.Tardiff JC, Factor SM, Tompkins BD, Hewett TE, Palmer BM, Moore RL. A truncated cardiac troponin T molecule in transgenic mice suggests multiple cellular mechanisms for familial hypertrophic cardiomyopathy. J Clin Invest. 1998;101:2800–11. doi: 10.1172/JCI2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yang Q, Sanbe A, Osinska H, Hewett TE, Klevitsky R, Robbins J. In vivo modeling of myosin binding protein C familial hypertrophic cardiomyopathy. Circ Res. 1999;85:841–7. doi: 10.1161/01.res.85.9.841. [DOI] [PubMed] [Google Scholar]
- 110.Spindler M, Saupe KW, Christe ME, Sweeney HL, Seidman CE, Seidman JG, et al. Diastolic dysfunction and altered energetics in the alphaMHC403/+ mouse model of familial hypertrophic cardiomyopathy. J Clin Invest. 1998;101:1775–83. doi: 10.1172/JCI1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Berul CI, Christe ME, Aronovitz MJ, Maguire CT, Seidman CE, Seidman JG, et al. Familial hypertrophic cardiomyopathy mice display gender differences in electrophysiological abnormalities. J Interv Card Electrophysiol. 1998;2:7–14. doi: 10.1023/a:1009700404218. [DOI] [PubMed] [Google Scholar]
- 112.Berul CI, Christe ME, Aronovitz MJ, Seidman CE, Seidman JG, Mendelsohn ME. Electrophysiological abnormalities and arrhythmias in alpha MHC mutant familial hypertrophic cardiomyopathy mice. J Clin Invest. 1997;99:570–6. doi: 10.1172/JCI119197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nagueh SF, Lakkis NM, Middleton KJ, Spencer WH, III, Zoghbi WA, Quinones MA. Doppler estimation of left ventricular filling pressures in patients with hypertrophic cardiomyopathy. Circulation. 1999;99:254–61. doi: 10.1161/01.cir.99.2.254. [DOI] [PubMed] [Google Scholar]
- 114.Swynghedauw B. Developmental and functional adaptation of contractile proteins in cardiac and skeletal muscles. Physiol Rev. 1986;66:710–71. doi: 10.1152/physrev.1986.66.3.710. [DOI] [PubMed] [Google Scholar]
- 115.Geisterfer-Lowrance AA, Christe M, Conner DA, Ingwall JS, Schoen FJ, Seidman CE, Seidman JG. A mouse model of familial hypertrophic cardiomyopathy. Science. 1996;272:731–4. doi: 10.1126/science.272.5262.731. [DOI] [PubMed] [Google Scholar]
- 116.Fatkin D, McConnell BK, Mudd JO, Semsarian C, Moskowitz IG, Schoen FJ, et al. An abnormal Ca(2+) response in mutant sarcomere protein-mediated familial hypertrophic cardiomyopathy. J Clin Invest. 2000;106:1351–9. doi: 10.1172/JCI11093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Janssen PM, Zeitz O, Keweloh B, Siegel U, Maier LS, Barckhausen P, et al. Influence of cyclosporine A on contractile function, calcium handling, and energetics in isolated human and rabbit myocardium [see comments] Cardiovasc Res. 2000;47:99–107. doi: 10.1016/s0008-6363(00)00052-3. [DOI] [PubMed] [Google Scholar]
- 118.Nagueh SF, Kopelen HA, Lim DS, Zoghbi WA, Quinones MA, Roberts R, et al. Tissue Doppler imaging consistently detects myocardial contraction and relaxation abnormalities, irrespective of cardiac hypertrophy, in a transgenic rabbit model of human hypertrophic cardiomyopathy. Circulation. 2000;102:1346–50. doi: 10.1161/01.cir.102.12.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rottbauer W, Gautel M, Zehelein J, Labeit S, Franz WM, Fischer C, et al. Novel splice donor site mutation in the cardiac myosin-binding protein-C gene in familial hypertrophic cardiomyopathy. Characterization Of cardiac transcript and protein. J Clin Invest. 1997;100:475–82. doi: 10.1172/JCI119555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jones WK, Grupp IL, Doetschman T, Grupp G, Osinska H, Hewett TE, et al. Ablation of the murine alpha myosin heavy chain gene leads to dosage effects and functional deficits in the heart. J Clin Invest. 1996;98:1906–17. doi: 10.1172/JCI118992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Blanchard EM, Iizuka K, Christe M, Conner DA, Geisterfer-Lowrance A, Schoen FJ, et al. Targeted ablation of the murine alpha-tropomyosin gene. Circ Res. 1997;81:1005–10. doi: 10.1161/01.res.81.6.1005. [DOI] [PubMed] [Google Scholar]
- 122.Rethinasamy P, Muthuchamy M, Hewett T, Boivin G, Wolska BM, Evans C, et al. Molecular and physiological effects of alpha-tropomyosin ablation in the mouse. Circ Res. 1998;82:116–23. doi: 10.1161/01.res.82.1.116. [DOI] [PubMed] [Google Scholar]
- 123.Nagueh SF, Bachinski L, Meyer D, Hill R, Zoghbi WA, Tam JW, et al. Tissue Doppler imaging consistently detects myocardial abnormalities in patients with familial hypertrophic cardiomyopathy and provides a novel means for an early diagnosis prior to an independent of hypertrophy. Circulation. 2001;104:128–30. doi: 10.1161/01.cir.104.2.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Marian AJ. Pathogenesis of diverse clinical and pathological phenotypes in hypertrophic cardiomyopathy. Lancet. 2000;355:58–60. doi: 10.1016/s0140-6736(99)06187-5. [DOI] [PubMed] [Google Scholar]
- 125.Assayag P, Carre F, Chevalier B, Delcayre C, Mansier P, Swynghedauw B. Compensated cardiac hypertrophy: arrhythmogenicity and the new myocardial phenotype. I. Fibrosis Cardiovasc Res. 1997;34:439–44. doi: 10.1016/s0008-6363(97)00073-4. [DOI] [PubMed] [Google Scholar]
- 126.Lim DS, Lutucuta S, Bachireddy P, Youker K, Evans A, Entman M, et al. Angiotensin II blockade reverses myocardial fibrosis in a transgenic mouse model of human hypertrophic cardiomyopathy. Circulation. 2001;103:789–91. doi: 10.1161/01.cir.103.6.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Oi S, Haneda T, Osaki J, Kashiwagi Y, Nakamura Y, Kawabe J, Kikuchi K. Lovastatin prevents angiotensin II-induced cardiac hypertrophy in cultured neonatal rat heart cells. Eur J Pharmacol. 1999;376:139–48. doi: 10.1016/s0014-2999(99)00282-4. [DOI] [PubMed] [Google Scholar]
- 128.Park HJ, Galper JB. 3-Hydroxy-3-methylglutaryl CoA reductase inhibitors up-regulate transforming growth factor-beta signaling in cultured heart cells via inhibition of geranylgeranylation of RhoA GTPase. Proc Natl Acad Sci USA. 1999;96:11525–30. doi: 10.1073/pnas.96.20.11525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Su SF, Hsiao CL, Chu CW, Lee BC, Lee TM. Effects of pravastatin on left ventricular mass in patients with hyperlipidemia and essential hypertension. Am J Cardiol. 2000;86:514–8. doi: 10.1016/s0002-9149(00)01004-3. [DOI] [PubMed] [Google Scholar]
- 130.Dorn GW. Calcineurin inhibition in hypertrophy: back from the dead! Circulation. 2001;104:9–11. doi: 10.1161/01.cir.104.1.9. [DOI] [PubMed] [Google Scholar]
- 131.Fatkin D, Christe ME, Aristizabal O, McConnell BK, Srinivasan S, Schoen FJ, et al. Neonatal cardiomyopathy in mice homozygous for the Arg403Gln mutation in the alpha cardiac myosin heavy chain gene. J Clin Invest. 1999;103:147–53. doi: 10.1172/JCI4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bevilacqua LM, Maguire CT, Seidman JG, Seidman CE, Berul CI. QT dispersion in alpha-myosin heavy-chain familial hypertrophic cardiomyopathy mice. Pediatr Res. 1999;45:643–7. doi: 10.1203/00006450-199905010-00005. [DOI] [PubMed] [Google Scholar]
- 133.Vikstrom KL, Bohlmeyer T, Factor SM, Leinwand LA. Hypertrophy, pathology, and molecular markers of cardiac pathogenesis. Circ Res. 1998;82:773–8. doi: 10.1161/01.res.82.7.773. [DOI] [PubMed] [Google Scholar]
- 134.Vikstrom KL, Factor SM, Leinwand LA. Mice expressing mutant myosin heavy chains are a model for familial hypertrophic cardiomyopathy. Mol Med. 1996;2:556–67. [PMC free article] [PubMed] [Google Scholar]
- 135.Evans CC, Pena JR, Phillips RM, Muthuchamy M, Wieczorek DF, Solaro RJ, et al. Altered hemodynamics in transgenic mice harboring mutant tropomyosin linked to hypertrophic cardiomyopathy. Am J Physiol Heart Circ Physiol. 2000;279:H2414–23. doi: 10.1152/ajpheart.2000.279.5.H2414. [DOI] [PubMed] [Google Scholar]
- 136.McConnell BK, Jones KA, Fatkin D, Arroyo LH, Lee RT, Aristizabal O, et al. Dilated cardiomyopathy in homozygous myosin-binding protein-C mutant mice. J Clin Invest. 1999;104:1235–44. doi: 10.1172/JCI7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sanbe A, Nelson D, Gulick J, Setser E, Osinska H, Wang X, et al. In vivo analysis of an essential myosin light chain mutation linked to familial hypertrophic cardiomyopathy. Circ Res. 2000;87:296–302. doi: 10.1161/01.res.87.4.296. [DOI] [PubMed] [Google Scholar]
- 138.James J, Zhang Y, Osinska H, Sanbe A, Klevitsky R, Hewett TE, Robbins J. Transgenic modeling of a cardiac troponin I mutation linked to familial hypertrophic cardiomyopathy. Circ Res. 2000;87:805–11. doi: 10.1161/01.res.87.9.805. [DOI] [PubMed] [Google Scholar]