Abstract
In nematodes, the ETS-family transcription factor ast-1 regulates multiple genes comprising the dopamine (DA) neuron phenotype, including biosynthetic enzymes and transporters. ETS transcription factors are hypothesized to play a similar role in vertebrates, and based on its expression in the adult mouse midbrain, Etv5/ERM has been proposed as a regulator of DA gene expression in the substantia nigra (SN) and ventral tegmental area (VTA). Here we show that Etv5 expression is not detectable until postnatal stages in the midbrain, well after development of the DA system, and that Etv5 knock-out and control mice show comparable tyrosine hydroxylase and dopamine transporter expression in the embryonic and adult midbrain. Other known members of the ETS family do not have expression patterns that are consistent with a role in DA gene regulation in the SN/VTA. These findings suggest that the ETS factors, while required for the generation of the DA phenotype in nematodes, do not play such a role in the mouse midbrain.
Introduction
Dopaminergic pathways have received extensive attention due to their roles in psychiatric and neurological disorders, reward mechanisms, and addiction. The DA neurons mediating these pathways reside in the substantia nigra (SN) and ventral tegmental area (VTA), of the ventral midbrain, and the recent identification of transcription factors regulating the embryonic development of these neurons represents a major advance in this field (Ang, 2006; Smidt and Burbach, 2007). In recent work using Caenorhabditis elegans as a model for DA neuron development, it was demonstrated that a transcription factor of the ETS family, encoded by the axon steering defect-1 (ast-1) gene, is a terminal selector gene for DA cell fate, although it is not necessary for the generation or survival of these neurons (Flames and Hobert, 2009). In ast-1-null mutants, five key dopamine pathway genes showed greatly reduced expression, including dat-1 (dopamine transporter, DAT), bas-1 (aromatic amino acid decarboxylase, AADC) cat-1 (vesicular monoamine transporter, VMAT) cat-2 (tyrosine hydroxylase), and cat-4 (GTP cyclohydrolase). In addition, misexpression of ast-1 induced ectopic expression of dat-1 and cat-2 reporter genes in a restricted subset of neurons, demonstrating that it is sufficient for cell-autonomous expression of DA markers in some contexts. Ast-1 appears to exert its effects on multiple genes through a conserved regulatory motif (Flames and Hobert, 2009; Spitzer, 2009).
In mammals, there are >20 known members of the ETS family, and these have been classified based on domain structure and homology within the ETS domain (Oikawa and Yamada, 2003). Ast-1 is a member of the fli subfamily of ETS proteins, and its closest vertebrate homologues are Fli-1 and Erg (Schmid et al., 2006). These factors have been identified as translocation breakpoints in human malignancies, and for their roles in hematopoiesis (Kruse et al., 2009). However, lack of expression in the brain indicates that they are not good candidates for regulating the development or maintenance of DA neurons. Instead, the principal vertebrate ETS factors with tissue-specific expression in the nervous system are those of the PEA3 class, including Etv1 (Er81), Etv4 (Pea3, E1AF) and Etv5 (ERM). Etv5 is expressed in the adult mouse SN/VTA, and it has also been shown that the tyrosine hydroxylase (TH) promoter can be activated by Etv5 in vitro (Flames and Hobert, 2009). For these reasons Etv5 has been proposed as a regulator of DA neuron differentiation in the midbrain.
Here we show that Etv5 mRNA is not detectable in the SN/VTA until postnatal day 7, well after the onset of the expression of DA pathway genes. DA markers including tyrosine hydroxylase (TH), the dopamine transporter (DAT, Slc6a3), and the vesicular monoamine transporter 2 (Vmat2, Slc18a2) are expressed normally in the embryonic and adult midbrain of Etv5 knock-out (KO) mice and in DA neuron target fields in the basal forebrain. Together these results indicate that Etv5 is not required for DA neuron development. In addition, a review of the expression patterns of other murine ETS factors in available gene expression databases does not reveal obvious candidates for a functional ast-1 homolog in mammals.
Materials and Methods
Mice, genotyping, timed matings.
129Sv/Ev wild-type (WT) mice were bred and maintained as described previously (Chen et al., 2005). Etv5−/− mice on a 129Sv/Ev background were produced by interbreeding Etv5+/− mice, and the pups were genotyped as described previously (Chen et al., 2005). Noon on the day of the detection of a mucous plug was designated embryonic day 0.5 (E0.5). All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of California, San Diego, and conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Immunofluorescence and in situ hybridization.
Tissue for immunofluorescence and in situ hybridization (ISH) was fixed by immersion in 4% paraformaldehyde in PBS for 1–3 h depending on the embryonic stage (E16.5 and earlier), or by cardiac perfusion for postnatal stages and adult mice (Quina et al., 2009). Cryostat sections at 20 μm were used for both techniques. Primary antibodies used included rabbit anti-TH (Millipore); rat anti-DAT (Millipore); rabbit anti-Nurr1 (Santa Cruz Biotechnology); and rabbit anti-vesicular monoamine transporter 2 (VMAT2, Millipore); further details appear in supplemental Table 1, available at www.jneurosci.org as supplemental material. Secondary antibodies conjugated to Alexa series fluorophores were obtained from Invitrogen. ISH was performed as described previously (Eng et al., 2004). The plasmids encoding the probes used for Etv1, Etv4 and Etv5, PDEF and ELF1 are described in supplemental Table 1, available at www.jneurosci.org as supplemental material.
Results
To begin to understand the roles of ETS factors in DA development, we examined the developing brain for expression of the PEA3-class factors by ISH beginning at E11.5. Etv1 and Etv4 have known roles in sensory and motor neuron differentiation (Arber et al., 2000; Ladle and Frank, 2002; Vrieseling and Arber, 2006). Etv1 knock-out mice have an apparent reduction in TH expression in the olfactory bulb (Flames and Hobert, 2009), but recent work has shown that loss of Etv1 does not affect olfactory expression of other markers of the DA pathway (Cave et al., 2010). Consistent with these prior reports and available database information, we did not observe Etv1 or Etv4 expression in the ventral midbrain at any stage examined from E11.5 to adult (Data not shown; http://www.genepaint.org/; http://developingmouse.brain-map.org/).
Etv5 mutant mice develop muscle weakness and show functional neuromuscular transmission defects (Hippenmeyer et al., 2007), and Etv5 is also required for spermatogenesis (Chen et al., 2005; Tyagi et al., 2009), but it has no known role in the CNS. To begin to understand the role of Etv5 in DA neuron development, we first examined its expression in the adult mouse. ISH revealed expression of Etv5 in the adult SN/VTA, as well as in the medial part of the cerebral cortex (supplemental Fig. 1, available at www.jneurosci.org as supplemental material), that is consistent with gene expression atlas data (http://mouse.brain-map.org/).
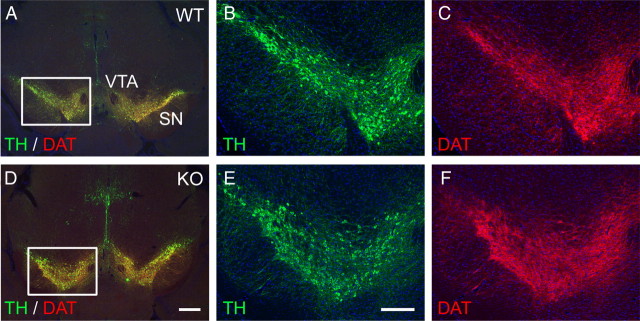
To evaluate the role of Etv5 in midbrain DA neuron differentiation, we first compared the morphology of the tegmental DA nuclei, the expression of the DA pathway genes, and the projections of these neurons to the striatum in Etv5−/− embryos and controls. Antibodies to TH, DAT, and Vmat2 were used to identify dopamine neurons and their projections. As expected, in the E16.5 mouse brain these markers revealed immunoreactive neurons in the SN pars compacta (SNc) and the VTA (Fig. 1). The expression patterns of all three markers were indistinguishable in Etv5−/− embryos and wild type controls. Loss of Etv5 also did not alter the expression of Nurr1, an essential transcription factor for DA neuron development (Fig. 1G,H).
Figure 1.
Expression of DA pathway genes in Etv5 knock-out embryos. Coronal sections of E16.5 embryos were examined for the expression of DA pathway genes. Immunofluorescence for TH (A, B), DAT (C, D), and VMAT2 (E, F) were similar in Etv5−/− (KO) embryos and control (WT) mice. The expression pattern of the regulator of DA differentiation Nurr1 was also indistinguishable in Etv5−/− embryos and controls (G, H). Scale bar, 80 μm.
Although Etv5 expression has been reported in the embryonic midbrain (Flames and Hobert, 2009), details of its ontogeny and localization have not been described. To determine whether the timing of Etv5 expression was consistent with a role in initiating DA pathway gene expression, we examined Etv5 expression from mid-gestation through postnatal development. Etv5 mRNA was undetectable in the ventral midbrain at E14.5, E16.5 and P3 (Fig. 2A–D and data not shown), and was first detected at low levels in the SN at P7, with increased signal at P14 (Fig. 2E–G). Expression of Etv5 in the cerebral cortex at P3 and in the lung at E14.5 and E16.5 served as positive controls for Etv5 detection at these stages (data not shown).
Figure 2.
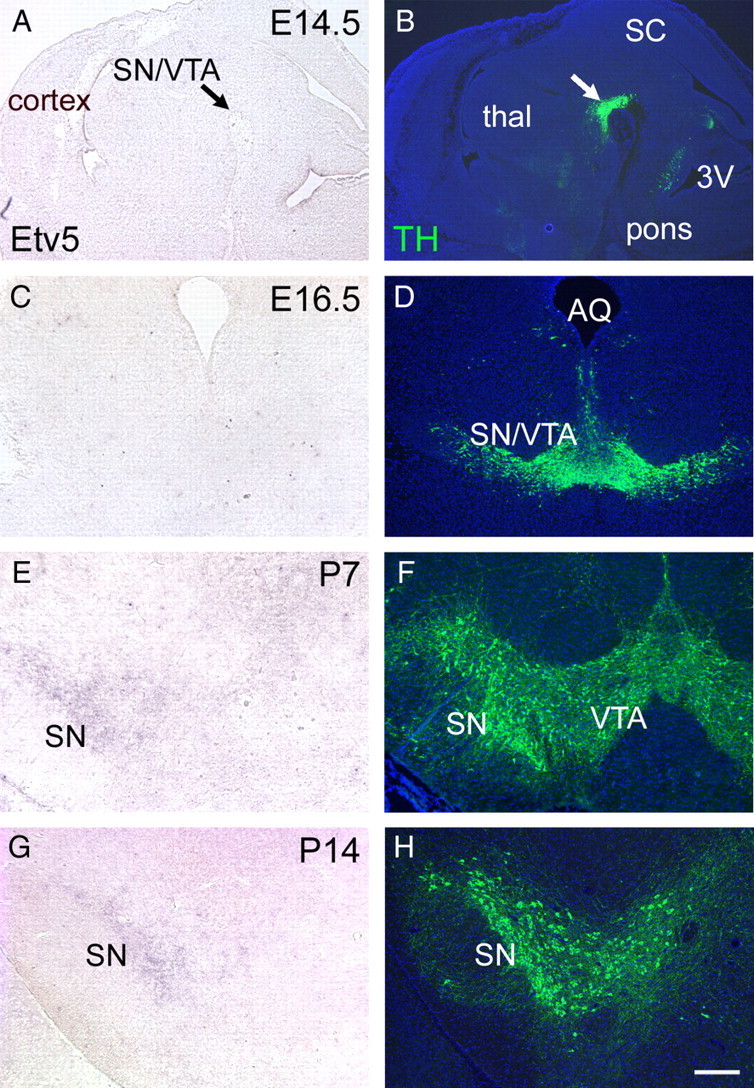
A, C, E, G, Ontogeny of Etv5 expression in the ventral midbrain. Etv5 expression was examined by ISH in sagittal sections at E14.5 (A), and in coronal sections at E16.5, P7, and P14 (C, E, G). B, D, F, H, Immunofluorescence for TH in adjacent sections was used to localize DA neurons in the ventral midbrain. Arrows in A and B indicate the location of differentiating SN/VTA neurons. Etv5 expression in the SNc was first detected at P7. 3V, third ventricle. SC, Superior colliculus; thal, thalamus. Scale bar, 250 μm.
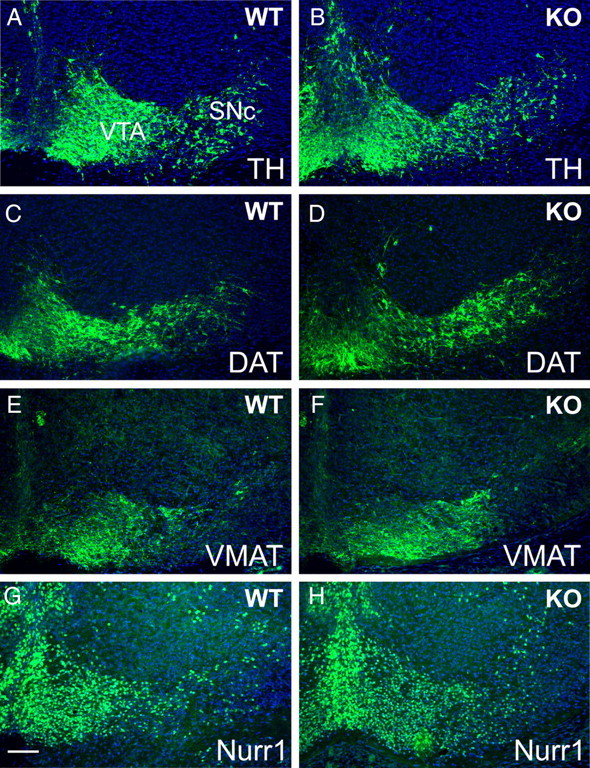
In C. elegans, ast-1 is required for the maintenance of the DA neuron phenotype as well as its initiation (Flames and Hobert, 2009). Because of this observation, combined with the late onset of Etv5 expression in mice, we considered that Etv5 might have a role in the maintenance of midbrain DA gene expression. To test this possibility, we examined the expression of DA pathway genes in adult Etv5−/− mice, and found that the expression of TH and DAT in the ventral midbrain is indistinguishable from controls (Fig. 3). In addition, TH-immunoreactive fibers in the striatum, which receives DA input from the ventral midbrain, appeared normal in the Etv5 knock-outs (supplemental Fig. S2, available at www.jneurosci.org as supplemental material), indicating that Etv5 is not needed for maintenance of the anatomical connection between the SN and the striatum. Together these results exclude Etv5 as a key regulator of midbrain DA pathway gene expression.
Figure 3.
A–F, Expression of DA markers in the ventral midbrain of adult Etv5−/− mice. Coronal sections of the brains of adult control (WT, A–C) and Etv5−/− mice (KO, D–F) were examined by immunofluorescence for the expression of TH and DAT. These markers are highly coexpressed in the DA neurons and their fibers, and are unchanged in the Etv5 knock-out. Inset boxes in A and D show the location of the enlarged areas in B, C and E, F. Scale bars: (in D) A, D, 500 μm; (in E) B, C, E, F, 250 μm.
Although a role for Etv5 in midbrain DA gene expression is effectively excluded by these results, the possibility remains that other vertebrate ETS genes could mediate DA differentiation. The plausibility of this hypothesis is underscored by the key role of the ETS gene Fev/Pet-1 in the differentiation of hindbrain serotonergic neurons, although this factor is known to have no role in DA neuron development (Hendricks et al., 2003). For this reason, we examined the expression of six other members of the ETS family with reported expression in CNS: Etv1, Etv4, Ets2, Fev, Spdef, and Elk1. None of these were detectable in the region of the mouse ventral midbrain defined by TH expression in E16.5 embryos (data not shown). The recent availability of gene expression database information for the mouse embryo (Visel et al., 2004; Allen Institute for Brain Science, 2009; http://developingmouse.brain-map.org) also allowed a more exhaustive search for expression of members of this gene family at E11.5, E13.5, E14.5, and E16.5. Because these efforts to characterize developmental gene expression have emphasized transcription factors, it was possible to examine the expression patterns of all 26 members of the ETS family for which spliced transcripts have been identified in the mouse (Oikawa and Yamada, 2003; Hollenhorst et al., 2007). The available database information did not reveal expression of any member of this class in differentiating DA neurons (supplemental Table S2, supplemental Figs. S3–S6, available at www.jneurosci.org as supplemental material).
Discussion
The development of midbrain DA neurons in vertebrates requires a complex combination of transcriptional regulators and diffusible signals to control both the acquisition and maintenance of a neurotransmitter-specific phenotype (Smidt and Burbach, 2007). Gain- and loss-of-function studies suggest that some of the key transcription factors required for the midbrain DA differentiation program, such as Otx2, Lmx1a/b, Msx1/2, Ngn2 and Mash1, Pitx3 and Nurr1 are likely to act sequentially and combinatorially to promote the mature midbrain DA phenotype (Andersson et al., 2006; Ang, 2006; Kele et al., 2006). It remains possible that Etv5 interacts with this gene expression program at a late phase of development to determine some aspect of the mature DA phenotype. However, the present study, combined with recent work demonstrating that Etv1 is not essential for the expression of most DA markers in olfactory neurons (Cave et al., 2010), largely excludes the ETS genes as master regulators of the DA phenotype in mice, and suggests that the fundamental mechanisms of DA differentiation in nematodes and vertebrates are in this way distinct.
Footnotes
This work was supported in part by Department of Veterans Affairs MERIT funding, and National Institutes of Health Awards HD33442, MH065496, and NS064933 (E.E.T.). E.E.T. is a National Alliance for Research on Schizophrenia and Depression Investigator. We thank Dr. Rex Hess, Dr. Paul Cooke, and Daryl Meling of the University of Illinois and Dr. Ken Murphy of Washington University in St. Louis for extensive assistance with Etv5/ERM knock-out mice.
References
- Allen Institute for Brain Science. Allen developing mouse brain atlas. Seattle, WA: Allen Institute for Brain Science; 2009. [Google Scholar]
- Andersson E, Tryggvason U, Deng Q, Friling S, Alekseenko Z, Robert B, Perlmann T, Ericson J. Identification of intrinsic determinants of midbrain dopamine neurons. Cell. 2006;124:393–405. doi: 10.1016/j.cell.2005.10.037. [DOI] [PubMed] [Google Scholar]
- Ang SL. Transcriptional control of midbrain dopaminergic neuron development. Development. 2006;133:3499–3506. doi: 10.1242/dev.02501. [DOI] [PubMed] [Google Scholar]
- Arber S, Ladle DR, Lin JH, Frank E, Jessell TM. ETS gene Er81 controls the formation of functional connections between group Ia sensory afferents and motor neurons. Cell. 2000;101:485–498. doi: 10.1016/s0092-8674(00)80859-4. [DOI] [PubMed] [Google Scholar]
- Cave JW, Akiba Y, Banerjee K, Bhosle S, Berlin R, Baker H. Differential regulation of dopaminergic gene expression by er81. J Neurosci. 2010;30:4717–4724. doi: 10.1523/JNEUROSCI.0419-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ouyang W, Grigura V, Zhou Q, Carnes K, Lim H, Zhao GQ, Arber S, Kurpios N, Murphy TL, Cheng AM, Hassell JA, Chandrashekar V, Hofmann MC, Hess RA, Murphy KM. ERM is required for transcriptional control of the spermatogonial stem cell niche. Nature. 2005;436:1030–1034. doi: 10.1038/nature03894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng SR, Lanier J, Fedtsova N, Turner EE. Coordinated regulation of gene expression by Brn3a in developing sensory ganglia. Development. 2004;131:3859–3870. doi: 10.1242/dev.01260. [DOI] [PubMed] [Google Scholar]
- Flames N, Hobert O. Gene regulatory logic of dopamine neuron differentiation. Nature. 2009;458:885–889. doi: 10.1038/nature07929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks TJ, Fyodorov DV, Wegman LJ, Lelutiu NB, Pehek EA, Yamamoto B, Silver J, Weeber EJ, Sweatt JD, Deneris ES. Pet-1 ETS gene plays a critical role in 5-HT neuron development and is required for normal anxiety-like and aggressive behavior. Neuron. 2003;37:233–247. doi: 10.1016/s0896-6273(02)01167-4. [DOI] [PubMed] [Google Scholar]
- Hippenmeyer S, Huber RM, Ladle DR, Murphy K, Arber S. ETS transcription factor Erm controls subsynaptic gene expression in skeletal muscles. Neuron. 2007;55:726–740. doi: 10.1016/j.neuron.2007.07.028. [DOI] [PubMed] [Google Scholar]
- Hollenhorst PC, Shah AA, Hopkins C, Graves BJ. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 2007;21:1882–1894. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kele J, Simplicio N, Ferri AL, Mira H, Guillemot F, Arenas E, Ang SL. Neurogenin 2 is required for the development of ventral midbrain dopaminergic neurons. Development. 2006;133:495–505. doi: 10.1242/dev.02223. [DOI] [PubMed] [Google Scholar]
- Kruse EA, Loughran SJ, Baldwin TM, Josefsson EC, Ellis S, Watson DK, Nurden P, Metcalf D, Hilton DJ, Alexander WS, Kile BT. Dual requirement for the ETS transcription factors Fli-1 and Erg in hematopoietic stem cells and the megakaryocyte lineage. Proc Natl Acad Sci U S A. 2009;106:13814–13819. doi: 10.1073/pnas.0906556106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladle DR, Frank E. The role of the ETS gene PEA3 in the development of motor and sensory neurons. Physiol Behav. 2002;77:571–576. doi: 10.1016/s0031-9384(02)00907-1. [DOI] [PubMed] [Google Scholar]
- Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- Quina LA, Wang S, Ng L, Turner EE. Brn3a and Nurr1 mediate a gene regulatory pathway for habenula development. J Neurosci. 2009;29:14309–14322. doi: 10.1523/JNEUROSCI.2430-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid C, Schwarz V, Hutter H. AST-1, a novel ETS-box transcription factor, controls axon guidance and pharynx development in C. elegans. Dev Biol. 2006;293:403–413. doi: 10.1016/j.ydbio.2006.02.042. [DOI] [PubMed] [Google Scholar]
- Smidt MP, Burbach JP. How to make a mesodiencephalic dopaminergic neuron. Nat Rev Neurosci. 2007;8:21–32. doi: 10.1038/nrn2039. [DOI] [PubMed] [Google Scholar]
- Spitzer NC. Neuroscience: a bar code for differentiation. Nature. 2009;458:843–844. doi: 10.1038/458843a. [DOI] [PubMed] [Google Scholar]
- Tyagi G, Carnes K, Morrow C, Kostereva NV, Ekman GC, Meling DD, Hostetler C, Griswold M, Murphy KM, Hess RA, Hofmann MC, Cooke PS. Loss of Etv5 decreases proliferation and RET levels in neonatal mouse testicular germ cells and causes an abnormal first wave of spermatogenesis. Biol Reprod. 2009;81:258–266. doi: 10.1095/biolreprod.108.075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Thaller C, Eichele G. GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 2004;32:D552–D556. doi: 10.1093/nar/gkh029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieseling E, Arber S. Target-induced transcriptional control of dendritic patterning and connectivity in motor neurons by the ETS gene Pea3. Cell. 2006;127:1439–1452. doi: 10.1016/j.cell.2006.10.042. [DOI] [PubMed] [Google Scholar]