Abstract
While whole genome resequencing remains expensive, genomic partitioning provides an affordable means of targeting sequence efforts towards regions of high interest. There are several competitive methods for targeted capture; these include molecular inversion probes, microdroplet-segregated multiplex PCR, and on-array or in-solution capture-by-hybridization. Enrichment of the human exome by array hybridization has been successfully applied to pinpoint the causative allele of Mendelian disorders. This protocol focuses on the application of Agilent 1M arrays for capture-by-hybridization and sequencing on the Illumina platform, though the library preparation method may be adaptable to other vendor’s array platforms and sequencing technologies.
Keywords: Resequencing, exome, hybridization, targeted enrichment
Unit Introduction
The identification of sequence-level variation in individuals is of immense utility in genetics, and is of particular interest in the discovery of disease genes and specific mutant alleles. Generating complete genome sequences is increasingly feasible on a large-scale with the introduction of “second-generation” sequencers, which are able to generate sequence data in a cost-effective, massively parallel fashion. However, although the cost of sequencing has dropped dramatically, it is still not low enough for whole-genome resequencing to be done routinely by many laboratories, even on a small-scale. However, for many projects, there is significant interest using new sequencing platforms to perform variation discovery in targeted subsets of the genome. A variety of methods to accomplish “genome partitioning” or “targeted capture” have been described (9), and include several that are based on hybridization (1–6) and others that include enzymatic activity to confer specificity (7, 8).
Here we describe the protocol that we have applied to enrich for all protein-coding regions in the human genome (the “exome”), using hybridization on microarrays. The basic protocol has two main components: construction of a shotgun sequencing library, and enrichment of that library for sequences of interest via hybridization to a programmable microarray.
Strategic Planning
This protocol largely relies on standard molecular biology techniques; however, special attention is required to limit cross-contamination between samples. Large amounts (~20 μg) of shotgun library are produced for each sample, which can easily contaminate gloves and workspaces. If samples are being prepared in parallel, careful segregation of different sample libraries is required.
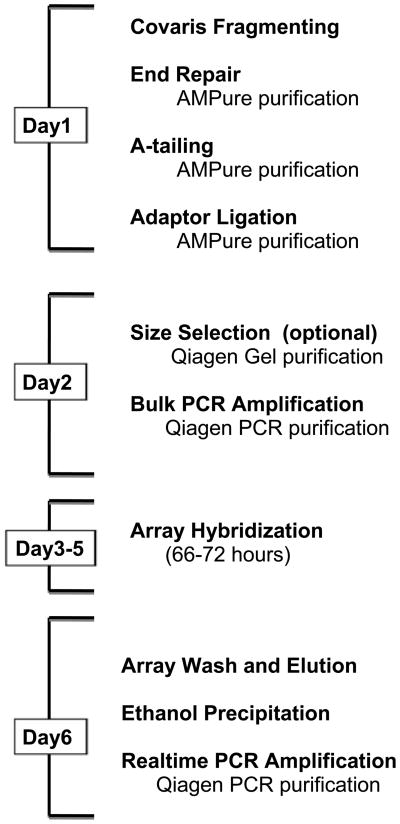
The flowchart shown in Figure 1 provides a suggested daily schedule for this protocol. Several breakpoints are indicated in the main protocol to aid in scheduling.
Figure 1.
Basic Flowchart for Genomic Enrichment by Array Hybridization
Basic Protocol Materials List
Solutions and Reagents
5 μg of genomic DNA
1X Tris-EDTA, pH 8.0 (e.g., Fisher BioReagents, BP2473-1)
10X Next End-Repair Buffer (NEB)
Next End-Repair Enzyme Mix (NEB)
Agencourt AMPure XP
10X PCR buffer w/15mM MgCl2 (Applied Biosystems N8080160)
dATP (100mM)
Taq (5U/μl)
T4 ligase (e.g., Enzymatics L603-HC-L)
10X T4 ligase buffer (e.g., Enzymatics L603-HC-L)
Certified low-range ultra agarose (e.g., Biorad)
10 mg/ml ethidium bromide
1X TAE buffer (0.04M Tris-acetate pH 8, 1mM EDTA)
5X gel loading buffer (e.g., QIAgen 239901)
100 bp DNA ladder (e.g.,NEB N3231)
Qiaquick Gel Extraction kit (Qiagen)
Hot Start Phusion polymerase (2U/μl) (Finnzymes)
5X Phusion buffer (Finnzymes)
25 mM dNTPs
Qiaquick PCR Purification kit (Qiagen)
10X Oligo aCGH/ChIP-on-Chip Blocking Agent (Agilent)
2X Hi-RPM Hybridization Buffer (Agilent)
Cot1 human DNA (1mg/ml)
DNA 1000 Bioanalyzer kit (Agilent)
DNA High Sensitivity Bioanalyzer kit (Agilent)
Oligo aCGH/ChIP-on-chip Hybridization Kit (Agilent)
Special Consumables and Equipment
1M DNA Capture Microarray (Agilent, PN: G3358A, AMADID: 027616).
DNA Microarray Hybridization Chamber (Agilent)
DNA Microarray Hybridization Oven (Agilent)
Secure Seal Tabs (Grace Bio Labs)
Basic Protocol: Genomic Enrichment by Array Hybridization
Design of Capture Arrays
Capture arrays can be designed against any set of genomic subsequences of interest, including the entire exome, with the caveat that repetitive regions of the genome are likely to be problematic. Programmable arrays are commercially available, e.g. arrays with densities as high as 1 million features per array and individual oligo lengths of up to 60 bp can be ordered from Agilent. Guidelines that have been successful for us include the following. In general, hybridization probes should be designed to be evenly spaced across each target region. Other general probe design constraints include: (1) to be relatively unique, such that the average occurrence of each 15-mer in the probe sequence is less than 10−8, (2) to be between 20 and 60 bases in length, with preference for longer probes, and (3) to have a calculated melting temperature (Tm) ≥69 °C, with preference for higher Tm values. The protocol described here was evaluated on array designs that followed these guidelines.
Prepare Working Solutions of Primers, Adaptors and Blocking Oligos
Protocol
Resuspend the oligos listed in Table 1 with 1x TE buffer to a final concentration of 100 μM.
For oligos EXOME_BULK_ FWD (100μM) and EXOME_BULK_ REV (100μM) do a 1:10 dilution to create a 10μM in 100 μl stock of each oligo.
To prepare a 50 μM stock solution of the paired-end adaptor, combine equal volumes of Exome_Adapt_Hi (100 μM) and Exome_Adapt_Lo (100 μM) in a strip tube.
Denature the mixture in a thermal cycler at 95°C for 5 minutes. Turn the thermal cycler off and allow the mixture to anneal while slowly cooling to room temperature (30–45 minutes).
Table 1.
Oligonucleotide sequences
| EXOME_BULK_ FWD | ACA CTC TTT CCC TAC ACG ACG* C |
| EXOME_BULK_ REV | CTC GGC ATT CCT GCT GAA C*C |
| EXOME_BLOCK_F | CAA GCA GAA GAC GGC ATA CGA GAT CGG TCT CGG CAT TCC TGC TGA ACC GCT CTT CCG ATC* T |
| EXOME_BLOCK_R | AAT GAT ACG GCG ACC ACC GAG ATC TAC ACT CTT TCC CTA CAC GAC GCT CTT CCG ATC* T |
| EXOME_BLOCK_F_rev | AGA TCG GAA GAG CGT CGT GTA GGG AAA GAG TGT |
| EXOME_BLOCK_R_rev | AGA TCG GAA GAG CGG TTC AGC AGG AAT GCC GAG |
| EXOME_SEQ_AMP_FWD | CAA GCA GAA GAC GGC ATA CGA GAT CGG TCT CGG CAT TCC TGC TGA ACC GCT CTT CCG ATC* T |
| EXOME_SEQ_AMP_REV | AAT GAT ACG GCG ACC ACC GAG ATC TAC ACT CTT TCC CTA CAC GAC GCT CTT CCG ATC* T |
| EXOME_ADAPT_HI | ACA CTC TTT CCC TAC ACG ACG CTC TTC CGA TC*T |
| EXOME_ADAPT_LO | /5Phos/GAT CGG AAG AGC GGT TCA GCA GGA ATG CCG AG |
Fragment Genomic DNA with Covaris
Materials required
Covaris (E-series or S-series)
Covaris microtubes
1x TE buffer
5 μg input genomic DNA
Protocol
Each sample requires 5 μg of high quality gDNA as input. Check the concentration of the input sample DNA by Picogreen or Nanodrop assays.
Dilute 5 μg of gDNA in total volume of 85 μL in 1x TE buffer.
Load samples into Covaris tubes, recording its positions in the Covaris tube plate. Ensure there are no air bubbles in the bottom of the tubes after loading the samples. In the case of air bubbles, gently tap the plate to bring the solution to the bottom of the tube.
-
Shear the samples on the Covaris under the following settings:
Duty Cycler: 20% Intensity: 5 Cycles/Burst: 200 Time: 180 seconds
Transfer the samples to a half skirted 300 μL 96 well plate and centrifuge. There is no cleanup after this step.
End Repair Fragmented DNA
Materials required
10X Next End-Repair Buffer (NEB)
Next End-Repair Enzyme Mix (NEB)
Agencourt AMPure XP
Thermal cycler
Magnetic plate
½ skirted 300 μl PCR plate
Protocol
Prepare a master mix based on Table 2, scaling the volumes for the number of samples being prepared.
Aliquot 15 μL of master mix into each well containing 85 μL of fragmented DNA, mix and spin down. Each well will contain 100 μL total volume.
React on thermal cycler for 30 minutes at 20°C.
Transfer 180 μL of room temperature AMPure beads to each well containing end-repaired DNA sample, mix well and incubate for 5 minutes at room temperature.
Place the reaction plate onto a magnet plate for 2 minutes to separate beads from the solution. Visually confirm that the beads have moved to the side of the plate. Aspirate the 280 μL of clear solution from the reaction plate and discard.
Dispense 200 μl of 70% ethanol to each well of the reaction plate and incubate for 30 seconds at room temperature. Aspirate out the ethanol and discard. Repeat for a total of two washes.
Take the reaction plate off the magnet plate, add 45 μl of ultrapure water to each well and mix. Place the reaction plate onto the magnet plate for 1 minute to separate the beads. Transfer the 45 μL of supernatant to a new standard 96 well plate.
The DNA is now sheared, end repaired and purified. This is a good stopping point if you need to pause the experiment. Store at −20°C.
Table 2.
Master Mix for End Repair (per sample)
| Component | Volume |
|---|---|
| 10X Next End-Repair Buffer (NEB) | 10 μl |
| Next End-Repair Enzyme Mix (NEB) | 5 μl |
| TOTAL | 15 μl |
A-Tail DNA
Materials required
10X PCR buffer w/15mM MgCl2
dATP (100mM)
Taq (5U/μL)
Agencourt AMPure XP
Thermal cycler
Magnetic plate
Protocol
Prepare a master mix based on Table 3, scaling the volumes for the number of samples being prepared.
Aliquot 6 μL of master mix to each well containing end-repaired DNA, mix and spin down. The final volume will be 50 μL in each well.
React on the thermal cycler at 70°C for 20 minutes.
Transfer 90 μL of room temperature AMPure beads to each well containing the A-tailed DNA sample, mix well and incubate for 5 minutes at room temperature.
Place the reaction plate onto a magnet plate for 2 minutes to separate beads from the solution. Visually confirm that the beads have moved to the side of the plate. Aspirate the 140 μL of clear solution from the reaction plate and discard.
Dispense 200 μL of 70% ethanol to each well of the reaction plate and incubate for 30 seconds at room temperature. Aspirate out the ethanol and discard. Repeat for a total of two washes.
Take the reaction plate off the magnet plate, add 37 μL of ultrapure water to each well and mix. Place the reaction plate onto the magnet plate for 1 minute to separate the beads. Transfer the 37 μL of supernatant to a new standard 96 well PCR plate.
Perform a picogreen assay to quantify DNA. Samples must be at least 20 ng/μL.
Table 3.
Master Mix for A-Tail (per sample)
| Component | Volume |
|---|---|
| 10X PCR buffer w/15mM MgCl2 | 5 μl |
| dATP (100mM) | 0.5 μl |
| Taq (5U/ul) | 0.5 μl |
| TOTAL | 6 μl |
Ligate Paired-end Adaptors to DNA
Materials required
T4 ligase (e.g. NEB, Enzymatics)
10X T4 ligase buffer (e.g. NEB, Enzymatics)
Annealed paired-end adaptor (50μM)
Agencourt AMPure XP
Thermal cycler
Magnetic plate
Protocol
Prepare a master mix based on Table 4, scaling the volumes for the number of samples being prepared.
Aliquot 15 μl of master mix to each well containing A-tailed sample, mix and spin down. Each well will contain 50 μL.
React at room temperature for 15 minutes.
Transfer 90 μL of room-temperature AMPure beads to each well containing the adaptor ligated DNA sample, mix well and incubate for 5 minutes at room temperature.
Place the reaction plate onto a magnet plate for 2 minutes to separate beads from the solution. Visually confirm that the beads have moved to the side of the plate. Aspirate the 140 μL of clear solution from the reaction plate and discard.
Dispense 200 μl of 70% ethanol to each well of the reaction plate and incubate for 30 seconds at room temperature. Aspirate out the ethanol and discard. Repeat for a total of two washes.
-
Take the reaction plate off the magnet plate, add 32 μl of ultrapure water to each well and mix. Place the reaction plate onto the magnet plate for 1 minute to separate the beads. Transfer the 32 μL of supernatant to a new standard 96 well plate.
The DNA is now sheared, end repaired, A-tailed, adaptor ligated and purified. This is a good stopping point if you need to pause the experiment. Store at −20°C.
Table 4.
Adaptor Ligation Master Mix (per sample):
| Component | Volume |
|---|---|
| Annealed paired-end adaptor (50μM) | 5 μl |
| 10X T4 Ligation Buffer | 5 μl |
| T4 ligase (400 U/μL) | 5 μl |
| TOTAL | 15 μl |
Size Select 250–350bp DNA
Special considerations
Size selection is optional. Performing size selection will result in a narrower insert size range, while performing a gel-free protocol will produce a more complex final library.
Materials required
Certified low-range ultra agarose (e.g. Biorad)
Comb: 1mm(d) × mm (w) × 10mm (h) (to accommodate 40uL sample/well)
10 mg/ml ethidium bromide
1x TAE buffer (0.04M Tris-acetate pH 8, 1mM EDTA)
5X gel loading buffer
100 bp DNA ladder
UV light box
Microcentrifuge
Razor blades
15mL Tubes
Clear plastic film
Isopropanol
Qiaquick Gel Extraction kit (Qiagen)
Prepare a 2% low melting agarose gel in a final volume of 200 ml of 1x TAE buffer. (200 ml TAE, 4 g Agarose. Four samples can fit on one gel.)
Microwave for about 2 minutes or until agarose has fully dissolved.
Add 8 μl of EtBr (0.4 μg/ml final concentration), pour gel onto the casting tray and place the wide comb in the upper section.
Once the gel has solidified, place the gel onto the gel box and pour fresh 1x TAE buffer until the gel is covered.
Dispense 10 μl of 4x loading buffer to each 30 ul of DNA sample.
For each sample, load 40 μl of sample, skip a well, then load ladder. Make sure the samples are loaded asymmetrically on the gel so you can distinguish left from right on the gel image.
Run electrophoresis at 100 V for 120 minutes.
Place agarose gel on saran wrap and view the gel on the UV light box.
For each sample, using the 100 bp ladder as reference, cut the region of interest between 250–350bp. Use a clean razor blade while cutting each sample and place each slice in a separate 15ml tube.
Weigh each gel slice and add 4x the volume of Qiagen Buffer QG to the sample (e.g. 400 mg slice, add 1600 μl Buffer QG). Allow the gel to melt at room temperature for 15 minutes vortexing frequently. Ensure the gel is fully dissolved before proceeding.
While gel is melting, heat 120 ul of Qiagen Buffer EB per sample to 65°C.
Add 1 gel volume of isopropanol to the gel solution and mix (e.g. 400mg slice, 400 μl isopropanol).
If the slice weighed more than 400 mg, split the solution across two columns. If the volume is greater than 750 μl, perform multiple centrifugations. Load the solution to the Qiagen column and centrifuge for 1 minute.
Discard flow-through and add 500 μl Buffer QG to each column. Centrifuge 1 minute and discard flow-through.
Add 750 μl Buffer PE to each column. Let the column stand for 2 minutes before centrifuging. Centrifuge 1 minute and discard flow-through. Centrifuge again for 1 minute.
Place the column in a clean labeled 1.5 ml micro-centrifuge tube. Add 50 μl of Qiagen Buffer EB that has been heated to 65°C to each column. Let the column sit for 1 minute before centrifuging for 1 minute.
Amplify shotgun library with bulk PCR
Materials Required
Hot Start Phusion polymerase (2U/μl) (Finnzymes)
5X Phusion buffer (Finnzymes)
25 mM dNTPs
EXOME_BULK_ FWD (100μM)
EXOME_BULK_ REV(100μM)
Qiaquick PCR Purification kit (Qiagen)
Thermal Cycler
Microcentrifuge
Protocol
Dilute each size-selected sample to 100 μL with EB.
Make the master mix described in Table 5, scaling as necessary for the number of samples being captured.
Aliquot 45.7 μL of the master mix to the no-template control well, and add 4.3 μL of water to that well.
Add 1100 μL master mix to the 100 μL of sample from step 1, mix and aliquot 50 μL across 23 wells.
-
Amplify the sample in a thermal cycler with the following program:
98°C, 0:30
98°C, 0:10
65°C, 0:30
72°C, 0:60
Go to Step b, 17 cycles
72°C, 2:00
16°C, forever
Pool the reaction volumes from each sample well into a 15 ml tube. Add 6 ml of Buffer PB to each tube.
Aliquot 750 μl of the sample/PB mixture into each of four Qiagen QIAquik columns. Centrifuge the column for 30 seconds and discard eluant. Repeat until all the solution has been loaded.
Aliquot 750 μl Buffer PE to each column. Centrifuge for 30 seconds and discard eluant. Centrifuge for an additional 2 minutes to remove all traces of Buffer PE.
Transfer the tube to a clean 1.5 ml tube. Aliquot 30 μl Buffer EB to each column. Centrifuge for 30 seconds.
Pool each 32 μl aliquot from the sample for a total volume of 128 μl.
Table 5.
Master mix for Pre-array bulk Amplification (per sample)
| Number of reactions (23 DNA, 1 no template control) | 24 |
| Total volume with DNA template | 1200 μl |
| Component | Volume |
| Water | 823.2 μl |
| 5x Phusion buffer (5x → 1x) | 240 μl |
| 25mM dNTPs [25mM → 200μM] | 9.6 μl |
| EXOME_BULK_ FWD [100μM → 500nM] | 6 μl |
| EXOME_BULK_ REV [100μM → 500nM] | 6 μl |
| Phusion HotStart (2U/μl → 0.02U/μl) | 12 μl |
| TOTAL | 1100 μl |
Quality Checkpoint – Bioanalyzer assay to confirm sample size and concentration
Materials Required
Agilent DNA 1000 kit
Agilent Bioanalyzer
Protocol
-
Run the sample on the Agilent DNA 1000 bioanalyzer.
Twelve samples can be run at one time. Given that the sample’s size selection was 250–350 bp, the bioanalyzer’s electropherograms should have a peak that averages at 300 bp. Look for unligated adaptor bands around 50 base pairs and for overamplification humps at a size range higher than the size selection (e.g. 500bp).
-
Calculate the mass of DNA in the sample by multiplying the concentration acquired from the bioanalyzer by the total elution volume from the previous step(128 μl). The concentration of the sample must be at least 5 μg to continue.
If quantifying using a nanodrop instead of the bioanalyzer or a pico-green assays aim for 15–20 μg of DNA.
Array hybridization
Materials Required
EXOME_BLOCK_F (100 μM)
EXOME_BLOCK_R (100 μM)
EXOME_BLOCK_F_rev (100 μM)
EXOME_BLOCK_R_rev (100 μM)
10X Oligo aCGH/ChIP-on-Chip Blocking Agent (Agilent)
2X Hi-RPM Hybridization Buffer (Agilent)
Cot1 human DNA (1mg/ml)
Thermal cycler
1M Exome Agilent Microarray
Gaskets (Agilent)
Chambers (Agilent)
Oven (Agilent)
Protocol
Make the master mix described in Table 6 scaling as necessary for the number of samples being captured.
Add a minimum of 5 μg of template DNA in 120 μl of water to the master mix for array hybridization. The ideal loading mass is 20 μg of library.
Split the volume of 520 μl into 4 wells of 130μl each.
-
Run in a thermal cycler using the following protocol:
95°C, 3:00
-
37°C, 30:00
Keep the sample in the thermal cycler at 37°C until it is ready to be loaded on the chip.
Assemble the gasket onto the chamber.
Transfer hybridizing solution to gasket pipetting sample down the middle of the gasket. Ensure that no sample spillover occurs, and pipette slowly to minimize bubbles.
Load the array chip to the chamber ensuring the numerical barcode is up and the Agilent label is down.
Assemble chamber, verify the bubble is mobile and place it in rotating oven at 65°C for 66–72 hours.
Table 6.
Master mix for array hybridization (per sample)
| Component | Volume |
|---|---|
| EXOME_BLOCK_F (100 μM) | 3.6 μl |
| EXOME_BLOCK_R (100 μM) | 3.6 μl |
| EXOME_BLOCK_F_rev (100 μM) | 3.6 μl |
| EXOME_BLOCK_R_rev (100 μM) | 3.6 μl |
| Cot1 human DNA (1mg/ml) | 50 μl |
| 10X Blocking Agent (Agilent) | 52 μl |
| 2X Hybridization Buffer (Agilent) | 260 μl |
| TOTAL | 400 μl |
Wash Array and Elute Captured DNA
Materials Required
Wash Buffer 1 (Agilent)
Wash Buffer 2 (Agilent)
15 ml tubes
Secure Seal Gaskets
Secure Seal dots
Elution buffer
Shaker
Tweezers
Heat block (with thermometer)
Water bath
Protocol
Set up for each array
-
1. Set heat block to 95°C and water bath to 37°C.
If using a heat block that can accommodate tubes, partially fill the wells with water.
2. Fill 2x 1.5 ml tubes with Elution Buffer and place on heat block at 95°C.
3. Fill 2x 50ml tubes of Wash Buffer 1 (Agilent).
4. Fill 1x 50ml tube of Wash Buffer 2 (Agilent) and place on water bath at 37°C.
5. Label elution tubes. There are two elutions and each requires two 1.5 ml tubes.
Array Wash
6. Disassemble array chamber.
7. Submerge array and gasket in Agilent Wash Buffer 1. Quickly pry the array/gasket assembly apart with tweezers and transfer the array into the second tube of Agilent Wash Buffer 1. Shake the tube at room temperature for 10 minutes.
8. Move array into Wash Buffer 2 (at 37°C) for 5 minutes.
Elution
9. Remove array from Agilent Wash Buffer 2.
10. Add Secure Chamber against the array’s Agilent side, sealing the edges by sliding the array on the heat block.
11. Seal the port farthest from the Agilent label with a Secure Seal dot.
12. Fill the array with the preheated 95°C EB (approximately 800 μL, prevent leaks and bubbles).
13. Seal the second port with a Secure Seal dot (leave overhang to facilitate removal).
14. Place on heat block at 95°C for 5 minutes.
15. Remove the Secure Seal and aspirate eluant into 2 tubes (approximately 400 μL each).
16. Refill Secure Chamber, re-tab Secure Seal, and repeat Steps 10–15 for Elution 2.
Ethanol Precipitation
Materials Required
100% cold ethanol
70% cold ethanol
Glycogen (5mg/ml)
Ammonium acetate NH4OAC (7.5 M)
4°C Microcentrifuge
Speed Vacuum
Protocol
Add the solutions described in Table 7 to each tube. Each tube will have an approximate final volume of 1.5ml.
Place tubes at −80°C for 90 minutes to overnight.
Spin in a 4°C mini-centrifuge at maximum speed for 30 minutes.
Remove supernatant.
Add 750 μl of 75% cold ethanol and spin at maximum speed for 5 minutes.
Remove supernatant (750 μl) and repeat Step 5.
Vacuum concentrate samples at 30°C for 15 minutes or until dried.
Resuspend pellet in 10μL EB for 30 minutes, vortex and spin down.
Pool both elution tubes into one (20 μL per sample/elution).
Freeze elution 2 as a back up sample and continue with elution 1.
Table 7.
Ethanol Precipitation Recipe (per elution)
| Component | Volume |
|---|---|
| 1/10 vol NH4OAC (7.5 M) | 40 μL |
| 1/100 vol glycogen (5mg/ml) | 4 μL |
| 2.5 vol 100% cold ethanol | 1000 μL |
| TOTAL | 1044 μL |
Amplify Captured DNA by Real-time PCR
Material Required
EXOME_SEQ_ FWD (10μM)
EXOME_SEQ_ REV (10μM)
2x iTaq Supermix
Agencourt AMPure XP
Real Time Thermal Cycler (e.g. Bio Rad)
Magentic Plate
Prepare a master mix based on Table 8, scaling the volumes for the number of samples being prepared plus a no template control.
Aliquot 30 μL of master mix into each a well containing 20 μL of water for the no template control.
Combine the master mix with the DNA sample, and aliquot to wells. Each well will contain 50 μL of solution.
-
Run samples in a RT-PCR thermal cycler with the following program:
95°C, 5:00
95°C, 0:30
55°C, 2:00
72°C, 2:00
Read plate
72°C, 0:15
Go to Step b, 29 cycles
72°C, 10:00
16°C, forever
-
Watch the amplification profile and remove samples from the instrument when the exponential curve reaches a plateau.
This program is designed so that a sample can be taken out mid-run immediately after a plate read. Step f allows the application to pause the run to take the sample out before cycling around and denaturing the sample. This program is designed to a total of 30 cycles to allow all possible amplification scenarios. Elution 1 samples should peak around cycle 15 and elution 2 samples around cycle 20.
Transfer 90 μL of room temperature AMPure beads to each well containing the RT amplified DNA sample, mix well and incubate for 5 minutes at room temperature.
Place the reaction plate onto a magnet plate for 2 minutes to separate beads from the solution. Visually confirm that the beads have moved to the side of the plate. Aspirate the 140 μL of clear solution from the reaction plate and discard.
Dispense 200 μL of 70% ethanol to each well of the reaction plate and incubate for 30 seconds at room temperature. Aspirate out the ethanol and discard. Repeat for a total of two washes.
Take the reaction plate off the magnet plate, add 32 μL of ultrapure water to each well and mix. Place the reaction plate onto the magnet plate for 1 minute to separate the beads. Transfer the 32 μL of supernatant to a new standard 96 well PCR plate.
Table 8.
Master mix for Pre-sequencing Amplification (per sample)
| Component | Volume |
|---|---|
| Water | 3 μL |
| 2x iTaq Supermix | 25 μL |
| EXOME_SEQ_ FWD [10μM →200nM] | 1 μL |
| EXOME_SEQ_ REV [10μM →200nM] | 1 μL |
| TOTAL | 30 μL |
Quality Control Assay – Bioanalyzer for sample size and concentration
-
Run the sample on the Agilent DNA 1000 bioanalyzer.
Note the average base pair size, the concentration of the sample as it will be important for sequencing calculations. Also look for adaptor and overamplification artifacts.
Add 3 μL 1% Tween to the finished library. The sample is now ready for sequencing.
Commentary
Background Information
With the development of next-generation sequencing platforms, the per-base cost of sequencing has dropped by several orders of magnitude. Several different capture strategies exist that allow for enrichment of mammalian genomes for regions of interest. These methods can be broadly divided into those that rely on an enzymatic step to achieve specificity (i.e. molecular inversion probes, multiplex PCR), and those that rely purely on hybridization to oligos complementary to sequences of interest. Hybridization-based methods can be further divided into in-solution and on-array methods. Performance parameters of relevance include input DNA requirements, capture uniformity, specificity, allelic bias, maintenance of sample complexity, scalability and cost (9).
The success of this array capture protocol relies on a strategy that carries forward the entire sample. Unlike traditional molecular biology protocols, where aliquots of sample are loaded to PCR, for example, the entire reaction volume is used for all steps to maintain final library complexity. In general, this protocol could be adapted to work with alternative sequencing methods by modifying the adaptor and PCR primer sequences, and with alternative capture reagents, by adjusting hybridization conditions to reflect the volume and temperature needs of different platforms.
Critical Parameters
Start input DNA
Currently 5μg is the lowest gDNA input recommended. If 10 μg is available, better results may be obtained using this higher amount. The protocol for 10 μg would be the same, with the exception of end repair. The end repair reaction is optimized for 5 μg; doubling the input DNA would require doubling the end repair reaction.
Reagent Purity – (Reagent Aliquots and Cross contamination)
Reagent purity and avoiding cross contamination are crucial factors of this protocol. Aliquot all reagents to single-use volumes and use PCR grade water.
There are multiple amplifications steps in this protocol and micrograms of DNA for a given sample are passed from one step to the next. When capturing multiple samples at once, samples must be handled carefully. Leave blank wells on agarose gels and between samples on plates where possible.
Magnetic Beads vs. Qiagen Columns
AMPure magnetic beads and Qiagen columns can be used interchangeably as purification steps without a significant difference in concentration yield. In a high-throughput setting, beads are cost- and time-efficient. However, magnetic beads require a minimum elution volume of 30μl. Since AMPure purification calls for 1.8μl of beads per 1μl of reaction, a 70 μl reaction is the maximum volume that can be completed in a standard PCR plate. Half skirted 300 μl plates are a solution; however, half skirted plates cannot be gripped or moved on some robotic platforms.
Adaptor Contamination
Achieving a robust T4 ligase reaction in the adaptor ligation is a key step. Unligated adaptors can be carried through to sequencing, reducing useful data output. If adaptor bands remain present in the final sample, titrating the adaptor ligation reaction can be helpful.
The blockers used during the array hybridization are designed with the purpose of blocking adaptor and primer sequences, and their inclusion has been demonstrated to be critical. We speculate that they prevent common adaptor sequences of different fragments in the genomic library from annealing to each other, i.e. “daisy-chaining”, which would be predicted to reduce specificity.
Array Handling
When eluting samples off of arrays controlling the heat block to 95°C with a thermometer is crucial, especially if using a digital heat block with a built in thermometer. If using blocks for micro-centrifuge tubes or strip-tubes, partially fill the wells with distilled water to create steam contact with the array.
Overamplification – RT PCR Machine with Pause Feature vs. Thermocycler
The PCR amplifications are key steps in the protocol that determine the sequence complexity of the samples. This protocol employs a “no molecule left behind strategy”, in which the entire sample mixture is carried forward into each PCR to ensure maximal library complexity. Overamplifying the sample will result in an over representation of some sequences. This artifact may create a bias in the library, thus necessitating more sequencing then would be otherwise required. Overamplification also makes it more difficult to quantify samples, as some fraction of the amplified product remains single stranded.
Running samples on a real-time PCR machine offers the advantage of visually verifying amplification based on individual sample curves. When the sample curve hits a plateau, the sample must be taken out at the end of that cycle. Real-time PCR machines with a pause feature where the lid can be opened, allowing removal of the sample mid-run. Some real-time machines have a delayed real-time analysis and will not be useful for stopping a sample from overamplification.
Samples that are captured in parallel through under the same conditions should behave within a few cycles from each other. Once the number of cycles is optimized for a specific input gDNA extraction method, a standard thermal cycler can replace the RT-PCR machine. When using 5 μg of gDNA the amplification should peak, on average, at 15 cycles.
Bioanalyzer vs. Gel and Quantification
Analysis of the library by Bioanalyzer allows direct determination of the DNA library insert size range and concentration, and is the recommended quality control method. However, a combination of gel electrophoresis and DNA quantitation (eg Nanodrop or Picogreen) methods could be used to give a similar, albeit less precise, quality control result.
Troubleshooting
The key failure points in this protocol are loss of DNA during the library preparation steps, and poor recovery during array elution. Care should be taken to ensure that elutions off AMPure or Qiagen columns are performed carefully. Spot-checking DNA concentrations throughout the protocol can help identify areas of loss. During array elution, it is important to heat the array and elution buffer to 95°C. Many heat blocks do not maintain temperatures evenly over long periods of time; frequent verification of the temperature will ensure full denaturation of the sample from the array. Small bubbles should be visible during the elution; turning and tapping the array during elution steps will ensure the entire surface is reached by the elution buffer. Care should be taken to avoid touching the active surface (Agilent label side) with fingertips or pipette tips. Any contact will result in the loss of DNA captured by probes in the affected region.
Anticipated Results
Assuming a starting input of 5 μg high-quality genomic DNA, the shotgun library preparation (Fragmentation – Bulk PCR) should produce 10–40 μg of library for hybridization to the array. The final PCR should produce ~30 ng/μL of product, which should be sufficient for multiple sequencing lanes.
Acknowledgments
Support for the development of this protocol was provided in part by grants from the NHLBI (5R01HL094976 to JS and DAN) and NHGRI (5R21HG004749 to JS).
References
- 1.Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X, Richmond TA, Middle CM, Rodesch MJ, Packard CJ, Weinstock GM, Gibbs RA. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4:903–5. doi: 10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- 2.Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, Gabriel S, Jaffe DB, Lander ES, Nusbaum C. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–9. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hodges E, Rooks M, Xuan Z, Bhattacharjee A, Benjamin Gordon D, Brizuela L, Richard McCombie W, Hannon GJ. Hybrid selection of discrete genomic intervals on custom-designed microarrays for massively parallel sequencing. Nat Protoc. 2009;4:960–74. doi: 10.1038/nprot.2009.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hodges E, Smith AD, Kendall J, Xuan Z, Ravi K, Rooks M, Zhang MQ, Ye K, Bhattacharjee A, Brizuela L, McCombie WR, Wigler M, Hannon GJ, Hicks JB. High definition profiling of mammalian DNA methylation by array capture and single molecule bisulfite sequencing. Genome Res. 2009;19:1593–605. doi: 10.1101/gr.095190.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hodges E, Xuan Z, Balija V, Kramer M, Molla MN, Smith SW, Middle CM, Rodesch MJ, Albert TJ, Hannon GJ, McCombie WR. Genome-wide in situ exon capture for selective resequencing. Nat Genet. 2007;39:1522–7. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- 6.Okou DT, Steinberg KM, Middle C, Cutler DJ, Albert TJ, Zwick ME. Microarray-based genomic selection for high-throughput resequencing. Nat Methods. 2007;4:907–9. doi: 10.1038/nmeth1109. [DOI] [PubMed] [Google Scholar]
- 7.Porreca GJ, Zhang K, Li JB, Xie B, Austin D, Vassallo SL, LeProust EM, Peck BJ, Emig CJ, Dahl F, Gao Y, Church GM, Shendure J. Multiplex amplification of large sets of human exons. Nat Methods. 2007;4:931–6. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- 8.Turner EH, Lee C, Ng SB, Nickerson DA, Shendure J. Massively parallel exon capture and library-free resequencing across 16 genomes. Nat Methods. 2009;6:315–6. doi: 10.1038/nmeth.f.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turner EH, Ng SB, Nickerson DA, Shendure J. Methods for genomic partitioning. Annu Rev Genomics Hum Genet. 2009;10:263–84. doi: 10.1146/annurev-genom-082908-150112. [DOI] [PubMed] [Google Scholar]