Abstract
The balance between cell cycle progression and apoptosis is important for both surveillance against genomic defects and responses to drugs that arrest the cell cycle. In this report, we show that the level of the human anti-apoptotic protein Mcl-1 is regulated during the cell cycle and peaks at mitosis. Mcl-1 is phosphorylated at two sites in mitosis, Ser64 and Thr92. Phosphorylation of Thr92 by cyclin-dependent kinase 1 (CDK1)–cyclin B1 initiates degradation of Mcl-1 in cells arrested in mitosis by microtubule poisons. Mcl-1 destruction during mitotic arrest requires proteasome activity and is dependent on Cdc20/Fizzy, which mediates recognition of mitotic substrates by the anaphase-promoting complex/cyclosome (APC/C) E3 ubiquitin ligase. Stabilisation of Mcl-1 during mitotic arrest by mutation of either Thr92 or a D-box destruction motif inhibits the induction of apoptosis by microtubule poisons. Thus, phosphorylation of Mcl-1 by CDK1–cyclin B1 and its APC/CCdc20-mediated destruction initiates apoptosis if a cell fails to resolve mitosis. Regulation of apoptosis, therefore, is linked intrinsically to progression through mitosis and is governed by a temporal mechanism that distinguishes between normal mitosis and prolonged mitotic arrest.
Keywords: apoptosis, Cdc20, cyclin-dependent kinase, Mcl-1, mitosis
Introduction
The fidelity of the cell cycle is monitored by checkpoint mechanisms that arrest cell cycle progression if a crucial process has not been completed successfully (Hartwell, 1992). During mitosis, chromosome segregation is restrained by the mitotic or spindle assembly checkpoint (SAC) until all kinetochores are properly attached to polar microtubules (Musacchio and Salmon, 2007). Otherwise, mitosis has the potential to result in chromosome missegregation and aneuploidy, which may promote tumour development in metazoans (Holland and Cleveland, 2009). If mitosis is not resolved successfully, then initiation of cell death through the process of apoptosis can kill a defective cell. The balance between these fates is likely to be important in determining the cellular response to anti-cancer drugs such as taxanes and vinca alkaloids, which disrupt spindle assembly, cause prolonged SAC activation and initiate apoptosis (Rieder and Maiato, 2004; Gascoigne and Taylor, 2009). The molecular mechanisms that determine this balance are still poorly understood. In particular, it is not clear how mechanisms controlling the initiation of apoptosis distinguish between normal mitosis and prolonged mitotic arrest (Clarke and Allan, 2009).
The SAC restrains progression through mitosis by inhibition of the ubiquitin ligase activity of the anaphase-promoting complex/cyclosome (APC/C) towards certain substrates (Acquaviva and Pines, 2006). These substrates include securin, which controls sister chromatid separation (Nasmyth et al, 2000), and cyclin B1, the regulatory subunit of the mitosis-promoting factor, cyclin-dependent kinase 1 (CDK1)–cyclin B1 protein kinase (Acquaviva and Pines, 2006). The SAC restrains the APC/C by inhibiting the function of Cdc20 (Fizzy), which has a critical role in the recognition of mitotic substrates by the APC/C and also activates its ubiquitin ligase activity (Yu, 2007). When proper attachment of microtubules to kinetochores is accomplished, the SAC is satisfied and the ubiquitin ligase activity of APC/C initiates the polyubiquitination and degradation of structural and regulatory proteins through the proteasome (Musacchio and Salmon, 2007). Securin degradation releases separase, which cleaves cohesin molecules that hold sister chromatids together, causing chromatid disjunction (Nasmyth et al, 2000). Degradation of cyclin B1 and other regulators causes irreversible exit from mitosis (Pines, 2006). However, the degradation of some other mitotic substrates of the APC/CCdc20, such as cyclin A, is not prevented by the SAC, and cyclin A destruction begins immediately after nuclear envelope breakdown at prometaphase (den Elzen and Pines, 2001; Geley et al, 2001).
Initiation of apoptosis in response to many stimuli, including microtubule poisons, requires the cysteine protease, caspase-9 (Hakem et al, 1998; Kuida et al, 1998; O'Connor et al, 2002; Janssen et al, 2007). Caspase-9 activation is induced by the release of cytochrome c from mitochondria, a process that is tightly controlled by pro-apoptotic and anti-apoptotic proteins that contain domains related to Bcl-2 (Youle and Strasser, 2008). Once activated, caspase-9 cleaves and activates the effector caspases-3 and -7, which target a variety of cellular components to dismantle a cell and present the fragments for phagocytosis (Taylor et al, 2008). We have shown that apoptosis is temporarily restrained during mitotic arrest through phosphorylation of caspase-9 by CDK1–cyclin B1. Abolition of caspase-9 phosphorylation by mutation of the inhibitory phosphorylation site, Thr125, accelerates the induction of downstream caspase-3 activation and apoptosis (Allan and Clarke, 2007). These results suggest that prolonged mitotic arrest causes activation of the mitochondrial apoptotic pathway upstream of caspase-9, which eventually initiates apoptosis when caspase-9 is dephosphorylated or the threshold to initiate downstream caspase activation is overcome (Allan and Clarke, 2008).
Regulation of apoptosis during mitotic arrest is unlikely to be controlled by transcriptional induction, although it could be affected by the widespread shutdown of transcription during mitosis (Blagosklonny, 2007). Apoptotic regulators might also be controlled at the translational level (Marash et al, 2008) or post-translationally through their modification or degradation. We hypothesised that apoptosis could be initiated during mitotic arrest by the timed degradation of an inhibitor of apoptosis that acts upstream of caspase-9. Good candidates are anti-apoptotic proteins of the Bcl-2 family, in particular Mcl-1 (Kozopas et al, 1993), which is relatively unstable (Nijhawan et al, 2003). Mcl-1 has a critical function in the control of apoptosis induced by many stimuli, including UV irradiation (Nijhawan et al, 2003), and is over-expressed in some human cancers (Michels et al, 2005). In interphase cells, Mcl-1 is degraded in response to cellular stresses by an ubiquitin–proteasome-mediated mechanism that involves the E3 ubiquitin ligases, Mule/ARF-BP1 (Zhong et al, 2005) and SCFβTrCP (Ding et al, 2007), which are opposed by the deubiquitinase USP9X (Schwickart et al, 2010). Phosphorylation of Mcl-1 at Ser159 by GSK-3 in response to growth factor (IL-3) withdrawl promotes Mcl-1 ubiquitination and degradation (Maurer et al, 2006). Recognition of Mcl-1 by GSK-3 requires a priming kinase, ERK (Domina et al, 2004; Ding et al, 2008) or JNK (Inoshita et al, 2002; Morel et al, 2009), which targets Thr163. Mcl-1 has also been reported to be phosphorylated in mitosis (Domina et al, 2004; De Biasio et al, 2007), but the regulation of Mcl-1 stability during the cell cycle and its possible function in controlling apoptosis during mitotic arrest have been unclear.
Here, we show that the initiation of apoptosis during a prolonged mitotic arrest is determined by Mcl-1 instability, which is controlled by a mechanism distinct from that operating in interphase. Proteasome-dependent destruction of Mcl-1 during mitotic arrest requires phosphorylation of a critical site, Thr92, by CDK1–cyclin B1 and is mediated by APC/CCdc20. Stabilisation of Mcl-1 by mutation of either Thr92 or a putative destruction box (D-box) makes cells resistant to apoptosis induced by prolonged mitotic arrest. This work identifies a direct link between the regulation of mitosis and the temporal control of apoptosis that is determined by the differential timing of substrate destruction through APC/CCdc20.
Results
Mcl-1 protein levels are regulated during the cell cycle
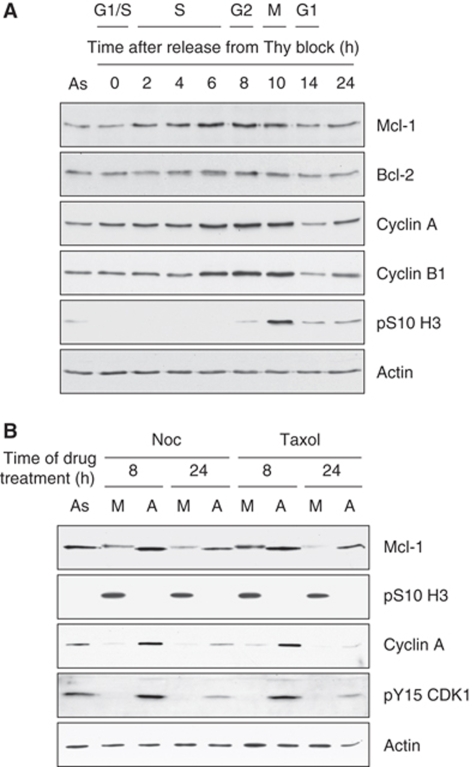
To study the regulation of the level of Mcl-1 protein during the cell cycle, human osteosarcoma U2OS cells were synchronised at the G1/S boundary using a double thymidine block, then released into the cell cycle. Cells reached mitosis at around 10 h, as determined by mitotic phosphorylation of histone H3 at Ser10 (Figure 1A). We found that Mcl-1 protein levels increased during S phase and G2, peaked in mitosis and then returned to baseline levels in G1, mirroring the cell cycle profiles of the mitotic cyclins A and B1. In contrast, other apoptotic regulators such as Bcl-2 (Figure 1A) and Bcl-xL (data not shown) did not show regulation of their levels during the cell cycle. These results suggested a specific function for Mcl-1 in the regulation of apoptosis during mitosis. Indeed, ablation of Mcl-1 by siRNA promoted apoptosis in U2OS cells treated with nocodazole, which inhibits microtubule polymerisation and arrests cells in mitosis (Supplementary Figure 1).
Figure 1.
Mcl-1 protein levels are regulated through the cell cycle. (A) Mcl-1 levels increase from G1 through to mitosis. Human U2OS cells were synchronised at the G1/S boundary by double thymidine block then released into the cell cycle. Samples were analysed by SDS–PAGE and immunoblotting using the specified antibodies at the times shown after release from the block. The predominant cell cycle phase at each time was confirmed by FACS analysis in parallel experiments (data not shown) as in Bennett and Clarke (2006). (B) Mcl-1 protein levels decrease in response to anti-mitotic drugs. Human U2OS cells growing in asynchronous (As) culture were treated with 200 ng/ml nocodazole (Noc) or 0.5 μM taxol for 8 or 24 h. Floating, mitotic cells (M) were separated from adherent, interphase cells (A) at each time and samples were analysed by immunoblotting using the specified antibodies.
In U2OS cells treated with nocodazole or taxol (which stabilises microtubules and also arrests cells in mitosis), there was a decline in Mcl-1 levels by 8 h in rounded-up, mitotically arrested cells (Figure 1B). The remaining Mcl-1 protein had reduced mobility on SDS–PAGE, indicating a post-translational modification, most likely phosphorylation. After 24 h, Mcl-1 was almost completely lost in rounded-up cells, particularly with taxol treatment. Mcl-1 was not modified and levels were more stable in drug-treated, adherent interphase cells, which were distinguished by the presence of cyclin A, inhibitory phosphorylation of CDK1 at Tyr15 and the lack of mitotic histone H3 phosphorylation. Thus, the effect of the drugs on Mcl-1 was due to their induction of mitotic arrest and not some other stress response.
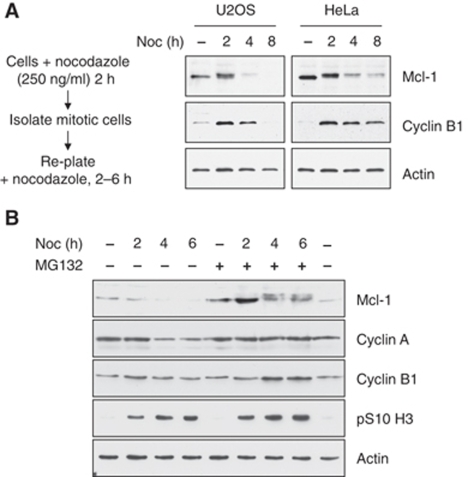
Since cells in an initially asynchronous culture treated with microtubule poisons will be arrested in mitosis for widely differing times depending on which stage of the cell cycle they are in when the treatment is started, we used a different protocol to study in more detail the temporal regulation of Mcl-1 during a mitotic arrest (Figure 2). Rounded-up mitotic cells were first collected after 2 h treatment with nocodazole, then re-plated in nocodazole-containing media such that the cells are near-synchronously arrested in mitosis. We confirmed that 75% or more of the cells collected by this protocol were mitotic by analysis of the phosphorylation of histone H3 at serine 10 using flow cytometry (Supplementary Figure 2). In either U2OS or HeLa cells arrested for up to 2 h, Mcl-1 was still present and mostly upshifted (suggesting it was phosphorylated), but at 4 h, the majority had disappeared and Mcl-1 was almost completely absent at 8 h (Figure 2A). In contrast, the levels of Bcl-2 and Bcl-xL were unchanged during the arrest. Although both of these proteins were highly phosphorylated, as indicated by a reduction in their mobility on SDS–PAGE, consistent with earlier reports (Scatena et al, 1998; Du et al, 2005; Terrano et al, 2010), this also did not change during the period of the arrest (Supplementary Figure 3).
Figure 2.
Mcl-1 is degraded by a proteasome-dependent mechanism after 2 h of mitotic arrest. (A) Loss of Mcl-1 in human cells arrested in mitosis. U2OS and HeLa cells were synchronised in mitotic arrest using the protocol shown. Samples incubated for a total of 2, 4 or 8 h in 250 ng/ml nocodazole were analysed by immunoblotting. (B) Loss of Mcl-1 during mitotic arrest is inhibited by the proteasome inhibitor, MG132. Cells were treated with 10 μM MG132 for 2 h before arrest in mitosis as in (A). Samples were immunoblotted with the antibodies shown.
The loss of Mcl-1 in U2OS cells arrested in mitosis for 4 h was not affected by the caspase inhibitor Z-VAD-FMK (Supplementary Figure 4), showing that it was not a consequence of apoptosis, but it was inhibited by MG132, a proteasome inhibitor. Mcl-1 that had reduced mobility on SDS–PAGE (and, therefore, was putatively phosphorylated) was present in MG132-treated cells even after 6 h nocodazole treatment (Figure 2B). As expected, MG132 also stabilised the levels of cyclin A, which is otherwise degraded in cells arrested in mitosis (den Elzen and Pines, 2001; Geley et al, 2001). MG132 also enhanced the amount of cyclin B1 in cells arrested for a prolonged period in mitosis by treatment with nocodazole for 4–6 h. This effect is probably due to inhibition of the slow degradation of cyclin B1 that occurs during mitotic arrest (Brito and Rieder, 2006).
Mcl-1 is phosphorylated at two sites in mitosis
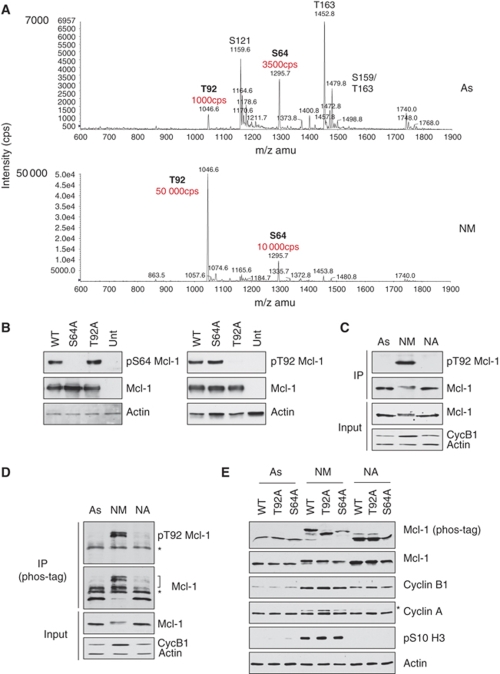
To characterise the cell cycle-dependent phosphorylation of Mcl-1, the Flag-tagged protein was expressed in HeLa cells, which were then treated with nocodazole for 3 h. Flag-Mcl-1 was immunoprecipitated from asynchronous or mitotic nocodazole-treated cells and analysed by mass spectroscopy of tryptic peptides (Figure 3A). Flag-Mcl-1 was phosphorylated at five sites in asynchronous cells: Ser64, Thr92, Ser121, Ser159 and Thr163. Ser159 was only found in conjunction with Thr163, consistent with a requirement for priming phosphorylation at Thr163 for phosphorylation of Ser159 by GSK-3 (Maurer et al, 2006; Morel et al, 2009). Ser121 has been described as a site phosphorylated together with Thr163 by JNK in response to oxidative stress (Inoshita et al, 2002), whereas Ser64 has been identified as phosphorylated in cells during G2 and/or mitosis (Kobayashi et al, 2007). Thr92 is phosphorylated by ERK in vitro (Ding et al, 2008), but its phosphorylation in cells has not been described earlier. Of these five sites, only Ser64 and Thr92 were detected in cells arrested in mitosis by nocodazole. Phosphorylation of the tryptic peptide-containing Ser64 increased about three-fold in the mitotic sample, whereas the phosphorylated Thr92 peptide increased 50-fold and was five times more abundant than the Ser64 peptide, suggesting that Thr92 is the major site in Mcl-1 that is phosphorylated specifically in mitosis (Figure 3A).
Figure 3.
Mcl-1 is phosphorylated at two sites in mitosis. (A) Flag-Mcl-1 was immunoprecipitated from HeLa cells growing in asynchronous culture (As) or treated with nocodazole for 3 h, followed by collection of the rounded-up, mitotic cells (NM). Tryptic peptides were analysed by mass spectrometry. The relative abundance of the phosphopeptides containing the Thr92 and Ser64 sites are highlighted. (B) Specificity of antibodies generated against Mcl-1 phosphorylation sites. HeLa cells were transiently transfected with either wild-type Mcl-1 (WT) or the phosphorylation site mutants S64A or T92A. After treatment with 250 ng/ml nocodazole for 2 h, samples were analysed by SDS–PAGE and immunoblotting with antibodies raised either against the phosphorylated Ser64 (left) or Thr92 (right) sites. (C, D) Mcl-1 is phosphorylated at Thr92 in mitosis. Endogenous Mcl-1 was immunoprecipitated from asynchronous (As) HeLa cells or after treatment with 100 ng/ml nocodazole for 2 h before separation of mitotic (NM) and adherent cells (NA). Cell lysates (Input) were analysed by conventional SDS–PAGE. Immunoprecipitates were analysed by conventional SDS–PAGE (C) or Phos-tag SDS–PAGE (D) and immunoblotted for Mcl-1 phosphorylated at Thr92 or total Mcl-1. Phosphorylated Mcl-1 is indicated by a bracket. *Indicates the heavy chain of the precipitating antibody. (E) Mcl-1 is highly phosphorylated at Thr92 and Ser64 during mitotic arrest. U2OS cells stably transfected with either wild-type Mcl-1 (WT), T92A or S64A mutants were untreated (As) or treated for 2 h with nocodazole before separation of mitotic (NM) and adherent cells (NA). Cell lysates were analysed by conventional SDS–PAGE or Phos-tag SDS–PAGE as indicated and immunoblotted with the specified antibodies. *Indicates a band corresponding to cyclin B1 caused by reprobing of the same blot used to identify cyclin B1.
We generated polyclonal antibodies against phosphopeptides containing either the Ser64 or Thr92 sites and validated their specificity (Supplementary Figure 5). The antibodies recognised Flag-Mcl-1 from mitotically arrested HeLa cells when phosphorylated at Ser64 or Thr92, respectively, as recognition of Flag-Mcl-1 was abolished by mutation of the specific residue to alanine. Phosphorylation of Thr92 was not affected by mutation of Ser64 and vice versa, indicating that each site was phosphorylated independently (Figure 3B). We confirmed that Mcl-1 was phosphorylated at Ser64 and Thr92 during mitosis in the absence of microtubule poisons using U2OS cells that were stably transfected with Flag-Mcl-1 and synchronised in the cell cycle (Supplementary Figure 6). Furthermore, we were able to show that immunoprecipitated endogenous Mcl-1 was phosphorylated at Thr92 in both mitotically arrested cells and in untreated mitotic cells, but not in interphase cells (Figure 3C; Supplementary Figure 7). Figure 3D shows that endogenous Mcl-1 immunoprecipitated from mitotically arrested cells migrated as three phosphorylated forms on a Phos-tag gel, which specifically retards the migration of phosphoproteins (Kinoshita et al, 2006). The upper two bands were detected by the pT92 Mcl-1 antibody and accounted for ∼50% of the total Mcl-1. This indicates that Mcl-1 is phosphorylated to a high level at Thr92 in mitotically arrested cells. Analysis of stably transfected Mcl-1 proteins on a Phos-tag gel (Figure 3E) showed that mutation of T92 or S64 to alanine produced distinct single bands that migrated in retarded positions, showing that both residues are highly phosphorylated under these conditions.
CDK1–cyclin B1 phosphorylates Mcl-1 at Thr92 directly and is required for phosphorylation at Ser64 in mitosis
To identify the kinase(s) that phosphorylate(s) Mcl-1 at Thr92 and Ser64, we used a cell-free system derived from mitotic HeLa cells (Hutchins et al, 2004). Recombinant Mcl-1-His6 was phosphorylated at Thr92 and Ser64 (detected by the specific antibodies) in nocodazole-treated mitotic cell extracts. Mitotic Mcl-1 phosphorylation at both sites was inhibited by either of the CDK inhibitors, roscovitine or purvalanol A (Figure 4A), but not by the MEK1/2 inhibitor, UO126, which blocks the ERK1/2 MAPK pathway, nor the p38 inhibitor, SB203580. SP600125 also partially inhibited phosphorylation at these sites; this reagent inhibits JNK, but also other kinases, including CDKs (Bain et al, 2003). As the mitotic extracts contained cyclin B1, but not cyclin A, these results are consistent with a dependency on CDK1–cyclin B1, the major mitotic CDK, for the phosphorylation of Mcl-1 at Thr92 and Ser64.
Figure 4.
Thr92 of Mcl-1 is phosphorylated directly by CDK1–cyclin B1, whereas Ser64 phosphorylation is dependent on CDK1, but may be indirect. (A) The CDK inhibitors roscovitine and purvalanol A abrogate phosphorylation of Thr92 and Ser64. Mcl-1-His6 was incubated for 1 h with extracts derived from HeLa cells either in asynchronous culture (As) or arrested in mitosis with nocodazole (N). Samples were analysed by SDS–PAGE and immunoblotting with the specified antibodies. (B) Depletion of CDK1–cyclin B1 from mitotic HeLa cell extracts inhibits phosphorylation of Mcl-1-His6 at Thr92 and Ser64. Mcl-1-His6 was incubated for 1 h with extracts depleted of CDK1–cyclin B1 by p13Suc1 beads. Purified recombinant CDK1–cyclin B1 was added back to depleted extracts to restore kinase activity. (C) Purified recombinant CDK1–cyclinB1 directly phosphorylates Mcl-1-His6 at Thr92, but not at Ser64. Mcl-1-His6 was incubated for 1 h with CDK1–cyclin B1 or mitotic extract made from nocodazole-treated cells. (D) Active CDK1 immunoprecipitated from HeLa cells arrested in mitosis with nocodazole (Noc) phosphorylates Mcl-1-His6 on Thr92 (lower panels). Mock immunoprecipitations were performed with an isotype-matched control antibody. Total cell lysates (Input) used for the precipitations are shown in the upper panels.
Depletion of CDK–cyclin complexes from mitotic extracts using p13suc1 beads resulted in inhibition of phosphorylation at Thr92 and Ser64 (Figure 4B). Similarly, the phosphorylation of Mcl-1 at Thr92 and Ser64 was abrogated in mitotic extracts depleted using anti-cyclin B1 antibodies (data not shown). In p13suc1-depleted extracts, addition of active recombinant CDK1–cyclin B1 fully restored phosphorylation of Mcl-1 at Thr92, and partially restored phosphorylation at Ser64 (Figure 4B). Incubation of Mcl-1-His6 with purified recombinant CDK1–cyclin B1 in the absence of mitotic extracts confirmed that Thr92 was phosphorylated directly by this kinase, whereas Ser64 was not (Figure 4C). This was corroborated when CDK1 immunoprecipitated from mitotic HeLa cells was incubated with Mcl-1-His6, resulting in phosphorylation of Thr92 (Figure 4D). Thus, CDK1–cyclin B1 is directly responsible for phosphorylation of Mcl-1 at Thr92. Ser64 phosphorylation is dependent on CDK1–cyclin B1, but requires an additional component of the mitotic extracts.
Phosphorylation of Thr92 is required for the mitotic degradation of Mcl-1
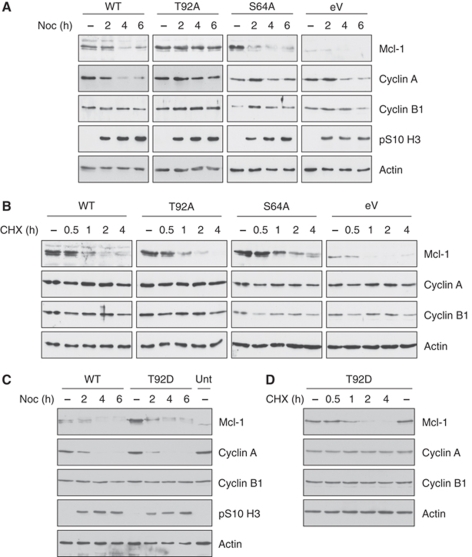
To investigate the function of the mitotic phosphorylation of Mcl-1, we established cell lines derived from U2OS cells that were stably transfected with Flag-tagged wild-type Mcl-1 (U2-Mcl-1-WT), or alanine mutants of Thr92 (U2-Mcl-1-T92A) or Ser64 (U2-Mcl-1-S64A). These cells were selected because they express similarly low levels of transfected Mcl-1. Cells were also derived that were transfected with an empty vector control (U2-eV). As before, we treated the cells with nocodazole for 2 h and re-plated the mitotic cells for a further 2 or 4 h (Figure 5A). The transfected Flag-Mcl-1 in U2-Mcl-1-WT was degraded in the mitotic block with a similar time course to the endogenous Mcl-1 protein in U2-eV cells. Flag-Mcl-1-S64A was also unstable in mitotically arrested U2-Mcl-1-S64A cells. In contrast, the degradation of Flag-Mcl-1 T92A was blocked in the U2-Mcl-1-T92A cells, showing that the phosphorylation of Thr92 is required specifically for the mitotic degradation of Mcl-1. Identical results were obtained with different clones of each cell type. Mutation of either Thr92 or Ser64 to alanine did not affect the degradation of Flag-Mcl-1 in interphase cells treated with the protein synthesis inhibitor cycloheximide (Figure 5B). In contrast to T92A, mutation of Thr92 to aspartate, which is predicted to mimic the phosphorylated form, did not stabilise Mcl-1 during mitotic arrest (Figure 5C). The T92D protein was also degraded like the wild-type protein in interphase cells treated with cycloheximide (Figure 5D). The destruction of Mcl-1 during mitotic arrest is, therefore, specifically dependent on Thr92 phosphorylation and is mediated by a mechanism different to that operating in interphase.
Figure 5.
Phosphorylation of Thr92 is required for mitotic destruction of Mcl-1. (A) Mcl-1 T92A is stable in cells arrested in mitosis. U2OS cells stably transfected with either wild-type Mcl-1 (WT), T92A or S64A mutants, or empty vector (eV) were treated for 2 h with nocodazole. Mitotic cells were then isolated and re-plated for a further 2 or 4 h so that cells were arrested in mitosis for a total of 2, 4 or 6 h. (B) The T92A mutant is degraded like wild type and S64A Mcl-1 in interphase. Time course after addition of 40 μg/ml cycloheximide (CHX) to asynchronous cell cultures. (C) Mcl-1 T92D phospho-mimetic mutant is degraded like wild-type Mcl-1 during mitotic arrest. U2OS cells stably transfected with either wild-type Mcl-1 (WT) or T92D mutant were treated as in (A). (D) The T92D mutant is degraded like wild-type Mcl-1 in interphase. Time course after addition of 40 μg/ml cycloheximide (CHX) to asynchronous cell cultures.
The degradation of cyclin A was also delayed in U2-Mcl-1-T92A cells during mitotic arrest (Figure 5A). The level of cyclin B1 in mitotically arrested U2-Mcl-1-T92A cells was also elevated and stabilised compared with U2-Mcl-1-WT, U2-Mcl-1-S64A and U2-eV cells. These results indicate that the mechanism of Mcl-1 destruction is closely related to that of the mitotic cyclins, and suggest that Mcl-1 competes for the same degradation machinery. Strong over-expression of Flag-Mcl-1 by transient transfection resulted in stabilisation of even the WT protein during a prolonged mitotic arrest (data not shown), suggesting that a component of the degradation machinery is limiting under these conditions.
Mcl-1 degradation during mitotic arrest requires Cdc20 and APC3
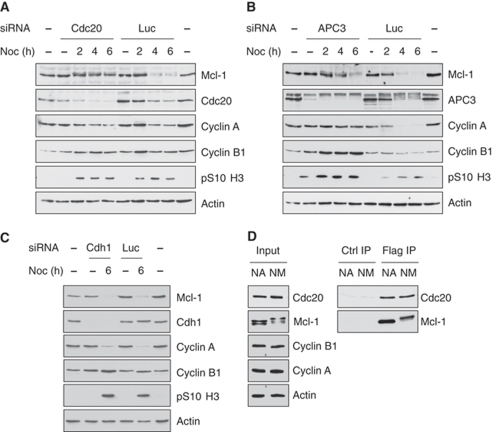
The degradation of mitotic cyclins and other mitotic regulators is mediated by Cdc20 (Fizzy) and the E3 ubiquitin ligase complex APC/C (Yu, 2007). To test the possible function of Cdc20 in the degradation of Mcl-1 during mitotic arrest, we depleted Cdc20 using siRNA in U2OS cells prior to the mitotic arrest (Figure 6A). Cdc20 itself is unstable during mitotic arrest (Nilsson et al, 2008) and the level of Cdc20 was further reduced with 2 h of nocodazole treatment: in control cells, Cdc20 was reduced to a low, basal level by 4 h in nocodazole and was barely detectable in cells treated with the siRNA after this time. The reduction in Cdc20 levels by siRNA completely inhibited the mitotic degradation of endogenous Mcl-1, and the stabilised protein migrated entirely in upshifted forms, whereas the level of Mcl-1 in asynchronous cells was not affected by Cdc20 depletion. Despite the strong reduction of Cdc20 in cells treated with the siRNA, the degradation of cyclin A during mitotic arrest was delayed, but not prevented. This is consistent with the work by Wolthuis et al (2008) showing that a residual level of Cdc20 is sufficient for cyclin A degradation in mitosis. Similarly, depletion of APC3 (CDC27), an APC/C component, which recruits the activators Cdc20 and Cdh1 (Fizzy related) to the complex (Vodermaier et al, 2003), also stabilised Mcl-1 as well as cyclin A and cyclin B1 during mitotic arrest (Figure 6B). In contrast, depletion of Cdh1 (Figure 6C) or Mule (data not shown) failed to stabilise Mcl-1. We found that Cdc20 co-precipitated with Flag-Mcl-1 from both interphase and mitotically arrested cells, indicating that their interaction is independent of mitotic Mcl-1 phosphoryation (Figure 6D). The association of Mcl-1 with Cdc20 is similar to the association between cyclin A and Cdc20, which is also observed in both interphase and mitosis (Wolthuis et al, 2008).
Figure 6.
Destruction of Mcl-1 during mitotic arrest is mediated by Cdc20. (A) Cdc20 is required for Mcl-1 destruction in cells arrested in mitosis. Cdc20 was depleted from U2OS cells using siRNA, then cells were synchronised in a mitotic arrest by treatment with nocodazole for 2 h followed by collection of rounded-up mitotic cells and further treatment with nocodazole for a total of 4 or 6 h. Cells treated with a luciferase siRNA (Luc) were used as a control. (B) APC3 is required for Mcl-1 destruction in cells arrested in mitosis. APC3 was depleted from U2OS cells using siRNA, then cells were synchronised as in (A). Luciferase siRNA (Luc) was used as a control. (C) Depletion of Cdh1 does not prevent Mcl-1 degradation during mitotic arrest. Cdh1 was depleted from U2OS cells using siRNA, then cells were synchronised in mitotic arrest by treatment with nocodazole for 2 h followed by collection of rounded-up mitotic cells and further treatment with nocodazole for a total of 6 h. (D) Cdc20 interacts with Mcl-1 in interphase and mitosis. U2-Mcl-1-WT cells were synchronised by a single thymidine block followed by release for 9 h before addition of nocodazole (100 ng/ml) and MG132 (10 μM) for 3 h. Flag-Mcl-1 was precipitated from mitotic (NM) and adherent (NA) cells using Flag-agarose and immunoblotted for Cdc20 and Mcl-1; HA-agarose was used as a control (Ctrl) for the precipitation (right panels). Cell lysates (Input) used for the precipitations are shown in the left panels.
Mitotic destruction of Mcl-1 requires a putative D-box
Cdc20 recognises proteins through destruction box (D-box) motifs. We scanned the primary sequence of Mcl-1 for a potential D-box and found one region (residues 207–215) with 6 out of 9 residues identical to the D-box of securin (Figure 7A). Within this motif, two residues, an arginine and a leucine in the minimal consensus RxxL, are also conserved in D-boxes from cyclin A and cyclin B, and these residues are essential for mitotic destruction of cyclin B (Glotzer et al, 1991). We also noticed that Mcl-1 carries an isoleucine-arginine motif at its C-terminus, similar to the C-terminal I/M-R motifs found in Cdc20, Cdh1, APC10 and Nek2A (Figure 7B). This motif is required for their interaction with the APC/C through binding to APC3 (Hayes et al, 2006; Kimata et al, 2008). To test the possible function of these motifs in the mitotic destruction of Mcl-1, we generated stable clones of U2OS cells that express a D-box mutant (R207A/L210A) or a mutant lacking the C-terminal isoleucine-arginine motif (ΔIR). We found that the D-box mutant was completely stable during nocodazole-induced mitotic arrest, whereas the ΔIR protein was unstable (Figure 7C). We also confirmed that both the D-box and ΔIR mutants were unstable like the wild type in interphase cells (Figure 7D). The stabilisation of the D-box mutant was not due to a defect in Thr92 phosphorylation, since both the D-box and ΔIR mutants were phosphorylated at this site in mitotically arrested cells (Figure 7E). Interestingly, the mitotic destruction of both cyclin A and cyclin B1 was still slightly delayed in the D-box mutant cells, suggesting that stabilised Mcl-1 interferes with cyclin destruction through a mechanism other than just competition for Cdc20 through its binding to the D-box.
Figure 7.
Mcl-1 contains a D-box motif that is required for its degradation during mitotic arrest. (A) Sequence alignment of putative Mcl-1 D-box motif compared with those from cyclin A, cyclin B1 and securin. The two residues in Mcl-1 mutated to alanine to yield the Mcl-1 D-box mutant (R207A/L210A) are indicated (arrows). (B) Sequence alignment of the C-terminal regions of Cdc20, Cdh1, APC10, Nek2A and Mcl-1. The I/M-R motif is underlined. (C) Mcl-1 D-box mutant is stable in cells arrested in mitosis. U2OS cells stably transfected with either wild-type Mcl-1 (WT), D-box or IR-deletion (ΔIR) mutants were treated for 2 h with nocodazole. Mitotic cells were then isolated and re-plated for a further 2 or 4 h so that cells were arrested in mitosis for a total of 2, 4 or 6 h. (D) Mcl-1 D-box and IR-deletion (ΔIR) mutants are degraded like wild-type Mcl-1 in interphase. Time course after addition of 40 μg/ml cycloheximide (CHX) to asynchronous cell cultures. (E) Mcl-1 D-box and IR-deletion (ΔIR) mutants are phosphorylated at Thr92 like wild-type Mcl-1 during mitotic arrest. U2OS cells stably transfected with either wild-type Mcl-1 (WT), D-box or IR-deletion (ΔIR) mutants were treated for 2 h with nocodazole before collection of mitotic cells (Noc) or left untreated (As). Flag-Mcl-1 was precipitated from cell lysates using Flag-agarose and immunoblotted for Mcl-1 phosphorylated at Thr92 or total Mcl-1. Cell lysates (Input) used for the precipitations are shown in the right panels.
Stabilisation of Mcl-1 during mitotic arrest inhibits apoptosis
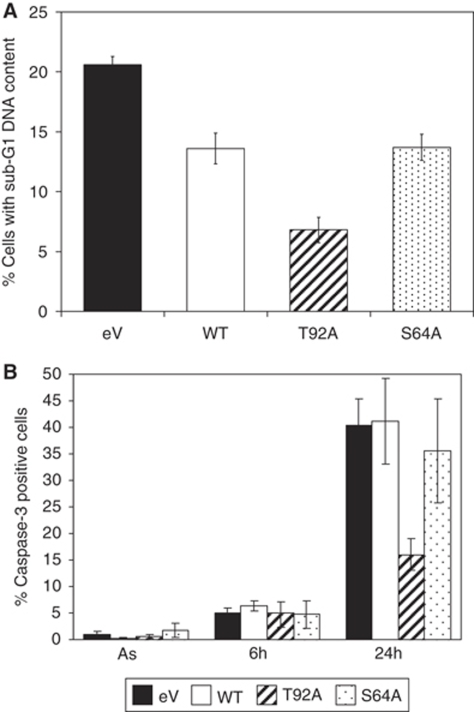
To test the possible function of Mcl-1 degradation during mitotic arrest in determining the sensitivity of cells to the induction of apoptosis, we analysed the response of U2-Mcl-1-WT, U2-Mcl-1-T92A, U2-Mcl-1-S64A and U2-eV cells to prolonged exposure to nocodazole. Analysis of apoptosis by measuring sub-G1 (<2N) DNA content using flow cytometry showed that cells expressing the T92A mutant were significantly more resistant to the induction of apoptosis than cells expressing either WT or S64A Mcl-1 (Figure 8A; Supplementary Figure 8). Analysis by immunofluorescence of individual cells showed that the number of apoptotic cells with active caspase-3 was reduced by ∼60% in U2-Mcl-1-T92A cells (Figure 8B). We also found that stabilisation of Mcl-1 through mutation of the D-box reduced sensitivity to nocodazole-induced apoptosis, whereas the ΔIR mutant had no inhibitory effect, consistent with its inability to prevent Mcl-1 destruction (Supplementary Figure 9). Cells expressing Mcl-1 T92A or the D-box mutant were protected against apoptosis to the same extent (Supplementary Figure 10). These results show that Thr92 phosphorylation and the subsequent destruction of Mcl-1 determines the sensitivity of cells to apoptosis induced by prolonged mitotic arrest.
Figure 8.
Abolition of Mcl-1 Thr92 phosphorylation inhibits apoptosis induced from a mitotic arrest. (A) U2OS cells stably transfected with Mcl-1 (WT, T92A or S64A) or empty vector cells (eV) were treated with nocodazole (250 ng/ml) for 45 h. Cells were analysed for sub-G1 DNA content by flow cytometry (±s.e.m., n=3). Statistical analyses were performed using a two-tailed unpaired Student's t-test. The percentage of cells with sub-G1 DNA content in WT, T92A or S64A cells was significantly different from the empty vector control (P<0.01) and T92A was significantly reduced compared with WT or S64A (P<0.02). (B) U2OS cells stably transfected with Mcl-1 (WT, T92A or S64A) or empty vector cells (eV) were treated with nocodazole (100 ng/ml) for times indicated. Percentage of cells with active caspase-3 is shown (±s.e.m., n=4). After 24 h, the percentage of caspase-3 positive cells was significantly reduced in T92A cells compared with the empty vector control (P<0.01) and WT (P<0.03).
Mcl-1 destruction precedes caspase-9 dephosphorylation during mitotic arrest
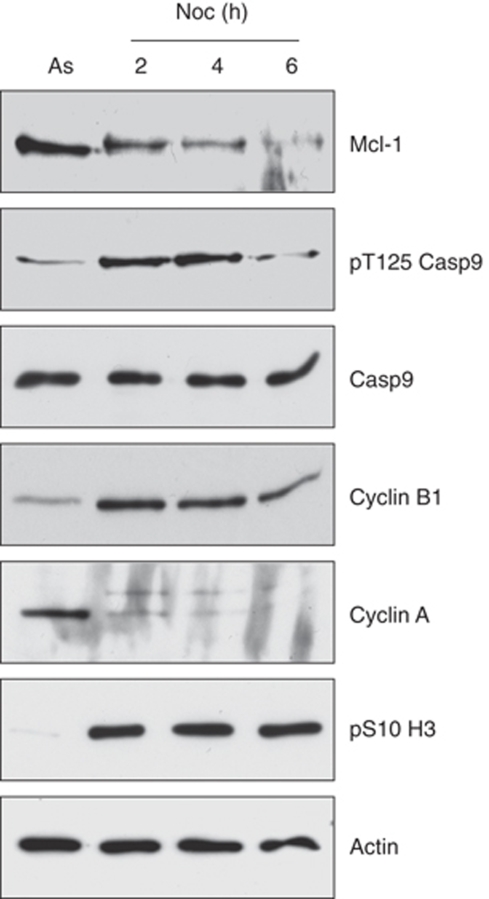
In contrast to the phosphorylation of Mcl-1, which initiates its destruction and promotes apoptosis, we have earlier shown that phosphorylation of caspase-9 at an inhibitory site (Thr125) by CDK1–cyclin B1 restrains the onset of apoptosis during mitotic arrest (Allan and Clarke, 2007). When compared with the levels of Mcl-1 in nocodazole-arrested mitotic cells, we found that the phosphorylation of caspase-9 at Thr125 was maintained until after the destruction of Mcl-1 had begun (Figure 9). Eventually, when cyclin B1 levels declined, caspase-9 was dephosphorylated, which would permit its activation. Thus, there is a temporal series of events that sequentially removes the brakes on the apoptotic pathway during prolonged mitotic arrest.
Figure 9.
The relative timing of Mcl-1 degradation and caspase-9 Thr125 dephosphorylation during mitotic arrest. U2.C9-C287A cells, a derivative of U2OS cells, which over-express an inactive form of caspase-9 (Allan and Clarke, 2007), were synchronised in mitotic arrest with nocodazole for 2 h. Mitotic cells were then re-plated in nocodazole-containing medium for a further 2 or 4 h. Cell lysates were immunoblotted for Mcl-1, caspase-9 phosphorylated at Thr125 and total caspase-9.
Discussion
In this report, we have identified a mechanism for the control of cell death by apoptosis during the cell division cycle and in response to mitotic arrest. We have shown that phosphorylation of the anti-apoptotic regulator Mcl-1 at Thr92 by CDK1–cyclin B1, the major mitosis-promoting factor, causes the complete destruction of Mcl-1 by an APC/CCdc20- and proteasome-dependent mechanism after a prolonged arrest in mitosis and, subsequently, induces apoptosis. Mcl-1 is also partially phosphorylated at Thr92 in a normal mitosis, but the reduction in Mcl-1 levels is insufficient to trigger apoptosis. Thus, the post-translational regulation and destruction of Mcl-1 provides a timing mechanism to distinguish a short delay in mitosis from a prolonged mitotic arrest caused by an unresolved problem with spindle assembly (Figure 10).
Figure 10.
Model for the regulation of caspase activation and apoptosis by Mcl-1 during mitosis and in response to mitotic arrest. Activation of CDK1–cyclin B1 through its dephosphorylation by Cdc25 causes entry into mitosis. Inhibitory phosphorylation of caspase-9 by CDK1–cyclin B1 inhibits apoptosis, whereas coincident phosphorylation of Mcl-1 by CDK1–cyclin B1 initiates its destruction after a delay. Cyclin B1 destruction and inactivation of CDK1 are restrained by the spindle assembly checkpoint (SAC) until proper kinetochore attachments are established. We propose that the loss of Mcl-1 drops below the threshold required to restrain the activation of an apoptotic signal after prolonged mitotic arrest, for instance in response to microtubule poisons. The timing of caspase activation depends on the relative rates of Mcl-1 destruction and caspase-9 dephosphorylation. The latter is determined by the balance between phosphatase activity and CDK1–cyclin B1 activity, which may decline even during mitotic arrest because of the slow degradation of cyclin B1. Whether apoptosis occurs in mitosis or after exit from mitosis depends on the relative timing of effector caspase activation and cyclin B1 destruction below the threshold required to hold a cell in mitosis.
This model of the temporal control of apoptosis during mitotic arrest is consistent with recent work from Bekier et al (2009), which showed that, when the length of mitotic arrest in HeLa-derived cells is controlled with an Aurora kinase inhibitor, prolonged arrest resulted in subsequent cell death, whereas a shorter arrest produced variable fates. Furthermore, Huang et al (2009) have shown that cells arrested in mitosis for an extended period by depletion of Cdc20 show increased sensitivity to microtubule poisons. This indicates that it is not the activity of the SAC per se, but rather the protracted arrest that promotes apoptosis. Under these conditions, cyclin B1 proteolysis was delayed, but not completely inhibited (Huang et al, 2009). We would expect the Cdc20-dependent destruction of Mcl-1 to be delayed similarly, and initiation of apoptosis could occur when the level of Mcl-1 eventually drops below a protective threshold. Alternatively, a pro-apoptotic signal might be generated that overcomes inhibition by Mcl-1 after a highly extended mitotic arrest in cells depleted of Cdc20. Nevertheless, in cells expressing Cdc20, we show that the regulation of mitotic Mcl-1 degradation has a significant role in determining the sensitivity of cells to drugs that interfere with mitosis.
Other recent work has shown that destruction of Mcl-1 in asynchronous cells through knockdown of the deubiquitinase USP9X sensitises cells to ABT-737, a pro-apoptotic BH3-domain mimetic that inhibits Bcl-2 and Bcl-xL, but not Mcl-1 (Schwickart et al, 2010). Here, we have shown that drugs that arrest cells in mitosis also cause the complete loss of Mcl-1. Although Bcl-2 and Bcl-xL are stable under these conditions, they are highly phosphorylated and probably inhibited (Terrano et al, 2010). Thus, mitotically arrested cells may be particularly sensitive to the loss of Mcl-1 even without the pharmacological inhibition of Bcl-2 and Bcl-xL.
The delay between entry into mitosis and the complete destruction of Mcl-1 might be accomplished if the rate of Mcl-1 phosphorylation by CDK1–cyclin B1 is slow compared with other substrates of this kinase and, therefore, only occurs to a high level during a prolonged arrest. However, the fact that Thr92 phosphorylation can be observed in mitotic cells suggests that there is a delay between phosphorylation of this site and destruction of the protein. In contrast, phosphorylation of Mcl-1 at Ser159 by GSK-3 in interphase is almost undetectable without stabilisation of Mcl-1 by a proteasome inhibitor (Maurer et al, 2006). The dependency of mitotic Mcl-1 destruction on the phosphorylation of Thr92 is unusual for an APC/CCdc20-directed mechanism. We find that the association of Mcl-1 with Cdc20 occurs in interphase before CDK1–cyclin B1 activation, indicating that their interaction is not controlled by mitotic Thr92 phosphorylation. This suggests that a subsequent step, such as recognition of the Cdc20-Mcl-1 complex by APC/C, requires Thr92 phosphorylation and is rate limiting. Alternatively, Thr92 phosphorylation might inhibit the interaction of an opposing deubiquitinase, allowing Mcl-1 polyubiquitination and degradation.
The possible function of the other mitotic phosphorylation site in Mcl-1, Ser64, is less clear. Although this site is highly phosphorylated in mitotically arrested cells, it is also phosphorylated to a lower level in interphase cells, consistent with it being targeted by a kinase other than CDK1–cyclin B1. Furthermore, we found that mutation of this site does not stabilise Mcl-1 either in interphase or during mitotic arrest, consistent with an earlier report (Kobayashi et al, 2007).
The apparent dominant inhibitory effect of stabilised Mcl-1 on the Cdc20-dependent destruction of cyclins might be explained through the sequestration of Cdc20 by Mcl-1. However, the ability of the D-box mutant of Mcl-1 to still delay cyclin destruction indicates that it does not work only through this mechanism and we, therefore, do not exclude a more direct inhibitory effect of stabilised Mcl-1 on the APC/C. These results suggest the intriguing possibility that the pathological over-expression of Mcl-1 in certain cancers could result not only in resistance to apoptosis, but also in the generation of mitotic defects through disruption of the functions of APC/CCdc20.
Regulation of the Cdc20-dependent destruction of mitotic substrates is differentially inhibited by the SAC (Pines, 2006). Cyclin B1 proteolysis is strongly inhibited, although there is evidence for the slow degradation of cyclin B1 even in cells in which the SAC is not satisfied (Brito and Rieder, 2006; Gascoigne and Taylor, 2008; Wolthuis et al, 2008). In contrast, cyclin A is rapidly degraded on entry into prometaphase and its destruction is not prevented by the SAC (den Elzen and Pines, 2001; Geley et al, 2001), even though its degradation still requires Cdc20 (Wolthuis et al, 2008). Interestingly, cyclin A interacts stably with Cdc20 even before mitosis, like Mcl-1, but in contrast to cyclin B1 (Wolthuis et al, 2008). Another mitotic APC/CCdc20 substrate, Nek2A, is degraded during mitotic arrest by a mechanism that allows the Cdc20-independent interaction of Nek2A with the APC/C, although APC/C activity towards Nek2A is still stimulated by Cdc20 (Hames et al, 2001; Hayes et al, 2006; Kimata et al, 2008). Sensitivity to the SAC might, therefore, be dependent on a requirement for Cdc20 to target a substrate to the APC/C, but an earlier stable interaction between Cdc20 and the substrate could protect against this. However, although the interaction of Mcl-1 with Cdc20 is maintained during mitotic arrest, we find that Mcl-1 destruction is more sensitive to the total level of Cdc20 than cyclin A destruction. This suggests that Mcl-1 differs from cyclin A in its mode of regulation by the APC/CCdc20.
Association of apoptotic BH3-domain proteins Noxa and Bim with Mcl-1 is not affected by Thr92 or Ser64 phosphorylation (Supplementary Figure 11), but the destruction of Mcl-1 after a prolonged mitotic arrest could release them to initiate apoptosome formation and caspase-9 activation, leading to caspase-3 activation and eventually apoptotic DNA fragmentation. The appearance of apoptotic characteristics takes several hours after Mcl-1 degradation (Figure 8B). This lag may be due not only to the time taken to assemble the apoptosome, but also to the requirement to overcome the downstream threshold set by inhibitory phosphorylation of caspase-9 at Thr125 by CDK1–cyclin B1 (Allan and Clarke, 2007). Consistent with this proposal, we find that the phosphorylation of caspase-9 at Thr125 is maintained during mitotic arrest until after Mcl-1 destruction has been initiated (Figure 9), and abolition of the phosphorylation of caspase-9 at this site by mutation accelerates apoptosis under these conditions (Allan and Clarke, 2007). The level of caspase-9 phosphorylation is likely to be determined by both CDK1–cyclin B1 activity, which could diminish with the slow degradation of cyclin B1 during a prolonged mitotic arrest (Brito and Rieder, 2006; Gascoigne and Taylor, 2008; Wolthuis et al, 2008), and by the opposing phosphatase activity. If a cell slips through the mitotic checkpoint and exits mitosis, a process that can be accelerated by caspase activity in a feedback mechanism (Clarke and Allan, 2009), then inactivation of CDK1–cyclin B1 and dephosphorylation of caspase-9 would amplify the apoptotic pathway. In the event that a cell slipping out of mitosis still contains elevated levels of Mcl-1 because of its over-expression or slow degradation, then the cell could survive and proliferate despite mitotic aberrations. Thus, the fate of a cell arrested in mitosis by a microtubule poison might be determined by the relative rates of Mcl-1 destruction, cyclin B1 destruction and caspase-9 dephosphorylation (Figure 10).
Recent studies in which cell fates in response to microtubule poisons were determined for a number of cell lines by analysis of individual cells have shown that there is considerable variation in responses, both between cell lines and between individual cells of a particular line (Gascoigne and Taylor, 2008; Shi et al, 2008; Brito and Rieder, 2009). The study by Gascoigne and Taylor (2008) also showed a relationship between the rate of cyclin B1 destruction during mitotic arrest and subsequent cell fate. We propose that the efficiency of Mcl-1 destruction and the timing of caspase-9 dephosphorylation are likely to determine the response of an individual cell to the disruption of mitosis. The rates of these processes, which are both influenced by the activity of CDK1–cyclin B1, may differ between cells. Our identification of Mcl-1 as a critical target for CDK1–cyclin B1 further emphasises the central function of this protein kinase in both driving mitosis and controlling apoptosis during this phase of the cell cycle.
Materials and methods
Antibodies and reagents
The following antibodies were used for immunoblotting according to standard protocols: Mcl-1 (mouse monoclonal), cyclin A, APC3 (BD Pharmingen); Mcl-1 (rabbit polyclonal), Bcl-2, Cdc20, cyclin B1, CDK1, CDK1 phospho-Tyr15 (Santa Cruz Biotechnology); Cdh1 (Thermo Fisher Scientific); actin (Sigma); histone H3 phospho-Ser10 (Millipore); total caspase-9 (Chemicon). Rabbit polyclonal phospho-specific antibodies were generated against Mcl-1 phosphopeptides (pT92 Mcl-1, EVPDVTAT*PARLLFF and pS64 Mcl-1, GGSAGAS*PPSTLT, where * represents the phosphorylated residue) (Moravian Biotechnology, Brno, Czech Republic). Phospho-specific antibodies were affinity purified by two rounds of negative selection against non-phosphorylated peptide followed by one round of positive selection with phosphorylated peptide (Supplementary Figure 5). Generation of the phospho-Thr125 caspase-9 antibody was described earlier (Allan et al, 2003). Cleaved caspase-3 used for immunofluorescence was from Cell Signalling.
Cells
HeLa (OHIO) and U2OS (HTB96) were obtained from Cell Services, Cancer Research UK London Research Institute (Lincoln's Inn Fields). To generate stable clones of U2OS cells expressing Mcl-1 [wild-type, T92A, S64A, T92D, D-box (R207A/L210A) or ΔIR (C-terminal 2 amino acids, IR, deleted)], U2OS cells were co-transfected with Mcl-1 subcloned into pCMV-Flag (MRC Protein Phosphorylation Unit, Dundee) and pcDNA3. Single clones were selected in G418 (800 μg/ml, Calbiochem). Cell culture conditions were described earlier (Allan et al, 2003).
Cell treatments
Cells were synchronised at the G1/S boundary by double thymidine block, which involved incubation with 2 mM thymidine for 16 h, followed by an 8 h release into normal medium before the addition of 2 mM thymidine for a further 16 h. To arrest cells in mitosis, cells were treated with the specified concentration of nocodazole or taxol for the times given. To synchronise cells in a mitotic arrest, rounded-up cells were collected after treatment with nocodazole for 2 h and re-plated in the same nocodazole-containing medium. For proteasome inhibition studies, cells were incubated with 10 μM MG132 (Calbiochem) for 2 h before further treatments. To study the stability of proteins in interphase, cells were treated with 40 μg/ml cycloheximide (Calbiochem).
Identification of phosphorylation sites
Flag-Mcl was immunoprecipitated from HeLa cells, either asynchronous or treated with nocodazole for 3 h (mitotic), using Flag-agarose (Sigma). Samples were analysed by electrophoresis through a NuPAGE 4–12% Bis–Tris gel (Invitrogen). Flag-Mcl-1 bands were excised and digested overnight at 30°C with trypsin. Peptides were analysed by liquid chromatography mass spectrometry using a 4000 Q-Trap system (Applied Biosystems) by Dr Nick Morrice of the MRC Protein Phosphorylation Unit, University of Dundee.
Immunoprecipitation
For Mcl-1 precipitations, cells were lysed in IP buffer (20 mM Tris [pH 7.5], 150 mM NaCl, 10% glycerol, 1% Triton X-100, 2 mM EDTA, 1 mM Na3VO4, 50 mM NaF, 5 mM β-glycerophosphate, 1 mM PMSF, 1 μg/ml each pepstatin, leupeptin and aprotinin, 0.07% 2-mercaptoethanol, 1 μM okadaic acid). A total of 1–2 mg of lysate was incubated with 2 μg/ml α-Mcl-1 antibody (mouse monoclonal, BD Pharmingen) at 4°C for 2 h and then with 15 μl protein G sepharose, 4°C, 1 h. Precipitates were washed three times for 5 min before addition of SDS–gel-loading buffer. For Flag-tagged precipitations, cells were lysed in buffer (as IP buffer above, except glycerol, Na3VO4, 2-mercaptoethanol and okadaic acid were omitted). A total of 700 μg lysate was incubated with 15 μl Flag-agarose at 4°C for 10 h. Precipitates were washed three times for 5 min before incubation in SDS–gel-loading buffer at 37°C for 3 min; 2-mercaptoethanol was added to the supernatants and the samples were boiled for 3 min.
Analysis of protein phosphorylation by Phos-tag SDS–PAGE
Protein samples were analysed on 10% SDS–PAGE with 30 μM Phos-tag acrylamide (Phos-tag, Hiroshima, Japan) added to the resolving gel. This reagent specifically retards phosphorylated proteins through its Mn2+-dependent interaction with phosphomonoester dianions bound to Ser, Thr and Tyr residues (Kinoshita et al, 2006). After electrophoresis, gels were soaked in standard transfer buffer with 1 mM EDTA at room temperature for 10 min, followed by another 10 min incubation in transfer buffer without EDTA.
Mitotic cell extracts and in vitro kinase assays
Human HeLa cell extracts were prepared as described by Hutchins et al (2004). For western analysis of Mcl-1 phosphorylation in extracts, 500 ng Mcl-1-His6 (expressed in E. coli from a pET21 vector and purified by nickel–agarose affinity chromatography) was incubated with 10 μg extract for 60 min at 30°C in kinase buffer (50 mM Tris–HCl [pH 7.5], 10 mM MgCl2 and 10 μM DTT) plus 100 μM ATP and kinase inhibitor where indicated. Kinase inhibitors were used at the following concentrations: 10 μM roscovitine, 2 μM purvalanol A, 10 μM UO126, 10 μM SB203580 and 10 μM SP600125. Samples were analysed by SDS–PAGE and immunoblotting. For p13suc1 depletion of CDK/cyclin complexes, 500 μg HeLa cell extract was depleted by two rounds of incubation for 60 min at 4°C with 20 μl p13suc1-agarose beads. To restore CDK1–cyclin B1 activity to depleted extracts, 1 μl containing the indicated amount of purified active recombinant CDK1–cyclin B1 (Millipore) was added to give a total volume of 10 μl per incubation.
Immunoprecipitated kinase assays
Asynchronous or mitotic HeLa cells treated with nocodazole were lysed in 50 mM Tris–HCl [pH 8.0], 150 mM NaCl, 1% Triton X-100, 2 mM EDTA, 20 mM EGTA, 1 mM Na3V04, 50 mM NaF, 5 mM β-glycerophosphate, 0.1 mM PMSF, 1 μg/ml each of aprotinin, leupeptin and pepstatin. For CDK1 immunoprecipitations, 1 mg cell lysate was incubated for 60 min at 4°C with 1 μg anti-CDK1 mouse monoclonal antibody, then 25 μl protein A beads were added and the incubation continued for an additional 60 min at 4°C. Precipitated samples were washed three times in lysis buffer, three times in kinase buffer containing 5 mM EGTA, then incubated with 500 ng Mcl-1-His6 in kinase buffer containing 100 μM ATP for 60 min at 30°C.
RNA interference
siRNA duplexes were directed against Mcl-1 (5′-GGACUUUUAGAUUUAGUGATT-3′; Adams and Cooper, 2007), Cdh1 (5′-AAUGAGAAGUCUCCCAGUCAG-3′; Bashir et al, 2004), Luciferase (5′-CGUACGCGGAAUACUUCGATT-3′) (MWG, Ebersberg, Germany), APC3 (APC3-1, 5′-GGAAAUAGCCGAGAGGUAA-3′; APC3-2, 5′-CAAAAGAGCCUUAGUUUAA-3′; Dharmacon) or Cdc20 (ON-TARGETplus SMARTpool siRNA Cdc20, L-003225; Dharmacon). For Mcl-1 or Cdc20 knockdown, cells were transfected with 10 nM duplex and siLentFect (Bio-Rad) for 5 h according to the manufacturer's instructions, then trypsinised into fresh media and cultured for 43 h before further treatment. To knockdown APC3, cells were transfected with 50 nM each siRNA and LipofectAmine 2000 (Invitrogen) for 24 h followed by a second transfection for 48 h before further treatment. For Cdh1 knockdown, 50 nM duplex was transfected with LipofectAmine 2000 for 48 h before further treatment.
Flow cytometry
For analysis of sub-G1 DNA content, cells were fixed in 70% ethanol and incubated on ice for 2 h. Samples were washed twice in phosphate-citrate buffer (192 mM disodium phosphate, 4 mM citric acid, pH 7.8) and resuspended in PBS containing 100 μg/ml RNase A and 50 μg/ml propidium iodide. DNA content was analysed by flow cytometry using a FACScan machine (Becton Dickinson). At least 10 000 cells were analysed per sample.
Immunofluorescence
Cells were fixed on poly-L-lysine-coated coverslips with 3.7% formaldehyde, permeabilised with 0.2% Triton X-100 in PBS for 2 min, and blocked in PBS plus 3% BSA at room temperature for 30 min. Samples were incubated overnight at 4°C with anti-cleaved caspase-3 (1:200), then Alexa-Fluor 488-conjugated anti-rabbit (1:200; Invitrogen) antibodies at room temperature for 30 min. DNA was detected by staining with ToPro (1:1000; Invitrogen).
Supplementary Material
Acknowledgments
We thank Nick Morrice of the MRC Protein Phosphorylation Unit, University of Dundee for phosphopeptide analysis. This work was funded by Cancer Research UK, Association for International Cancer Research, Biotechnology and Biological Sciences Research Council and Tenovus Scotland. MEH was supported by a Medical Research Council Studentship and PRC was a Royal Society-Wolfson Research Merit Awardee.
Footnotes
The authors declare that they have no conflict of interest.
References
- Acquaviva C, Pines J (2006) The anaphase-promoting complex/cyclosome: APC/C. J Cell Sci 119: 2401–2404 [DOI] [PubMed] [Google Scholar]
- Adams KW, Cooper GM (2007) Rapid turnover of mcl-1 couples translation to cell survival and apoptosis. J Biol Chem 282: 6192–6200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan LA, Clarke PR (2007) Phosphorylation of caspase-9 by CDK1/cyclin B1 protects mitotic cells against apoptosis. Mol Cell 26: 301–310 [DOI] [PubMed] [Google Scholar]
- Allan LA, Clarke PR (2008) A mechanism coupling cell division and the control of apoptosis. SEB Exp Biol Ser 59: 257–265 [PubMed] [Google Scholar]
- Allan LA, Morrice N, Brady S, Magee G, Pathak S, Clarke PR (2003) Inhibition of caspase-9 through phosphorylation at Thr 125 by ERK MAPK. Nat Cell Biol 5: 647–654 [DOI] [PubMed] [Google Scholar]
- Bain J, McLauchlan H, Elliott M, Cohen P (2003) The specificities of protein kinase inhibitors: an update. Biochem J 371: 199–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir T, Dorrello NV, Amador V, Guardavaccaro D, Pagano M (2004) Control of the SCF(Skp2-Cks1) ubiquitin ligase by the APC/C(Cdh1) ubiquitin ligase. Nature 428: 190–193 [DOI] [PubMed] [Google Scholar]
- Bekier ME, Fischbach R, Lee J, Taylor WR (2009) Length of mitotic arrest induced by microtubule-stabilizing drugs determines cell death after mitotic exit. Mol Cancer Ther 8: 1646–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett LN, Clarke PR (2006) Regulation of claspin degradation by the ubiquitin-proteasome pathway during the cell cycle and in response to ATR-dependent checkpoint activation. FEBS Lett 580: 4176–4181 [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV (2007) Mitotic arrest and cell fate: why and how mitotic inhibition of transcription drives mutually exclusive events. Cell Cycle 6: 70–74 [DOI] [PubMed] [Google Scholar]
- Brito DA, Rieder CL (2006) Mitotic checkpoint slippage in humans occurs via cyclin B destruction in the presence of an active checkpoint. Curr Biol 16: 1194–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito DA, Rieder CL (2009) The ability to survive mitosis in the presence of microtubule poisons differs significantly between human nontransformed (RPE-1) and cancer (U2OS, HeLa) cells. Cell Motil Cytoskeleton 66: 437–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke PR, Allan LA (2009) Cell-cycle control in the face of damage—a matter of life or death. Trends Cell Biol 19: 89–98 [DOI] [PubMed] [Google Scholar]
- De Biasio A, Vrana JA, Zhou P, Qian L, Bieszczad CK, Braley KE, Domina AM, Weintraub SJ, Neveu JM, Lane WS, Craig RW (2007) N-terminal truncation of antiapoptotic MCL1, but not G2/M-induced phosphorylation, is associated with stabilization and abundant expression in tumor cells. J Biol Chem 282: 23919–23936 [DOI] [PubMed] [Google Scholar]
- den Elzen N, Pines J (2001) Cyclin A is destroyed in prometaphase and can delay chromosome alignment and anaphase. J Cell Biol 153: 121–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, He X, Hsu JM, Xia W, Chen CT, Li LY, Lee DF, Liu JC, Zhong Q, Wang X, Hung MC (2007) Degradation of Mcl-1 by beta-TrCP mediates glycogen synthase kinase 3-induced tumor suppression and chemosensitization. Mol Cell Biol 27: 4006–4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Huo L, Yang JY, Xia W, Wei Y, Liao Y, Chang CJ, Yang Y, Lai CC, Lee DF, Yen CJ, Chen YJ, Hsu JM, Kuo HP, Lin CY, Tsai FJ, Li LY, Tsai CH, Hung MC (2008) Down-regulation of myeloid cell leukemia-1 through inhibiting Erk/Pin 1 pathway by sorafenib facilitates chemosensitization in breast cancer. Cancer Res 68: 6109–6117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domina AM, Vrana JA, Gregory MA, Hann SR, Craig RW (2004) MCL1 is phosphorylated in the PEST region and stabilized upon ERK activation in viable cells, and at additional sites with cytotoxic okadaic acid or taxol. Oncogene 23: 5301–5315 [DOI] [PubMed] [Google Scholar]
- Du L, Lyle CS, Chambers TC (2005) Characterization of vinblastine-induced Bcl-xL and Bcl-2 phosphorylation: evidence for a novel protein kinase and a coordinated phosphorylation/dephosphorylation cycle associated with apoptosis induction. Oncogene 24: 107–117 [DOI] [PubMed] [Google Scholar]
- Gascoigne KE, Taylor SS (2008) Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell 14: 111–122 [DOI] [PubMed] [Google Scholar]
- Gascoigne KE, Taylor SS (2009) How do anti-mitotic drugs kill cancer cells? J Cell Sci 122: 2579–2585 [DOI] [PubMed] [Google Scholar]
- Geley S, Kramer E, Gieffers C, Gannon J, Peters JM, Hunt T (2001) Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J Cell Biol 153: 137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW (1991) Cyclin is degraded by the ubiquitin pathway. Nature 349: 132–138 [DOI] [PubMed] [Google Scholar]
- Hakem R, Hakem A, Duncan GS, Henderson JT, Woo M, Soengas MS, Elia A, de la Pompa JL, Kagi D, Khoo W, Potter J, Yoshida R, Kaufman SA, Lowe SW, Penninger JM, Mak TW (1998) Differential requirement for caspase 9 in apoptotic pathways in vivo. Cell 94: 339–352 [DOI] [PubMed] [Google Scholar]
- Hames RS, Wattam SL, Yamano H, Bacchieri R, Fry AM (2001) APC/C-mediated destruction of the centrosomal kinase Nek2A occurs in early mitosis and depends upon a cyclin A-type D-box. EMBO J 20: 7117–7127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L (1992) Defects in a cell cycle checkpoint may be responsible for the genomic instability of cancer cells. Cell 71: 543–546 [DOI] [PubMed] [Google Scholar]
- Hayes MJ, Kimata Y, Wattam SL, Lindon C, Mao G, Yamano H, Fry AM (2006) Early mitotic degradation of Nek2A depends on Cdc20-independent interaction with the APC/C. Nat Cell Biol 8: 607–614 [DOI] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW (2009) Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol 10: 478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HC, Shi J, Orth JD, Mitchison TJ (2009) Evidence that mitotic exit is a better cancer therapeutic target than spindle assembly. Cancer Cell 16: 347–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins JR, Moore WJ, Hood FE, Wilson JS, Andrews PD, Swedlow JR, Clarke PR (2004) Phosphorylation regulates the dynamic interaction of RCC1 with chromosomes during mitosis. Curr Biol 14: 1099–1104 [DOI] [PubMed] [Google Scholar]
- Inoshita S, Takeda K, Hatai T, Terada Y, Sano M, Hata J, Umezawa A, Ichijo H (2002) Phosphorylation and inactivation of myeloid cell leukemia 1 by JNK in response to oxidative stress. J Biol Chem 277: 43730–43734 [DOI] [PubMed] [Google Scholar]
- Janssen K, Pohlmann S, Janicke RU, Schulze-Osthoff K, Fischer U (2007) Apaf-1 and caspase-9 deficiency prevents apoptosis in a Bax-controlled pathway and promotes clonogenic survival during paclitaxel treatment. Blood 110: 3662–3672 [DOI] [PubMed] [Google Scholar]
- Kimata Y, Baxter JE, Fry AM, Yamano H (2008) A role for the Fizzy/Cdc20 family of proteins in activation of the APC/C distinct from substrate recruitment. Mol Cell 32: 576–583 [DOI] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T (2006) Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics 5: 749–757 [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Lee SH, Meng XW, Mott JL, Bronk SF, Werneburg NW, Craig RW, Kaufmann SH, Gores GJ (2007) Serine 64 phosphorylation enhances the antiapoptotic function of Mcl-1. J Biol Chem 282: 18407–18417 [DOI] [PubMed] [Google Scholar]
- Kozopas KM, Yang T, Buchan HL, Zhou P, Craig RW (1993) MCL1, a gene expressed in programmed myeloid cell differentiation, has sequence similarity to BCL2. Proc Natl Acad Sci USA 90: 3516–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuida K, Haydar TF, Kuan CY, Gu Y, Taya C, Karasuyama H, Su MS, Rakic P, Flavell RA (1998) Reduced apoptosis and cytochrome c-mediated caspase activation in mice lacking caspase 9. Cell 94: 325–337 [DOI] [PubMed] [Google Scholar]
- Marash L, Liberman N, Henis-Korenblit S, Sivan G, Reem E, Elroy-Stein O, Kimchi A (2008) DAP5 promotes cap-independent translation of Bcl-2 and CDK1 to facilitate cell survival during mitosis. Mol Cell 30: 447–459 [DOI] [PubMed] [Google Scholar]
- Maurer U, Charvet C, Wagman AS, Dejardin E, Green DR (2006) Glycogen synthase kinase-3 regulates mitochondrial outer membrane permeabilization and apoptosis by destabilization of MCL-1. Mol Cell 21: 749–760 [DOI] [PubMed] [Google Scholar]
- Michels J, Johnson PW, Packham G (2005) Mcl-1. Int J Biochem Cell Biol 37: 267–271 [DOI] [PubMed] [Google Scholar]
- Morel C, Carlson SM, White FM, Davis RJ (2009) Mcl-1 integrates the opposing actions of signaling pathways that mediate survival and apoptosis. Mol Cell Biol 29: 3845–3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED (2007) The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 8: 379–393 [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Peters JM, Uhlmann F (2000) Splitting the chromosome: cutting the ties that bind sister chromatids. Science 288: 1379–1385 [DOI] [PubMed] [Google Scholar]
- Nijhawan D, Fang M, Traer E, Zhong Q, Gao W, Du F, Wang X (2003) Elimination of Mcl-1 is required for the initiation of apoptosis following ultraviolet irradiation. Genes Dev 17: 1475–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson J, Yekezare M, Minshull J, Pines J (2008) The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat Cell Biol 10: 1411–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor DS, Wall NR, Porter AC, Altieri DC (2002) A p34(cdc2) survival checkpoint in cancer. Cancer Cell 2: 43–54 [DOI] [PubMed] [Google Scholar]
- Pines J (2006) Mitosis: a matter of getting rid of the right protein at the right time. Trends Cell Biol 16: 55–63 [DOI] [PubMed] [Google Scholar]
- Rieder CL, Maiato H (2004) Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev Cell 7: 637–651 [DOI] [PubMed] [Google Scholar]
- Scatena CD, Stewart ZA, Mays D, Tang LJ, Keefer CJ, Leach SD, Pietenpol JA (1998) Mitotic phosphorylation of Bcl-2 during normal cell cycle progression and Taxol-induced growth arrest. J Biol Chem 273: 30777–30784 [DOI] [PubMed] [Google Scholar]
- Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, Maecker H, O'Rourke K, Bazan F, Eastham-Anderson J, Yue P, Dornan D, Huang DC, Dixit VM (2010) Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature 463: 103–107 [DOI] [PubMed] [Google Scholar]
- Shi J, Orth JD, Mitchison T (2008) Cell type variation in responses to antimitotic drugs that target microtubules and kinesin-5. Cancer Res 68: 3269–3276 [DOI] [PubMed] [Google Scholar]
- Taylor RC, Cullen SP, Martin SJ (2008) Apoptosis: controlled demolition at the cellular level. Nat Rev Mol Cell Biol 9: 231–241 [DOI] [PubMed] [Google Scholar]
- Terrano DT, Upreti M, Chambers TC (2010) Cyclin-dependent kinase 1-mediated Bcl-xL/Bcl-2 phosphorylation acts as a functional link coupling mitotic arrest and apoptosis. Mol Cell Biol 30: 640–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodermaier HC, Gieffers C, Maurer-Stroh S, Eisenhaber F, Peters JM (2003) TPR subunits of the anaphase-promoting complex mediate binding to the activator protein CDH1. Curr Biol 13: 1459–1468 [DOI] [PubMed] [Google Scholar]
- Wolthuis R, Clay-Farrace L, van Zon W, Yekezare M, Koop L, Ogink J, Medema R, Pines J (2008) Cdc20 and Cks direct the spindle checkpoint-independent destruction of cyclin A. Mol Cell 30: 290–302 [DOI] [PubMed] [Google Scholar]
- Youle RJ, Strasser A (2008) The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol 9: 47–59 [DOI] [PubMed] [Google Scholar]
- Yu H (2007) Cdc20: a WD40 activator for a cell cycle degradation machine. Mol Cell 27: 3–16 [DOI] [PubMed] [Google Scholar]
- Zhong Q, Gao W, Du F, Wang X (2005) Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell 121: 1085–1095 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.