Abstract
Non-cell-autonomous effect of mutant proteins expressed in glia has been implicated in several neurodegenerative disorders, whereas molecules mediating the toxicity are currently not known. We identified a novel molecule named multiple α-helix protein located at ER (Maxer) downregulated by mutant ataxin-1 (Atx1) in Bergmann glia. Maxer is an endoplasmic reticulum (ER) membrane protein interacting with CDK5RAP3. Maxer anchors CDK5RAP3 to the ER and inhibits its function of Cyclin D1 transcription repression in the nucleus. The loss of Maxer eventually induces cell accumulation at G1 phase. It was also shown that mutant Atx1 represses Maxer and inhibits proliferation of Bergmann glia in vitro. Consistently, Bergmann glia are reduced in the cerebellum of mutant Atx1 knockin mice before onset. Glutamate-aspartate transporter reduction in Bergmann glia by mutant Atx1 and vulnerability of Purkinje cell to glutamate are both strengthened by Maxer knockdown in Bergmann glia, whereas Maxer overexpression rescues them. Collectively, these results suggest that the reduction of Maxer mediates functional deficiency of Bergmann glia, and might contribute to the non-cell-autonomous pathology of SCA1.
Keywords: ataxin-1, Bergmann glia, Maxer, non-cell-autonomous pathology, SCA1
Introduction
Effects of the non-neuronal expression of mutant proteins have been implicated in the pathology of neurodegenerative disorders including ALS (Di Giorgio et al, 2007; Nagai et al, 2007; Yamanaka et al, 2008a, 2008b), Huntington's disease (Shin et al, 2005; Lievens et al, 2008; Tamura et al, 2009) and spinocerebellar ataxias (Kretzschmar et al, 2005; Custer et al, 2006; Tamura et al, 2009). Astrocytes and microglia are suspected to have a central function in the ALS pathology (Lobsiger and Cleveland, 2007). Glutamate toxicity mediated by Bergmann glia was proposed in the SCA7 pathology by a recent report (Custer et al, 2006), whereas details of the non-cell-autonomous pathology in neurodegenerative disorders remain largely unknown. Glial endoplasmic reticulum (ER) stress and subsequent hypofunction of glutamate uptake as well as secretion of toxic signals including superoxide are suggested (reviewed by Ilieva et al, 2009). However, molecules mediating the non-cell-autonomous pathology in glia and neurons are not completely determined, and additional mechanisms in glia and neurons are also possible.
In a mouse model of spinocerebellar ataxia type 1 (SCA1), expression of mutant ataxin-1 (Atx1) for the first 3 weeks after birth remarkably influences the symptoms observed later in adulthood (Serra et al, 2006). It corresponds well to the critical period for cerebellar development when external granule cells and Bergmann glia are actively proliferating to form tissue architecture (Basco et al, 1977; Shiga et al, 1983; Goldowitz and Hamre, 1998). In the SCA7 mouse model, ataxin-7 (Atx7) expression in Bergmann glia that is located adjacent to Purkinje cells also induces neurodegeneration of Purkinje cells through perturbation of reuptake of glutamate by Bergmann glia (Custer et al, 2006).
The results in different SCA models suggest that proliferation and/or dysfunction of Bergmann glia might have an intriguing function in the fate of Purkinje cells in adulthood. Although functions of Bergmann glia in developing and mature brains have not been elucidated completely, they are considered to guide neuronal migration with their radial fibres (Yamada and Watanabe, 2002) similar to radial glia in the cerebral cortex. Bergmann glia are also suspected to act as stem cells because they express a stem cell marker Sox2 (Sottile et al, 2006) and have been shown to proliferate (Gaiano and Fishell, 2002; Yamada and Watanabe, 2002). The earlier study used the gfa2 enhancer to drive Atx7 (Custer et al, 2006), which is more active in proliferating glia, hence Bergmann glia proliferation, as well as their glutamate uptake, might be involved in the pathology.
Beyond the function of protein synthesis, ER was recently reported to have a critical function in the cell cycle regulation (Fearon and Cohen-Fix, 2008). For instance, G1 cyclin Cln3 is retained and bound to ER at early G1 phase, whereas it relocates to the nucleus at late G1 phase and contributes to initiation of S phase (Verges et al, 2007). SCAPER, a novel ER protein, is shown to regulate G1/S transition and M phase through interaction with cyclin A/Cdk2 at multiple phases of the cell cycle (Tsang et al, 2007).
From expression profiling analyses with primary neurons expressing mutant polyglutamine proteins (Tagawa et al, 2007), we unexpectedly found an ER protein, designated as multiple α-helix protein located at ER (Maxer) that has a critical function in G1/S transition and regulates cell proliferation. Maxer interacts with CDK5RAP3 (also known as LZAP and C53), a putative tumour suppressor (Wang et al, 2007), regulates subcellular localization of CDK5RAP3, and affects the expression level of Cyclin D1 through CDK5RAP3. Maxer is expressed at a high level in Bergmann glia of the cerebellum from perinatal period to adulthood. Maxer co-expression and RNAi-mediated knockdown rescuers and enhances, respectively, mutant Atx1-induced pathologies of primary Bergmann glia and Purkinje cells. In keeping with this, SCA1 model mice exhibited reduction of Bergmann glia that precedes decrease of Purkinje cells. Collectively, these results indicate that Maxer is a new type of cell cycle regulator at ER-like SCAPER, and suggest that Maxer might be involved in the SCA1 pathology through functional deficiency of Bergmann glia.
Result
Identification of a novel gene, Maxer
In the earlier study, we performed the expression profiling analysis with gene chips to investigate the molecular mechanism of neurodegenerative diseases (Tagawa et al, 2007). In parallel, we performed the proteome analysis for the same purpose (Qi et al, 2007). Consequently, we identified several candidate genes including Hsp70 and HtrA2/Omi that might influence cell type-specific vulnerability in polyglutamine (polyQ) diseases (Tagawa et al, 2007; Inagaki et al, 2008) and several molecules that could commonly affect pathologies (Qi et al, 2007).
Through gene expression profiling with microarrays, we found KIAA0776, which was downregulated by mutant Atx1 expression in primary culture of cerebellar neurons from rat P7 pups, which inevitably includes Bergmann glia in addition to granular and Purkinje cells even after the treatment of Ara-C. Database searches by standardbasic local alignment and search tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and PredictProtein program (Rost et al, 2004) revealed that Maxer is an evolutionally conserved protein highly rich in α-helix structure (Figure 1A and B). The orthologues are conserved in mammals (identity to human NM_015323: 90%, mouse NM_026194: 95%) and in Drosophila melanogaster (NM_141433: 33%), Anopheles gambiae (XM_317778: 33%), Caenorhabditis elegans (NM_068689: 22%), Oryza sativa (NM_001061011: 29%) and Arabidopsis thaliana (CAB90949: 24%), suggesting that Maxer possesses a critical biological function. Bioinformatics analysis by PredictProtein (Rost et al, 2004) predicted that Maxer contains a nuclear localization signal (NLS) and a transmembrane domain (Figure 1A and B). Earlier protein phosphorylation omics studies (Beausoleil et al, 2004; Olsen et al, 2006) also suggested that Maxer possesses two phosphorylation sites (Figure 1A). However, Maxer is a highly unique protein without any homologue in mammalian databases. We named it Maxer because it possesses multiple α-helixes and locates at ER as shown later.
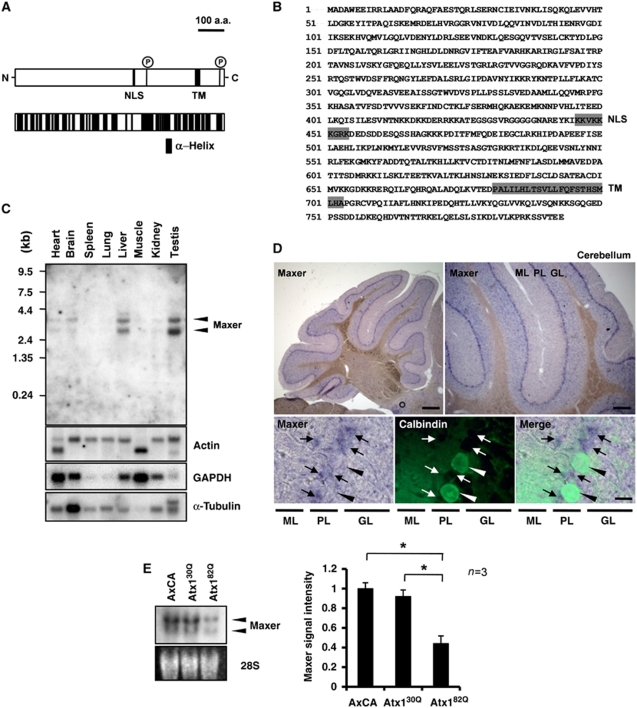
Figure 1.
Identification and tissue expression of Maxer. (A) Structure of Maxer protein. NLS, nuclear localization signal; TM, transmembrane domain; P, possible phosphorylation site. Lower scheme shows multiple α-helix (black). (B) Amino-acid sequence of rat Maxer. (C) Tissue expression profiles of Maxer mRNA in adult rat. Northern blot with full-length rat Maxer cDNA. (D) In situ hybridization analysis of Maxer at 8 weeks. Maxer is expressed at Purkinje cell layer (upper panels, purple). Lower panels are higher magnifications of co-staining with anti-calbindin antibody (green, arrowheads). Maxer is expressed in calbindin-negative cells (arrows), but not in calbindin-positive Purkinje cells. ML, molecular layer; PL, Purkinje cell layer; GL; granule cell layer. Bar: 200 μm (upper left), 100 μm (upper right) and 10 μm (lower). (E) Northern blot analysis of primary cerebellar culture prepared at P7. Cells were infected with AxCA (empty vector)-, AxCA-Atx130Q (Atx130Q)- or AxCA-Atx182Q (Atx182Q)-expressing adenovirus vectors. Maxer was reduced by mutant Atx1. The error bars represent s.e.m. n=3, *P<0.01, Student's t-test.
To determine the tissue expression pattern of Maxer mRNA, we first performed northern blot analysis and found its expression in brain, heart, liver and testis (Figure 1C). Among various regions of the brain, Maxer expression was high in the cerebellum (data not shown). At 8 weeks after birth, in situ hybridization showed a high expression at Purkinje cell layer, whereas the signals of Maxer did not merge with that of calbindin (Figure 1D). Double staining with anti-Maxer, whose characterization will be described later (Figure 2), and anti-calbindin antibodies also excluded Maxer expression in Purkinje cells (Supplementary Figure S1A). Consistent with our earlier microarray analysis (Tagawa et al, 2007), northern blot analysis with primary culture of cerebellar neurons at P7 showed that mutant Atx1 reduced Maxer (Figure 1E).
Figure 2.
Maxer is expressed in neural stem cells and Bergmann glia. (A) Western blot analysis of Maxer with C15-01D anti-Maxer antibody. C6, P19 and HeLa cells as well as primary cerebellar glia expressed Maxer endogenously (left). Maxer reduction in mutant Atx1-infected primary cerebellar culture (right upper) and in mutant Atx1-KI mouse cerebellum (right lower). (B) Immunohistochemistry of E17 cerebral cortex with anti-Maxer antibody. Cytoplasmic Maxer merged with GLAST (upper and middle panels). Sox2 was expressed in the nuclei of Maxer-positive cells (lower panels). VZ, ventricular zone. Bar: 100 μm. (C) Immunohistochemistry of cerebellum with anti-Maxer antibody at P7, the time point used for our original microarray analysis (Tagawa et al, 2007). Maxer merged with GLAST in Bergmann glia. EGL, external granule cell layer; PL, Purkinje cell layer; IGL, internal granule cell layer. Bar: 20 μm. (D) Maxer was reduced in Bergmann glia of mutant Atx1-KI mice (16 weeks). ML, molecular layer; PL, Purkinje cell layer; GL, granule cell layer. Bar: 20 μm. (E) Immunohistochemistry of cerebellum at 16 weeks. Purkinje cell nuclei (arrow) and Sox2-positive Bergmann glia nuclei (arrowheads) were stained with Atx1 (anti-ataxin-1 antibody 11NQ) and DAPI. Bar: 20 μm. (F) PCNA-positive cells expressed Maxer at P7 (arrows). Bar: 20 μm.
Maxer is expressed in Bergmann glia and neural stem cells
To analyse protein expression patterns, we generated six antibodies against different regions of Maxer. Among them, the antibody against a C-terminal sequence of Maxer (C15-01D) detected specifically the Maxer protein (Figure 2A). The Maxer antibody detected endogenous Maxer in C6, P19 and HeLa cells in addition to cerebellar tissue (Figure 2A). We used this antibody to confirm that Maxer protein is reduced in Atx1-infected primary cerebellar neuron culture and in the cerebellum of Atx1-KI mice (Figure 2A). These experiments consistently support the fidelity of the C15-01D antibody.
With C15-01D, we examined the cell-type specificity of Maxer protein expression in embryonic, perinatal and adult brains. In the mouse embryonic brain at E17, Maxer was detected in Sox2-positive neural stem cells and in glutamate-aspartate transporter (GLAST)-positive radial glia (Figure 2B). In perinatal brain, Maxer was highly expressed in GLAST-positive Bergmann glia of the cerebellum (Figure 2C) possessing a morphological character similar to radial glia. Maxer and Sox2, another Bergmann glia marker protein, were also localized to identical cells (Supplementary Figure S1B). Collectively, Maxer is expressed in Bergmann glia, but not Purkinje cells (Figure 1D; Supplementary Figure S1A). Maxer protein continues to be expressed in GFAP-positive Bergmann glia of adult brain at 16 weeks (Figure 2D). These results reconfirmed the expression pattern of Maxer observed by in situ hybridization.
In agreement with the result of primary culture (Figure 1E), Maxer protein was reduced in Bergmann glia of mutant Atx1-KI mice (Figure 2C and D). To illustrate the relevance of Maxer reduction to Atx1, we tested Atx1 expression in Bergmann glia. Double staining of Atx1 and Sox2 confirmed that Atx1 was expressed in Bergmann glia in addition to Purkinje cells (Figure 2E). In addition, the double staining with anti-GFAP and Atx1 antibodies reconfirmed that Atx1 was expressed both in Bergmann glia and Purkinje cells (Supplementary Figure S1C).
As several reports showed perinatal proliferation of Bergmann glia (Basco et al, 1977; Shiga et al, 1983), we also tested with a proliferation marker, PCNA, whether Maxer-positive cells proliferate or not. PCNA-positive cells expressed Maxer in their cytoplasm (Figure 2F), supporting proliferation of Maxer-positive cells at 7 days after birth.
Maxer is a novel ER-associated protein
To investigate the molecular function of Maxer, we investigated the subcellular localization of Maxer by using Maxer–EGFP fusion protein. Fluorescent imaging of EGFP–Maxer showed ER-like distribution in HeLa cells that merged with an ER marker, ER-DsRed, at the confocal microscopic level (Figure 3A). In this case, no EGFP signal bled through fluorescence to the DsRed image in single transfection of Maxer–EGFP, and vice versa (Figure 3A). EGFP–Maxer was found in the membrane fraction, whereas the truncated form (EGFP–Maxer 1–683) lacking the transmembrane domain was shifted to the cytoplasm (Figure 3B). Further to support its localization at ER membrane, we performed gradient subcellular fractionation with OptiPrep (Figure 3C). Maxer was co-fractionated with an ER membrane protein, Calnexin, but not with Golgi or mitochondria proteins (Figure 3C). Endogenous Maxer protein detected by Maxer antibody in immunocytochemistry also confirmed Maxer localization to ER (Figure 3D). Although Maxer possesses NLS, the sequence was functional only when the transmembrane domain was deleted (Supplementary Figure S2).
Figure 3.
Maxer is a novel ER protein. (A) Confocal images show colocalization of Maxer and ER-DsRed in HeLa cells. Higher magnifications are shown from the fourth to sixth lines. Maxer 1–711 is colocalized with ER-DsRed, whereas Maxer 1–683 showed diffuse distribution. Single transfection of Maxer or ER-DsRed did not make signals that bleed through fluorescence. Bar: 10 or 1 μm in higher magnification. (B) Deletion constructs of Maxer–EGFP (left). Maxer and Maxer 1–711 were enriched in cellular membrane fraction of HeLa cells, whereas Maxer 1–683 was enriched in cytosolic fraction (right); α-tubulin and calnexin were used as cytosolic and membrane fraction marker, respectively. (C) Density gradient fractionation showed Maxer enrichment in ER, but not golgi or mitochondria. Calnexin (ER), golgi 58 K (golgi) and Tom70 (mitochondria) were used as organelle markers. (D) Confocal images showed colocalization of endogenous Maxer and ER-DsRed. Bar: 10 μm.
Maxer interacts with CDK5RAP3
To elucidate the molecular function of Maxer, we further screened binding partners of Maxer using an interactome database of Drosophila published earlier (Giot et al, 2003). One candidate was cyclin E, which is known to be a component of the Rb signalling pathway. Phosphorylation of Rb by cyclin E-Cdk2 and cyclin D-Cdk4/6 dissociates E2F to separate from Rb, and the released E2F promotes entry to S phase through transcription of target genes (Sherr and McCormick, 2002). However, we could not detect interaction between Maxer and cyclin E in immunoprecipitation (Figure 4A) and excluded cyclin E from candidate.
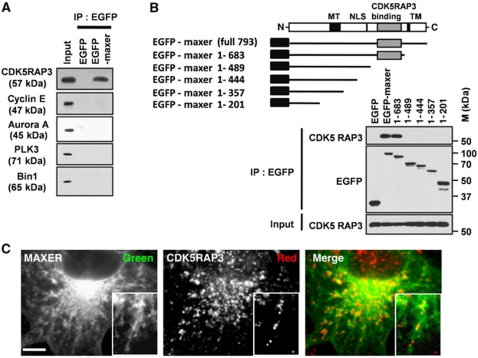
Figure 4.
Maxer interacts with CDK5RAP3. (A) Cell lysates from EGFP or EGFP–Maxer-expressing HeLa cells are subjected to precipitation by anti-EGFP antibody and blotted by indicated antibodies. (B) Deletion constructs of Maxer–EGFP were used for immunoprecipitation. The binding domain to CDK5RAP3 was shaded. (C) Confocal images showed colocalization of EGFP–Maxer and CDK5RAP3. Bar: 5 μm.
Another candidate was CDK5RAP3. CDK5RAP3 was identified as a putative tumour suppressor protein (Wang et al, 2007). Interestingly, CDK5RAP3 overexpression induces G1 arrest in cell lines (Wang et al, 2006). The interactome data suggested that Maxer binds to CDK5RAP3 (Ewing et al, 2007). As expected, CDK5RAP3 was efficiently co-immunoprecipitated with EGFP–Maxer, whereas other cell cycle proteins were not (Figure 4A). Specificity of the interaction was further confirmed by deletion analysis (Figure 4B). Carboxyl terminal region (490–683 a.a.) of Maxer was essential for the interaction with CDK5RAP3 (Figure 4B), whereas the binding domain does not share any homology to the other protein–protein interaction module. Confocal microscopic analysis also showed colocalization of Maxer with endogenous CDK5RAP3 in the cytoplasm (Figure 4C).
Maxer regulates G1/S transition through CDK5RAP3
Earlier biochemical studies suggested that CDK5RAP3 is distributed both in the nucleus and cytoplasm (Jiang et al, 2005; Wang et al, 2006, 2007). As overexpression of CDK5RAP3 in the nucleus induces G1 arrest (Wang et al, 2006), Maxer might change the subcellular localization of CDK5RAP3 and affect cell cycle.
To test this hypothesis, we used Maxer-shRNA-DsRed expressing IRES-intercalated Maxer-shRNA and DsRed from the same enhancer/promoter. We confirmed that transfected cells expressing DsRed showed reduction of endogenous Maxer protein (Figure 5A). Maxer knockdown by shRNA shifted CDK5RAP3 from the cytoplasm to nucleus (Figure 5B), and the shift was confirmed by quantitative analysis with >100 cells (Figure 5C). Consistently, western blot analysis with nuclear and cytoplasmic fractions prepared from total cells supported the Maxer knockdown-induced nuclear shift of CDK5RAP3 (Figure 5D) despite dilution of the effect by non-transfected cells. As CDK5RAP3 binds and inhibits RelA, a component of NFκB (Wang et al, 2007), and NFκB regulates transcription of cyclin D1 (Cao et al, 2001; Karin et al, 2002), Maxer could affect the expression of cyclin D1. Consistently, western blot analysis showed reduction of cyclin D1 by Maxer-shRNA (Figure 5E). The transfection efficiencies of non-silencing shRNA and Maxer-shRNA were equivalent (60 and 62%) in these experiments (Supplementary Figure S3A).
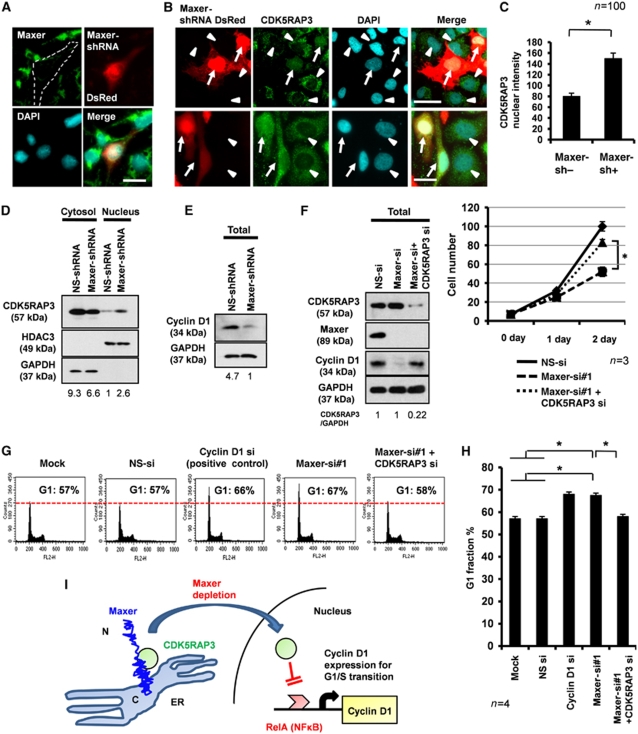
Figure 5.
Maxer regulates nuclear translocation of CDK5RAP3. (A) Endogenous Maxer was reduced in Maxer-shRNA/DsRed-transfected HeLa cell (dotted). Maxer-shRNA and DsRed are expressed from a plasmid. Bar: 20 μm. (B) Transfection of Maxer-shRNA/DsRed increased nuclear CDK5RAP3 in HeLa cells. Arrow: shRNA-transfected cells, arrowhead: non-transfected cells. Bar: 20 μm. (C) Quantification of CDK5RAP3 intensity in the nuclei by MetaMorph. Mean+s.d. n=100, *P<0.01. Three independent experiments showed the similar result. (D) Western blot analysis shows the increase of CDK5RAP3 in the nuclear fraction of Maxer-shRNA-transfected cells. GAPDH and HDAC3 were cytoplasmic and nuclear marker, respectively. The ratio below the panel indicates CDK5RAP3 intensity in comparison with non-silencing (NS) shRNA. (E) Cyclin D1 expression level was reduced in Maxer-shRNA-transfected cells. The ratio indicates GAPDH-corrected signal intensity of cyclin D1. (F) C6 glioma cells were transfected with NS siRNA, Maxer-siRNA#1 or Maxer-siRNA + CDK5RAP3-siRNA. Their effects on protein expression (western blots, left) and on cell proliferation (graph, right) were simultaneously analysed. Co-transfection of CDK5RAP3-siRNA rescued cyclin D1 and proliferation suppressed by Maxer-siRNA#1. Error bar: s.e.m. n=3, *P<0.01. The ratio indicates GAPDH-corrected intensity of CDK5RAP3. (G) C6 glioma cells were transfected with siRNAs indicated above the panels. Fluorescence-activated cell sorter (FACS) analysis was performed at 72 h after transfection. Histograms with 10 000 cells indicated G1 accumulation by Maxer knockdown. Cyclin D1-siRNA was a positive control. Co-transfection of CDK5RAP3-siRNA rescued the G1 accumulation by Maxer-siRNA#1. (H) G1 per cent in multiple FACS analyses was statistically analysed. Error bar: s.d. n=4, *P<0.01. (I) A hypothetical scheme of molecular function of Maxer to regulate G1/S transition through CDK5RAP3 and cyclin D1.
These results prompted us to hypothesize that suppression of Maxer permits nuclear translocation of CDK5RAP3 to inhibit cyclin D1 and induce G1 accumulation. Consequently, we then analysed the effects of Maxer on proliferation and cell cycle. Simultaneously, we tested the rescue effect of CDK5RAP3. For these purposes, we designed siRNAs against rat Maxer. Two siRNAs against rat Maxer were designed. Maxer-siRNA#1 suppressed Maxer in C6 glioma cells more efficiently than siRNA#2, as shown with northern blot analysis (Supplementary Figure S4A), western blot analysis (data not shown), immunocytochemistry (Supplementary Figure S4B) and RT–PCR (Supplementary Figure S4C). We confirmed that transfection efficiency was similar among non-silencing siRNA (100%), Maxer-siRNA#1 (100%) and Maxer-siRNA#2 (100%), by using non-silencing siRNA Alexa488 as an internal control (Supplementary Figure S3B). Therefore, we used siRNA#1 and analysed physiological functions of Maxer in the following experiments.
As expected, Maxer-siRNA suppressed proliferation of C6 glioma cells, and the knockdown of CDK5RAP3 reactivated proliferation (Figure 5F). Maxer knockdown clearly induced G1 accumulation such as the positive control of cyclin D1 knockdown (Figure 5G). Again, the knockdown of CDK5RAP3 rescued the G1 accumulation (Figure 5G). No effect was observed with negative controls such as mock treatment or transfection of non-silencing siRNA (Figure 5G). The increase of G1 fraction by Maxer knockdown and its rescue by CDK5RAP3 knockdown were statistically confirmed by multiple experiments (Figure 5H). Collectively, our results support the scheme that suppression of Maxer permits nuclear translocation of CDK5RAP3, inhibits cyclin D1 expression and induces G1 accumulation (Figure 5I).
Mutant Atx1 represses proliferation and GLAST expression of Bergmann glia through repression of Maxer
As Maxer regulating cell proliferation is expressed in Bergmann glia, it is plausible that mutant Atx1-induced reduction of Maxer inhibits proliferation of Bergmann glia during development and perinatal period. As expected, immunohistochemistry with two Bergmann glia markers, Sox2 and Sox9, revealed that Bergmann glia were actually decreased in mutant Atx1-KI mice (B6.129S-Atxn1tm1Hzo: Watase et al, 2002) at 1 week (Figure 6A and B; Supplementary Figure S5A and B). The reduction of Sox2 was reconfirmed by western blot analysis (Figure 6C). The Bergmann glia reduction (Figure 6A–C) preceded the reduction of Purkinje cells starting at 19 weeks (Watase et al, 2002) (Figure 6B; Supplementary Figure S5B). The reduction was not due to the increased cell death of Bergmann glia because Tunel staining of P7 cerebellar tissue did not show apoptosis at Purkinje cell layer (Supplementary Figure S6A).
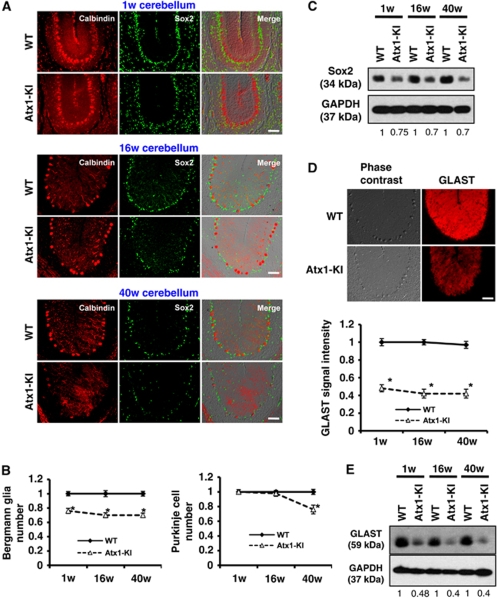
Figure 6.
Reduction of Bergmann glia and GLAST in Atx1-KI mice. (A) Bergmann glia were reduced in the cerebellum of mutant Atx1-KI mice. Bergmann glia and Purkinje cells were immunostained with anti-Sox2 antibody and anti-calbindin antibody, respectively. Corresponding cerebellar folia from three different ages (1, 16 and 40 weeks) were analysed. Bar: 50 μm. (B) Quantification of Bergman glia number (left) and Purkinje cell number (right) of three different ages (1, 16 and 40 weeks). Error bar: s.e.m. n=4, *P<0.01, Student's t-test. (C) Western blot analysis with anti-Sox2 antibody to quantify Bergmann glia. The ratio indicates GAPDH-corrected signal intensity of Sox2. (D) Mutant Atx1 expression reduced GLAST in KI mice. Cerebella from WT and Atx1-KI were stained with anti-GLAST antibody (upper). GLAST signal intensity was quantified (lower). The error bars represent s.e.m. n=4 mice, *P<0.01, Student's t-test. Bar: 50 μm. (E) Western blot analysis of GLAST expression in WT and Atx1-KI cerebellum.
As GLAST in the cerebellum is mainly expressed by Bergmann glia, we re-examined the expression of GLAST in mutant Atx1-KI mice by immunohistochemistry. As expected, GLAST was reduced from 1 week after birth (Figure 6D) in consistence with the reduced number of Bergmann glia (Figure 6B). The reduction of GLAST was also confirmed by western blot analysis (Figure 6E). However, both immunohistochemistry and western blot analyses showed that the reduction of GLAST was larger than that of Sox2 or Sox9, suggesting that GLAST per cell was also reduced.
In addition to GLAST, we performed similar analyses with glutamate transporter 1 (GLT1), another glial protein involved in glutamate reuptake. We found reduction of GLT1 signals in the cerebellar cortex of Atx1-kock in mice from 1 week after birth (Supplementary Figure S7A), which was confirmed by western blot (Supplementary Figure S7B).
The reduction of Bergmann glia and GLAST and GLT1 in the SCA1 pathology shown in this study (Figure 6; Supplementary Figure S7) are consistent with the proposed mechanism in the SCA7 pathology in which reduction of GLAST in Bergmann glia leads to the increase of local glutamate concentration around neurons and the excitatory toxicity for neurons (Custer et al, 2006). Therefore, similar pathology mediated by Bergmann glia might be shared by multiple polyQ degenerations including SCA1 and SCA7.
Maxer is involved in the Bergmann glia-mediated pathology of SCA1
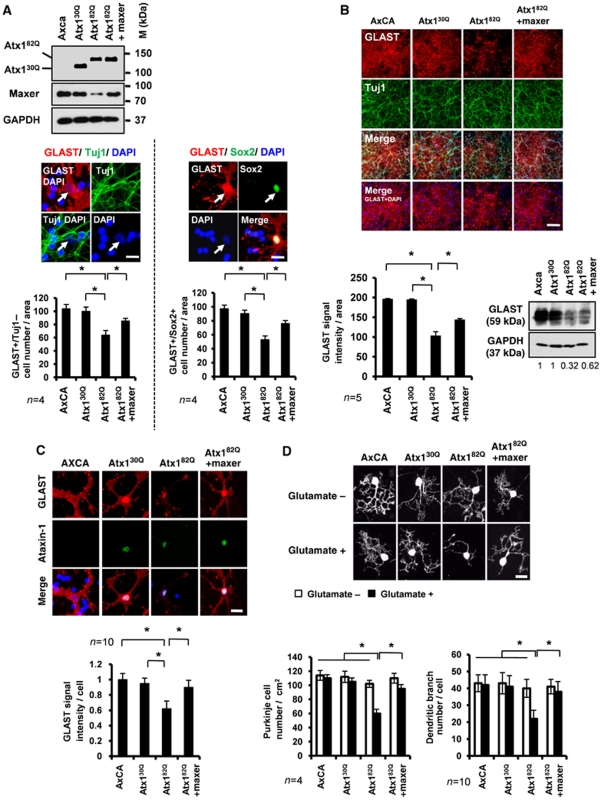
Further to test the involvement of Maxer in the SCA1 pathology, we performed two types of experiment. First, we tested whether Maxer overexpression rescues the SCA1 pathology. Second, we examined whether Maxer knockdown accelerates the SCA1 pathology.
In the first line of experiment, we evaluated the effect of Maxer on proliferation of Bergmann glia in primary culture (Figure 7A), GLAST expression in Bergmann glia (Figure 7B and C) and glutamate toxicity on Purkinje cells in co-culture (Figure 7D). All the experiments were performed with adenovirus vectors (AxCA: empty, Atx130Q, Atx182Q, Maxer), and their infection efficiencies into primary cerebellar cells were checked using an EGFP-expressing adenovirus vector (AxCA–EGFP) as an internal control (Supplementary Figure S3C) and by Atx1 expression (Supplementary Figure S3D).
Figure 7.
Maxer rescues Purkinje cells from glutamate toxicity through Bergmann glia. (A) Rescue experiment using Maxer-expressing adenovirus. Primary Bergmann glia were prepared at P7 and infected with AxCA (empty vector)-, AxCA-Atx130Q (Atx130Q)-, AxCA-Atx182Q (Atx182Q)- or AxCA-Maxer (Maxer)-expressing adenovirus vectors. Cell lysates were immunobloted with indicated antibodies. Atx1 expression levels were almost equivalent (upper). Bergmann glia were identified either by GLAST/Tuj1 staining (GLAST+/Tuj1−cells) or by GLAST/Sox2 staining (GLAST+/Sox2+cells) (middle). Bergmann glia number was quantified (lower). Mutant Atx1 suppressed proliferation of Bergmann glia, but co-expression of Maxer rescued the proliferation in primary culture. The error bars represent s.e.m. n=4, *P<0.01, Student's t-test. Bar: 10 μm. (B) Co-expression of Maxer rescued suppression of GLAST by Atx182Q. Primary cerebellar cell culture was prepared at P7 and infected with indicated adenovirus vectors as in (A). Cells were immunostained with anti-GLAST and anti-Tuj1 antibodies (upper). The signal intensities of GLAST were quantified (lower left). In parallel, cell lysates were blotted with anti-GLAST antibody (lower right). The error bars represent s.e.m. n=5, *P<0.01, Student's t-test. Bar: 100 μm. (C) Co-expression of Maxer rescued suppression of GLAST by Atx182Q at a single cell level. Primary cerebellar cell culture after infection was stained with anti-GLAST and anti-ataxin-1 antibodies (upper). GLAST signal intensities were quantified at a single cell level (lower). The error bars represent s.d. n=10, *P<0.05, Student's t-test. Bar: 10 μm. (D) Bergmann glia–Purkinje cell co-culture was exposed to 50 μM of glutamate. Cells were infected with indicated adenovirus vectors. Purkinje cell number and Purkinje branch complexity were significantly reduced when Bergmann glia were infected with Atx182Q and rescued with co-expression of Maxer (lower). Purkinje cells were visualized by anti-calbindin antibody (upper: representative cells). For Purkinje cell number, the error bars represent s.e.m. n=4, *P<0.05, Student's t-test. For Purkinje branch complexity, the error bars represent s.d. n=10, *P<0.05, Student's t-test. Four independent experiments showed the similar results. Bar: 10 μm.
We used two types of triple staining, GLAST/Tuj1/DAPI and GLAST/Sox2/DAPI, for identification of Bergmann glia. In both the methods, Bergmann glia were easily identified as cells possessing large but weakly stained nuclei in DAPI. During the primary culture for 7 days, Bergmann glia prepared at P7 proliferate to two-folds (data not shown). Mutant Atx1 (Atx182Q) expression suppressed proliferation of Bergmann glia in primary culture and decreased the number of Bergmann glia at day 7 (Figure 7A), and the suppressed proliferation was not due to the increase of apoptosis (Supplementary Figure S6B). Maxer re-expression rescued the Atx182Q-induced proliferation block (Figure 7A). In these experiments, we also confirmed that expression of Atx130Q or Atx182Q in Bergmann glia was almost equivalent (Supplementary Figure S4D). Western blot and immunocytochemistry analyses reconfirmed that Maxer rescued suppression of GLAST by Atx182Q (Figure 7B). We performed similar experiments with GLT1, and observed the rescue effect of Maxer on GLT1 expression in mixed culture of primary cerebellar cells (Supplementary Figure S7C).
In addition, we investigated the effects of Atx182Q and Maxer at a single cell level of Bergmann glia. As expected from the results of Atx1-KI mice (Figure 6B–E), Atx182Q reduced the GLAST signal intensities at a single Bergmann glia level, and Maxer co-expression rescued the reduction of GLAST per cell (Figure 7C). Furthermore, Atx182Q increased vulnerability of Purkinje cells to glutamate in the co-culture with Bergmann glia, as the number of Purkinje cells and their dendritic branching were decreased by Atx182Q. Again, Maxer co-expression rescued the glutamate toxicity to Purkinje cell (Figure 7D). We could not find the similar rescue effects on Atx182Q-induced phenotypes when we co-expressed α-tubulin as a negative control (data not shown).
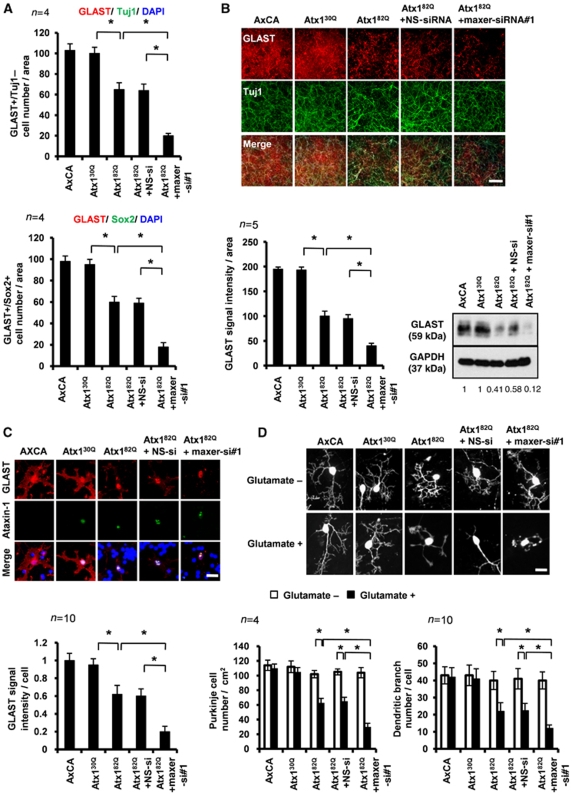
In the second line of the experiment, we tested whether Maxer knockdown accelerates SCA1 pathology. Maxer knockdown enhanced suppressive effect of Atx182Q on proliferation of GLAST+/Tuj1− or GLAST+/Sox2+ Bergmann glia in primary culture (Figure 8A). Maxer-specific siRNA enhanced reduction of GLAST by Atx182Q in co-culture of primary cerebellar neurons and glia (Figure 8B). The GLAST reduction by Maxer-siRNA was also observed at a single Bergmann glia cell level (Figure 8C). The transfection efficiencies of the control and Maxer siRNAs were confirmed to be similar by using siRNA Alexa488 as an internal control (Supplementary Figure S3E). Purkinje cells in co-culture obviously became more vulnerable to glutamate, judging from the number of Purkinje cells in co-culture and their dendritic branching number (Figure 8D). These enhancing effects on the Atx182Q-induced phenotypes were not observed with non-silencing siRNA (Figure 8A–D).
Figure 8.
Maxer knockdown accelerates glutamate toxicity on Purkinje cells. (A) Co-expression of Maxer-RNAi enhanced suppression of Bergmann glia proliferation by Atx182Q. Primary culture of cerebellar cells (P7) were infected with adenovirus vectors or transfected with Maxer siRNA#1 or NS siRNA. Bergmann glia were identified either by GLAST/Tuj1 staining (GLAST+/Tuj1−cells) or by GLAST/Sox2 staining (GLAST+/Sox2+cells) 4 days after the transfection. The error bars represent s.e.m. n=4, *P<0.01, Student's t-test. (B) Co-expression of Maxer-RNAi further suppressed Atx182Q-induced suppression of GLAST. Primary cerebellar culture cells (P7) were prepared as in (A) and double stained with anti-GLAST antibody and anti-Tuj1 antibody (upper). GLAST intensities were quantified (lower left). Simultaneously, cell lysates were blotted with anti-GLAST antibody (lower right). The error bars represent s.e.m. n=5, *P<0.01, Student's t-test. Bar: 100 μm. (C) Co-expression of Maxer-RNAi enhanced Atx182Q-induced suppression of GLAST at a single cell level. Primary cerebellar culture cells were prepared and double stained with anti-GLAST antibody and anti-Atx1 antibody (upper). GLAST signal intensity was quantified at a single cell level (lower). The error bars represent s.d. n=10, *P<0.05, Student's t-test. Three different experiments showed the similar result. Bar: 10 μm. (D) Bergmann glia–Purkinje cell co-culture was exposed to 50 μM of glutamate. Purkinje cell number and Purkinje branch complexity were significantly reduced when Bergmann glia were infected with Atx182Q (lower). Co-expression of Maxer-siRNA further suppressed the Purkinje cell number and its branch complexity (lower). Purkinje cells were visualized with anti-calbindin antibody (upper: representative cells). For Purkinje cell number, the error bars represent s.e.m. n=4, *P<0.05, Student's t-test. For Purkinje branch complexity, the error bars represent s.d. n=10, *P<0.05, Student's t-test. Four independent experiments showed the similar results. Bar: 10 μm.
Finally, we tested whether the phenotypes only by Maxer knockdown mimics the phenotypes by mutant Atx1. We confirmed suppression of Bergmann glia proliferation (Supplementary Figure S8A), reduction of GLAST (Supplementary Figure S8B and C) and increased vulnerability to glutamate (Supplementary Figure S8D), supporting that Maxer is the downstream effector of mutant Atx1 in the SCA1 Bergmann glia pathology.
Discussion
In this study, we identified a novel ER membrane protein that affects G1/S transition through regulation of nuclear translocation of CDK5RAP3. Maxer belongs to a new category of ER proteins that regulate the cell cycle (reviewed by Fearon and Cohen-Fix, 2008). As far as we know, Maxer is the third molecule of this category after Cln3 and SCAPER (Fearon and Cohen-Fix, 2008). Translocation of G1 cyclin Cln3 from ER to nucleus regulates initiation of S phase (Verges et al, 2007), whereas the molecule anchoring Cln3 is not yet known. SCAPER retains cyclin A in the cytoplasm (Tsang et al, 2007). Depletion of SCAPER decreases the cytoplasmic pool of cyclin A and delays G1/S transition, whereas ectopic expression of SCAPER inhibits M phase (Tsang et al, 2007). The function of Maxer seems homologous to that of SCAPER controlling nuclear translocation of a cell cycle regulatory protein. Meanwhile, the specific effect of Maxer on G1/S transition is different from that of SCAPER affecting both G1/S transition and M phase (Tsang et al, 2007).
Maxer was reduced in Bergmann glia of mutant Atx1-KI mice (Figure 2D). Bergmann glia morphologically mimic radial glia that act as stem cells in the cerebral cortex. Thus, Bergmann glia have been suspected to be actually stem cells in the cerebellum (Gaiano and Fishell, 2002; Alcock et al, 2007), although no direct proof has been reported. They express stem cell marker proteins such as Sox 1 and Sox2 (Sottile et al, 2006) and share the Notch-mediated differentiation mechanism with neural stem cells (Eiraku et al, 2005). Most importantly, Bergmann glia proliferate until the second postnatal week (Basco et al, 1977; Shiga et al, 1983; Gaiano and Fishell, 2002; Yamada and Watanabe, 2002), and this time point corresponds well to the critical period for the SCA1 pathology reported by the Orr group (Serra et al, 2006). As Bergmann glia remain in the adult brain, in addition to the developmental proliferation, a low level of proliferation after birth might be necessary to keep a certain cell number sufficient for their function in the adult cerebellum, just similar to hippocampal neural progenitor cells that keep proliferation at a low level in adulthood.
It is widely accepted that cell-autonomous effect of mutant Atx1 in Purkinje cells has a central function in SCA1 pathology. Molecular functions of Atx1 have been intensively analysed and its interactions with Capicua (Lam et al, 2006) and RBM17 (Lim et al, 2008) have been shown to have critical functions in SCA1 pathology. These molecules are suggested to be involved in splicing such as PQBP1, another candidate molecule in SCA1 pathology that interacts with a splicing factor, U5-15kD (Waragai et al, 2000; Zhang et al, 2000; Okazawa et al, 2002). It is well known that splicing is coupled closely to transcription. Thus, either transcriptional or post-transcriptional control of certain target genes is implicated in SCA1 pathology.
On the other hand, the significance of Bergmann glia in the cerebellar degeneration pathology was found in the SCA7 pathology by the La Spada group (Custer et al, 2006). Their impressive data suggested that dysfunction of Bergmann glia might impair glutamate reuptake and induce excitatory toxicity. Our results support their hypothesis, as well as further expanding the Bergmann glia hypothesis to another type of SCA, SCA1. Moreover, we identified a novel molecule, Maxer, which could be involved in the non-cell-autonomous pathway.
Regarding the reduction of GLAST, we found the discrepancy between reduction of Bergmann glia number and GLAST signal intensity (Figure 6D). We also found in WB that reduction of GLAST is higher than reduction of Sox2 (Figure 6C and E). Figure 7C supported that GLAST signal/Bergmann glia was decreased. Thus, both Bergmann glia cell number and GLAST expression per Bergmann glia contributed to the decrease of GLAST by Maxer reduction. The Atx1-induced Maxer reduction and the rescue effect of Maxer against Atx1 neurotoxicity suggest that Maxer could be a downstream effector of mutant Atx1. However, definite proof requires the generation of Bergmann glia-specific conditional KO mice of Maxer and Bergmann glia-specific transgenic mice of Maxer and for their genetic effect on Atx1-KI mice.
Considering that CDK5RAP3 is a binding partner of p35, an essential activator of CDK5 (Ching et al, 2000) and that CDK5 is involved in the terminal cell cycle exit, differentiation and migration of neural stem cells in developmental brain (Cicero and Herrup, 2005; Kawauchi et al, 2006), the function of Maxer and CDK5RAP3 in neuronal differentiation would be worth testing. Especially in the Atx1-KI mice, Maxer's function in neural stem cells should be carefully estimated, as downregulation of Maxer is not significant in pre-differentiated neural stem cells in comparison with that in Bergmann glia (data not shown).
Finally, although it is not clear at this moment how mutant Atx1 reduces Maxer, molecular functions of Atx1-interacting molecules (Waragai et al, 2000; Zhang et al, 2000; Okazawa et al, 2002; Lam et al, 2006; Lim et al, 2008) might suggest that Maxer is one of the target genes of Atx1 whose transcriptional or post-transcriptional regulation is affected by mutant Atx1.
In conclusion, we discovered a novel type of ER membrane protein regulating cell cycle through G1/S transition, which might participate in the SCA1 pathology through its functions in Bergmann glia.
Materials and methods
RNA and cDNA preparation
Total RNA was extracted from the primary neurons of Wistar rat cerebellum, ICR mouse embryo (E14) primary neural stem cells and Drosophila embryos by Trizol reagent (Invitrogen, Carlsbad, CA). The synthesis of cDNA by reverse transcription was performed using an LA PCR kit version 2.1 (Takara, Tokyo, Japan) and an oligo-dT primer.
Maxer cloning and plasmid construction
For cloning of full-length rat Maxer, the primers of MaxerF (5′-CCGCTCGAGTGATGGCGGACGCCTGG-3′) and MaxerR (5′-CGGAATTCGCTACTCCTCTGTCACAGAT-3′) were used. For generation of deletion construct of Maxer, the following primers were also used: Maxer489R (5′-CGGAATTCGCTAGTGTTTCCTTAAGCAACCTTCAA-3′), Maxer444R (5′-CGGAATTCGCTACTTGTATTCTCTGGCATTGCC-3′), Maxer357R (5′-CGGAATTCGCTAGACTGTGGCTGACGCATGCTT-3′), Maxer201R (5′-CGGAATTCGCTAGGCCGGGTAATAGCACTG-3′) and Maxer202F (5′-CCGCTCGAGTGGCAGTGAACTCTTTGGTTTCAAAATA-3′). The Maxer cDNAs were digested by XhoI and EcoR I, and cloned into XhoI and EcoR I site of pEGFP-C1 (Clontech). All constructs were verified by sequencing. For generating shRNA against rat or human Maxer, pSIREN-DNR-DsRed-Express (Clontech) shRNA expression vector was used. The inserted sequence was derived from rMaxer siRNA#1 and hMaxer siRNA (see below).
Cerebellar cell culture, neural stem cell culture and Purkinje cell culture
Cerebellar glia were prepared from Wistar rat pups at postnatal day 7 (SLC, Shizuoka, Japan). Cells were cultured in DMEM (GIBCO) containing 25 mM D-glucose, 4 mM L-glutamine, 25 mM KCl and 25 μg/ml gentamycin with 10% foetal bovine serum.
Neural stem cells were isolated from the cerebral cortex of ICR mouse embryo (E14). Dissected cerebral cortex was digested at 37°C with 0.0625% trypsin (GIBCO) and 100 μg/ml DNAse (Roche) to dissociate the cells. Tissue was treated with 350 μg/ml Ovomucoid (Sigma) to inhibit trypsin activity and passed through a 70 μm cell strainer (BD). NSCs were cultured in a DMEM/F12 (GIBCO), basic fibroblast growth factor (20 ng/ml, Promega), epidermal growth factor (20 ng/ml, Promega) and B27 supplement (GIBCO). NSCs at 3–5 passages were used for experiments.
Purkinje cells were cultured as described earlier (Tabata et al, 2000). Briefly, cerebella from E18 embryo were incubated in 0.0125% trypsin (GIBCO) at 33°C for 15 min. Dissociated cells were cultured on poly-L-ornithine-coated dish in DMEM/F12 (GIBCO) supplemented with B27(GIBCO), L-glutamine(1.4 mM) and 10% FBS.
Culture of cell lines, transfections and RNAi sequences
C6 glioma and HeLa cells were maintained at 37°C, 5% CO2 in DMEM (Sigma) supplemented with 10% FBS. P19 cells were in MEM (GIBCO) supplemented with 10% FBS. Transfections of plasmids and siRNAs were performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. siRNAs against Maxer (rat, human, mouse) and CDK5RAP3 were purchased from Qiagen (Valencia, CA). They are abbreviated in this study as rMaxer siRNA#1 (target sequence, TTGCAAGAAAGTGGTCAAGTA), rMaxer siRNA#2 (target sequence, CAAGAAGAGGTTTCAAATCTA), hMaxer siRNA (target sequence, TCCGTGGACTATTCAGTGCTA), mMaxer siRNA (target sequence, CTGCATTGAGATTGTGAATAA) and CDK5RAP3 siRNA (target sequence, AAGATTAATTTCCATAATAAA), respectively. As the control, negative control siRNA (Qiagen) UUCUCCGAACGUGUCACGUdTdT was used.
In situ hybridization
Free-floating sections were rinsed with PBS and fixed with 4% PFA 15 min. Then, sections were permeabilized with 0.3% Triton X-100 in PBS for 15 min, washed with PBS for 5 min and treated with Proteinase K (10 μg/ml in 0.1M Tris–HCl; 50 mM EDTA, pH 8.0) for 30 min at 37°C. Sections were rinsed twice with 0.75% glycine in PBS, acetylated with 0.25% acetic anhydride in 0.1 M triethanolamine for 15 min and rinsed twice with PBS. Prehybridization was performed in hybridization buffer [50% deionized formamide (dFA), 2% Blocking Reagent (Roche Diagnostics), 5 × SSC (1 × SSC consists of 0.88% NaCl and 0.44% Na3C6H5O3 2H2O), 0.1% N-lauroylsarcosine and 0.1% sodium lauryl sulphate] at 60°C for 2 h. Sections were transferred to new hybridization buffer containing DIG-labelled antisense RNA probes and incubated at 60°C overnight. They were washed twice in 2 × SSC, 50% dFA, 0.1% N-lauroylsarcosine at 50°C for 15 min, rinsed with RNase A buffer (10 mM Tris–HCl; 10 mM EDTA; 0.5M NaCl pH 8.0) and incubated with 20 μg/ml RNase A in RNase A buffer at 37°C for 20 min. The sections were washed twice in 2 × SSC; 0.1% N-lauroylsarcosine, for 15 min, twice in 0.2 × SSC; 0.1% N-lauroylsarcosine, for 20 min at 37°C, rinsed with TS7.5 buffer (0.1 M Tris–HCl; 0.15 M NaCl pH 7.5) and incubated in 1% Blocking Reagent in TS7.5 for 2 h at room temperature. After blocking, sections were transferred to anti-DIG antibody (1:2000, Roche Diagnostics) in 1% blocking reagent. After overnight antibody incubation, sections were washed three times with TS7.5 containing 0.1% Tween 20 for 20 min, rinsed in TS9.5 buffer (0.1 M Tris–HCl; 0.1 M NaCl; 50 mM MgCl2 pH 9.5) and incubated in NBT/BCIP (Roche Diagnostics) solution until adequate colour development. Digoxigenin-labelled probes for Maxer were synthesized from a 750 bp fragment of Maxer cDNA (nucleotides 1–750 from the initiation codon). Control experiments were carried out, and no signals were observed.
Quantitative analysis of signal intensity in images
Signal intensity in immunostaining was calculated by using MetaMorph software (Universal Imaging Corporation, Downingtown). The fluorescence signal intensities per area (μm2) of molecular layer (for immunohistochemistry; Figure 8D; Supplementary Figure 7A), total visual field (for immunocytochemistry; Figures 7B, 8B; Supplementary Figure 7B) or cytoplasm (for single cell intensity; Figures 7C and 8C) were measured. To exclude background fluorescence, we measured signal intensity of 10 randomly selected non-cell existing areas of each sample, and their mean value was subtracted from the cellular fluorescence signals.
Immunoprecipitation
Hela cells (80% confluency in 10 cm dish) were transfected with pEGFP-C1, pEGFP-C1–Maxer and serial of pEGFP-C1–Maxer deletion constructs by Lippofectamin 2000 (Invitrogen). The cells were harvested, incubated in TNE buffer (10 mM Tris–HCl at pH 7.8, 10% NP-40, 0.15 M NaCl, 1 mM EDTA) at 4°C for 1 h in a rotator, and centrifuged at 17 400 g for 20 min. The supernatant was preincubated with protein G–Sepharose (Amersham Biosciences) for 2 h at 4°C and centrifuged. The supernatant was incubated either with anti-GFP antibodies (1:200; Clontech) overnight and with protein G–Sepharose for 2 h. Protein G beads were collected by centrifugation at 2000 g for 5 min, washed five times with TNE buffer. Bound proteins were eluted in sample buffer, separated on SDS–PAGE, and blotted with antibodies.
Fluorescence-activated cell sorter analysis
At 72 h after transfection, cells were harvested and fixed with 70% ethanol in PBS at −20°C overnight. Cells were washed and labelled with propidium iodide (Sigma) at 40 μg/ml for 30 min at 37°C. For each analysis, 10 000 gated cells were collected by FACSCalibur system and analysed with the CELLQuest program (Becton Dickinson).
Immunocytochemistry and immunohistochemistry
Cultured cells were fixed in 4% paraformaldehyde (prepared in PB) at room temperature for 15 min. After fixation, cells were treated with 0.1% Triton X-100 in PBS for 10 min, blocked with PBS containing 5% skimmed milk for 30 min at room temperature and incubated with primary antibodies diluted in the blocking buffer. The primary antibodies were anti-α-tubulin antibody DM1A (1:1000; Sigma-Aldrich), anti-CDK5RAP3 antibody (1:200; Bethyl), anti-Atx1 antibody H-21 (1:200; Santa Cruz), anti-GLAST (1:500; Chemicon), anti-GFAP antibody GA-5 Cy3 conjugated (1:3000; Sigma-Aldrich) and anti-MAP2 HM-2 (1:500; Sigma-Aldrich).
To prepare cortical sections, mouse embryos or adult brains were dissected, and fixed in 4% paraformaldehyde for overnight at 4°C. The fixed samples were gradually soaked in 30% sucrose as a final concentration before embedded in OCT compound or paraffin. A total of 10 μm of sections were prepared. Mutant Atx1-KI mice (B6.129S-Atxn1tm1Hzo; Sca1154Q/2Q), which had been made by one of the co-authors in the Zoghbi's laboratory (Watase et al, 2002), were generous gift from Professor Huda Y Zoghbi (Baylor College of Medicine). Immunohistochemical staining of Maxer by anti-Maxer antibody C15-01D (1:20) was performed by TSA Plus Fluorescence Systems (PerkinElmer) according to the manufacturer's protocol. The following primary antibodies were also used: anti-GLAST antibody (1:2500; Chemicon), anti-GLT1 antibody (1:100; Matsugami et al, 2006), anti-Atx1 antibody 11NQ (1:100; Watase et al, 2002), anti-GFAP antibody GA-5 Cy3-conjugated (1:3000; Sigma-Aldrich), anti-Sox2 Y17 antibody (1:200; Santa Cruz), anti-PCNA antibody (1:2000; SIGMA) and anti-NeuN antibody A-60 (1:500; Chemicon). The secondary antibodies were Cy3-labelled anti-rabbit and anti-goat IgG antibody (Jackson Immuno-Rearch), anti-rabbit IgG goat Alexa 488 antibody (Molecular Probes), anti-mouse IgG goat Alexa 546 and 488 antibody (Molecular Probes), and Anti-guinea pig IgG goat Alexa 568 antibody (Molecular Probes).
Images were acquired using an LSM510 Meta confocal microscope (Zeiss).
Quantitative RT–PCR
Quantitative PCR analyses were performed with 7300 Real-time PCR system (Applied Biosystems). CG1104 expression was analysed by forward primer (5′-GGGTAGTGACTGGGACGAGATC-3′), reverse primer (5′-ACGTGAGCTGCGCCTTCT-3′) and Taqman probe (5′-AGCGCTTGGCCGCCGACTT-3′); actin5C level was measured by forward primer (5′-CCGAGCGCGGTTACTCTTT-3′), reverse primer (5′-CAACATAGCACAGCTTCTCCTTGAT-3′) and Taqman probe (5′-CCGCTGAGCGTGAAATCGTCCGT-3′) as a control.
Anti-Maxer antibody
C15-01D was raised against the C-terminal region of Maxer by immunizing a rabbit with the peptide C-KDLVLKSRKSSVTEE (amino acids 779–793) conjugated to the carrier protein by cysteine.
Adenovirus vector
The cosmid of rat Maxer-pAxCA was transfected into 293 cells by the calcium–phosphate method using the digested DNA of adenoviruses. After the cells died, the medium was recovered as the virus solution. We then rechecked the construction of the adenovirus vectors through PCR and confirmed that the E1A protein was deleted and that the insert was maintained correctly. After the check, we amplified the adenoviruses two to three times. Adenovirus, AxCA-Atx130Q and -Atx182Q, were constructed as described earlier (Tagawa et al, 2007). The adenovirus vectors contain the full-length Atx1 protein. The vector was used to infect cerebellar primary culture cells and neural stem cells at a multiplicity of infection of 100.
Western blot analysis
Western blot analysis was performed as described earlier (Tagawa et al, 2007). Briefly, whole cells were dissolved in 62.5 mM Tris–HCl, pH 6.8, 2% (w/v) SDS, 2.5% (v/v) 2-mercaptoethanol, 5% (v/v) glycerin and 0.0025% (w/v) bromophenol blue. Samples were separated by SDS–PAGE, transferred onto Immobilon-P Transfer Membrane (Millipore) through a semidry method, blocked by 5% milk in TBS with Tween 20 (TBST) (10 mM Tris/Cl, pH 8.0, 150 mM NaCl, 0.05% Tween 20). The filters were incubated with each primary antibody for overnight at 4°C. Primary antibodies used were anti-Maxer antibody C15-01D (1:100), anti-α-tubulin antibody DM1A (1:1000; Sigma-Aldrich), anti-CDK5RAP3 antibody (1:200; Abnova), anti-GFP antibody JL-8 (1:1000; Clontech), anti-cyclin E1 HE247 (1:200; Santa Cruz), anti-cyclin D1 antibody C-20 (1:10 000; Santa Cruz), anti-HDAC3 antibody (1:500; Upstate), anti-Dynein IC(74-1) antibody (1:1000; Santa Cruz), anti-Aurora-A 35C1 (1:2000; Sigma-Aldrich), anti-Bin1 (1:300; Upstate), anti-GLAST (1:300; Chemicon), anti-GLT1 antibody (1:1000; Matsugami et al, 2006), anti-Atx1 antibody H-21 (1:500; Santa Cruz) and anti-GAPDH antibody 6C5 (1:5000; Chemicon). The filters were then incubated with the corresponding horseradish peroxidase-conjugated second antibody at a 1:3000 dilution for 1 h at room temperature in 5% milk/TBST. Finally, the target molecules were visualized through an enhanced chemiluminescence western blotting detection system (Amersham Biosciences, GE Health Care Biosciences, Hong Kong).
Subcellular fractionation
Hela cells were incubated in the same volume of hypotonic buffer (20 mM HEPES, pH 7.9, 10 mM KCl, 2 mM MgCl2, 0.3% Nonidet P-40, 1 mM dithiothreitol, 1 mM EDTA and protease Inhibitor Cocktail (Roche Diagnostics)) and placed on ice for 20 min. The extracts were centrifuged at 15 000 g for 20 min at 4°C. The supernatants were collected as the cytoplasmic fraction, and pellets were collected as nuclear fraction. For membrane fractionation, Hela cells were incubated in hypotonic lysis buffer (10 mM Tris, pH 8.0, 10 mM NaCl, 3 mM MgCl2 and 1 mM EGTA with a protease Inhibitor Cocktail) for 10 min, followed by 20 strokes of a Dounce B homogenizer. The lysates were spun down for 5 min at 500 g. The supernatant was spun at 100 000 g for 60 min, and the pellet was collected as membrane fraction.
Northern blot analysis
RNAs were subjected to electrophoresis using an MOPS/formaldehyde gel. Separated RNAs were blotted to Hybond-N (GE Healthcare) and fixed by UV cross-linking (120 000 μJ/cm2). Full-length cDNAs of rat Maxer, mouse Maxer, rat α-tubulin1A and rat GAPDH were digested from plasmids and radiolabelled by a random primer DNA labelling kit (Takara). 32P-labelled probes were hybridized to nylon membrane at 42°C overnight with shaking.
Statistical analysis
Statistical analysis was performed using Student's t-test. Error bars indicate s.e.m. or s.d.
Supplementary Material
Acknowledgments
This work was supported by grants to HO from Japan Science Technology Agency (PRESTO and CREST, JST, Japan) and from Ministry of Education, Culture, Sports, Science and Technology of Japan (16390249, 16650076, 18390254, 18650097, Research on Pathomechanisms of Brain Disorders: 17025017, 18023014, 20023011). We are grateful to Professor Huda Y Zoghbi (Baylor College of Medicine) for providing mutant Atx1-KI (Sca1154Q/2Q) mice, Professor Erich E Wanker (Max-Delbrück Center for Molecular Medicine) for kind support and Mrs Tayoko Tajima for her excellent technical support. We thank Dr Sam Barclay (Imperial College of London, Medical School) for editing the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alcock J, Scotting P, Sottile V (2007) Bergmann glia as putative stem cells of the mature cerebellum. Med Hypotheses 69: 341–345 [DOI] [PubMed] [Google Scholar]
- Basco E, Hajos F, Fulop Z (1977) Proliferation of Bergmann-glia in the developing rat cerebellum. Anat Embryol (Berl) 151: 219–222 [DOI] [PubMed] [Google Scholar]
- Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP (2004) Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci USA 101: 12130–12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Bonizzi G, Seagroves TN, Greten FR, Johnson R, Schmidt EV, Karin M (2001) IKKalpha provides an essential link between RANK signaling and cyclin D1 expression during mammary gland development. Cell 107: 763–775 [DOI] [PubMed] [Google Scholar]
- Ching YP, Qi Z, Wang JH (2000) Cloning of three novel neuronal Cdk5 activator binding proteins. Gene 242: 285–294 [DOI] [PubMed] [Google Scholar]
- Cicero S, Herrup K (2005) Cyclin-dependent kinase 5 is essential for neuronal cell cycle arrest and differentiation. J Neurosci 25: 9658–9668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custer SK, Garden GA, Gill N, Rueb U, Libby RT, Schultz C, Guyenet SJ, Deller T, Westrum LE, Sopher BL, La Spada AR (2006) Bergmann glia expression of polyglutamine-expanded ataxin-7 produces neurodegeneration by impairing glutamate transport. Nat Neurosci 9: 1302–1311 [DOI] [PubMed] [Google Scholar]
- Di Giorgio FP, Carrasco MA, Siao MC, Maniatis T, Eggan K (2007) Non-cell autonomous effect of glia on motor neurons in an embryonic stem cell-based ALS model. Nat Neurosci 10: 608–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiraku M, Tohgo A, Ono K, Kaneko M, Fujishima K, Hirano T, Kengaku M (2005) DNER acts as a neuron-specific Notch ligand during Bergmann glial development. Nat Neurosci 8: 873–880 [DOI] [PubMed] [Google Scholar]
- Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O'Connor L, Li M, Taylor R, Dharsee M, Ho Y, Heilbut A, Moore L, Zhang S, Ornatsky O, Bukhman YV, Ethier M, Sheng Y et al. (2007) Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol 3: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon P, Cohen-Fix O (2008) The endoplasmic reticulum takes center stage in cell cycle regulation. Sci Signal 1: pe4. [DOI] [PubMed] [Google Scholar]
- Gaiano N, Fishell G (2002) The role of notch in promoting glial and neural stem cell fates. Annu Rev Neurosci 25: 471–490 [DOI] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M et al. (2003) A protein interaction map of Drosophila melanogaster. Science 302: 1727–1736 [DOI] [PubMed] [Google Scholar]
- Goldowitz D, Hamre K (1998) The cells and molecules that make a cerebellum. Trends Neurosci 21: 375–382 [DOI] [PubMed] [Google Scholar]
- Ilieva H, Polymenidou M, Cleveland DW (2009) Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J Cell Biol 187: 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki R, Tagawa K, Qi ML, Enokido Y, Ito H, Tamura T, Shimizu S, Oyanagi K, Arai N, Kanazawa I, Wanker EE, Okazawa H (2008) Omi / HtrA2 is relevant to the selective vulnerability of striatal neurons in Huntington's disease. Eur J Neurosci 28: 30–40 [DOI] [PubMed] [Google Scholar]
- Jiang H, Luo S, Li H (2005) Cdk5 activator-binding protein C53 regulates apoptosis induced by genotoxic stress via modulating the G2/M DNA damage checkpoint. J Biol Chem 280: 20651–20659 [DOI] [PubMed] [Google Scholar]
- Karin M, Cao Y, Greten FR, Li ZW (2002) NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer 2: 301–310 [DOI] [PubMed] [Google Scholar]
- Kawauchi T, Chihama K, Nabeshima Y, Hoshino M (2006) Cdk5 phosphorylates and stabilizes p27kip1 contributing to actin organization and cortical neuronal migration. Nat Cell Biol 8: 17–26 [DOI] [PubMed] [Google Scholar]
- Kretzschmar D, Tschape J, Bettencourt Da Cruz A, Asan E, Poeck B, Strauss R, Pflugfelder GO (2005) Glial and neuronal expression of polyglutamine proteins induce behavioral changes and aggregate formation in Drosophila. Glia 49: 59–72 [DOI] [PubMed] [Google Scholar]
- Lam YC, Bowman AB, Jafar-Nejad P, Lim J, Richman R, Fryer JD, Hyun ED, Duvick LA, Orr HT, Botas J, Zoghbi HY (2006) ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell 127: 1335–1347 [DOI] [PubMed] [Google Scholar]
- Lievens JC, Iche M, Laval M, Faivre-Sarrailh C, Birman S (2008) AKT-sensitive or insensitive pathways of toxicity in glial cells and neurons in Drosophila models of Huntington's disease. Hum Mol Genet 17: 882–894 [DOI] [PubMed] [Google Scholar]
- Lim J, Crespo-Barreto J, Jafar-Nejad P, Bowman AB, Richman R, Hill DE, Orr HT, Zoghbi HY (2008) Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature 452: 713–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobsiger CS, Cleveland DW (2007) Glial cells as intrinsic components of non-cell-autonomous neurodegenerative disease. Nat Neurosci 10: 1355–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsugami TR, Tanemura K, Mieda M, Nakatomi R, Yamada K, Kondo T, Ogawa M, Obata K, Watanabe M, Hashikawa T, Tanaka K (2006) From the cover: indispensability of the glutamate transporters GLAST and GLT1 to brain development. Proc Natl Acad Sci USA 103: 12161–12166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai M, Re DB, Nagata T, Chalazonitis A, Jessell TM, Wichterle H, Przedborski S (2007) Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat Neurosci 10: 615–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazawa H, Rich T, Chang A, Lin X, Waragai M, Kajikawa M, Enokido Y, Komuro A, Kato S, Shibata M, Hatanaka H, Mouradian MM, Sudol M, Kanazawa I (2002) Interaction between mutant ataxin-1 and PQBP-1 affects transcription and cell death. Neuron 34: 701–713 [DOI] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127: 635–648 [DOI] [PubMed] [Google Scholar]
- Qi ML, Tagawa K, Enokido Y, Yoshimura N, Wada Y, Watase K, Ishiura S, Kanazawa I, Botas J, Saitoe M, Wanker EE, Okazawa H (2007) Proteome analysis of soluble nuclear proteins reveals that HMGB1/2 suppress genotoxic stress in polyglutamine diseases. Nat Cell Biol 9: 402–414 [DOI] [PubMed] [Google Scholar]
- Rost B, Yachdav G, Liu J (2004) The PredictProtein server. Nucleic Acids Res 32: W321–W326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra HG, Duvick L, Zu T, Carlson K, Stevens S, Jorgensen N, Lysholm A, Burright E, Zoghbi HY, Clark HB, Andresen JM, Orr HT (2006) RORalpha-mediated Purkinje cell development determines disease severity in adult SCA1 mice. Cell 127: 697–708 [DOI] [PubMed] [Google Scholar]
- Sherr CJ, McCormick F (2002) The RB and p53 pathways in cancer. Cancer Cell 2: 103–112 [DOI] [PubMed] [Google Scholar]
- Shiga T, Ichikawa M, Hirata Y (1983) Spatial and temporal pattern of postnatal proliferation of Bergmann glial cells in rat cerebellum: an autoradiographic study. Anat Embryol (Berl) 167: 203–211 [DOI] [PubMed] [Google Scholar]
- Shin JY, Fang ZH, Yu ZX, Wang CE, Li SH, Li XJ (2005) Expression of mutant huntingtin in glial cells contributes to neuronal excitotoxicity. J Cell Biol 171: 1001–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sottile V, Li M, Scotting PJ (2006) Stem cell marker expression in the Bergmann glia population of the adult mouse brain. Brain Res 1099: 8–17 [DOI] [PubMed] [Google Scholar]
- Tabata T, Sawada S, Araki K, Bono Y, Furuya S, Kano M (2000) A reliable method for culture of dissociated mouse cerebellar cells enriched for Purkinje neurons. J Neurosci Methods 104: 45–53 [DOI] [PubMed] [Google Scholar]
- Tagawa K, Marubuchi S, Qi ML, Enokido Y, Tamura T, Inagaki R, Murata M, Kanazawa I, Wanker EE, Okazawa H (2007) The induction levels of heat shock protein 70 differentiate the vulnerabilities to mutant huntingtin among neuronal subtypes. J Neurosci 27: 868–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T, Sone M, Yamashita M, Wanker EE, Okazawa H (2009) Glial cell lineage expression of mutant ataxin-1 and huntingtin induces developmental and late-onset neuronal pathologies in Drosophila models. PLoS One 4: e4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang WY, Wang L, Chen Z, Sanchez I, Dynlacht BD (2007) SCAPER, a novel cyclin A-interacting protein that regulates cell cycle progression. J Cell Biol 178: 621–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verges E, Colomina N, Gari E, Gallego C, Aldea M (2007) Cyclin Cln3 is retained at the ER and released by the J chaperone Ydj1 in late G1 to trigger cell cycle entry. Mol Cell 26: 649–662 [DOI] [PubMed] [Google Scholar]
- Wang J, An H, Mayo MW, Baldwin AS, Yarbrough WG (2007) LZAP, a putative tumor suppressor, selectively inhibits NF-kappaB. Cancer Cell 12: 239–251 [DOI] [PubMed] [Google Scholar]
- Wang J, He X, Luo Y, Yarbrough WG (2006) A novel ARF-binding protein (LZAP) alters ARF regulation of HDM2. Biochem J 393: 489–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waragai M, Junn E, Kajikawa M, Takeuchi S, Kanazawa I, Shibata M, Mouradian MM, Okazawa H (2000) PQBP-1/Npw38, a nuclear protein binding to the polyglutamine tract, interacts with U5-15kD/dim1p via the carboxyl-terminal domain. Biochem Biophys Res Commun 273: 592–595 [DOI] [PubMed] [Google Scholar]
- Watase K, Weeber EJ, Xu B, Antalffy B, Yuva-Paylor L, Hashimoto K, Kano M, Atkinson R, Sun Y, Armstrong DL, Sweatt JD, Orr HT, Paylor R, Zoghbi HY (2002) A long CAG repeat in the mouse Sca1 locus replicates SCA1 features and reveals the impact of protein solubility on selective neurodegeneration. Neuron 34: 905–919 [DOI] [PubMed] [Google Scholar]
- Yamada K, Watanabe M (2002) Cytodifferentiation of Bergmann glia and its relationship with Purkinje cells. Anat Sci Int 77: 94–108 [DOI] [PubMed] [Google Scholar]
- Yamanaka K, Boillee S, Roberts EA, Garcia ML, McAlonis-Downes M, Mikse OR, Cleveland DW, Goldstein LS (2008a) Mutant SOD1 in cell types other than motor neurons and oligodendrocytes accelerates onset of disease in ALS mice. Proc Natl Acad Sci USA 105: 7594–7599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K, Chun SJ, Boillee S, Fujimori-Tonou N, Yamashita H, Gutmann DH, Takahashi R, Misawa H, Cleveland DW (2008b) Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat Neurosci 11: 251–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lindblom T, Chang A, Sudol M, Sluder AE, Golemis EA (2000) Evidence that dim1 associates with proteins involved in pre-mRNA splicing, and delineation of residues essential for dim1 interactions with hnRNP F and Npw38/PQBP-1. Gene 257: 33–43 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.