Abstract
Integral membrane proteins of the inner nuclear membrane (INM) are inserted into the endoplasmic reticulum membrane during their biogenesis and are then targeted to their final destination. We have used human SUN2 to delineate features that are required for INM targeting and have identified multiple elements that collectively contribute to the efficient localization of SUN2 to the nuclear envelope (NE). One such targeting element is a classical nuclear localization signal (cNLS) present in the N-terminal, nucleoplasmic domain of SUN2. A second motif proximal to the cNLS is a cluster of arginines that serves coatomer-mediated retrieval of SUN2 from the Golgi. Unexpectedly, also the C-terminal, lumenal SUN domain of SUN2 supports NE localization, showing that targeting elements are not limited to cytoplasmic or transmembrane domains of INM proteins. Together, SUN2 represents the first mammalian INM protein relying on a functional cNLS, a Golgi retrieval signal and a perinuclear domain to mediate targeting to the INM.
Keywords: coatomer, inner nuclear membrane, nuclear localization signal, nuclear transport, SUN domain
Introduction
The nuclear envelope (NE) consists of two closely juxtaposed lipid bilayers termed outer and inner nuclear membrane (ONM and INM). These are fused at numerous sites, generating NE perforations occupied by large marcomolecular assemblies called nuclear pore complexes (NPCs) (Tran and Wente, 2006). NPCs serve as diffusion barrier and mediate receptor-facilitated transport of macromolecules between the nucleus and the cytoplasm. Although INM and ONM are connected at the sites of pore insertion, both membranes possess a distinct membrane protein composition. In metazoan cells, the INM is tightly associated with the nuclear lamina, an intermediate filament network composed of nuclear lamins, and many INM proteins are lamin interaction partners (Worman and Courvalin, 2000; Stewart et al, 2007). The ONM is an extension of the rough endoplasmic reticulum (ER) membrane. These contiguous membranes share a common set of integral membrane proteins. Some membrane proteins, however, are ONM specific and are retained at the ONM by interaction with INM proteins in the perinuclear space (PNS).
Like ER and ONM proteins, newly synthesized integral INM proteins are inserted into the ER membrane. INM-destined proteins diffuse laterally through the ER membrane system and the ONM, where they encounter NPCs. To traverse NPCs, INM proteins move along the pore membrane, likely passing peripheral channel-like openings of the NPC by a poorly defined mechanism. Only a dozen INM-specific proteins were known in mammalian cells until recently when proteomic approaches identified many novel INM proteins (Schirmer and Gerace, 2005). To date, we know a little <100 INM-resident integral membrane proteins. Many of them connect to chromatin, the nuclear lamina or both. Those associated with lamins and linked to genetic diseases have been studied in particular detail (Burke and Stewart, 2002; Gruenbaum et al, 2005).
The specific targeting of proteins to the INM has long been explained solely by the ‘diffusion–retention model' (Smith and Blobel, 1993; Soullam and Worman, 1995; Holmer and Worman, 2001). According to this model, sorting of INM proteins relies on their diffusion from the membrane insertion site in the ER along the pore membrane to the INM followed by their association with nuclear binding partners. Evidence for this model has been obtained through the analysis of several INM proteins such as LAP1, LBR, MAN1, LAP2 and emerin (Powell and Burke, 1990; Smith and Blobel, 1993; Soullam and Worman, 1993, 1995; Ellenberg et al, 1997; Yang et al, 1997; Furukawa et al, 1998; Ostlund et al, 1999; Tsuchiya et al, 1999; Wu et al, 2002). Notably, the diffusion–retention model does not invoke any energy-dependent step in the targeting pathway. However, INM localization of a diffusible INM reporter protein has been shown to be compromised by ATP depletion without affecting its mobility in the peripheral ER, suggesting that an energy-dependent step might help INM proteins during NPC passage (Ohba et al, 2004).
The universality of the diffusion–retention model has further been challenged by the concept of receptor-mediated translocation of certain yeast INM proteins through NPCs (King et al, 2006). The yeast INM proteins Heh1p and Heh2p possess classical nuclear localization signals (cNLSs), normally directing import of soluble nuclear transport cargo. Mutation of the NLS sequence in Heh2 causes its mislocalization to the ER. INM targeting of the Heh proteins requires both the heterodimeric NLS import receptor formed by karyopherin (kap) α and β1 and a functional RanGTPase system. Thus, mechanistically, import of the Heh proteins shares features with facilitated transport of soluble cargo through the NPC. It is currently unclear if NLS-dependent INM protein sorting is limited to yeast. Notably, NLS-like sequences can be identified in a significant number of mammalian INM proteins by computer prediction, yet direct evidence for the functionality of any of these NLSs is lacking (Lusk et al, 2007).
Sorting of INM proteins has also been explained by the existence of INM sorting motifs (INM-SMs), first discovered in baculovirus-derived membrane proteins (Braunagel et al, 2004). INM-SMs are characterized by positively charged amino acids located 4–8 amino acids from the nucleoplasmic face of the transmembrane domain. These INM-SMs are found in many INM proteins from different species (Braunagel et al, 2004). INM-SMs are recognized co-translationally by a truncated, membrane-associated isoform of karyopherin α referred to as importin-α (Impα)-16/KPNA-4-16 (Saksena et al, 2006; Braunagel et al, 2007). Impα-16 has been suggested to facilitate accumulation of newly synthesized INM proteins at the nuclear periphery (Braunagel et al, 2007) as well as their subsequent transport to the INM (Braunagel et al, 2009).
To elucidate the mechanisms that underlie integral INM protein sorting in mammalian cells, we have used human SUN2 as a model protein for analysis of sequence elements that contribute to its INM targeting. SUN2 belongs to the family of conserved SUN domain containing transmembrane proteins that are part of so-called LINC complexes involved in connecting the NE to the cytoskeleton (Hodzic et al, 2004; Crisp et al, 2006; Tzur et al, 2006; Stewart et al, 2007; Starr, 2009). Mammals encode for at least five different SUN proteins, of which SUN1 and SUN2 are known to localize specifically to the INM (Hodzic et al, 2004; Padmakumar et al, 2005; Crisp et al, 2006; Haque et al, 2006; Hasan et al, 2006). The N-termini of SUN1 and SUN2 extend into the nucleoplasm, whereas their conserved C-terminal SUN domains reside in the PNS. Within the PNS, the SUN domains associate with short, lumenal peptides of KASH domain containing membrane proteins of the ONM to form a molecular bridge linking the ONM and INM (Padmakumar et al, 2005; Crisp et al, 2006; McGee et al, 2006). SUN2 forms stable homodimers through coiled-coil regions adjacent to its SUN domain, but may also heteroligomerize with SUN1 to some extent (Wang et al, 2006; Lu et al, 2008). Our analysis reveals that INM targeting of SUN2 is a robust process that relies on several sorting elements present in its N-terminal, nucleoplasmic domain as well as in its C-terminal, lumenal region.
Results
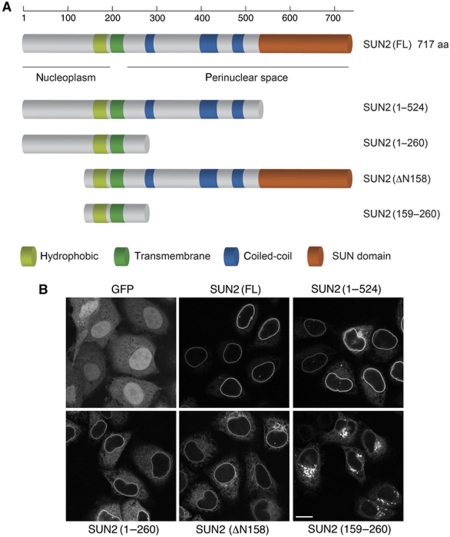
To characterize the regions of SUN2 required for its localization to the NE, we expressed truncated versions of SUN2 fused to GFP at their C-termini in HeLa cells (Figure 1A). Similar to endogenous SUN2, full-length SUN2–GFP was efficiently targeted to the nuclear rim (Figure 1B). Deletion of most of the N-terminal, nucleoplasmic domain (ΔN158) or of the C-terminal, lumenal domain (1–260) did not abolish NE targeting. N- or C-terminal deletion led to some accumulation of these SUN2 fragments in the membrane system of the ER, and for SUN2(ΔN158) also in the Golgi, suggesting that targeting of SUN2 to the INM may rely on contributions of both its N- and C-terminal domains. In agreement, concomitant deletion of both the N-terminal 158 aa and the C-terminal, lumenal domain in SUN2(159–260) caused a loss of NE signal and a pronounced localization of this SUN2 fragment in the Golgi.
Figure 1.
Deletion of the N- or C-terminal domain of SUN2 results in NE targeting defects. (A) Schematic representation of SUN2 and its deletion constructs. The conserved SUN domain is depicted in orange, the coiled coils in blue, the transmembrane segment in dark green and additional hydrophobic stretches in light green. (B) HeLa cells were transiently transfected with GFP, SUN2–GFP, SUN2(1–524)–GFP, SUN2(1–260)–GFP, SUN2(ΔN158)–GFP or SUN2(159–260)–GFP; 24 h post-transfection, cells were fixed and analysed by confocal fluorescence microscopy. Scale bar is 15 μm.
The nucleoplasmic domain of SUN2 binds the Impα/Impβ heterodimer
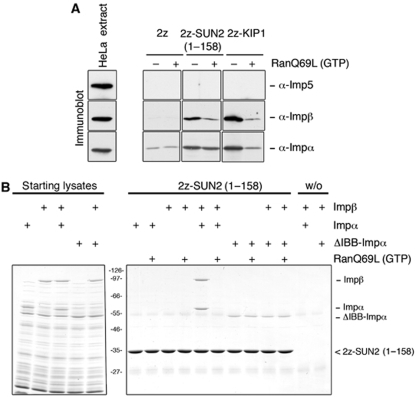
Given that its removal affected NE localization of SUN2, the N-terminal fragment of SUN2 might either constitute a domain involved in nuclear retention or contain a nuclear import signal. As active transport of yeast transmembrane proteins to the INM depends on the karyopherin family member Kap95, we tested whether the N-terminal domain of SUN2 can interact with the metazoan homologue of Kap95p termed importin β (Impβ). We expressed and purified recombinant SUN2(1–158) fused to an N-terminal protein A tag (2z-tag) from bacteria and used it for pull-down experiments with HeLa cell extracts (Figure 2A). As a positive control, we analysed binding of Impβ to 2z-KIP1, a cyclin-dependent kinase inhibitor known to be imported into the nucleus by use of a classical, basic NLS (Sekimoto et al, 2004). Binding to 2z alone served as a negative control. Western blot analysis revealed that Impβ was recruited to both 2z-SUN2(1–158) and 2z-Kip1 from HeLa cell extract. Another importin, Imp5, that recognizes a different type of basic NLS (Jakel and Gorlich, 1998) was not bound.
Figure 2.
The N-terminal domain of SUN2 binds the nuclear transport receptor heterodimer Impα/Impβ. (A) HeLa cell extract was incubated with 1.5 μM of recombinant, purified 2z, 2z-SUN2(1–158) or 2z-KIP1 in the absence or presence of 2.5 μM RanQ69L(GTP). 2z-tagged proteins and associated factors were retrieved from the reaction mixtures with IgG sepharose. Bound proteins were eluted and separated by 8% SDS–PAGE followed by immunoblotting using antibodies directed to human Impβ, Impα (RCH1) and Imp5. The load in the extract lanes corresponds to 0.1% of the input and the load for the bound fractions corresponds to 1/12 of the total. (B) 2z-SUN2(1–158) directly binds the Impα/Impβ heterodimer; 1.5 μM recombinant, purified 2z-SUN2(1–158) was mixed with the indicated purified factors (Impα, Impβ, ΔIBBImpα; 0.4 μM each) in E. coli lysate (starting lysates) in the absence or presence of 2.5 μM RanQ69L(GTP). For control reactions, 2z-SUN2(1–158) was omitted (w/o). Bound proteins were retrieved from the reaction mixtures with IgG sepharose, eluted and separated by 8% SDS–PAGE followed by colloidal Coomassie staining. Load for the starting material corresponds to 1% of the total and load for the bound fraction corresponds to 20% of the total.
The binding of importins to their nuclear transport cargo is known to be sensitive to the addition of RanGTP, which causes substrate release from nuclear import receptors. The association of Impβ with 2z-SUN2(1–158) was indeed abolished in the presence of RanQ69L(GTP) (Figure 2A), a GTPase-deficient Ran mutant locked in the GTP-bound form, indicating that the interaction satisfies this criterion for an importin–substrate interaction.
Impβ can bind import substrates either directly or in conjunction with import adaptor proteins such as Impα, which recognizes basic NLSs. Immunoblot analysis of 2z-SUN2(1–158)- and 2z-Kip1-bound fractions revealed that the abundant Impα isoform RCH1 was associated with both proteins suggesting that the N-terminal domain of SUN2 recruits the heterodimeric NLS import receptor complex composed of Impα and Impβ.
To test whether the interaction of Impα and Impβ with SUN2(1–158) was direct, we performed binding experiments using purified transport factors expressed in Escherichia coli. Recombinant Impβ bound efficiently to 2z-SUN2(1–158) in the presence of Impα (RCH1, hsImpα2), but not if Impα was omitted from the binding reaction or if an Impα mutant lacking its N-terminal Impβ-binding domain (ΔIBB) was used (Figure 2B). The binding of recombinant Impβ to 2z-SUN2(1–158) was prevented by the addition of RanQ69L(GTP). Note that Impα was only strongly recruited in the presence of Impβ because of the auto-inhibitory effect of Impα's IBB domain on NLS binding. In contrast, the ΔIBB mutant of Impα bound to 2z-SUN2(1–158) in the absence of Impβ. This direct binding between Impα and the N-terminal, nucleoplasmic part of SUN2 suggested that SUN2 contains a cNLS that could, in principle, support nuclear import of SUN2.
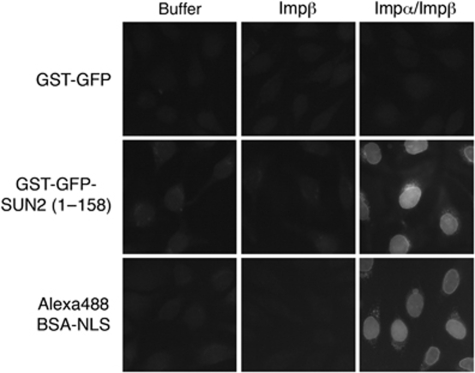
Nuclear import of soluble nuclear proteins can be investigated in an in vitro nuclear import assay using digitonin semi-permeabilized HeLa cells (Adam et al, 1990) and fluorescently labelled import cargo in conjunction with purified nuclear transport factors. To study nuclear import of the soluble N-terminal domain of SUN2, a GST–GFP fusion of SUN2(1–158) was generated. Nuclear import of a fluorescent BSA-NLS conjugate and GST–GFP served as positive and negative controls, respectively. Both GST–GFP–SUN2(1–158) and BSA-NLS were efficiently imported into the nuclei of the semi-permeabilized cells in the presence of both Impα and Impβ, but not in control reactions containing Impβ only (Figure 3). Collectively, these data suggest that the nucleoplasmic domain of SUN2 contains a functional nuclear import signal recognized by Impα/Impβ.
Figure 3.
The N-terminal domain of SUN2 is imported by Impα/Impβ in vitro. Nuclear import of recombinant, purified GST–GFP, GST–GFP–SUN2(1–158) (1.5 μM each) or an Alexa488-labelled BSA-NLS conjugate (0.75 μM) into the nuclei of semi-permeabilized HeLa cells was performed in the presence of Ran and energy mix in either buffer alone, in the presence of Impβ (0.5 μM) or of both Impα (0.6 μM)/Impβ (0.5 μM). After import, samples were fixed and analysed by wide-field fluorescence microscopy. Note that import of the BSA-NLS conjugate was more efficient because of the presence of multiple NLS peptides per BSA molecule; pictures were taken at half of the exposure time that was used for GST–GFP and GST–GFP–SUN2(1–158).
Mapping the NLS in SUN2
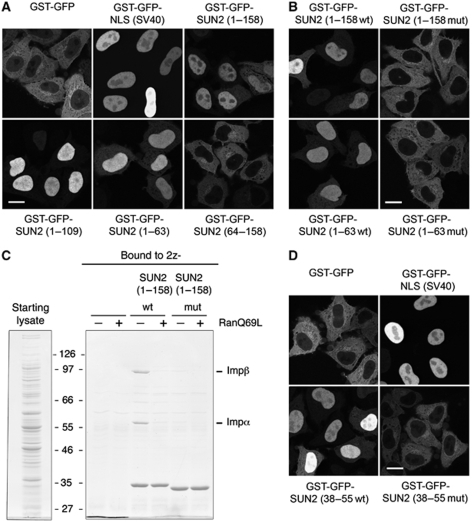
To map the NLS within the N-terminal 158 aa of SUN2, additional truncation mutants were generated and expressed as GST–GFP fusion proteins in HeLa cells (Figure 4A). Whereas GST–GFP–SUN2(1–63), GST–GFP–SUN2(1–109) and GST–GFP–SUN2(1–158) all displayed efficient nuclear accumulation, GST–GFP–SUN2(64–158) was excluded from nuclei, suggesting that the first 63 aa of SUN2 carry the NLS.
Figure 4.
The N-terminal domain of SUN2 contains a functional NLS. (A) The N-terminal domain of SUN2 can target a heterologous protein to the nucleus. HeLa cells were transfected with the indicated derivatives of GST–GFP; 24 h post-transfection, cells were fixed and analysed by confocal fluorescence microscopy. Scale bar is 15 μm. (B) Mutation of the SUN2–NLS abolishes nuclear import of the N-terminal domain in vivo. HeLa cells were transfected with the wild type or NLS mutant versions of GST–GFP–SUN2(1–158) and GST–GFP–SUN2(1–63); 24 h after transfection, cells were fixed and analysed by confocal fluorescence microscopy. (C) Mutation of the SUN2–NLS impairs Impα/Impβ binding. 1.5 μM recombinant, purified 2z, 2z-SUN2(1–158wt) or 2z-SUN2(1–158mut) were mixed with Impα and Impβ in E. coli lysate (starting lysates) in the absence or presence of RanQ69L(GTP) as in Figure 2B. Bound proteins were retrieved from the reaction mixtures with IgG sepharose, eluted and separated by 8% SDS–PAGE followed by colloidal Coomassie staining. (D) Amino acids 38–55 of human SUN2 function as NLS. HeLa cells were transfected with GST–GFP, GST–GFP fused to the SV40-NLS, GST–GFP–SUN2(38–55wt) or GST–GFP–SUN2(38–55mut); 24 h after transfection, cells were fixed and analysed by confocal fluorescence microscopy.
The consensus motif of a classical NLS is defined by a stretch of basic amino acids (5–20 residues) that is sufficient to direct a heterologous protein to the cell nucleus. Simple, monopartite cNLSs contain just a single cluster of lysine and arginine residues, but more complex cNLSs as exemplified by the bipartite NLS in nucleoplasmin also exist (Dingwall and Laskey, 1991). Both types of cNLSs are bound by Impα in an extended conformation (Cook et al, 2007). Inspection of the N-terminus of SUN2 (Supplementary Figure S1) revealed a cluster of basic residues between amino acids 41 and 52 for which no secondary structure is predicted using PSIPRED. To test whether this basic cluster contributes to NE targeting of SUN2, basic residues within this stretch, namely R41, K44, R45, K46, R51 and R52, were mutated to alanines in the context of both GST–GFP–SUN2(1–63) and GST–GFP–SUN2(1–158). When expressed in HeLa cells, both mutants, GST–GFP–SUN2(1–63mut) and GST–GFP–SUN2(1–158mut), failed to accumulate in nuclei and displayed a cytoplasmic localization pattern (Figure 4B). Note that mutation of the three central amino acids namely K44A, R45A, K46A were sufficient to abolish nuclear import (data not shown). This import defect of the NLS mutant constructs is consistent with the loss of Impα/Impβ binding in vitro (Figure 4C). The 2z-SUN2(1–158mut) protein used in this in vitro binding experiment could be purified efficiently after expression in E. coli suggesting that the mutations do not induce protein misfolding and aggregation.
To test whether the NLS of SUN2 can function in an isolated manner, we fused the putative SUN2–NLS to GST–GFP. Whereas GST–GFP alone was predominantly cytoplasmic, GST–GFP–SUN2(38–55) localized to the nucleus (Figure 4D). Nuclear import of this reporter was dependent on the functionality of the NLS, as the NLS mutant was cytoplasmic. Thus, SUN2 contains a bona fide NLS between aa 38 and 55.
Contribution of the NLS to NE localization of SUN2
Having identified an NLS in the N-terminal domain of SUN2, we next addressed whether the NLS contributes to NE targeting in the context of the full-length protein. SUN2–GFP and SUN2–GFP(NLS-mut) were transiently expressed in HeLa cells. To our surprise, we only observed a subtle effect of the NLS mutation, as SUN2–GFP(NLS-mut) was efficiently targeted to the NE and only showed minor mislocalization to the ER network in comparison with wild-type SUN2–GFP (Supplementary Figure S2A and B). To obtain a quantitative readout for this difference, we measured the ratio of GFP fluorescence between NE and ER. This quantification showed a minor, but significant difference between wild type and mutant SUN2–GFP. As SUN2 is known to form homodimers (Wang et al, 2006), we reasoned that formation of mixed homodimers between endogenous and exogenous SUN2 might overcome NLS requirement of the mutant for INM targeting. Therefore, we repeated our localization analysis after depletion of endogenous SUN2 by RNA interference (RNAi). Still, only a subtle contribution of the NLS to NE localization of SUN2 could be found (Supplementary Figures S2A, B, C and S3).
To exclude that we might have overlooked an accumulation of the NLS mutant at the ONM, we compared antibody accessibility of an N-terminal epitope of SUN2 by immunofluorescence after differential permeabilization of cells. After fixation, cells were either permeabilized only with a low concentration of digitonin that fails to solubilize the nuclear membrane, or additionally treated with a mixture of Triton X-100 (TX) and SDS that allows access of the antibodies to the nuclear interior. With this differential permeabilization protocol, antibody accessibility to an epitope exposed to the cytoplasm or nuclear interior is evaluated, as verified by nuclear laminA/C staining. This experiment confirmed that both wild-type SUN2–GFP and SUN2–GFP(NLS-mut) efficiently reached the INM (Supplementary Figure S2D). Thus, the NLS in the N-terminal domain of SUN2 does not strongly contribute to NE localization in the context of the full-length protein, indicating that either the NLS is of minimal importance for INM targeting or that an additional targeting signal exists.
Identification of a coatomer-binding motif in SUN2
To elucidate whether other sequence elements in SUN2 override the requirement for the NLS, we constructed a series of additional truncation mutants. We created a C-terminal deletion of SUN2, SUN2(1–260), to study NE localization in the absence of signals provided by the entire C-terminal region.
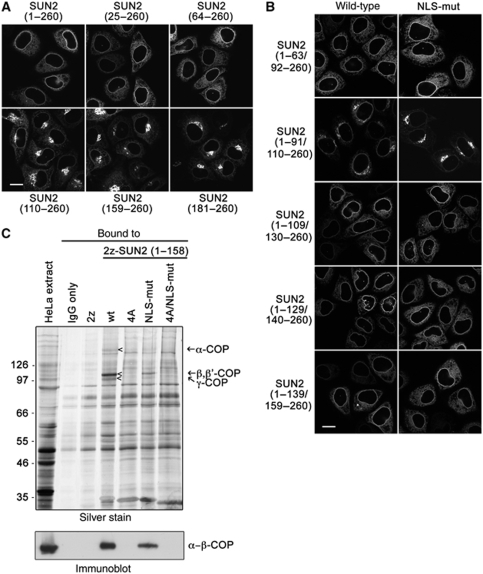
SUN2(1–260) was enriched at the NE, but also localized significantly to the ER network (Figures 1A and 5A). Deletion of the N-terminal 24 or 63 aa did not grossly change this localization pattern. In contrast, the lack of the first 109 aa caused a decrease of nuclear rim signal and a striking accumulation of the construct in the Golgi, as verified by colocalization with the Golgi marker protein giantin (Supplementary Figure S4). Removal of additional 49 or 71 aa (constructs SUN2(159–260) and SUN2(181–260), respectively) did not further strengthen the phenotype, suggesting that deletion of an element in between aa 64 and 109 is causing mislocalization of the protein from the NE/ER to the Golgi complex.
Figure 5.
SUN2 contains an Arg-based ER retrieval signal recognized by COPI (coatomer). (A) Deletion of the N-terminal 109 aa of SUN2(1–260) leads to its mislocalization to the Golgi. HeLa cells were transiently transfected with the indicated derivatives of SUN2(1–260)–GFP; 24 h post-transfection, cells were fixed and analysed by confocal fluorescence microscopy. Scale bar is 15 μm. (B) NE targeting of SUN2(1–260) depends on its NLS and amino acids 92–109 of SUN2. HeLa cells were transfected with the indicated internal deletion mutants of either wild type or the NLS mutant of SUN2(1–260)–GFP. Cells were fixed 24 h after transfection and analysed by confocal fluorescence microscopy. (C) The N-terminal domain of SUN2 interacts with the COPI complex depending on the presence of a 4 Arg cluster. HeLa cell extract was incubated with 1.5 μM of recombinant, purified 2z or 2z-SUN2(1–158) wild type and its mutant derivatives: NLS-mut, 4A (102RRRR105 to AAAA), 4A/NLS-mut. 2z-tagged proteins and associated factors were retrieved from the reaction mixtures with IgG sepharose. Bound proteins were eluted and separated by 8% SDS–PAGE followed by silver staining (upper) or immunoblotting (lower) using antibodies directed to β-COP. The load in the extract lanes corresponds to 0.25% of the input and the load for the bound fractions corresponds to 1/6 of the total. Note that the MS analysis of the proteins migrating at the indicated positions (arrow heads) led to the identification of coatomer subunits (arrows).
A series of internal deletions within this region was used to further narrow the responsible sequence element. Deletion of aa 92–109 (SUN2(1–91/110–260)) was sufficient to cause Golgi accumulation. Strikingly, when this deletion was combined with the N-terminal NLS mutation, the resulting double mutant SUN2(1–91/110–260mut) strongly accumulated in the Golgi and was barely visible at the NE (Figure 5B). This result suggested that the NLS together with a motif located between aa 92 and 109 ensures NE targeting of SUN2(1–260).
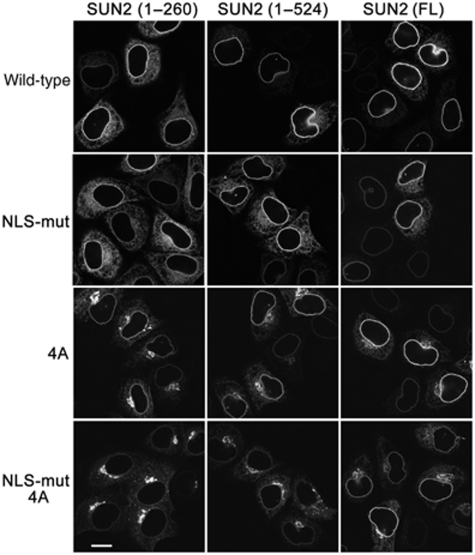
Within the region comprising aa 92–109, we spotted a conserved cluster of 3–4 Arg residues (Supplementary Figure S1). We changed these four arginines (aa 102–105) of human SUN2 into alanines, in the backbones of SUN2(1–260)–GFP, SUN2(1–524)–GFP and SUN2(FL)–GFP. In all cases, the 4A mutations gave rise to accumulation of a fraction of SUN2 in the Golgi, an effect that was most striking for SUN2(1–260)4A and SUN2(1–524)4A (Figure 6).
Figure 6.
The NLS and the ER retrieval signal together determine NE localization of SUN2 fragments lacking the SUN domain. HeLa cells were transfected with the indicated derivatives of SUN2(1–260)–GFP, SUN2(1–524)–GFP and SUN2(FL)–GFP. 4A stands for mutation of 102RRRR105 to AAAA. Cells were fixed 24 h after transfection and analysed by confocal fluorescence microscopy. Scale bar is 15 μm.
Retrieval of many membrane proteins from the Golgi to the ER relies on the recognition of ER localization signals by the coat protein complex I (COPI, coatomer). The identified arginine cluster in SUN2 resembles known Arg-based ER localization signals earlier characterized in multimeric membrane proteins (for review see Michelsen et al, 2005). To test whether the N-terminal domain of SUN2 can associate with coatomer dependent on the presence of the 4R motif, we performed pull-down experiments from HeLa cell extracts using wild-type 2z-SUN2(1–158) and 2z-SUN2(1–158)4A as baits. COPI was efficiently recruited to wild-type 2z-SUN2(1–158) as judged both by mass spectrometric analysis of bands from Coomassie stained gels (identified subunits were α-COP, β-COP, β′-COP, γ-COP, δ-COP) and by immunoblotting using β-COP-specific antibodies. When the 4R motif was mutated (2z-SUN2(1–158)4A), COPI association was lost (Figure 5C). In contrast, 2z-SUN2(1–158) harbouring the NLS mutation still bound the coatomer complex, albeit with reduced efficiency when compared with the wild-type construct. Together, these data show that the 4R motif represents a Golgi retrieval signal.
To verify that the NLS and the 4R motif together ensure NE localization of SUN2, we next combined the mutations in both elements. Importantly, the GFP fluorescence at the nuclear rim was significantly diminished by the combined mutations in the background of SUN2(1–260), and also for SUN2(1–524), whereas the localization of full-length SUN2 was less affected (Figure 6).
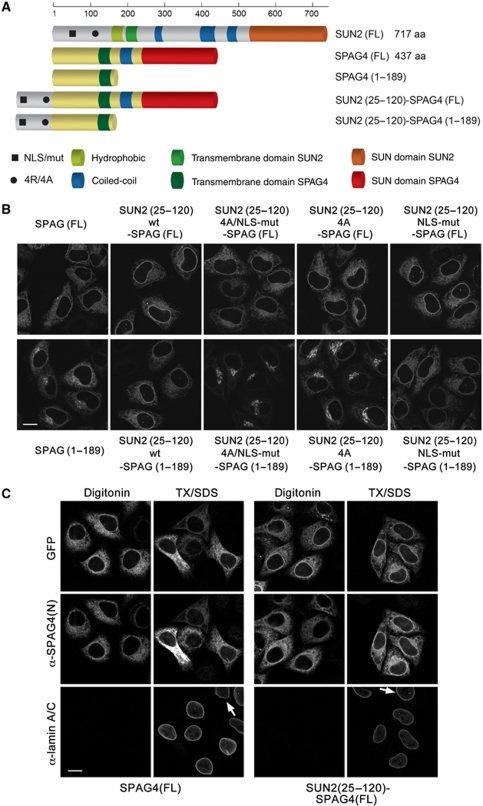
To test whether the identified sequence elements in the N-terminal domain of SUN2 can confer NE localization to a reporter membrane protein, we used a domain transfer approach to the related SUN protein SPAG4 (SUN4). Unlike SUN2–GFP, SPAG4–GFP localizes to the ER in HeLa cells (Hasan et al, 2006). We fused a SUN2 fragment (aa 25–120), which contains both the NLS and the 4R motif, to full-length SPAG4–GFP (Figure 7A). The resulting fusion protein was targeted to the NE and also showed residual ER localization (Figure 7B). To confirm that SUN2(25–120)–SPAG4(FL) had reached the INM, we performed antibody accessibility analysis using an antibody recognizing an epitope in the N-terminal domain of SPAG4 after differential detergent permeabilization of cells (Figure 7C). SPAG4(FL) served as negative control. This analysis verified that SUN2(25–120)–SPAG4(FL) was targeted to the INM as shown by the nuclear rim staining obtained by the anti-SPAG4 antibody upon full permeabilization of the NE with Triton/SDS.
Figure 7.
An N-terminal fragment of SUN2 comprising the NLS and the 4R motif can target a reporter membrane protein to the NE. (A) Schematic representation of SUN2, SPAG4 and their derivatives as in Figure 1. The black square and circle indicate the positions of the NLS and the 4R motif, respectively. (B) HeLa cells were transfected with the indicated C-terminally GFP-tagged derivatives of SPAG4(FL) or SPAG4(1–189). Cells were fixed 24 h after transfection and analysed by confocal fluorescence microscopy. Scale bar is 15 μm. (C) HeLa cells expressing C-terminal GFP fusions of SPAG4(FL) or SUN2(25–120)–SPAG4(FL) were fixed and permeabilized with either only digitonin or subsequently also with Triton X-100/SDS (TX/SDS). Immunostaining was performed using a mouse anti-laminA/C antibody detected by an Alexa633-labelled secondary antibody, and a rabbit anti-SPAG4(N) antibody detected by an Alexa568-labelled secondary antibody. Scans were taken by confocal fluorescence microscopy by sequential scanning of GFP, Alexa568 and Alexa633. Note that the SPAG4 antibody does not yield a signal in non-transfected cells (arrows).
NE accumulation of SUN2(25–120)–SPAG4(FL) was supported by both the NLS and the 4R motif, as a fusion protein bearing mutations in both signals showed a SPAG4-like ER localization pattern, whereas fusions bearing mutations in either the NLS or the 4R were only slightly compromised in NE localization (Figure 7B). Unlike SUN2, these SPAG4 derivatives were not found at the Golgi even when the 4R motif was mutated, suggesting that SPAG4 contains features that prevent Golgi accumulation. These features are contained in the C-terminal, lumenal part of SPAG4, as SPAG4(1–189)–GFP, which is membrane-bound, but lacks the lumenal domain, was localized to both the ER and the Golgi (Figure 7B). When the N-terminal fragment of SUN2 was attached to SPAG4(1–189)–GFP, we again observed an increase of the GFP signal at the nuclear rim and loss of Golgi localization. Mutation of the 4R motif resulted in a strongly increased Golgi accumulation and a residual localization at the NE that was lost upon the additional mutation of the NLS. These data show that the NLS and the 4R motif together promote NE localization of the SPAG4-derived reporter proteins. Moreover, we also transferred the wild type and mutant N-terminal fragments of SUN2 to the ER-resident form of cytochrome b5, confirming the results obtained for the SPAG4 fusions (Supplementary Figure S5).
The SUN domain of SUN2 contributes to NE localization
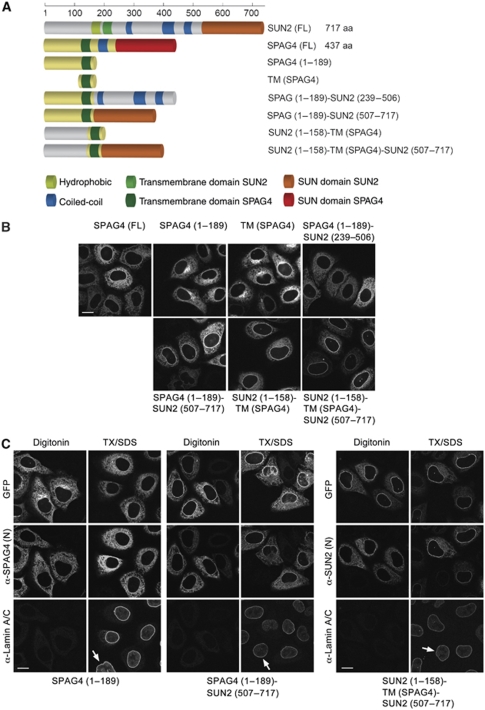
When comparing the effects of NLS and 4R mutations within full-length SUN2, SUN2(1–524) and SUN2(1–260) on subcellular localization (Figure 6), we observed that the presence of the C-terminal SUN domain in full-length SUN2 seemed to enhance NE localization. To directly test the function of the SUN domain of SUN2 in NE targeting, we attached it to SPAG4(1–189). The resulting protein fusion SPAG4(1–189)–SUN2(507–717) was still localized to the ER, but was additionally found enriched at the NE in most cells (Figure 8). This result shows that the SUN domain can indeed guide a reporter protein to the NE. In agreement with our earlier conclusions, NE targeting was further enhanced when the N-terminal domain of SPAG4 was replaced by the N-terminal domain of SUN2, yielding SUN2(1–158)–TM(SPAG4)–SUN2(507–717). In contrast, a fusion construct of SPAG4(1–189) and the lumenal coiled-coil domain of SUN2 did not support NE targeting.
Figure 8.
The SUN domain contributes to INM targeting. (A) Schematic representation of SUN2, SPAG4 and their derivatives as in Figure 7. (B) The SUN domain can target a reporter protein to the NE. HeLa cells were transiently transfected with SPAG4(FL)–GFP, SPAG4(1–189)–GFP, TM(SPAG4)–GFP (a SPAG4 fragment comprising the transmembrane domain of SPAG4 (aa 131–189)), or the indicated chimera of SPAG4 and SUN2; 24 h post-transfection, cells were fixed and analysed by confocal fluorescence microscopy. Scale bar is 15 μm. (C) The N-terminal domain of SUN2 and the SUN domain contribute to INM targeting. HeLa cells were transfected with the indicated GFP fusions and treated as in Figure 7C. Immunostaining was performed using a mouse anti-laminA/C (Alexa633-labelled secondary antibody) and rabbit anti-SPAG4(N) or anti-SUN2(N) antibodies recognizing epitopes in their respective N-terminal domains (Alexa568-labelled secondary antibody). Scans were taken by confocal fluorescence microscopy. Note that SPAG4 and endogenous SUN2 are not detected in non-transfected cells (arrows) under these conditions.
Again, we confirmed that the SPAG4(1–189)–SUN2(507–717) and SUN2(1–158)–TM(SPAG4)–SUN2(507–717)–GFP fusion proteins had reached the INM by antibody accessibility analysis (Figure 8C). Compared with the control SPAG4(1–189), both fusion constructs displayed obvious nuclear rim staining upon TX/SDS permeabilization, but not after digitonin treatment, suggesting that both proteins reached the INM. We also noted that SUN2(1–158)–TM(SPAG4)–SUN2(507–717) was strongly enriched at the INM, whereas SPAG4(1–189)–SUN2(507–717) was enriched to a lesser extent and also faintly appeared at the NE after digitonin permeabilization. Collectively, these data show that the N-terminal domain of SUN2, containing a classical NLS and the 4R motif, combined with the C-terminal SUN domain is sufficient to accomplish efficient targeting to the INM.
Discussion
We have analysed how the INM protein SUN2 is targeted to the NE and identified three features that jointly contribute to its NE localization. The first element is a basic segment (aa 38–52) in the N-terminal domain matching several criteria defining classical NLSs: (1) it is required for nuclear import of the soluble, N-terminal domain of SUN2, (2) similar to cNLSs, the SUN2–cNLS is recognized by the designated transport receptor dimer Impα/Impβ and (3) it can direct a heterologous protein into the nucleus. The second signal facilitating efficient NE localization of SUN2 is a cluster of four Arg residues at positions 102–105. This 4R motif functions as an ER localization signal that prevents accumulation of SUN2 in the Golgi complex by serving as a binding platform for the coatomer complex I (COPI). The third contribution to NE targeting of SUN2 stems from its C-terminal SUN domain.
The SUN domain contributes to INM targeting of SUN2
The SUN domain of SUN2 functions as a transferable NE targeting element (Figure 8). In the context of SUN2, the necessity of the SUN domain became most obvious when the two signals in the N-terminal domain, the cNLS and the 4R motif, were inactivated. SUN2 bearing mutations in both N-terminal signals still accumulated at the NE, but additional deletion of the SUN domain greatly reduced NE targeting (Figure 6). Thus, an interaction of the SUN domain with factors in the PNS may help retaining SUN2 at the NE, thereby contributing to its INM localization. Prime candidates for such interaction partners are the KASH proteins of the ONM. The interaction between lumenal SUN and KASH domains is well established and KASH proteins themselves are recruited to the ONM by interaction with SUN family members in various organisms (Starr and Han, 2002; Malone et al, 2003; Padmakumar et al, 2005; Crisp et al, 2006; Ketema et al, 2007; Kracklauer et al, 2007; Minn et al, 2009; Roux et al, 2009).
The identification of the SUN domain as one NE targeting element in SUN2 may suggest that the localization of SUN and KASH proteins is in part interdependent. Whereas KASH proteins seem to rely exclusively on the interaction of their KASH domains with SUN family members for NE targeting, SUN2 uses its N-terminal nucleoplasmic domain in addition to the lumenal SUN domain to reach its final destination in the INM. According to one possible scenario, the initial targeting of SUN2 to the INM is mediated by its N-terminal domain and occurs independently of KASH interaction of the C-terminal SUN domain. On arrival of SUN2 at the INM, SUN–KASH interactions may be established to stabilize NE localization. Anchorage of these SUN2–KASH complexes might be further supported by higher order structures built through the interaction of KASH proteins with cytoskeletal components in the cytoplasm. The use of a protein domain exposed to the PNS such as the SUN domain has not yet been described to support INM protein sorting and thus adds a new mechanistic facet to this targeting pathway.
An ER retrieval signal in SUN2
When the 4R motif in SUN2 is mutated, SUN2 is unable to interact with COPI in vitro and SUN2 derivatives tend to enrich in the Golgi. Golgi localization was most striking for constructs that contained a mutated NLS, showing that Golgi retrieval becomes crucial if efficient INM targeting of SUN2 is compromised. Our data indicate that there exist two possible routes for newly synthesized SUN2—one to the INM and one to the Golgi from where SUN2 is constantly retrieved to the ER, maintaining a pool of SUN2 deliverable to the INM.
The presence of an Arg-based ER localization signal in SUN2 was unexpected and to our knowledge, it is the first eukaryotic INM protein for which such a signal could be identified. Earlier, several virus-encoded membrane glycoproteins have been suggested to possess similar Arg-based motifs contributing to their ER/NE localization, although in those cases binding to COPI has not been investigated (Lee, 1999; Meyer and Radsak, 2000; Meyer et al, 2002). Interestingly, when inspecting the sequences of mammalian INM proteins, we could spot clusters of arginine residues in LBR (74–76), in emerin (44–46) and in LEM2 (130–132; 454–456), indicating that other INM proteins might also rely on the use of Golgi retrieval signals to support their efficient delivery to the INM.
Arg-based sorting signals have been mainly characterized in membrane protein complexes that are destined for the plasma membrane (Michelsen et al, 2005). In these multimeric complexes, the Arg-based signals function in keeping unassembled subunits in the ER, whereas the motifs are thought to be masked once the subunits are assembled. It remains to be seen if the recognition of the 4R signal in SUN2 is controlled by the assembly state of SUN2 into homodimers or higher order complexes.
But does the 4R motif also have a direct function in INM targeting of SUN2? The combined mutation of the NLS and the 4R motif in fusions with SPAG4 and cytochrome b5 that seem not to shuttle to the Golgi synergistically reduced NE targeting, suggesting that the 4R motif has a positive influence on NE localization independently of its function in Golgi retrieval. This could be explained by a contribution of the 4R motif to nuclear retention and/or receptor-mediated nuclear import. However, our FRAP analysis did not detect obvious changes in the diffusional mobility of SUN2 or SUN2(1–524) at the NE upon inactivation of the 4R motif (Supplementary Figure S6), indicating that other features are the main determinants of SUN2's dynamic properties. Further, the 4R to 4A mutant bound to Impα/Impβ in vitro (not shown) and the inactivation of the NLS in the N-terminal domain of SUN2 was sufficient to block nuclear import (Figure 4), suggesting that the 4R motif cannot function as an autonomous nuclear import signal. Thus, it is presently unclear how the 4R motif would directly support INM targeting. For SUN2 derivatives, the main phenotype of inactivating the 4R motif was the accumulation in the Golgi. These data are consistent with the assumption that a major function of the 4R motif lies in retrieving SUN2 from the Golgi to the ER by COPI-meditated retrograde transport in case it eventually escapes the ER network.
Contribution of the SUN2–NLS to NE localization
The presence of a cNLS in the N-terminal domain of SUN2 might indicate that karyopherin-mediated nuclear import of INM proteins is conserved from yeast to mammals. Although the mutation of the NLS impaired NE localization of full-length SUN2–GFP to only a minor extent, the combined mutations of the NLS and the 4R motif diminished NE enrichment of full-length SUN2 and almost completely prevented NE targeting of constructs lacking the SUN domain (Figure 6). These data speak in support of an NLS contribution to INM targeting of SUN2. Yet, it is important to consider whether the NLS does so by recruiting Impα/Impβ or by acting as a retention motif. For many NLSs, these two functions cannot be separated. NLSs can work both as transport receptor recognition motifs and retention signals, for instance, as part of DNA or RNA-binding domains (LaCasse and Lefebvre, 1995; Cokol et al, 2000). Therefore, the NLS may act as DNA-binding site in SUN2 and could, in principle, exert its effect by contributing to nuclear retention. However, mutation of the NLS did not significantly increase the diffusional mobility of SUN2 in the NE (Supplementary Figure S6).
So far, targeting of transmembrane proteins to the INM in higher eukaryotes has not implied the use of NLS sequences. Instead, many INM proteins are retained at the NE by interaction with chromatin and/or nuclear lamins (Worman and Courvalin, 2000). As SUN2 is known to interact with A-type lamins (Crisp et al, 2006; Haque et al, 2010), we have also tested whether SUN2 targeting to the NE is lamin-dependent. In agreement with the published data (Crisp et al, 2006; Schmitt et al, 2007; Haque et al, 2010), we did not observe changes in SUN2 localization on depletion of nuclear lamins by RNAi (not shown).
Direct evidence that the SUN2–cNLS or the earlier identified NLSs in the yeast INM proteins Heh1 and Heh2 contribute to NPC passage by recruiting Impα/Impβ is missing and difficult to obtain. Development of an in vitro nuclear transport assay reconstituting nuclear import of INM proteins may in the future provide an adequate experimental system to approach this question.
Still, some considerations on receptor-mediated import of INM proteins can be made based on our current knowledge of NPC structure and constraints for the size of INM proteins to be transported through the NPC. Several early studies revealed that nucleoplasmic domains of INM proteins should not exceed 60 kDa to allow for their NPC passage (Soullam and Worman, 1995; Ohba et al, 2004). In contrast, soluble nuclear transport cargo can be orders of magnitudes bigger, such as snRNPs or viral capsids. Is this size limitation valid only for INM proteins that lack an NLS or, in other words, does the presence of an NLS allow for a bigger nucleoplasmic domain in an INM protein? For SUN2, we observed that attaching two GFP moieties to the N-terminus of SUN2(1–260) strongly decreased NE localization and three consecutive GFPs prevented INM targeting (Supplementary Figure S7), suggesting that the SUN2 NLS is unable to overcome this size restriction.
Unlike soluble cargo, INM proteins must traverse the pore in close proximity to the pore membrane. Ultrastructural analysis of NPCs has revealed the existence of lateral channels of small size (approximately 9 nm in diameter; Beck et al, 2004) that may provide the structural explanation for the size limitation. On the basis of these current structural models, it is unclear if these lateral channels would be wide enough to accommodate Impα/Impβ heterodimers. Moreover, Impβ's function in cargo translocation should be linked to the binding of FG repeat containing nucleoporins (nups) and it remains to be seen which FG nups would come close enough to this translocation route to help pore passage. Notably, only non-FG nups such as GP210 (Ohba et al, 2004) in mammalian cells and Nup170 in yeast (King et al, 2006) have so far been implicated in translocation of INM proteins.
One possibility to reconcile these considerations with receptor-mediated transport of INM proteins could be that INM proteins extend their NLSs through sideward openings of the lateral channels to provide a handle for Impα/Impβ transport through the central FG meshwork. Nucleoporins such as yeast Nup170 or mammalian GP210 might be involved in ensuring structural arrangements of the NPC to allow for such a scenario. In future, a better understanding of overall NPC architecture might help to solve the questions of if and how the NPC can house a classical, receptor-mediated nuclear import pathway along the pore membrane.
It is noteworthy that a function of Impα in INM targeting could also be Impβ independent. Studies on targeting of INM proteins in insect cells have discovered INM-SMs that associate with a shortened Impα variant, Impα-16 (Saksena et al, 2006). Similar truncated Impα isoforms seem to exist in human cells (Braunagel et al, 2007). Notably, these Impα derivatives do not possess an IBB and must work independently of Impβ. Owing to their small size, they are good candidates for sorting chaperones that would fit through the lateral NPC channels. Also, SUN2 contains basic residues (RR205/206) resembling an INM-SM in close proximity to its transmembrane segment. Individual mutation of R205 or R206 has no effect on INM targeting (not shown). When both these residues are mutated to Ala, SUN2 is still targeted to the NE; however, the membrane orientation of the SUN domain is changed (Supplementary Figure S8), making it difficult to study the contribution of this potential sorting motif.
A role for NLS sequences of INM proteins in post-mitotic NE assembly?
Many vertebrate INM proteins contain predictable cNLSs (Lusk et al, 2007). But why do INM proteins such as SUN2 harbour NLS-like sequences with Impα/Impβ-binding ability? If it was not only for INM targeting during interphase, then another function for such NLSs could lie in the process of open mitosis. During open mitosis, INM proteins are partitioned into the membrane system of the ER (Ellenberg et al, 1997; Yang et al, 1997) from where they need to be sorted back to chromatin during nuclear assembly. Notably, about 50% of mammalian INM proteins carry basic, extralumenal domains and several INM proteins can directly bind to DNA (Ulbert et al, 2006). DNA binding of INM proteins seems instrumental in the process of NE reformation (Ulbert et al, 2006). As NLS sequences are often found in DNA-binding motifs, it is tempting to speculate that many more INM proteins might be able to directly interact with Impα/Impβ. Thus, in the mitotic cytoplasm, Impα/Impβ heterodimers could serve INM proteins as molecular chaperones to prevent undesired interactions through their exposed DNA-binding domains. During NE reformation, Impα/Impβ would be released from NLSs of INM proteins in the vicinity of chromatin, guided by the RanGTP gradient, allowing for a spatial control of DNA-binding activity. For one INM protein, the lamin B receptor, release of Impβ has already been suggested to control its chromatin reassociation during NE reformation (Ma et al, 2007).
On the basis of the identification of the NLS in SUN2 and the likely existence of more INM proteins carrying cNLSs in higher eukaryotes, it would not be surprising if more INM proteins benefited from possessing an NLS—for INM targeting during interphase and/or an unperturbed transit through mitosis.
Materials and methods
Molecular cloning
A full-length cDNA clone (pcDNA3.1 TOPO/V5-His SUN2) encoding for human SUN2 (Hodzic et al, 2004) was kindly provided by P Stahl (Washington University School of Medicine, St Louis). This plasmid served as PCR template for the generation of pEGFP–N3–SUN2(FL), which contains the SUN2 insert in the BglII/EcoRI sites, followed by the EGFP open reading frame. For all further subcloning, pEGFP–N3–SUN2(FL) was used as PCR or mutagenesis template, yielding the following pEGFP–N3–SUN2 derivatives (restriction sites used): (1–524) (EcoRI/BglII); (1–260) (HindIII/BamHI); (ΔN158) (SalI/EcoRI); (25–260), (64–260) (both PstI/BamHI); (110–260), (159–260), (181–260) (all SalI/BamHI); (1–63/92–260), (1–91/110–260), (1–109/130–260), (1–129/140–260), (1–139/159–260) (all BglII/SalI/BamHI). The NLS mutants of human SUN2 derivatives (NLS-mut: R41A, K44A, R45A, K46A, K51A, R52A) and the respective 4A mutants (R102A, R103A, R104A and R105A) were generated using the QuikChange Mutagenesis kit (Stratagene).
For the domain swap experiments, the SPAG4 coding region was amplified from HeLa cDNA and inserted into the EcoRI/BamHI sites of pEGFP–N3. pEGFP–N3–SPAG4(FL) served as PCR template for the generation of the following SPAG4 fragments and fusion proteins assembled in the pEGFP–N3 backbone (restriction sites used): SPAG4(1–189), SUN2(25–120)–SPAG4(1–189) (BglII/EcoRI/Sal); SUN2(25–120)–SPAG4(FL) (BglII/EcoRI/BamHl); TM(SPAG4)(131–189) (EcoRI/SalI); SPAG4(1–189)–SUN2(239–506), SPAG4(1–189)–SUN2(507–717) (EcoRI/SalI/BglII-BamHI); SUN2(1–158)–TM(SPAG4), (BglII/EcoRI/Sal); SUN2(1–158)–TM(SPAG4)–SUN2(507–717) (BglII/EcoRI/SalI/BglII-BamHI).
For expression of GST–GFP fusion proteins in HeLa cells, PCR fragments obtained on SUN2(FL) and mutant derivatives were subcloned into pK7–GST–GFP (Erkmann et al, 2005) using the BamHI-EcoRI sites, yielding the following C-terminal fusions: SUN2(1–158wt), SUN2(1–158mut), SUN2(1–109), SUN2(1–63wt), SUN2(1–63mut), SUN2(64–158). pK7–GST–GFP–SUN2 NLS (wt or mut, aa 38–55) were generated by annealing the respective sense/antisense primers and cloning into the BamHI-EcoRI sites. pK7–GST–GFP–NLS(SV40) has been described earlier (Erkmann et al, 2005).
For expression of proteins in E. coli, the respective PCR fragments were either cloned into the NcoI-BamHI sites of pQE60-2z (Kutay et al, 1997b) or of pQE60–GST–GFP (generated by subcloning GST–GFP as a BspHI/NcoI fragment into the NcoI site of pQE60 (Qiagen)), yielding the following constructs: (1) pQE60-2z derivatives: SUN2(1–158wt), SUN2(1–158mut), Kip1 (amplified from HeLa cell cDNA) and (2) pQE60–GST–GFP derivatives: SUN2(1–158wt), SUN2(1–158mut).
Antibodies
Antibodies against Impα2 (RCH1), Imp and Imp5 were kind gifts of D Görlich (MPI Göttingen, Germany). Anti-β-COP (ab2899) was purchased from Abcam, anti-laminA/C was from Novacastra. Antibodies to human SUN2 and SPAG4 were generated in rabbits using aa 1–158 of SUN2 and the peptide RPGSASSSRKHTPNFFSENC (aa 6–24 of SPAG4) as antigenes and affinity purified.
Recombinant protein expression
The expression and purification of human Impα (RCH1) and ΔIBB-Impα (ΔIBB-RCH1) (Kutay et al, 1997a), human Impβ (Kutay et al, 1997b), and RanQ69L (Izaurralde et al, 1997) has been described.
2z, 2z-Kip1, 2z-SUN2(1–158wt), 2z-SUN2(1–158mut), GST–GFP–SUN2(1–158wt) and GST–GFP were expressed in E. coli BLR(pREP4) at 20°C by induction with 0.5 mM isopropyl β-d-thiogalactoside. Cells were lysed by sonication in 50 mM Tris, pH 7.5, 700 mM NaCl, 3 mM MgCl2, 5% glycerol, 2 mM 2-mercaptoethanol. The lysate was cleared by ultracentrifugation, passed over Ni-NTA agarose (Qiagen) and eluted with 400 mM imidazole in lysis buffer. Peak fractions were pooled and buffer exchanged to 50 mM Tris, pH 7.5, 350 mM NaCl, 3 mM MgCl2, 250 mM sucrose for 2z, 2z-Kip1, 2z-SUN2(1–158wt), 2z-SUN2(1–158mut) and to 50 mM Hepes pH 7.5, 150 mM potassium acetate, 5 mM Mg acetate for GST–GFP–SUN2(1–158wt) and GST–GFP.
Pull-down experiments
HeLa cell extract was prepared as described in Kutay et al (1998). A total of 500 μl of HeLa cell extract (adjusted to 50 mM Tris, pH 7.5, 225 mM potassium acetate, 2 mM MgCl2) was supplemented with purified 2z, 2z-Kip1, 2z-SUN2(1–158wt) (1.5 μM each) and, if indicated, RanQ69L(GTP) (2.5 μM) was added. Samples were incubated on ice for 4 h. Then, 12.5 μl of IgG sepharose was added and samples were gently mixed for 45 min. Beads were washed three times in 1.5 ml of binding buffer. Bound proteins were eluted with 1.5 M MgCl2, 50 mM Tris, pH 7.5, precipitated with isopropanol, and dissolved in SDS sample buffer.
For binding experiments using purified factors, each sample contained 225 μl of E. coli lysate in 50 mM Tris, pH 7.5, 230 mM potassium acetate, 2 mM MgCl2 supplemented with purified recombinant transport receptors Impα (RCH1), ΔIBB-Impα (ΔIBB-RCH1), Impβ to 0.4 μM (starting lysates). Then, 2z-tagged proteins (2z, 2z-Kip1, 2z-SUN2(1–158wt) or 2z-SUN2(1–158mut); 1.5 μM each), and, where indicated, RanQ69L (2.5 μM) were added. Samples were incubated on ice for 2 h and further processed as described above. Elution was with SDS sample buffer to which DTT was added after the elution step.
Cell culture and transient transfections
HeLa cells were maintained in DMEM containing 10% FCS and penicillin/streptomycin at 37°C, 5% CO2. Transient transfections were performed using FuGene transfection reagent (Roche). Cells were fixed 20–36 h after transfection with 4% PFA for 10 min. After washing with PBS, cover slips were mounted in VectaShield (VectorLabs) for microscopic analysis.
In vitro nuclear import assay
Alexa488 (Molecular Probes) labelling of BSA-NLS conjugates was performed according to (Gorlich et al, 1994). In vitro transport reactions were performed essentially as described (Adam et al, 1990; Gorlich et al, 1994). HeLa cells were grown on coverslips and permeabilized with PB buffer (20 mM Hepes, pH 7.5, 110 mM potassium acetate, 5 mM magnesium acetate, 0.5 mM EGTA, 250 mM sucrose) containing 40 μg/ml digitonin. The 20 μl import mixtures in 50 mM Hepes, pH 7.5, 80 mM potassium acetate, 5 mM magnesium acetate, 250 mM sucrose contained 0.75 or 1.5 μM import substrate (Alexa488-BSA-NLS and GST–GFP, GST–GFP–hSUN2(1–158wt), respectively), and, where indicated, Impβ (0.5 μM) and Impα (0.6 μM). All samples also contained an energy regenerating system (Gorlich et al, 1994) and Ran mix (Kutay et al, 1997a). Import was allowed to proceed for 20 min at room temperature. Then, coverslips were washed once with PB, cells were fixed with 3% PFA, washed in PBS and mounted for microscopy.
Microscopy
For immunofluorescence analysis, cells were fixed in 4% PFA and washed with PBS. Permeabilization was either performed with 0.001% digitonin for 14 min at 4°C only, or for additional 5 min using a mixture of 0.1% Triton X-100 and 0.02% SDS at RT. Immunostaining was performed as described earlier (Zemp et al, 2009). Wide-field images were taken on an Olympus BX51 microscope connected to a DP 50 camera using an UPlanFl 40 × , NA 0,75 objective. Confocal microscopy was performed with a Leica TCS-SP2/AOBS microscope using an HCX Pl APO Ibd.Bl. 40 × , NA 1.25 or HCX Pl APO lbd.Bl. 63 × , NA 1.4 oil immersion lens or with a Leica TCS-SP1 microscope using a HCX Pl APO Ibd.Bl. 40 × , NA 1.25 or HCX Pl APO Ibd.Bl. 63 × , NA 1.32 oil immersion lens.
Supplementary Material
Acknowledgments
We thank C Ashiono for excellent technical assistance, Drs T Schwartz and M Beck for helpful discussions, Drs D Görlich, G Rabut and P Stahl for providing reagents, as well as Drs A Smith and E Laurell for critical reading of the paper. Our former colleagues Drs S Hasan and R Koller-Eichhorn are thanked for the generation of some DNA constructs. Imaging was performed on instruments of the ETH LMC facility. This work was supported by Boehringer Ingelheim PhD fellowships to Y Turgay and A Rothballer and a Swiss National Science Foundation grant (31003A-118053) to UK.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adam SA, Marr RS, Gerace L (1990) Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol 111: 807–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Forster F, Ecke M, Plitzko JM, Melchior F, Gerisch G, Baumeister W, Medalia O (2004) Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science 306: 1387–1390 [DOI] [PubMed] [Google Scholar]
- Braunagel SC, Cox V, Summers MD (2009) Baculovirus data suggest a common but multifaceted pathway for sorting proteins to the inner nuclear membrane. J Virol 83: 1280–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunagel SC, Williamson ST, Ding Q, Wu X, Summers MD (2007) Early sorting of inner nuclear membrane proteins is conserved. Proc Natl Acad Sci USA 104: 9307–9312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunagel SC, Williamson ST, Saksena S, Zhong Z, Russell WK, Russell DH, Summers MD (2004) Trafficking of ODV-E66 is mediated via a sorting motif and other viral proteins: facilitated trafficking to the inner nuclear membrane. Proc Natl Acad Sci USA 101: 8372–8377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke B, Stewart CL (2002) Life at the edge: the nuclear envelope and human disease. Nat Rev Mol Cell Biol 3: 575–585 [DOI] [PubMed] [Google Scholar]
- Cokol M, Nair R, Rost B (2000) Finding nuclear localization signals. EMBO Rep 1: 411–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook A, Bono F, Jinek M, Conti E (2007) Structural biology of nucleocytoplasmic transport. Annu Rev Biochem 76: 647–671 [DOI] [PubMed] [Google Scholar]
- Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D (2006) Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol 172: 41–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C, Laskey RA (1991) Nuclear targeting sequences—a consensus? Trends Biochem Sci 16: 478–481 [DOI] [PubMed] [Google Scholar]
- Ellenberg J, Siggia ED, Moreira JE, Smith CL, Presley JF, Worman HJ, Lippincott-Schwartz J (1997) Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J Cell Biol 138: 1193–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkmann JA, Wagner EJ, Dong J, Zhang Y, Kutay U, Marzluff WF (2005) Nuclear import of the stem-loop binding protein and localization during the cell cycle. Mol Biol Cell 16: 2960–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K, Fritze CE, Gerace L (1998) The major nuclear envelope targeting domain of LAP2 coincides with its lamin binding region but is distinct from its chromatin interaction domain. J Biol Chem 273: 4213–4219 [DOI] [PubMed] [Google Scholar]
- Gorlich D, Prehn S, Laskey RA, Hartmann E (1994) Isolation of a protein that is essential for the first step of nuclear protein import. Cell 79: 767–778 [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL (2005) The nuclear lamina comes of age. Nat Rev Mol Cell Biol 6: 21–31 [DOI] [PubMed] [Google Scholar]
- Haque F, Lloyd DJ, Smallwood DT, Dent CL, Shanahan CM, Fry AM, Trembath RC, Shackleton S (2006) SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol Cell Biol 26: 3738–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque F, Mazzeo D, Patel JT, Smallwood DT, Ellis JA, Shannahan CM, Shackleton S (2010) Mammalian SUN protein networks at the inner nuclear membrane and their role in laminopathy disease processes. J Biol Chem 285: 3487–3498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan S, Guttinger S, Muhlhausser P, Anderegg F, Burgler S, Kutay U (2006) Nuclear envelope localization of human UNC84A does not require nuclear lamins. FEBS Lett 580: 1263–1268 [DOI] [PubMed] [Google Scholar]
- Hodzic DM, Yeater DB, Bengtsson L, Otto H, Stahl PD (2004) Sun2 is a novel mammalian inner nuclear membrane protein. J Biol Chem 279: 25805–25812 [DOI] [PubMed] [Google Scholar]
- Holmer L, Worman HJ (2001) Inner nuclear membrane proteins: functions and targeting. Cell Mol Life Sci 58: 1741–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Kutay U, von Kobbe C, Mattaj IW, Gorlich D (1997) The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J 16: 6535–6547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakel S, Gorlich D (1998) Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J 17: 4491–4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketema M, Wilhelmsen K, Kuikman I, Janssen H, Hodzic D, Sonnenberg A (2007) Requirements for the localization of nesprin-3 at the nuclear envelope and its interaction with plectin. J Cell Sci 120: 3384–3394 [DOI] [PubMed] [Google Scholar]
- King MC, Lusk CP, Blobel G (2006) Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature 442: 1003–1007 [DOI] [PubMed] [Google Scholar]
- Kracklauer MP, Banks SM, Xie X, Wu Y, Fischer JA (2007) Drosophila klaroid encodes a SUN domain protein required for Klarsicht localization to the nuclear envelope and nuclear migration in the eye. Fly (Austin) 1: 75–85 [DOI] [PubMed] [Google Scholar]
- Kutay U, Bischoff FR, Kostka S, Kraft R, Gorlich D (1997a) Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell 90: 1061–1071 [DOI] [PubMed] [Google Scholar]
- Kutay U, Izaurralde E, Bischoff FR, Mattaj IW, Gorlich D (1997b) Dominant-negative mutants of importin-beta block multiple pathways of import and export through the nuclear pore complex. EMBO J 16: 1153–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Lipowsky G, Izaurralde E, Bischoff FR, Schwarzmaier P, Hartmann E, Gorlich D (1998) Identification of a tRNA-specific nuclear export receptor. Mol Cell 1: 359–369 [DOI] [PubMed] [Google Scholar]
- LaCasse EC, Lefebvre YA (1995) Nuclear localization signals overlap DNA- or RNA-binding domains in nucleic acid-binding proteins. Nucleic Acids Res 23: 1647–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK (1999) Four consecutive arginine residues at positions 836–839 of EBV gp110 determine intracellular localization of gp110. Virology 264: 350–358 [DOI] [PubMed] [Google Scholar]
- Lu W, Gotzmann J, Sironi L, Jaeger VM, Schneider M, Luke Y, Uhlen M, Szigyarto CA, Brachner A, Ellenberg J, Foisner R, Noegel AA, Karakesisoglou I (2008) Sun1 forms immobile macromolecular assemblies at the nuclear envelope. Biochim Biophys Acta 1783: 2415–2426 [DOI] [PubMed] [Google Scholar]
- Lusk CP, Blobel G, King MC (2007) Highway to the inner nuclear membrane: rules for the road. Nat Rev Mol Cell Biol 8: 414–420 [DOI] [PubMed] [Google Scholar]
- Ma Y, Cai S, Lv Q, Jiang Q, Zhang Q, Sodmergen, Zhai Z, Zhang C (2007) Lamin B receptor plays a role in stimulating nuclear envelope production and targeting membrane vesicles to chromatin during nuclear envelope assembly through direct interaction with importin beta. J Cell Sci 120: 520–530 [DOI] [PubMed] [Google Scholar]
- Malone CJ, Misner L, Le Bot N, Tsai MC, Campbell JM, Ahringer J, White JG (2003) The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell 115: 825–836 [DOI] [PubMed] [Google Scholar]
- McGee MD, Rillo R, Anderson AS, Starr DA (2006) UNC-83 IS a KASH protein required for nuclear migration and is recruited to the outer nuclear membrane by a physical interaction with the SUN protein UNC-84. Mol Biol Cell 17: 1790–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer G, Gicklhorn D, Strive T, Radsak K, Eickmann M (2002) A three-residue signal confers localization of a reporter protein in the inner nuclear membrane. Biochem Biophys Res Commun 291: 966–971 [DOI] [PubMed] [Google Scholar]
- Meyer GA, Radsak KD (2000) Identification of a novel signal sequence that targets transmembrane proteins to the nuclear envelope inner membrane. J Biol Chem 275: 3857–3866 [DOI] [PubMed] [Google Scholar]
- Michelsen K, Yuan H, Schwappach B (2005) Hide and run. Arginine-based endoplasmic-reticulum-sorting motifs in the assembly of heteromultimeric membrane proteins. EMBO Rep 6: 717–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn IL, Rolls MM, Hanna-Rose W, Malone CJ (2009) SUN-1 and ZYG-12, mediators of centrosome-nucleus attachment, are a functional SUN/KASH pair in Caenorhabditis elegans. Mol Biol Cell 20: 4586–4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba T, Schirmer EC, Nishimoto T, Gerace L (2004) Energy- and temperature-dependent transport of integral proteins to the inner nuclear membrane via the nuclear pore. J Cell Biol 167: 1051–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostlund C, Ellenberg J, Hallberg E, Lippincott-Schwartz J, Worman HJ (1999) Intracellular trafficking of emerin, the Emery-Dreifuss muscular dystrophy protein. J Cell Sci 112: 1709–1719 [DOI] [PubMed] [Google Scholar]
- Padmakumar VC, Libotte T, Lu W, Zaim H, Abraham S, Noegel AA, Gotzmann J, Foisner R, Karakesisoglou I (2005) The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J Cell Sci 118: 3419–3430 [DOI] [PubMed] [Google Scholar]
- Powell L, Burke B (1990) Internuclear exchange of an inner nuclear membrane protein (p55) in heterokaryons: in vivo evidence for the interaction of p55 with the nuclear lamina. J Cell Biol 111: 2225–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KJ, Crisp ML, Liu Q, Kim D, Kozlov S, Stewart CL, Burke B (2009) Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc Natl Acad Sci USA 106: 2194–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksena S, Summers MD, Burks JK, Johnson AE, Braunagel SC (2006) Importin-alpha-16 is a translocon-associated protein involved in sorting membrane proteins to the nuclear envelope. Nat Struct Mol Biol 13: 500–508 [DOI] [PubMed] [Google Scholar]
- Schirmer EC, Gerace L (2005) The nuclear membrane proteome: extending the envelope. Trends Biochem Sci 30: 551–558 [DOI] [PubMed] [Google Scholar]
- Schmitt J, Benavente R, Hodzic D, Hoog C, Stewart CL, Alsheimer M (2007) Transmembrane protein Sun2 is involved in tethering mammalian meiotic telomeres to the nuclear envelope. Proc Natl Acad Sci USA 104: 7426–7431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimoto T, Fukumoto M, Yoneda Y (2004) 14-3-3 suppresses the nuclear localization of threonine 157-phosphorylated p27(Kip1). EMBO J 23: 1934–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Blobel G (1993) The first membrane spanning region of the lamin B receptor is sufficient for sorting to the inner nuclear membrane. J Cell Biol 120: 631–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soullam B, Worman HJ (1993) The amino-terminal domain of the lamin B receptor is a nuclear envelope targeting signal. J Cell Biol 120: 1093–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soullam B, Worman HJ (1995) Signals and structural features involved in integral membrane protein targeting to the inner nuclear membrane. J Cell Biol 130: 15–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA (2009) A nuclear-envelope bridge positions nuclei and moves chromosomes. J Cell Sci 122: 577–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Han M (2002) Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science 298: 406–409 [DOI] [PubMed] [Google Scholar]
- Stewart CL, Roux KJ, Burke B (2007) Blurring the boundary: the nuclear envelope extends its reach. Science 318: 1408–1412 [DOI] [PubMed] [Google Scholar]
- Tran EJ, Wente SR (2006) Dynamic nuclear pore complexes: life on the edge. Cell 125: 1041–1053 [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Hase A, Ogawa M, Yorifuji H, Arahata K (1999) Distinct regions specify the nuclear membrane targeting of emerin, the responsible protein for Emery-Dreifuss muscular dystrophy. Eur J Biochem 259: 859–865 [DOI] [PubMed] [Google Scholar]
- Tzur YB, Wilson KL, Gruenbaum Y (2006) SUN-domain proteins: ‘Velcro' that links the nucleoskeleton to the cytoskeleton. Nat Rev Mol Cell Biol 7: 782–788 [DOI] [PubMed] [Google Scholar]
- Ulbert S, Platani M, Boue S, Mattaj IW (2006) Direct membrane protein-DNA interactions required early in nuclear envelope assembly. J Cell Biol 173: 469–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Du X, Cai Z, Greene MI (2006) Characterization of the structures involved in localization of the SUN proteins to the nuclear envelope and the centrosome. DNA Cell Biol 25: 554–562 [DOI] [PubMed] [Google Scholar]
- Worman HJ, Courvalin JC (2000) The inner nuclear membrane. J Membr Biol 177: 1–11 [DOI] [PubMed] [Google Scholar]
- Wu W, Lin F, Worman HJ (2002) Intracellular trafficking of MAN1, an integral protein of the nuclear envelope inner membrane. J Cell Sci 115: 1361–1371 [DOI] [PubMed] [Google Scholar]
- Yang L, Guan T, Gerace L (1997) Integral membrane proteins of the nuclear envelope are dispersed throughout the endoplasmic reticulum during mitosis. J Cell Biol 137: 1199–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemp I, Wild T, O'Donohue MF, Wandrey F, Widmann B, Gleizes PE, Kutay U (2009) Distinct cytoplasmic maturation steps of 40S ribosomal subunit precursors require hRio2. J Cell Biol 185: 1167–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.