Abstract
Purpose of review
The review will integrate current knowledge of transcriptional circuits whose dysregulation leads to autoimmunity, neutropenia and leukemia.
Recent findings
Growth factor independent-1 is a transcriptional repressor with essential roles in controlling hematopoietic stem cell biology, myeloid and lymphoid differentiation and lymphocyte effector functions. Recent work has suggested that Gfi1 competes or collaborates with other transcription factors to modulate transcription programs and lineage decisions.
Summary
Gfi1 is central to several transcriptional circuits whose dysregulation leads to abnormal or malignant hematopoiesis. These functional relationships are conserved from Drosophila development. Such conserved pathways represent central oncogenic or “gatekeeper” pathways that are pivotal to understanding the process of cellular transformation, and illustrate key targets for clinical intervention.
Keywords: Growth factor independent-1, HoxA9, Pu.1, transcription, hematopoiesis, leukemia, severe congenital neutropenia
Introduction
Growth factor independent-1 (Gfi1) is a zinc-finger transcriptional repressor that was originally identified in an in vitro screen for Moloney murine leukemia virus (MMLV) insertion loci that could progress IL-2 dependent T-cell leukemias to IL-2 independent growth [1]. In fact, the Gfi1 locus is activated by multiple common insertion loci [1,2], which when considered together reveal Gfi1 as the most frequently activated gene in MMLV-induced lymphoid malignancies [3]. In contrast, Gfi1 is a rare target of MMLV-related retroviruses in myeloid malignancies, perhaps because Gfi1 suppresses myeloid transformation [4]. Similar to other MMLV targets, Gfi1 is a critical regulator of hematopoietic development and function (see reviews[5-7]). Moreover, Gfi1 functions extend to intestinal, neuroendocrine and sensory neural cell differentiation [8-10].
Gfi1 mediates transcriptional repression dependent upon DNA binding and a 20 amino-acid amino-terminal “SNAG” transcriptional repressor domain [11]. Gfi1 repression function is mediated by the recruitment of a small number of potent chromatin modifying enzymes such as ETO [12], histone deacetylases (HDAC) 1, 2 & 3[12,13], histone lysine methyltransferase G9A [13], lysine specific demethylase-1 (LSD1)/CoREST [14], and Ajuba [15]. While these cofactors function by different mechanisms, they share the ability to interact with Gfi1 on target specific DNA sequences to gene transcription. Recent work has suggested that Gfi1 DNA binding sites can overlap or occur in close proximity to those of HoxA9, Pu.1, or C/EBP proteins, and competition or collaboration between Gfi1 and these proteins induces different transcriptional outcomes [4,16,17]. The focus of this review will highlight Gfi1 as a central player in transcriptional circuits whose dysregulation leads to neutropenia and cancer.
Gfi1 in Hematopoietic Stem Cell Biology
Gfi1 is required for adult hematopoietic stem cell (HSC) quiescence [18,19]. Deregulation of the Gfi1 target gene p21cip1/waf[20] was posited as a partial mechanism to explain the loss of HSC quiescence [18,19]. However, Gfi1−/− and p21−/− display dissimilar lineage negative, Sca1+, cKit+(LSK) numbers, with Gfi1−/− stem/progenitors in active cell cycle leading to decreasing numbers of LSK while p21−/− induces a decrease in stem/progenitor G2-M phase [18,19,21-23]. In fact, recent studies have demonstrated that HSC quiescence is a p21-independent function of p53 [24]. Notably, while deregulated p21Waf1 is not sufficient to phenocopy p53−/− or Gfi1−/− HSC effects, it is formally possible that deregulation might still be a necessary precursor to p53−/− or Gfi1−/− HSC phenotypes.
Recently, p53 and the Ets transcription factor Mef/Elf4 have been implicated in controlling HSC maintenance, perhaps through Gfi1 (Fig 1) [24]. In the absence of Mef/Elf4, a known regulator of HSC [25], LSK display a 10-fold increase in Gfi1 mRNA [24]. Mef controls the expression of Mdm2, which regulates the stability of the tumor suppressor p53. In turn, p53 regulates multiple HSC functions including proliferation and apoptosis [26,27]. Interestingly, Mef−/− induced Gfi1 mRNA was entirely dependent upon p53, as p53−/−Mef−/− mice had the same 2.5 fold decrease in Gfi1 as p53−/− LSK [24]. In agreement with this, p53−/− HSC show a partial phenocopy of Gfi1−/− HSC: decreased LSK cells in G0, and increased proliferation of LSK. However, while Gfi1−/− LSK fail in competitive BM repopulation assays, p53−/− LSK are competitive [19,24]. This may reflect either hypomorphic levels of Gfi1 in p53−/− (as opposed to the complete absence of Gfi1 in Gfi1−/− mice) or a divorce between the previously welded concepts of Gfi1−/−HSC fitness defects and Gfi1−/−increased cell cycle. Whether p53 regulates Gfi1 outside of HSC is not known.
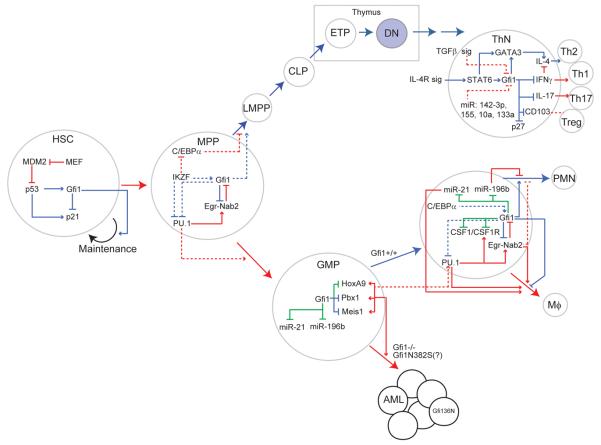
Figure 1.
Integrated view of Gfi1 transcriptional networks. Using a linear model of hematopoiesis, the published role of Gfi1 in each cell type is outlined by showing upstream activators and downstream targets of Gfi1. Solid lines: Direct targets, as confirmed by ChIP, EMSA or other physical binding data. Dotted lines: indirect targets, suggested by gene expression data. Green lines: relevant to SCN-related Gfi1N382S mutation. Blue lines: Gfi1 active circuit. Red lines: antagonism to Gfi1. The requirement for Gfi1 in DN T cells is shown with blue fill.
Independent of p53, the functional effects of hypomorphic Gfi1 levels remain to be directly proven. Kuo et al. recently demonstrated that Runx2 induces acute myeloid leukemia in cooperation with Cbfβ-SMMHC in mice, and that one effect of Runx2 forced expression was to repress Gfi1 expression [28]. In addition, Huh et al. demonstrated that Gfi1 transcript levels were low in patient myelodysplastic syndrome (MDS) samples, but whether this is due to low numbers of progenitors or a functional link to MDS is unknown [29]. We now know that Gfi1−/− myeloid progenitors are predisposed to transformation [4], but future work is needed to rigorously determine the biological sequelae of hypomorphic levels of Gfi1.
Gfi1 in Severe Congenital Neutropenia, Myeloid Development and Leukemia
Severe Congenital Neutropenia
Severe congenital neutropenia (SCN) is characterized by arrested granulocyte development and disappearance of mature neutrophils (<0.5 x 109/L)[30]. SCN is associated with mutations in ELANE[31,32], WAS [33], HAX1 [34], CSF3R [35] and GFI1[36]. Targeted sequencing of these candidate genes in Severe Congenital Neutropenia International Registry patient samples recently revealed not only additional GFI1N382S and GFI1K403R mutations, but also novel non-synomomous SNPs—GFI1L400F and GFI1R412X [37]. Notably, mutations found in genes associated with SCN have not modeled the biology of the syndrome when introduced into the murine homolog, with the exception of the Gfi1N382S mutant [38].
Previously, the lineage choice between neutrophil and macrophage differentiation was suggested to be controlled by antagonism between the Cebpα and Pu.1 transcription factors [39]. Since both factors are required at some level for control of neutrophil and macrophage genes, neither Cebpα nor Pu.1 may resolve the fate of mixed lineage cells by suppressing expression of the opposing factor. Instead, Laslo et al. hypothesized that mixed granulocyte-monocyte lineage might be resolved by direct reciprocal antagonism of Gfi1 (putatively downstream of Cebpα) versus the Egr1 and Egr2 transcription factors and the Egr-corepressor Nab2 (downstream of Pu.1). In this study, Gfi1 directly repressed Egr2 [40] during late stages of myeloid development. In separate work, the expression of Gfi1N382S in the 32D cell line induced the expression of Cebpε resulting in G-CSF-stimulated cell death instead of differentiation [41]. Thus, Gfi1 has been thought to repress Egr2 [40] and Cebpε [41] to control granulopoiesis. However, Zarebski et al. showed that Gfi1N382S (which blocked granulopoiesis) and wild type Gfi1 (which stimulated granulopoiesis) similarly affected the expression of Egr1, Egr2, Nab2 and Cebpε in primary Lin- cells [38]. Instead, Gfi1N382S selectively deregulated a subset of Gfi1 target genes including Csf1 and Csf1R. GFI1-mutant SCN patient CD34+ bone marrow cells displayed over 60 fold deregulated expression of CSF1 mRNA. This appears causative, because while expression of murine Gfi1N382S blocked granulopoiesis in murine Lin- bone marrow cells, genetic or antibody ablation of Csf1 expression restored granulopoiesis [38]. Next, Velu et al. defined microRNA (miR)-21 and miR196b as targets of Gfi1 repression and Gfi1N382S derepression [42]. Notably, Gfi1 regulates miR-21 and miR-196b in granulocytic-monocytic progenitors (GMP). Repression of these miRs is critical for allowing granulocytic differentiation as miR-21 functioned in a pro-monopoietic manner, while miR-196b exhibited anti-granulopoietic function in vitro (Fig. 1). Both miR were deregulated in a Gfi1N382S patient sample. Importantly, forced expression of miR-21 with miR-196b in Gfi1+/+ cells completely blocked G-CSF-stimulated granulopoiesis, phenocoping Gfi1N382S SCN patients [42]. The latter data illustrate microRNA as potent transducers of Gfi1 signaling; however, the relevant miR targets remain unknown.
Additional work linking Gfi1 to SCN-associated genes has demonstrated that Gfi1 regulates ELANE (encoding neutrophil elastase = NE) [36]. Recently, Salipante et al. discovered a novel protein, PFAAP5 (phosphonoformate immunoassociated protein-5), in a yeast-two hybrid screen linking both Gfi1 and NE. PFAAP5 was immunoprecipitated with both Gfi1 and NE and may bridge the molecules in the nucleus. In the presence of NE, PFAAP5 enhanced Gfi1's repression activity. Knockdown of PFAAP5 in human CD34+ cells lead to decreased myeloid colony formation but not total CFU as is the case with Gfi1 knockdown [43]. Future work is necessary to determine how NE and PFAAP5 control Gfi1 target genes, and whether SCN-associated NE mutant proteins induce defects in Gfi1 repression mechanisms.
Myeloid Development & Leukemia
The HoxA9 transcription factor is of critical interest in human acute myeloid leukemia (AML). However, in-depth molecular analysis of HoxA9 gene target regulation and evaluation of their requirement for HoxA9-mediated oncogenesis is limited to Pim1 [44], c-Myb [45] and Flt3 [46,47]. Only Pim1 [44] or c-Myb [45] impaired HoxA9 oncogenesis. Thus, the direct transcriptional network affected by endogenous HoxA9 (and therefore the HoxA9 mechanism of transformation) remains largely unknown.
Li-Kroeger et al. exploited Drosophila genetics to address a key mechanism of Hox factor function. Notably, the fly Hox factor complex is comprised of three proteins: (1) the Hox factor (Abd-A = Hox ortholog), (2) Extradenticle (Exd = Pbx ortholog) and (3) Homothorax (Hth = Meis ortholog). During analysis of the rhomboid (rho) gene cis-regulatory sequences, they made found: (1) Senseless (Sens = Gfi1 ortholog) binding sites can overlap Hox complex sites through the Sens “AATC” versus Exd “GATT” core sequence; (2) both Hox complexes and Sens can bind these sequences; and (3) transgenic-lacZ reporters, driven by one such sequence, faithfully recapitulated the expression of rho in vivo. Thus, the Hox complex competes with Sens for binding sites on target genes. If the Hox complex “wins”, the gene is activated. If the Sens protein “wins”, the gene is repressed [48].
In mammals, Horman et al. demonstrated that Gfi1 directly represses expression of HoxA9, Meis1 and Pbx1 during myelopoiesis [4]. Gfi1-mutant SCN patients and Gfi1−/− mice display increased numbers of myeloid progenitors. Importantly, the Gfi1−/− accumulation was dose-dependent upon alleles of HoxA9, genetically linking myeloid progenitor accumulation to this specific Gfi1 target gene. Epistasis experiments revealed a genetic relationship between Gfi1 and HoxA9−/− deficits in monopoeisis, which were corrected by Gfi1 haploinsufficiency; however, no such relationship was discerned between HoxA9 and Gfi1−/− granulopoietic defects. These studies reveal that progenitors blocked in differentiation do not accumulate by default. Instead, the data demonstrate that transcriptional networks regulating granulopoietic programming and progenitor programming are separable, but commonly regulated by Gfi1 [4].
The accumulation of progenitors and high levels of HoxA9 with loss of Gfi1 may explain why SCN patients are at increased risk for the induction of AML. Indeed, HoxA9 expression was elevated in an SCN patient carrying the Gfi1N382S mutation. Although the limited lifespan of Gfi1−/− mice precludes the induction of a slow-acting HoxA9-induced leukemia, the conditional Cre-mediated deletion of Gfi1 and induction of K-RasG12D expression induced a transplantable AML in 17 days [4]. These data provide the first incisive proof that Gfi1 functions to suppress myeloid progenitor transformation.
Furthermore, an association between Gfi1 and human AML has recently been suggested. Khandanpour et al. sequenced the DNA of AML patients and identified a GFI136S versus GFI136N single nucleotide polymorphism (SNP) [49] previously seen in an SCN patient with persistent tetraploid mosaicism in the bone marrow [50]. Despite the minor change in the amino acid structure (OH vs COOH), the SNP showed increased risk for disease (OR=1.6), but no effect was seen on survival or other clinical manifestations or AML subgroup. The exact mechanism remains to be determined. Global gene expression changes were not found in AML patients with the mutant allele compared to AML patients with wild type Gfi1 [49]. Future studies modeling this SNP in mice are expected to reveal specific Gfi1 target gene dysregulation.
In sum, Gfi1 is integral to multiple transcriptional programs regulating myeloid development: (1) the direct repression of target genes preventing neutrophil differentiation such as Spfi1, Egr2, Csf1, miR-21 and miR196b (the latter three are relevant to Gfi1N382S-induced SCN-associated defects), (2) the direct repression of HoxA9, Meis1 and Pbx1 in the transition to GMP. Putative target genes of Gfi1 and HoxA9 include Cxcr4, Cdkn2B, and Spfi1 [4,51]. Thus, the transcriptional sequelae of Gfi1 loss of function is amplified by the derepression of the activator of a subset of Gfi1 target genes (HoxA9); creating a feed forward loop. Future work is necessary to utilize Gfi1-HoxA9 and Pu.1-Gfi1 antagonism as tools to dissect myelopoiesis and reveal critical targets controlling myelopoiesis. Notably, the two circuits interact (Table 1) (Fig. 1).
Table 1.
Potential mechanisms of Gfi1–HoxA9/Pu.1 antagonism
| Mechanism | Effect |
|---|---|
| 1. Gfi1 binding to HoxA9, Meis1, Pbx1 promoters. |
Decreased HoxA9, Meis1 and Pbx1 protein expression and indirect repression of Hox- complex targets (putatively Spfi1) |
| 2. Gfi1 competing for HoxA9-Pbx1-Meis1 binding sites (not proven in mammals). |
Direct repression of a subset of HoxA9 target genes with composite Gfi1-HoxA9 binding sites. |
| 3. Gfi1 binding to Spfi1 promoter | Decreased Pu.1 protein |
| 4. Gfi1 binding to Csf1R promoter (a Pu.1 target) |
Decreased Csf1R protein |
| 5. Gfi1-Pu.1 protein-protein interaction | Repression of Pu.1 target genes (e.g. Csf1R) |
| 6. Gfi1-Pu.1 bind in close proximity on a subset of common target genes (not proven) |
Repression of a subset of Pu.1 target genes |
| 7. Pu.1 binding promoters of Egr-1, Egr-2, Nab2 |
Increased expression of Egr-1, Egr-2, Nab2 and subsequent repression of Gfi1 |
Gfi1 in Lymphoid Development and Function
T cell development
Gfi1 is required for T lymphopoiesis [52] as well as mature CD4+ and CD8+ T cell function and maintenance. Notably, while Gfi1 is required for naïve CD8+ (but not CD4+) T cell maintenance [53], Gfi1 appears to be essential for the CD4+ (but not CD8+) immune response [54]. Specifically, Gfi1 controls CD4+ T helper-2 (Th2) cell polarization, which requires expression of the IL-4-Stat6 target genes Gfi1 and Gata3 [55]. In human blood cells, GFI1 and GATA3 display significant concordant expression in CD4+ and CD8+ T cells as well as CD56+ natural killer (NK) cells [56]. Thus, the role of Gfi1 and Gata3 in cells outside the Th2 lineage needs to be explored more thoroughly. Within the Th2 lineage, Gfi1 and Gata3 biologically integrate the IL4 signal by enhancing proliferation and inhibiting apoptosis to expand presumptive Th2 cells [55]. Gfi1 physically associates with Gata3 preventing its ubiquitination and degradation [57]. One caveat to this explanation of Gfi1-Gata3 synergy is that the biological effects of Gfi1-Gata3 were dependent upon an intact Gfi1 SNAG repressor domain [55], and the role of SNAG associated proteins was not addressed.
Gfi1 controls Th2 lineage fidelity. Th2 cells express IL-4, -5 and -13, but not IFNγ. Gfi1−/− cells show lower levels of repressive marks at the IFNγ locus, and decreased activating histone modifications at the IL-5 promoter, suggesting that in conjunction with Gata3, Gfi1 may also control Th2 effector function [57]. Moreover, loss of Gfi1 leads to increased production of the hallmark cytokine of Th17 cells, IL-17α, as well as an increased plasticity in switching from Th2 to Th17 [58]. Furthermore, overexpression of Gfi1 prevented the formation of Th17 cells and IL-17α production, but not RORγt expression. This result was independently replicated by another group [59]. Both groups demonstrated that Gfi1 levels where repressed upon TGFβ signaling, a requisite of Th17 polarization [58,59]. Although Gfi1−/− cells display dramatic differences in levels of histone 3 lysine 4 (H3K4) trimethylation at the RORc locus, no direct Gfi1 binding was identified. In contrast, the Gfi1 cofactor, Lsd1, occupied an intergenic region of the IL-17 gene in Gfi1+/+ (but not in Gfi1−/−) Th2 primed cells. Thus, high levels of Gfi1 ensure the fidelity of Th2 polarization by repressing genes controlling alternative cell fates. Moreover, Th17 differentiation is initiated by a feed forward loop initiated by TGFβ-signal-mediated repression of Gfi1, which releases IL-17 from Gfi1-mediated repression and induces IL-17-stimulated Th17 polarization [58].
CD103 is an important E-cadherin binding integrin found on human intraepithelial lymphocytes and on naturally occurring T-regulatory cells with high suppressive activity [60,61]. Gfi1−/− cells lack CD103-locus-associated Lsd1, had higher levels of H3K4 trimethylation at the CD103 locus, and had increased ability to prime iTregs, especially Foxp3+, CD103+ iTregs [58]. To delineate the functional relevance of these findings, Zhu et al. used an experimental model of multiple sclerosis (MS). MS is thought to be driven by Th17 biology. Loss of Gfi1 leads to increased Th17 polarization. However, the conditional Gfi1 knockout mice showed delayed disease onset as well as decreased severity. This was likely due to the increased numbers of Foxp3+ CD103+ regulatory cells and their ability to modulate disease severity [58]. These findings may have some relevance to human disease in that 21 SNPs within the GFI-EVI5-RPL5-FAM69A locus were associated with MS in 732 patients. One SNP in the 17th intron of EVI5 had a significant association with disease (P=0.008, OR=1.29; 95% CI=1.08-1.54) [62]. Notably, MMLV insertion mutagenesis in the last intron of Evi5 activates murine Gfi1 expression [2], but it currently unclear whether the SNP functions similarly to affect human GFI1 expression.
B-lymphopoiesis
Gfi1−/− mice display profound deficits in B cell numbers due to a block in early lymphoid precursors [52]. IL-7 is critical for murine B lymphopoiesis. In CD8+ T cells, IL7 induces Gfi1 to antagonize GABPα activation of IL7Ra [63] and maintain naïve T cell pools [53], In B cells, Gfi1 loss was associated with a deficit in response to IL7 [64], providing the possibility that Gfi1 is a downstream effector of IL7 signaling. More recently, a transcriptional network was suggested to explain Gfi1−/− defective B lymphopoiesis [16]. Specifically, the “C/Ebpα-Gfi1 versus Egr2-Pu.1” circuit [40] was revised to posit that in multipotent progenitors (MPPs) the transcription factor Ikaros upregulates Gfi1 expression. In this “Ikaros-Gfi1 versus Egr2-Pu.1” circuit, Ikaros replaces Cebpα, but Gfi1 still represses Egr2 to prevent myeloid differentiation and allow lymphoid priming. Previously, Gfi1 was shown to physically bind Pu.1 protein and antagonize Pu.1 target gene activation [65]. Using Pu.1 ChIP-on-chip analysis, Spooner et al showed that 19% of Pu.1-bound regions contained putative binding sites for both Gfi1 and Pu.1 within 10bp of each other, including the promoter of the Pu.1-encoding gene (Sfpi1). Global Pu.1/Gfi1 binding was not proven, but the suggestion is that direct antagonism between Pu.1 and Gfi1 is possible. In agreement with this, haploinsufficiency of Sfpi1 on a Gfi1−/− null background, partially restored B cell potential [16]; however, this analysis does not delineate between Gfi1 repression of Sfpi1 versus direct Gfi1 antagonism of Pu.1 target genes.
Gfi1 is not expressed in mature unchallenged B cells. Interestingly, Gfi1, Egr1, Egr2 and Nab2 are coordinately upregulated subsequent to B cell receptor (BCR) ligation through an ERK-dependent tolerogenic signal [66]. Gfi1−/− B-cell-driven autoimmunity has been reported [67], but not confirmed [68]. Instead, the second group noted Gfi1−/− B cells display increased levels of serum IgG antibodies after antigen challenge. Gfi1 did not bind the promoters of any of the Iγ loci [68]. Instead, the authors posit a 32-fold increase in TGFβ1 transcript levels as causative; however, this only correlated to ~2-fold increase in TGFβ1 protein following lipopolysaccharide (LPS)-challenge [68]. Thus, tolerogenic BCR signaling induces Gfi1 which may control IgG expression; however, the functional ramifications of Gfi1 in mature B cells needs to be resolved.
Gfi1 in Lymphoid Leukemia
Dabrowska et al. demonstrated how MMLV insertion mutagenesis overcomes miR targeting to activate Gfi1 expression [69]. While insertion sites in the Gfi1 3′ UTR were known to lead to increased transcript levels [2,70], these insertions also truncate the Gfi1 transcript to eliminate binding sites for a number of miRs, including miR-155 and miR-10a [69]. Luciferase sensor assays confirmed targeting of Gfi1 3′UTR by the miR [69]. These new findings implicate miRs in controlling Gfi1 protein levels in both normal and oncogenic settings, and suggest future work to delineate proteosomal degradation [71,72] versus miR mechanisms to control Gfi1 levels. Interestingly, they also showed leukemia-specific Gfi1 mRNA alternative splicing, but did not document transcript abundance [69].
Using a yeast 2-hybrid screen, Gfi1 and Miz-1 (Myc-interacting zinc finger protein-1) were found to interact and form a complex with c-Myc [73]. This interaction was present in multiple cell lines and could repress target genes Cdkn2B (p15) and Cdkn1B (p27) in lucerifase assays. Increased expression of Cdkn2B (a putative HoxA9 target gene) was observed in Gfi1−/− bone marrow [73]. Upregulation of Cdkn2B may be augmented by deregulated expression of HoxA9, Meis1 and Pbx1 in Gfi1−/− cells [4]. The repression of Cdkn2B might explain the collaboration between Gfi1 and Myc in lymphoid malignancies, but this remains to be proven.
Hematopoietic-relevant lessons learned from Gfi1 in non-hematopoeitic tissues
While Gfi1 repression of target genes is well known, work from the Drosophila field suggests that Gfi1 may synergize with other transcription factors to activate target genes, and this may be relevant to humans with specific granule deficiency (SGD). Studies from three groups show that Drosophila Senseless can function as a potent transcriptional co-activator. In developing sensory precursor cells, the basic Helix-Loop-Helix (bHLH) factors Achaete, Scute, and Atonal stimulate gene expression by recruiting Sens through its zinc finger motifs [74,75]. Additionally, Sens stimulates the ability of the Orthodenticle homeodomain transcription factor to activate rhodopsin gene expression within photoreceptor cells [76]. In humans, Khanna-Gupta et al. demonstrated that a patient with SGD who was wild type for C/EBPε, had abnormally low levels of GFI1. She further showed that GFI1 and C/EBP proteins bind in close proximity in the promoter of the neutrophil collagenase promoter, and that cotransfection of GFI1 and C/EBPε activated a neutrophil-collagenase-promoter driven reporter [17]. While the mechanism of putative Gfi1 coactivator function is unknown, the effect appears to span evolution.
In a pathway similar to its Drosophila ortholog Senseless, Gfi1 is essential for the bHLH-initiated differentiation and survival of several solid organ cell types. A common mechanism is antagonism to Notch. The atonal homologs Atoh1 and Atoh7 function upstream of Gfi1 in the inner ear, intestine, and retina, respectively [8,10,77]. In the nervous system Gfi1 is required for differentiation and survival of the mechanosensory hair cells of the inner ear, and for survival of cochlear ganglion neurons and cerebellar purkinje cells [10,78]. In the small intestine and colon, Gfi1 is expressed in progenitor cells and directs appropriate differentiation of enteroendocrine, goblet and Paneth cells [8]. In the lung, Gfi1 functions downstream of the achaete/scute homolog Ascl1, and is required for pulmonary neuroendocrine cell differentiation [9]. In each of these contexts, Gfi1 functions in concert with its respective bHLH factor to oppose the activity of Notch, and thereby direct differentiation of a specific cell fate [78,79]. Future work is necessary to discern a functional relationship between Gfi1 and Notch signaling in hematopoietic cells.
Conclusion
Complex transcriptional circuits promoting differentiation and proliferation are intimately linked to Gfi1. Multiple disease states are observed with Gfi1 gain and loss of function such as severe congenital neutropenia, non-immune chronic idiopathic neutropenia of adults, specific granule deficiency and perhaps AML and T-ALL. However, large gaps in knowledge still surround Gfi1 critical targets and cofactors. Defining these targets and partners will potentiate methods to manipulate these proteins for clinical intervention.
Acknowledgments
Funding: NIH CA142601, HL079574, the Leukemia and Lymphoma Society of America, and Alex's Lemonade Stand.
REFERENCES
- 1.Gilks CB, Bear SE, Grimes HL, Tsichlis PN. Progression of interleukin-2 (IL-2)-dependent rat T cell lymphoma lines to IL-2-independent growth following activation of a gene (Gfi-1) encoding a novel zinc finger protein. Mol Cell Biol. 1993;13:1759–1768. doi: 10.1128/mcb.13.3.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scheijen B, Jonkers J, Acton D, Berns A. Characterization of pal-1, a common proviral insertion site in murine leukemia virus-induced lymphomas of c-myc and Pim-1 transgenic mice. J Virol. 1997;71:9–16. doi: 10.1128/jvi.71.1.9-16.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uren AG, Kool J, Matentzoglu K, de Ridder J, Mattison J, van Uitert M, Lagcher W, Sie D, Tanger E, Cox T, et al. Large-scale mutagenesis in p19(ARF)- and p53-deficient mice identifies cancer genes and their collaborative networks. Cell. 2008;133:727–741. doi: 10.1016/j.cell.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4**.Horman SR, Velu CS, Chaubey A, Bourdeau T, Zhu J, Paul WE, Gebelein B, Grimes HL. Gfi1 integrates progenitor versus granulocytic transcriptional programming. Blood. 2009;113:5466–5475. doi: 10.1182/blood-2008-09-179747. Gfi1 represses HoxA9, Meis1 and Pbx1 in myeloid progenitors. HoxA9 is required for Gfi1−/− myeloid progenitor accumulation. First definitive report proving that Gfi1−/− predisposes to myeloid leukemia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kazanjian A, Gross EA, Grimes HL. The growth factor independence-1 transcription factor: new functions and new insights. Crit Rev Oncol Hematol. 2006;59:85–97. doi: 10.1016/j.critrevonc.2006.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boztug K, Klein C. Novel genetic etiologies of severe congenital neutropenia. Curr Opin Immunol. 2009;21:472–480. doi: 10.1016/j.coi.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 7.Moroy T, Zeng H, Jin J, Schmid KW, Carpinteiro A, Gulbins E. The zinc finger protein and transcriptional repressor Gfi1 as a regulator of the innate immune response. Immunobiology. 2008;213:341–352. doi: 10.1016/j.imbio.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Shroyer NF, Wallis D, Venken KJ, Bellen HJ, Zoghbi HY. Gfi1 functions downstream of Math1 to control intestinal secretory cell subtype allocation and differentiation. Genes Dev. 2005;19:2412–2417. doi: 10.1101/gad.1353905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kazanjian A, Wallis D, Au N, Nigam R, Venken KJ, Cagle PT, Dickey BF, Bellen HJ, Gilks CB, Grimes HL. Growth factor independence-1 is expressed in primary human neuroendocrine lung carcinomas and mediates the differentiation of murine pulmonary neuroendocrine cells. Cancer Res. 2004;64:6874–6882. doi: 10.1158/0008-5472.CAN-04-0633. [DOI] [PubMed] [Google Scholar]
- 10.Wallis D, Hamblen M, Zhou Y, Venken KJ, Schumacher A, Grimes HL, Zoghbi HY, Orkin SH, Bellen HJ. The zinc finger transcription factor Gfi1, implicated in lymphomagenesis, is required for inner ear hair cell differentiation and survival. Development. 2003;130:221–232. doi: 10.1242/dev.00190. [DOI] [PubMed] [Google Scholar]
- 11.Grimes HL, Chan TO, Zweidler-McKay PA, Tong B, Tsichlis PN. The Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol Cell Biol. 1996;16:6263–6272. doi: 10.1128/mcb.16.11.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGhee L, Bryan J, Elliott L, Grimes HL, Kazanjian A, Davis JN, Meyers S. Gfi-1 attaches to the nuclear matrix, associates with ETO (MTG8) and histone deacetylase proteins, and represses transcription using a TSA-sensitive mechanism. J Cell Biochem. 2003;89:1005–1018. doi: 10.1002/jcb.10548. [DOI] [PubMed] [Google Scholar]
- 13.Duan Z, Zarebski A, Montoya-Durango D, Grimes HL, Horwitz M. Gfi1 coordinates epigenetic repression of p21Cip/WAF1 by recruitment of histone lysine methyltransferase G9a and histone deacetylase 1. Mol Cell Biol. 2005;25:10338–10351. doi: 10.1128/MCB.25.23.10338-10351.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saleque S, Kim J, Rooke HM, Orkin SH. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol Cell. 2007;27:562–572. doi: 10.1016/j.molcel.2007.06.039. [DOI] [PubMed] [Google Scholar]
- 15.Montoya-Durango DE, Velu CS, Kazanjian A, Rojas ME, Jay CM, Longmore GD, Grimes HL. Ajuba functions as a histone deacetylase-dependent co-repressor for autoregulation of the growth factor-independent-1 transcription factor. J Biol Chem. 2008;283:32056–32065. doi: 10.1074/jbc.M802320200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16**.Spooner CJ, Cheng JX, Pujadas E, Laslo P, Singh H. A recurrent network involving the transcription factors PU.1 and Gfi1 orchestrates innate and adaptive immune cell fates. Immunity. 2009;31:576–586. doi: 10.1016/j.immuni.2009.07.011. Revised transcriptional circuit from Laslo et al. replaces C/Ebpα with Ikaros (Ikaros-Gfi1 versus Egr2-Pu.1) delineates lymphoid versus myeloid cell fate in MPP. 20% of PU.1 bound regions contain Gfi1 consensus binding sites. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khanna-Gupta A, Sun H, Zibello T, Lee HM, Dahl R, Boxer LA, Berliner N. Growth factor independence-1 (Gfi-1) plays a role in mediating specific granule deficiency (SGD) in a patient lacking a gene-inactivating mutation in the C/EBPepsilon gene. Blood. 2007;109:4181–4190. doi: 10.1182/blood-2005-05-022004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng H, Yucel R, Kosan C, Klein-Hitpass L, Moroy T. Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J. 2004;23:4116–4125. doi: 10.1038/sj.emboj.7600419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hock H, Hamblen MJ, Rooke HM, Schindler JW, Saleque S, Fujiwara Y, Orkin SH. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature. 2004;431:1002–1007. doi: 10.1038/nature02994. [DOI] [PubMed] [Google Scholar]
- 20.Tong B, Grimes HL, Yang TY, Bear SE, Qin Z, Du K, El-Deiry WS, Tsichlis PN. The Gfi-1B proto-oncoprotein represses p21WAF1 and inhibits myeloid cell differentiation. Mol Cell Biol. 1998;18:2462–2473. doi: 10.1128/mcb.18.5.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng T, Rodrigues N, Shen H, Yang Y, Dombkowski D, Sykes M, Scadden DT. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science. 2000;287:1804–1808. doi: 10.1126/science.287.5459.1804. [DOI] [PubMed] [Google Scholar]
- 22.van Os R, de Haan G, Dykstra BJ. Hematopoietic stem cell quiescence: yet another role for p53. Cell Stem Cell. 2009;4:7–8. doi: 10.1016/j.stem.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 23*.Foudi A, Hochedlinger K, Van Buren D, Schindler JW, Jaenisch R, Carey V, Hock H. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat Biotechnol. 2009;27:84–90. doi: 10.1038/nbt.1517. p21−/− hematopoietic stem cells do not phenocopy Gfi1−/− defects. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24**.Liu Y, Elf SE, Miyata Y, Sashida G, Huang G, Di Giandomenico S, Lee JM, Deblasio A, Menendez S, Antipin J, et al. p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell. 2009;4:37–48. doi: 10.1016/j.stem.2008.11.006. p53 activates Gfi1 in HSCs. p53-mediated HSC maintenance is not dependent upon p21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lacorazza HD, Yamada T, Liu Y, Miyata Y, Sivina M, Nunes J, Nimer SD. The transcription factor MEF/ELF4 regulates the quiescence of primitive hematopoietic cells. Cancer Cell. 2006;9:175–187. doi: 10.1016/j.ccr.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 26.Shounan Y, Dolnikov A, MacKenzie KL, Miller M, Chan YY, Symonds G. Retroviral transduction of hematopoietic progenitor cells with mutant p53 promotes survival and proliferation, modifies differentiation potential and inhibits apoptosis. Leukemia. 1996;10:1619–1628. [PubMed] [Google Scholar]
- 27.TeKippe M, Harrison DE, Chen J. Expansion of hematopoietic stem cell phenotype and activity in Trp53-null mice. Exp Hematol. 2003;31:521–527. doi: 10.1016/s0301-472x(03)00072-9. [DOI] [PubMed] [Google Scholar]
- 28.Kuo YH, Zaidi SK, Gornostaeva S, Komori T, Stein GS, Castilla LH. Runx2 induces acute myeloid leukemia in cooperation with Cbfbeta-SMMHC in mice. Blood. 2009;113:3323–3332. doi: 10.1182/blood-2008-06-162248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huh HJ, Chae SL, Lee M, Hong KS, Mun YC, Seong CM, Chung WS, Huh JW. CD34, RAB20, PU.1 and GFI1 mRNA expression in myelodysplastic syndrome. Int J Lab Hematol. 2009;31:344–351. doi: 10.1111/j.1751-553X.2008.01056.x. [DOI] [PubMed] [Google Scholar]
- 30.Boxer LA. Severe congenital neutropenia: genetics and pathogenesis. Trans Am Clin Climatol Assoc. 2006;117:13–31. discussion 31-12. [PMC free article] [PubMed] [Google Scholar]
- 31.Dale DC, Person RE, Bolyard AA, Aprikyan AG, Bos C, Bonilla MA, Boxer LA, Kannourakis G, Zeidler C, Welte K, et al. Mutations in the gene encoding neutrophil elastase in congenital and cyclic neutropenia. Blood. 2000;96:2317–2322. [PubMed] [Google Scholar]
- 32.Ancliff PJ, Gale RE, Liesner R, Hann IM, Linch DC. Mutations in the ELA2 gene encoding neutrophil elastase are present in most patients with sporadic severe congenital neutropenia but only in some patients with the familial form of the disease. Blood. 2001;98:2645–2650. doi: 10.1182/blood.v98.9.2645. [DOI] [PubMed] [Google Scholar]
- 33.Devriendt K, Kim AS, Mathijs G, Frints SG, Schwartz M, Van Den Oord JJ, Verhoef GE, Boogaerts MA, Fryns JP, You D, et al. Constitutively activating mutation in WASP causes X-linked severe congenital neutropenia. Nat Genet. 2001;27:313–317. doi: 10.1038/85886. [DOI] [PubMed] [Google Scholar]
- 34.Klein C, Grudzien M, Appaswamy G, Germeshausen M, Sandrock I, Schaffer AA, Rathinam C, Boztug K, Schwinzer B, Rezaei N, et al. HAX1 deficiency causes autosomal recessive severe congenital neutropenia (Kostmann disease) Nat Genet. 2007;39:86–92. doi: 10.1038/ng1940. [DOI] [PubMed] [Google Scholar]
- 35.Dror Y, Ward AC, Touw IP, Freedman MH. Combined corticosteroid/granulocyte colony-stimulating factor (G-CSF) therapy in the treatment of severe congenital neutropenia unresponsive to G-CSF: Activated glucocorticoid receptors synergize with G-CSF signals. Exp Hematol. 2000;28:1381–1389. doi: 10.1016/s0301-472x(00)00544-0. [DOI] [PubMed] [Google Scholar]
- 36.Person RE, Li FQ, Duan Z, Benson KF, Wechsler J, Papadaki HA, Eliopoulos G, Kaufman C, Bertolone SJ, Nakamoto B, et al. Mutations in proto-oncogene GFI1 cause human neutropenia and target ELA2. Nat Genet. 2003;34:308–312. doi: 10.1038/ng1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37*.Xia J, Bolyard AA, Rodger E, Stein S, Aprikyan AA, Dale DC, Link DC. Prevalence of mutations in ELANE, GFI1, HAX1, SBDS, WAS and G6PC3 in patients with severe congenital neutropenia. Br J Haematol. 2009;147:535–542. doi: 10.1111/j.1365-2141.2009.07888.x. Novel patient mutations in Gfi1, as well as additional Gfi1N38S and Gfi1K403R mutations were associated with SCN. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zarebski A, Velu CS, Baktula AM, Bourdeau T, Horman SR, Basu S, Bertolone SJ, Horwitz M, Hildeman DA, Trent JO, et al. Mutations in growth factor independent-1 associated with human neutropenia block murine granulopoiesis through colony stimulating factor-1. Immunity. 2008;28:370–380. doi: 10.1016/j.immuni.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dahl R, Walsh JC, Lancki D, Laslo P, Iyer SR, Singh H, Simon MC. Regulation of macrophage and neutrophil cell fates by the PU.1:C/EBPalpha ratio and granulocyte colony-stimulating factor. Nat Immunol. 2003;4:1029–1036. doi: 10.1038/ni973. [DOI] [PubMed] [Google Scholar]
- 40.Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R, Gantner BN, Dinner AR, Singh H. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126:755–766. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- 41.Zhuang D, Qiu Y, Kogan SC, Dong F. Increased CCAAT enhancer-binding protein epsilon (C/EBPepsilon) expression and premature apoptosis in myeloid cells expressing Gfi-1 N382S mutant associated with severe congenital neutropenia. J Biol Chem. 2006;281:10745–10751. doi: 10.1074/jbc.M510924200. [DOI] [PubMed] [Google Scholar]
- 42**.Velu CS, Baktula AM, Grimes HL. Gfi1 regulates miR-21 and miR-196b to control myelopoiesis. Blood. 2009;113:4720–4728. doi: 10.1182/blood-2008-11-190215. Gfi1 controls the expression of microRNAs; whose deregulated expression phenocopies Gfi1−/− or Gfi1N382S blocks to granulopoiesis. It is the second report of microRNA synergy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43*.Salipante SJ, Rojas ME, Korkmaz B, Duan Z, Wechsler J, Benson KF, Person RE, Grimes HL, Horwitz MS. Contributions to neutropenia from PFAAP5 (N4BP2L2), a novel protein mediating transcriptional repressor cooperation between Gfi1 and neutrophil elastase. Mol Cell Biol. 2009;29:4394–4405. doi: 10.1128/MCB.00596-09. Links SCN relevant proteins Neutrophil Elastase and Gfi1 through PFAAP5 protein interaction. PFAAP5 knockdown affects granulopoiesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu YL, Passegue E, Fong S, Largman C, Lawrence HJ. Evidence that the Pim1 kinase gene is a direct target of HOXA9. Blood. 2007;109:4732–4738. doi: 10.1182/blood-2006-08-043356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hess JL, Bittner CB, Zeisig DT, Bach C, Fuchs U, Borkhardt A, Frampton J, Slany RK. c-Myb is an essential downstream target for homeobox-mediated transformation of hematopoietic cells. Blood. 2006;108:297–304. doi: 10.1182/blood-2005-12-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morgado E, Albouhair S, Lavau C. Flt3 is dispensable to the Hoxa9/Meis1 leukemogenic cooperation. Blood. 2007;109:4020–4022. doi: 10.1182/blood-2006-01-039586. [DOI] [PubMed] [Google Scholar]
- 47.Wang GG, Pasillas MP, Kamps MP. Meis1 programs transcription of FLT3 and cancer stem cell character, using a mechanism that requires interaction with Pbx and a novel function of the Meis1 C-terminus. Blood. 2005;106:254–264. doi: 10.1182/blood-2004-12-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li-Kroeger D, Witt LM, Grimes HL, Cook TA, Gebelein B. Hox and senseless antagonism functions as a molecular switch to regulate EGF secretion in the Drosophila PNS. Dev Cell. 2008;15:298–308. doi: 10.1016/j.devcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49**.Khandanpour C, Thiede C, Valk PJ, Sharif-Askari E, Nueckel H, Lohmann D, Horsthemke B, Siffert W, Neubauer A, Grzeschik KH, et al. A variant allele of Growth Factor Independence 1 (GFI1) is associated with acute myeloid leukemia. Blood. doi: 10.1182/blood-2009-08-239822. A GFI1 SNP encoding Gfi136N is more frequently found in AML. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hochberg JC, Miron PM, Hay BN, Woda BA, Wang SA, Richert-Przygonska M, Aprikyan AA, Newburger PE. Mosaic tetraploidy and transient GFI1 mutation in a patient with severe chronic neutropenia. Pediatr Blood Cancer. 2008;50:630–632. doi: 10.1002/pbc.21094. [DOI] [PubMed] [Google Scholar]
- 51.De La Luz Sierra M, Gasperini P, McCormick PJ, Zhu J, Tosato G. Transcription factor Gfi-1 induced by G-CSF is a negative regulator of CXCR4 in myeloid cells. Blood. 2007;110:2276–2285. doi: 10.1182/blood-2007-03-081448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yucel R, Karsunky H, Klein-Hitpass L, Moroy T. The transcriptional repressor Gfi1 affects development of early, uncommitted c-Kit+ T cell progenitors and CD4/CD8 lineage decision in the thymus. J Exp Med. 2003;197:831–844. doi: 10.1084/jem.20021417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park JH, Yu Q, Erman B, Appelbaum JS, Montoya-Durango D, Grimes HL, Singer A. Suppression of IL7Ralpha transcription by IL-7 and other prosurvival cytokines: a novel mechanism for maximizing IL-7-dependent T cell survival. Immunity. 2004;21:289–302. doi: 10.1016/j.immuni.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 54.Pargmann D, Yucel R, Kosan C, Saba I, Klein-Hitpass L, Schimmer S, Heyd F, Dittmer U, Moroy T. Differential impact of the transcriptional repressor Gfi1 on mature CD4+ and CD8+ T lymphocyte function. Eur J Immunol. 2007;37:3551–3563. doi: 10.1002/eji.200737130. [DOI] [PubMed] [Google Scholar]
- 55.Zhu J, Guo L, Min B, Watson CJ, Hu-Li J, Young HA, Tsichlis PN, Paul WE. Growth factor independent-1 induced by IL-4 regulates Th2 cell proliferation. Immunity. 2002;16:733–744. doi: 10.1016/s1074-7613(02)00317-5. [DOI] [PubMed] [Google Scholar]
- 56.Watkins NA, Gusnanto A, de Bono B, De S, Miranda-Saavedra D, Hardie DL, Angenent WG, Attwood AP, Ellis PD, Erber W, et al. A HaemAtlas: characterizing gene expression in differentiated human blood cells. Blood. 2009;113:e1–9. doi: 10.1182/blood-2008-06-162958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shinnakasu R, Yamashita M, Kuwahara M, Hosokawa H, Hasegawa A, Motohashi S, Nakayama T. Gfi1-mediated stabilization of GATA3 protein is required for Th2 cell differentiation. J Biol Chem. 2008;283:28216–28225. doi: 10.1074/jbc.M804174200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58**.Zhu J, Davidson TS, Wei G, Jankovic D, Cui K, Schones DE, Guo L, Zhao K, Shevach EM, Paul WE. Down-regulation of Gfi-1 expression by TGF-beta is important for differentiation of Th17 and CD103+ inducible regulatory T cells. J Exp Med. 2009;206:329–341. doi: 10.1084/jem.20081666. Gfi1 enforces the fidelity of Th2 lineage specification. Characterizes chromatin changes associated with Gfi1 loss of function that are depenedent on site-specific recruitment of the Gfi1 cofactor LSD1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ichiyama K, Hashimoto M, Sekiya T, Nakagawa R, Wakabayashi Y, Sugiyama Y, Komai K, Saba I, Moroy T, Yoshimura A. Gfi1 negatively regulates T(h)17 differentiation by inhibiting RORgammat activity. Int Immunol. 2009;21:881–889. doi: 10.1093/intimm/dxp054. [DOI] [PubMed] [Google Scholar]
- 60.Suffia IJ, Reckling SK, Piccirillo CA, Goldszmid RS, Belkaid Y. Infected site-restricted Foxp3+ natural regulatory T cells are specific for microbial antigens. J Exp Med. 2006;203:777–788. doi: 10.1084/jem.20052056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suffia I, Reckling SK, Salay G, Belkaid Y. A role for CD103 in the retention of CD4+CD25+ Treg and control of Leishmania major infection. J Immunol. 2005;174:5444–5455. doi: 10.4049/jimmunol.174.9.5444. [DOI] [PubMed] [Google Scholar]
- 62.Alcina A, Fernandez O, Gonzalez JR, Catala-Rabasa A, Fedetz M, Ndagire D, Leyva L, Guerrero M, Arnal C, Delgado C, et al. Tag-SNP analysis of the GFI1-EVI5-RPL5-FAM69 risk locus for multiple sclerosis. Eur J Hum Genet. doi: 10.1038/ejhg.2009.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chandele A, Joshi NS, Zhu J, Paul WE, Leonard WJ, Kaech SM. Formation of IL-7Ralphahigh and IL-7Ralphalow CD8 T cells during infection is regulated by the opposing functions of GABPalpha and Gfi-1. J Immunol. 2008;180:5309–5319. doi: 10.4049/jimmunol.180.8.5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rathinam C, Klein C. Transcriptional repressor Gfi1 integrates cytokine-receptor signals controlling B-cell differentiation. PLoS One. 2007;2:e306. doi: 10.1371/journal.pone.0000306. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 65.Dahl R, Iyer SR, Owens KS, Cuylear DD, Simon MC. The transcriptional repressor GFI-1 antagonizes PU.1 activity through protein-protein interaction. J Biol Chem. 2007;282:6473–6483. doi: 10.1074/jbc.M607613200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Glynne R, Ghandour G, Rayner J, Mack DH, Goodnow CC. B-lymphocyte quiescence, tolerance and activation as viewed by global gene expression profiling on microarrays. Immunol Rev. 2000;176:216–246. doi: 10.1034/j.1600-065x.2000.00614.x. [DOI] [PubMed] [Google Scholar]
- 67.Rathinam C, Lassmann H, Mengel M, Klein C. Transcription factor Gfi1 restricts B cell-mediated autoimmunity. J Immunol. 2008;181:6222–6229. doi: 10.4049/jimmunol.181.9.6222. [DOI] [PubMed] [Google Scholar]
- 68.Igwe E, Kosan C, Khandanpour C, Sharif-Askari E, Brune B, Moroy T. The zinc finger protein Gfi1 is implicated in the regulation of IgG2b production and the expression of Igamma2b germline transcripts. Eur J Immunol. 2008;38:3004–3014. doi: 10.1002/eji.200838251. [DOI] [PubMed] [Google Scholar]
- 69*.Dabrowska MJ, Dybkaer K, Johnsen HE, Wang B, Wabl M, Pedersen FS. Loss of MicroRNA targets in the 3′ untranslated region as a mechanism of retroviral insertional activation of growth factor independence 1. J Virol. 2009;83:8051–8061. doi: 10.1128/JVI.00427-09. Gfi1 is targeted by microRNA including miR-155 and miR10a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zornig M, Schmidt T, Karsunky H, Grzeschiczek A, Moroy T. Zinc finger protein GFI-1 cooperates with myc and pim-1 in T-cell lymphomagenesis by reducing the requirements for IL-2. Oncogene. 1996;12:1789–1801. [PubMed] [Google Scholar]
- 71.Marteijn JA, van der Meer LT, van Emst L, van Reijmersdal S, Wissink W, de Witte T, Jansen JH, Van der Reijden BA. Gfi1 ubiquitination and proteasomal degradation is inhibited by the ubiquitin ligase Triad1. Blood. 2007;110:3128–3135. doi: 10.1182/blood-2006-11-058602. [DOI] [PubMed] [Google Scholar]
- 72.Marteijn JA, van der Meer LT, Van Emst L, de Witte T, Jansen JH, van der Reijden BA. Diminished proteasomal degradation results in accumulation of Gfi1 protein in monocytes. Blood. 2007;109:100–108. doi: 10.1182/blood-2006-02-003590. [DOI] [PubMed] [Google Scholar]
- 73*.Basu S, Liu Q, Qiu Y, Dong F. Gfi-1 represses CDKN2B encoding p15INK4B through interaction with Miz-1. Proc Natl Acad Sci U S A. 2009;106:1433–1438. doi: 10.1073/pnas.0804863106. Gfi1 interacts with Miz-1 to complex with c-Myc and repress CDKi encoding genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Acar M, Jafar-Nejad H, Giagtzoglou N, Yallampalli S, David G, He Y, Delidakis C, Bellen HJ. Senseless physically interacts with proneural proteins and functions as a transcriptional co-activator. Development. 2006;133:1979–1989. doi: 10.1242/dev.02372. [DOI] [PubMed] [Google Scholar]
- 75.Powell LM, Deaton AM, Wear MA, Jarman AP. Specificity of Atonal and Scute bHLH factors: analysis of cognate E box binding sites and the influence of Senseless. Genes Cells. 2008;13:915–929. doi: 10.1111/j.1365-2443.2008.01217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xie B, Charlton-Perkins M, McDonald E, Gebelein B, Cook T. Senseless functions as a molecular switch for color photoreceptor differentiation in Drosophila. Development. 2007;134:4243–4253. doi: 10.1242/dev.012781. [DOI] [PubMed] [Google Scholar]
- 77.Yang Z, Ding K, Pan L, Deng M, Gan L. Math5 determines the competence state of retinal ganglion cell progenitors. Dev Biol. 2003;264:240–254. doi: 10.1016/j.ydbio.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 78.Tsuda H, Jafar-Nejad H, Patel AJ, Sun Y, Chen HK, Rose MF, Venken KJ, Botas J, Orr HT, Bellen HJ, et al. The AXH domain of Ataxin-1 mediates neurodegeneration through its interaction with Gfi-1/Senseless proteins. Cell. 2005;122:633–644. doi: 10.1016/j.cell.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 79.Jafar-Nejad H, Acar M, Nolo R, Lacin H, Pan H, Parkhurst SM, Bellen HJ. Senseless acts as a binary switch during sensory organ precursor selection. Genes Dev. 2003;17:2966–2978. doi: 10.1101/gad.1122403. [DOI] [PMC free article] [PubMed] [Google Scholar]