Summary
The co-repressor proteins SMRT and NCoR concentrate in specific subnuclear compartments and function with DNA-binding factors to inhibit transcription. To provide detailed mechanistic understanding of these activities, this study tested the hypothesis that functional interactions with transcription factors, such as the pituitary-gland-specific Pit-1 homeodomain protein, direct the subnuclear organization and activity of co-repressor complexes. Both SMRT and NCoR repressed Pit-1-dependent transcription, and NCoR was co-immunoprecipitated with Pit-1. Immunofluorescence experiments confirmed that endogenous NCoR is concentrated in small focal bodies and that incremental increases in fluorescent-protein-tagged NCoR expression lead to progressive increases in the size of these structures. In pituitary cells, the endogenous NCoR localized with endogenous Pit-1 and the co-expression of a fluorescent-protein-labeled Pit-1 redistributed both NCoR and SMRT into diffuse nucleoplasmic compartments that also contained histone deacetylase and chromatin. Automated image-analysis methods were applied to cell populations to characterize the reorganization of co-repressor proteins by Pit-1 and mutation analysis showed that Pit-1 DNA-binding activity was necessary for the reorganization of co-repressor proteins. These data support the hypothesis that spherical foci serve as co-repressor storage compartments, whereas Pit-1/co-repressor complexes interact with target genes in more widely dispersed subnuclear domains. The redistribution of co-repressor complexes by Pit-1 might represent an important mechanism by which transcription factors direct changes in cell-specific gene expression.
Keywords: Transcriptional regulation, Prolactin, Nuclear co-repressor, SMRT, Nuclear structure, Green fluorescent protein, Fluorescence microscopy, Pit-1
Introduction
Coactivator and co-repressor protein complexes control gene expression by modifying the acetylation states of histones and other transcriptional regulators (for reviews, see Orphanides and Reinberg, 2002; Rosenfeld and Glass, 2001). Through interactions with specific DNA elements, transcription factors recruit a balance of these opposing co-regulatory proteins to target gene promoter and enhancer regions, providing precise homeostatic control. These balanced interactions have been characterized for Pit-1, a homeodomain transcription factor that is required to express the gene encoding prolactin (PRL) in differentiated anterior pituitary cells (Scully et al., 2000; Xu et al., 1998; Zanger et al., 1999). Pit-1 recruits a coactivator complex containing histone acetyltransferase (HAT) proteins such as the CREB-binding protein (CBP/p300) to activate transcription (Xu et al., 1998; Zanger et al., 1999). Counterbalancing the function of CBP, Pit-1 also interacts with co-repressor proteins such as nuclear-receptor co-repressor (NCoR) (Scully et al., 2000; Xu et al., 1998). Both NCoR and the closely related silencing mediator of retinoid- and thyroid-hormone receptors (SMRT) protein recruit the histone deacetylase (HDAC) proteins, which modify chromatin structure and actively repress transcription (for reviews, see Jepsen and Rosenfeld, 2002; Rosenfeld and Glass, 2001). Similar complexes are used by many divergent classes of transcription factors (Bailey et al., 1999; Dhordain et al., 1997; Hong et al., 1997; Hu et al., 2001; Jimenez-Lara and Aranda, 1999; Kakizawa et al., 2001; Lavinsky et al., 1998; Lee et al., 2000), stressing the importance of this balanced recruitment mechanism for the regulation of gene expression.
Many observations indicate that transcription factors and co-regulatory proteins localize to particular subnuclear sites (Day et al., 1999; Downes et al., 2000; Enwright et al., 2003; Kim et al., 1996; Lamond and Earnshaw, 1998; Misteli, 2001b; Pombo et al., 1998; Schaufele et al., 2001; van Wijnen et al., 1993; Zeng et al., 1998). The recent characterization of many small spherical nuclear bodies including Cajal bodies (Gall et al., 1999), gems (Hebert and Matera, 2000) and promyelocytic leukemia (PML) bodies (Maul et al., 2000) illustrate the highly ordered intranuclear environment. In addition to biochemical and immunofluorescence methods, the spectral variants of the fluorescent proteins (FPs) have been used as genetically encoded markers to study these nuclear subcompartments in living cells (Patterson et al., 2001; van Roessel and Brand, 2002). The dynamic partitioning of these different subcompartments without intervening membranes provides evidence for self-organization of the proteins that form these structures (Misteli, 2001b). Moreover, the chromatin in the interphase nucleus is similarly organized into distinct domains, including chromosomal territories, interchromatin spaces and centromeric heterochromatin (Lamond and Earnshaw, 1998). Recent evidence indicates that genes are transcribed at specific intranuclear sites (Cook, 1999) and there are examples of genes that become spatially positioned in different subnuclear regions depending on their activation state (Andrulis et al., 1998; Belmont et al., 1999; Brown et al., 1999; Brown et al., 1997). This area of intense research is revealing how these remarkably organized and structurally complex microenvironments contribute to the regulation of gene expression.
Several studies have shown that SMRT and NCoR are concentrated with their HDAC partners in matrix-associated deacetylase (MAD) bodies (Dhordain et al., 1997; Downes et al., 2000; Li et al., 2000; Nagy et al., 1997; Ordentlich et al., 1999; Soderstrom et al., 1997; Wu et al., 2001). A pharmacological HDAC inhibitor disrupted these subnuclear bodies, suggesting that they are maintained by specific functional interactions (Downes et al., 2000). This concept is supported by the observation that the spherical co-repressor focal bodies are spatially distinct from several other subnuclear protein compartments (Ariumi et al., 2003; Downes et al., 2000; Wu et al., 2001). Using a quantitative imaging approach, we demonstrated that MAD-body formation by NCoR was directly related to expression level (Voss et al., 2004), indicating a highly regulated mechanism controlling focal-body formation or maintenance. This regulated enrichment of co-repressor complexes in specific subnuclear compartments might contribute to the control of gene expression by at least two different mechanisms: (1) compartments might concentrate active co-repressor complexes at sites of gene regulation; (2) compartments might assemble or store co-repressor complexes away from sites of active transcription.
In this study, we use a combination of biochemistry, immunofluorescence, live-cell microscopy and quantitative image analysis to investigate the mechanisms controlling the subnuclear organization and function of co-repressor complexes. The data presented here suggest that spherical intranuclear foci serve as co-repressor reservoirs that are in balance with the available interacting partner proteins. As Pit-1 interacts with the co-repressors to regulate transcription, it also disperses the co-repressor complexes into diffuse nucleoplasmic compartments containing high concentrations of chromatin. Importantly, this redistribution of the co-repressor requires a functional Pit-1 DNA-binding domain. These data support the hypothesis that interactions with limiting amounts of transcription factors control co-repressor positioning and function, and form an important mechanism by which transcription factors direct changes in cell-specific gene expression.
Materials and Methods
Plasmids, cDNAs and other constructs
The cDNAs encoding the human SMRT, mouse NCoR and GFP-NCoR and rat Pit-1 have been described previously (Day et al., 1990; Chen and Evans, 1995; Horlein et al., 1995; Voss et al., 2004). The Pit-1 W261C mutant was generated by site-directed mutagenesis (QuikChange, Stratagene, La Jolla, CA). These cDNAs and their derivatives, as well as the cDNA encoding human PML were inserted in frame to sequences encoding each of the indicated fluorescent proteins (FPs) as described previously (Schaufele et al., 2001). For the reporter gene experiments, the indicated regions of the 2.5 kb flanking sequence of the rat gene encoding PRL were linked to the firefly luciferase (luc) reporter gene as described (Day et al., 1998; Day and Maurer, 1989; Howard and Maurer, 1994). The −39 to +34 base rat prolactin (rPRL) minimal promoter was inserted into the promoterless pGL3 luc reporter vector (Promega, Madison WI) to generate the −39 PRL luc vector. Tandem copies of the 1P Pit-1 response element (RE) or the 3P Pit-1 RE (Day et al., 1998) were inserted upstream of −39 PRL luc to produce the 1P −39 PRL luc or 3P −39 PRL luc vectors. In addition, an enhancer-trap reporter vector was generated to evaluate the 1P RE activity in the context of a Rous sarcoma virus (RSV) promoter. The oligonucleotide containing two copies of the 1P RE was inserted upstream of the RSV promoter in the pGL3 luc vector to produce the 1P RSV luc vector. Construction of all expression and reporter vectors was confirmed by automated nucleotide sequencing.
Transfection of cell lines and reporter-gene assays
For transfection, mouse GHFT1-5 (Lew et al., 1993), mouse 3T3-L1 (ATCC CL-173), rat GH4ZR7 (Elsholtz et al., 1991) and human HeLa (ATCC CCL-2) cell lines were maintained as monolayer cultures in Dulbecco's modified Eagle's medium containing 10% fetal calf serum. The cells were harvested and transfected with the indicated plasmid DNA(s) by electroporation. Briefly, approximately 1×106 cells were resuspended in Ca2+/Mg2+-free Dulbecco's PBS and transferred into 0.2 cm gap electroporation cuvettes containing 10 μg of the indicated luc reporter gene and various concentrations of the indicated expression vector DNAs. The total amount of DNA was kept constant using the cytomegalovirus (CMV) green fluorescent protein (CMV-GFP) expression plasmid. This provided a control for the effects of both the co-transfected CMV promoter and the GFP The transfected cells were transferred to 35 mm dishes and maintained in culture. Extracts were prepared from the cells after 24 hours and Luc activity was determined as described by the manufacturer (Promega). Each experiment was performed a minimum of three times and Luc activity, corrected for total protein, was expressed as the mean±s.e.m. The statistical analysis [analysis of variance (ANOVA) followed by the Student-Newman-Keuls post-hoc test where warranted to compare multiple conditions] was performed using SPSS 11 software, with differences considered significant when P<0.05.
Immunoprecipitation and immunodetection
For immunoprecipitation, cells were transfected with expression vectors encoding combinations of hemagglutinin (HA)-tagged Pit-1, GFP-SMRT or GFP-NCoR, and lysed after 24 hours as previously described for the immunoprecipitation of co-repressor complexes (Downes et al., 2000). The lysates were incubated with agarose beads conjugated with anti-HA antibody (Santa Cruz Biotechnology). The beads were then washed and proteins were eluted with denaturing loading buffer (Invitrogen Life Technologies) and analysed by western blot. Immunodetection was performed using anti-GFP (Molecular Probes) or anti-Pit-1 [131 polyclonal serum (S. Rhodes, Indiana University-Purdue University, Indianapolis, IN)] primary antibody, horseradish-peroxidase-conjugated anti-rabbit secondary antibody (Pierce Biotechnology) and chemiluminescence reagent (Amersham Biosciences).
Live cell microscopy and immunocytochemistry
Cells transfected with the indicated plasmid DNA(s) encoding the FP fusion proteins were transferred to 35 mm dishes that contained a sterile 25 mm circular cover glass and maintained in culture as described above. On the following day, the cover glass with the monolayer of cells was transferred to a medium-filled chamber fitted to the microscope stage (Day et al., 2001). Where indicated, the living cells were stained with Hoechst 33342 (H33342; Molecular Probes) as described previously (Schaufele et al., 2001). Wide-field microscopy (WFM) was performed with an inverted Olympus IX-70 epifluorescence microscope equipped with a 60× water-immersion 1.2 NA objective and filters sets specific for each fluorophore (Chroma Technology). Grayscale images with no saturated pixels were obtained using a cooled digital interline camera (Orca-200, Hamamatsu). All images were collected at a similar gray-level intensity by controlling the excitation intensity with constant neutral density filtration and by varying the on-camera integration time (0.1-8.0 seconds). Indirect immunocytochemical (ICC) detection of endogenous NCoR and Pit-1 proteins in fixed cells was performed as reported previously (Voss et al., 2005). Reference images of standard fluorescent beads were acquired to monitor consistency of microscope performance for all quantitative imaging experiments. All image files were processed for presentation using ISee software (Inovision) and Canvas 8.0 software (Deneba)
Integrated image analysis of cell populations
Cell populations producing a fusion between yellow fluorescent protein (YFP) and NCoR (YFP-NCoR) or SMRT (YFP-SMRT), the unfused monomeric red fluorescent protein (mRFP), and the indicated blue fluorescent protein (BFP) fusion protein were co-transfected by electroporation as described above. On the following day, images of the living cells were acquired using an integrated imaging protocol for the unbiased selection of cells based on mRFP expression. The subcellular features defined by the other expressed fusion proteins in the selected cells were measured using automated quantitative image analysis as described earlier (Voss et al., 2004). The area and fluorescence intensity of each automatically selected focal-body region of interest (ROI) was measured and the center of the ROI was used to position a second rectangular ROI that measured the fluorescence intensity of the nucleoplasm surrounding the selected feature. An enrichment factor (EF) was then calculated as the ratio of the intensities in the two regions. All the measurements were exported to text files and linear regression analysis was performed using spreadsheet software (Microsoft Excel) to determine the relationship between the labeled protein expression levels and co-repressor subnuclear organization. SPSS 11 software was used to perform ANOVA and post-hoc tests for the statistical comparison of imaging data from multiple cell populations.
Results
NCoR is organized in subnuclear compartments
Earlier studies showed that co-repressor complexes were localized to distinct subnuclear focal bodies called MAD bodies (Dhordain et al., 1997; Downes et al., 2000; Li et al., 2000; Nagy et al., 1997; Ordentlich et al., 1999; Soderstrom et al., 1997; Wu et al., 2001). Here, ICC staining was used to determine the subnuclear distribution of the endogenous NCoR in fixed 3T3-L1 cells, which were counterstained with the DNA-binding dye H33342 to label tracts of heterochromatin preferentially in the mouse cells (Amirand et al., 1998; Miller et al., 1974; Schaufele et al., 2001). The specificity of staining was confirmed by analysis of cells stained with the secondary antibody alone, which exhibited no nuclear staining at the exposure times used here. The results showed that NCoR was distributed in a reticular pattern throughout the nucleus that contained small focal bodies (Fig. 1A). The ICC images were analysed using automated quantitative image analysis (Voss et al., 2004) to identify focal bodies formed by the endogenous protein (Fig. 1A, circles). The H33342 counterstaining showed that these focal bodies were in regions of the nucleus containing low levels of stained chromatin (Fig. 1A, profile). A similar pattern was observed for endogenous SMRT in the nuclei of HeLa adenocarcinoma cells (Wu et al., 2001) and in GHFT1-5 pre-somatolactotrope cells (data not shown), indicating the formation of higher-order subnuclear structures is a property shared by the endogenous co-repressor proteins.
Fig. 1.
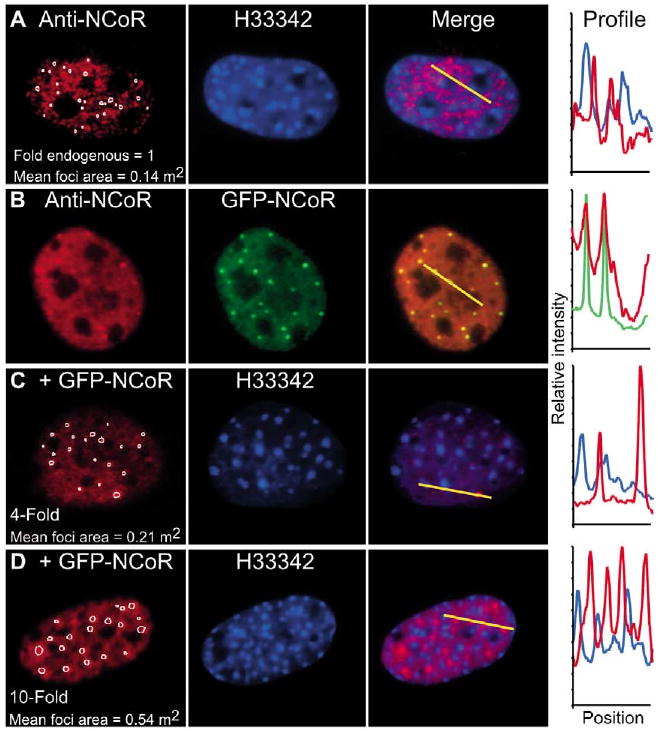
NCoR is localized to distinct subnuclear compartments. 3T3-L1 cells were fixed and subjected to ICC for detection of endogenous NCoR in (A) mock-transfected cells or (B-D) total NCoR in cells transfected with GFP-NCoR. Cells were stained with the H33342 chromatin dye. The profile plots (right) quantify the relative intensity of the fluorophores at the position along the yellow line in each overlay image. (B) The direct fluorescence signal from GFP-NCoR colocalized with anti-NCoR/Texas-Red immunofluorescence signal. (C,D) Representative images are labeled with the relative NCoR expression level based on the mean secondary-antibody/Texas-Red fluorescence intensity per nucleus. The computerized image-analysis algorithm automatically selected the foci (outlined in white) in each Texas-Red image. The mean area of the automatically selected foci is shown for each cell.
We have previously demonstrated that the formation of MAD bodies by exogenous NCoR in transfected cells is directly related to protein expression levels (Voss et al., 2004), consistent with the results of others (Soderstrom et al., 1997). Here, we demonstrate that the focal bodies formed by the endogenous NCoR and those formed by exogenous GFP-NCoR are the same bodies (Fig. 1). In parallel with the untransfected cells, ICC was performed on 3T3-L1 cells transfected with the plasmid encoding GFP-NCoR. The expressed protein was identified by green fluorescence, whereas total NCoR protein was measured using the Texas-Red-labeled antibody signal, allowing direct comparison of the relative NCoR expression levels (Fig. 1B). An automated image analysis algorithm was then used to identify and measure the characteristics of the focal bodies in the control and GFP-NCoR-expressing cells. As little as a fourfold increase in the total immunostaining signal within the nucleus resulted in a measurable increase in focal-body size (Fig. 1C). Further increase in the level of exogenous GFP-NCoR resulted in enlargement of the focal bodies (Fig. 1D). The profile plot analysis showed the same subnuclear positioning of GFP-NCoR relative to stained chromatin that was observed for the endogenous protein (Fig. 1A,C,D). Similar results were obtained using ICC analysis of endogenous SMRT and GFP-SMRT (data not shown). In summary, GFP-NCoR was localized with the endogenous protein and the size of the NCoR focal bodies was very sensitive to changes in relative protein expression level.
Co-repressor compartments recruit specific protein partners
To demonstrate that the MAD bodies formed by GFP-NCoR were different from other small nuclear compartments formed by proteins unrelated to the co-repressors, GFP-NCoR was co-expressed with PML fused to RFP (Day et al., 2001). PML is a component of a well-defined nuclear substructure called PML bodies (Zhong et al., 2000). The mouse pituitary GHFT1-5 cells co-transfected with plasmids encoding the FP-labeled NCoR and PML were counterstained with the chromatin dye H33342. Merger of the images revealed that the focal bodies formed by NCoR and the PML bodies localized to subnuclear regions separated by tracts of highly stained heterochromatin (Fig. 2A and overlay). The labeled PML bodies were frequently observed to be close to MAD bodies formed by NCoR, but these two nuclear proteins clearly occupied distinct compartments (Fig. 2A, overlay). These results support the view that the small nuclear bodies formed by NCoR are distinct from other subnuclear protein domains (Wu et al., 2001).
Fig. 2.
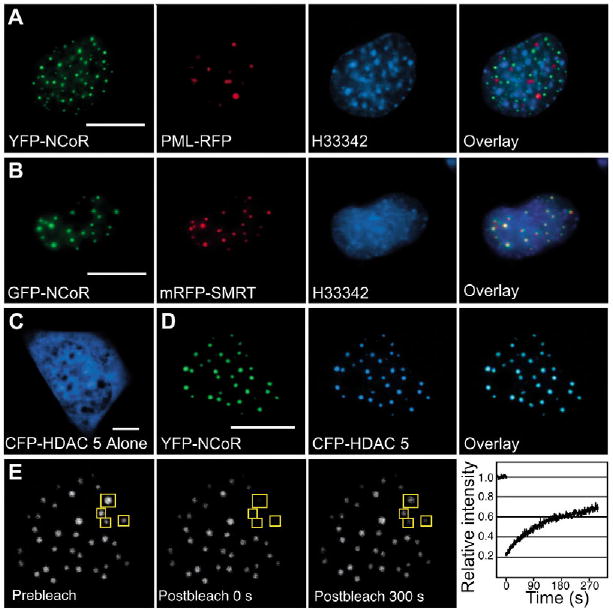
Co-repressors form specific intranuclear focal bodies. WFM images show living GHFT1-5 cells expressing: (A) both YFP-NCoR and RFP-PML, and stained briefly with the cell-permeant chromatin stain H33342; (B) both GFP-NCoR and mRFP-SMRT, and stained briefly with H33342; (C) CFP-HDAC-5 alone; or (D) YFP-NCoR and CFP-HDAC5 together. Each fluorescence channel is displayed separately and together in the overlay panel. Notice the different scale. Scale bars, 10 μm. (E) Living GHFT1-5 cells producing GFP-SMRT were subjected to FRAP analysis. The images show the nucleus of a cell taken at the same focal plane before selective photobleaching and at the indicated time points after. Fluorescence intensity in four foci was measured as indicated by the white square ROIs. The recovery plot shows the mean change in relative fluorescence intensity over a 300 second time frame, normalized to the prebleaching level for each ROI. Error bars denote s.e.m.
By contrast, images acquired of cells that co-expressed mRFP-SMRT and GFP-NCoR showed that these related co-repressor proteins occupied the same focal bodies (Fig. 2B and overlay). Earlier studies demonstrated that the MAD bodies formed by NCoR and SMRT also contain their HDAC partners, which function as part of the co-repressor complex to modify chromatin and repress transcription (Dhordain et al., 1997; Downes et al., 2000; Li et al., 2000; Nagy et al., 1997; Ordentlich et al., 1999; Soderstrom et al., 1997; Wu et al., 2001). We then examined the subcellular localization of HDAC5 to determine whether it was co-localized with NCoR. Some HDACs, including HDAC5, are known to shuttle between the cytoplasm and nucleus (McKinsey et al., 2000), and, when expressed in living GHFT1-5 cells, CFP-HDAC5 was distributed throughout the cell (Fig. 2C). However, when co-expressed with YFP-NCoR, there was redistribution of HDAC5 to the intranuclear foci occupied by NCoR (Fig. 2D and overlay). Equivalent results were obtained when CFP-HDAC5 was co-expressed with YFP-SMRT, but there was no association of the CFP-HDAC5 with the focal bodies formed by the co-expressed PML-RFP (data not shown). These living-cell observations are consistent with previous ICC studies showing that MAD bodies contain other members of the co-repressor complex and suggest a mechanism whereby specific protein interactions organize the co-repressor protein complex within specialized intranuclear compartments. To explore the dynamics of co-repressor foci, fluorescence recovery after photobleaching (FRAP) experiments were performed on GHFT1-5 cells producing YFP-SMRT. Following the photobleaching of YFP-SMRT in selected foci, the fluorescence intensity recovered to 70% in approximately 5 minutes (Fig. 2E,right). These results clearly demonstrate the dynamic exchange of co-repressor protein between the foci and nucleoplasm.
Co-repressor proteins functionally interact with Pit-1 to repress transcription
We next investigated the inhibition of Pit-1-dependent gene transcription by the co-repressors. Pit-1 functions during development to establish the pituitary somatolactotrope cell lineage, and is required for the transcription of both the growth hormone (GH)-encoding and PRL-encoding genes in these cells (Elsholtz et al., 1991). The pituitary somatolactotrope cell line GH4ZR7 expresses high levels of endogenous Pit-1 protein, which strongly activated the transfected rat PRL-promoter-fused luc reporter gene (−2.5 rPRL luc; Fig. 3A). Co-transfection with increasing amounts of an expression vector encoding SMRT resulted in a dose-dependent decrease in Luc activity, with greater than 80% inhibition of −2.5 rPRL Luc activity at the maximum amount of SMRT expression vector tested. Importantly, similar activity was observed for increasing amounts of expression vector encoding GFP-SMRT, demonstrating that the GFP fusion did not interfere with the PRL gene inhibitory activity of SMRT.
Fig. 3.
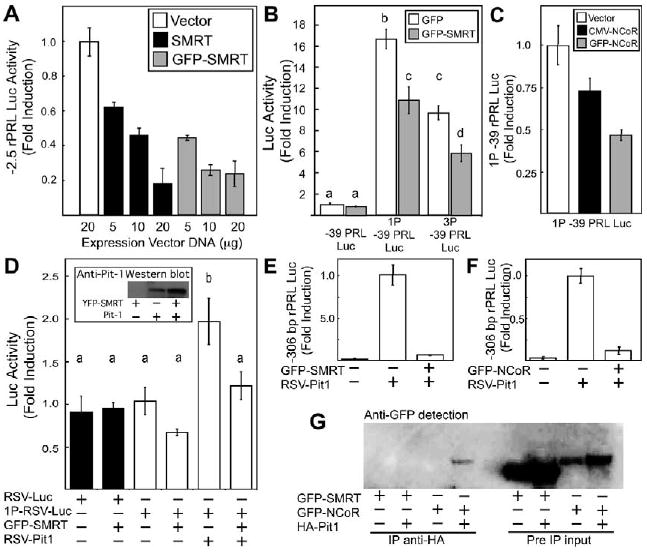
Co-repressors regulate Pit-1-dependent transcription. Luciferase reporter-gene transcription assays were performed using (A-C) GH4ZR7 pituitary cells or (D-F) non-pituitary HeLa cells. Cells were transfected with the indicated reporter vectors and expression vectors. The panels show the relative luciferase activity, corrected for total cellular protein in the lysates. Results are the means of measurements from at least three cultures and error bars denote the s.e.m. (B,D) Statistical analysis was performed with P<0.05 considered to be significant. The lower-case letters indicate sets of measurements that are statistically different. (D, insert) Western-blot analysis of HeLa cells transfected with the indicated expression vectors. (G) Lysates were prepared from cells transfected with the indicated expression vectors.
Immunoprecipitations were performed using an anti-HA agarose conjugate and anti-GFP antibody was used for western blot analysis.
The activity of the PRL promoter is dependent upon the binding of Pit-1 to multiple DNA REs (Ingraham et al., 1988). We evaluated the effect of co-repressor expression on transcription mediated by tandem copies of two different Pit-1 REs, each linked to the minimal −39 to +34 rPRL promoter linked to luc. The 1P or 3P −39 PRL luc constructs conferred a 16- and a 10-fold increase in reporter gene activity, respectively, when expressed in the pituitary GH4ZR7 cells (Fig. 3B). Where the expression of GFP alone or GFP-SMRT did not inhibit the basal activity of the minimal promoter, the activity of the 1P and 3P −39 PRL luc was significantly repressed by expression of GFP-SMRT compared with the control containing an equal amount of GFP expression vector (Fig. 3B). Similar to the results obtained with SMRT, we also observed that expression of full-length NCoR or GFP-NCoR repressed the activity of the 1P RE in GH4ZR7 somatolactotrope cells (Fig. 3C).
To determine whether the co-repressor activity observed in pituitary somatolactotrope cells was dependent upon Pit-1, PRL promoter activity was reconstituted in a heterologous cell line. Non-pituitary HeLa cells do not express crucial regulators of the PRL-encoding gene, including Pit-1, allowing dissection of Pit-1-dependent PRL promoter activation (Bradford et al., 1997; Ingraham et al., 1988; Nelsen et al., 1993; Walter et al., 1985). The expression of GFP-SMRT did not inhibit the RSV promoter in the HeLa cells (Fig. 3D), indicating that SMRT was not a global inhibitor of promoter activity in these cells. Linking the 1P Pit-1 RE to the RSV promoter (1P RSV luc) conferred responsiveness to Pit-1 when co-expressed in the HeLa cells (Fig. 3D). Western blotting showed that the expression of SMRT did not inhibit the expression of the RSV-Pit-1 plasmid (Fig. 3D, inset). Although the magnitude of the Pit-1-response was much less than that observed in GH4ZR7 cells (Fig. 3B), which probably reflects the high basal activity of the RSV promoter, the co-expression of GFP-SMRT led to a significant inhibition of the Pit-1-dependent reporter-gene activity (Fig. 3D). The rat PRL-encoding gene proximal promoter (−306 rPRL) contains four Pit-1 binding sites (Ingraham et al., 1988) and has very low basal activity in HeLa cells, but could be induced more than 600-fold by co-transfection with the RSV Pit-1 expression plasmid (Fig. 3E). The co-expression of GFP-SMRT with Pit-1 resulted in more than 90% inhibition of the Pit-1-dependent promoter activity, and a similar level of inhibition occurred with the co-expression of GFP-NCoR (Fig. 3F). Together, these reporter gene experiments indicated that both SMRT and NCoR inhibited Pit-1-dependent gene expression.
These results implied a functional interaction between Pit-1 and the co-repressor protein complex, and earlier studies indicated a direct physical interaction between NCoR and Pit-1 (Xu et al., 1998). We used co-immunoprecipitation (co-IP) to assess potential direct interactions between the FP-labeled co-repressor proteins and Pit-1 (Fig. 3G). Lysates were prepared from cells that co-expressed HA-Pit-1 and either GFP-NCoR or GFP-SMRT. The HA-Pit-1 expression in the cell lysates was confirmed by western blot analysis (data not shown). Following IP with an antibody specific for the HA epitope tag, western blotting with a GFP-specific antibody was used to detect co-IPed GFP fusion proteins. The results showed that GFP-NCoR, but not GFP-SMRT, was co-IPed with HA-Pit-1, despite similar levels of both proteins in the input lysate (Fig. 3G). This indicated stable interactions between GFP-NCoR and Pit-1, consistent with that previously reported for untagged NCoR (Xu et al., 1998). However, under the same conditions, an interaction with SMRT could not be detected. Given the equivalent activity of SMRT and NCoR in vivo, this was an unexpected result and might indicate a weaker physical association between SMRT and Pit-1 under the conditions used for the co-IP assay.
NCoR and Pit-1 co-localize in mouse pituitary somatotrope progenitor cells
We next examined whether NCoR co-localized with Pit-1 in mouse pituitary cells, which endogenously express both proteins. The mouse pituitary GHFT1-5 cell line was derived by targeted transformation of embryonic pituitary cells and has characteristics of the progenitor for the GH-secreting pituitary somatotrope cell lineage (Lew et al., 1993). The GHFT1-5 cells express Pit-1 at a much lower level than differentiated PRL and GH-secreting cell lines, including GH4ZR7 cells (Schaufele et al., 2001). ICC staining was used to compare the subnuclear distribution of the endogenous NCoR and Pit-1 in fixed GHFT1-5 cells, and the results demonstrated that the endogenous NCoR and Pit-1 were both distributed in a nuclear reticular pattern that contained very small focal structures (Fig. 4). The profile analysis shows a very high degree of co-localization of the immunostained proteins (Fig. 4). The specificity of staining was confirmed by analysis of cells stained with both secondary antibodies alone, each individual primary antibody with the opposite secondary antibody, or each primary with both secondary antibodies.
Fig. 4.
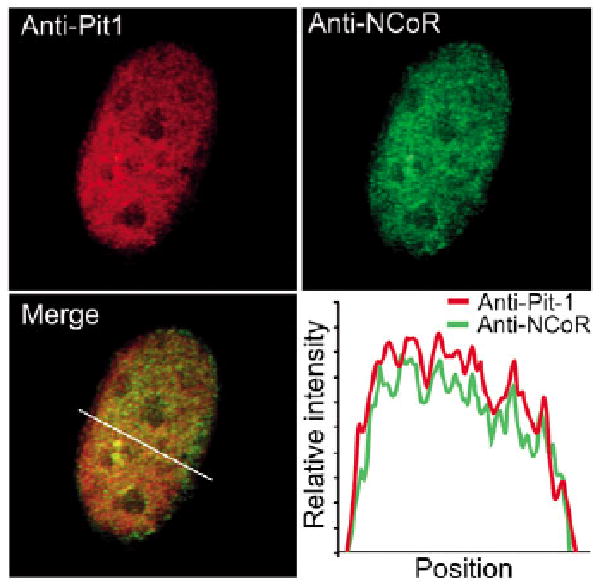
NCoR and Pit-1 are co-localized within the nucleus of mouse pituitary GHFT1-5 cells. Immunocytochemical staining was used to compare the subnuclear distribution of the endogenous NCoR and Pit-1 in fixed GHFT1-5 cells. GHFT1-5 cells probed with anti-Pit1 antibody followed by Texas-Red-conjugated anti-rabbit antibody, and anti-NCoR antibody followed by FITC-conjugated anti-goat antibody. The profile analysis shows the degree of co-localization of the immunostained proteins. The specificity of staining was confirmed by analysis of cells stained with both secondary antibodies alone, each individual primary antibody with the opposite secondary antibody, or each primary with both secondary antibodies.
Recruitment of GFP-NCoR by co-expressed BFP-Pit-1
We next determined whether the association of Pit-1 and NCoR could be detected in living cells. In mouse pituitary GHFT1-5 cells, GFP-Pit-1 was dispersed throughout the cell nucleus in a reticular pattern (Fig. 5A). The GFP-Pit-1 co-localized with regions of H33342-stained chromatin (Fig. 5A, overlay and profile), and we demonstrated earlier that this pattern is qualitatively indistinguishable from that of the endogenous Pit-1 (Enwright et al., 2003). When NCoR and Pit-1 were co-expressed as fusions to different FP color variants, there was reorganization of the GFP-NCoR to a distribution that co-localized with that of Pit-1 (Fig. 5B). The recruitment of GFP-NCoR by the co-expressed Pit-1 appeared to be specific because PML-RFP that was expressed in the same cell was not influenced by BFP-Pit-1 and remained localized to nuclear bodies. Importantly, the reorganized GFP-NCoR overlapped precisely with the web-like distribution of BFP-Pit-1 (Fig. 5B, profile), which could provide access of the co-repressor to the Pit-1-dependent genes.
Fig. 5.
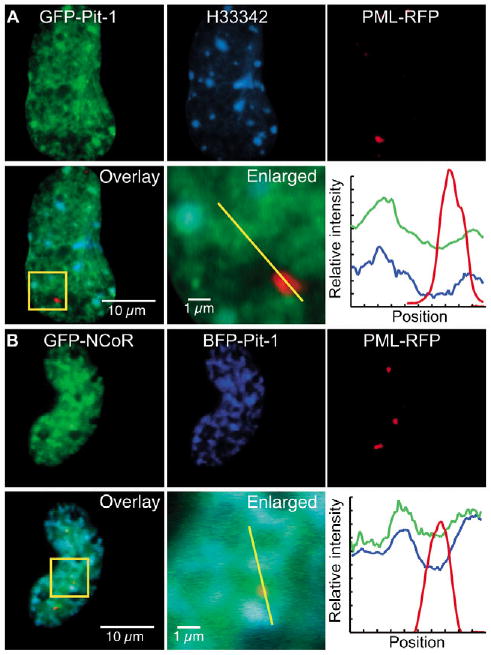
Pit-1 redistributes NCoR in the nucleus. (A) GHFT1-5 cells were transfected with GFP-Pit-1 and PML-RFP, and the living cells were stained briefly with H33342 DNA dye immediately before imaging. (B) GHFT1-5 cells were co-transfected with GFP-NCoR, BFP-Pit-1 and PML-RFP, and the living cells were imaged using WFM. The square ROI in the overlay image is enlarged to highlight the subnuclear protein organization. Cyan in the overlay images indicates colocalization of the green and blue channels. The profile plots display the relative intensity of the fluorophores at the positions along the yellow line in the overlay images. The labeled bars indicate scale.
Quantitative cell-population analysis confirms that Pit-1 reorganizes NCoR
The subnuclear organization of FP-NCoR in transfected cells can be heterogeneous (Voss et al., 2004) and the ability of Pit-1 to reorganize the co-expressed FP-NCoR was also variable. This is exemplified by the images of YFP-NCoR in randomly selected cells that co-express BFP-Pit-1 (Fig. 6A). Although YFP-NCoR was clearly dispersed in some cells (Fig. 6A, top), other cells had NCoR in a dispersed pattern with some small foci (Fig. 6A, middle) or in larger more distinct foci (Fig. 6A, bottom). The BFP-Pit-1 and YFP-NCoR were strongly co-localized to the dispersed compartments in these cells (Fig. 6A, profile). This heterogeneity in the subnuclear organization of NCoR creates a problem for image analysis, in which the interpretation of protein distribution in representative high-resolution microscopy images is subjective and might not accurately reflect the cell population.
Fig. 6.
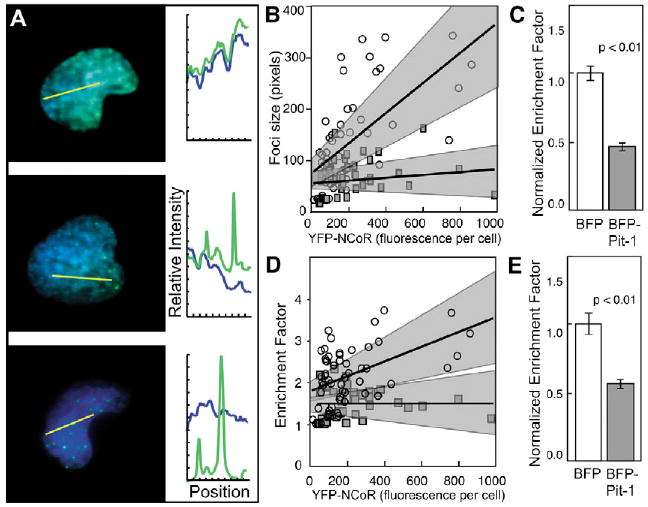
Pit-1 disperses NCoR foci in cell populations. Living cells were selected for WFM imaging using the mRFP signal, and subcellular ROIs were quantified using an automated computer algorithm to select ROI. The area and fluorescence intensity of each selected ROI were automatically measured, and the ratio of the surrounding region defines the enrichment factor (EF). (A) Overlay images of the YFP-NCoR and BFP-Pit-1 in three cells. The profile plots show the relative intensities of the fluorophores along the yellow line in each image. (B,D) Numerical results for 61 cells in the control population (open circles) and 48 cells in the experimental population (gray squares). Each point represents the mean data from a single cell. The relationships between YFP-NCoR expression level and (B) co-repressor focus size or (D) EF are shown. The black best-fit lines and 95% confidence intervals (surrounding gray regions) were calculated by linear regression for each population. The mean values of (C) focus size and (E) EF are shown normalized for expression level in each cell population.
To overcome this problem, we developed an integrated imaging protocol that uses the unbiased selection of transiently transfected cells based on the diffuse fluorescence from co-expressed mRFP, coupled with automated quantitative image analysis of the selected cells (Voss et al., 2004). Using this integrated approach, we examined the effect of Pit-1 on the subnuclear organization of NCoR by rigorous analysis of large populations of living cells. As a control, cells that co-expressed YFP-NCoR and unfused BFP were also analysed. The sampled population of control cells (61 cells) expressed YFP-NCoR over a 50-fold range and, based on the comparison with the immunofluorescence studies (Fig. 1), we estimate that the mean YFP-NCoR expression level was approximately 20 times that of the endogenous protein in 3T3-L1 cells. Graphical analysis of the population data showed that the cells with higher levels of YFP-NCoR expression organized the protein in larger focal bodies, and the best-fit line and narrow 95% confidence intervals illustrate this strong correlation (Fig. 6B). In striking contrast to this, the population of cells that co-expressed BFP-Pit-1 (48 cells) expressed YFP-NCoR over the same concentration range but had much reduced NCoR focal-body size, as indicated by the best-fit line with reduced slope and confidence intervals that do not overlap with those of the control population (Fig. 6B). This reduction in the mean NCoR focal-body size in the cells that co-expressed BFP-Pit-1 was verified by further statistical analysis of the data from these cell populations (Fig. 6C). The relative concentration of YFP-NCoR in the foci compared with the surrounding nucleoplasm was estimated by calculating the enrichment factor (EF), which is the ratio of the YFP-NCoR intensities in the two regions. The graphical analysis indicated that the EF of the NCoR foci was also reduced in the cell population that co-expressed BFP-Pit-1, indicating a more dispersed subnuclear organization (Fig. 6D). This was confirmed by analysis of the mean EF per cell within the two cell populations (Fig. 6E). This rigorous quantitative analysis of cell populations clearly demonstrated that, when they are co-expressed, BFP-Pit-1 reorganizes NCoR from focal bodies to a more widely distributed organization within the living cell nucleus.
Pit-1 also disperses the SMRT/HDAC co-repressor complex to a chromatin-enriched subnuclear pattern
NCoR and SMRT occupied the same focal bodies when co-expressed (Fig. 2B) and reporter-gene analysis indicated that both SMRT and NCoR inhibited Pit-1-dependent gene expression (Fig. 3E,F). However, biochemical analysis failed to detect an interaction between Pit-1 and SMRT (Fig. 3G). Because this could reflect a basic difference in Pit-1 interactions with the SMRT co-repressor complex, we next determined whether Pit-1 also reorganized SMRT in the nuclei of living cells. When they were co-expressed, BFP-Pit-1 was reorganized GFP-SMRT but not the PML bodies into a more dispersed distribution in which SMRT was localized with Pit-1 (Fig. 7A,B). The results in Fig. 7C further show that, when RFP-SMRT was recruited by GFP-Pit-1, both proteins were co-localized with the H33342-stained chromatin (Fig. 7C and profile). In addition, when Pit-1 reorganized SMRT, there was concomitant reorganization of the co-expressed HDAC-5 (Fig. 7D). This repositioning of multiple members of the co-repressor complex provides further support for the hypothesis that functional interactions with transcription factors direct the subnuclear organization and activity of co-repressor complexes.
Fig. 7.
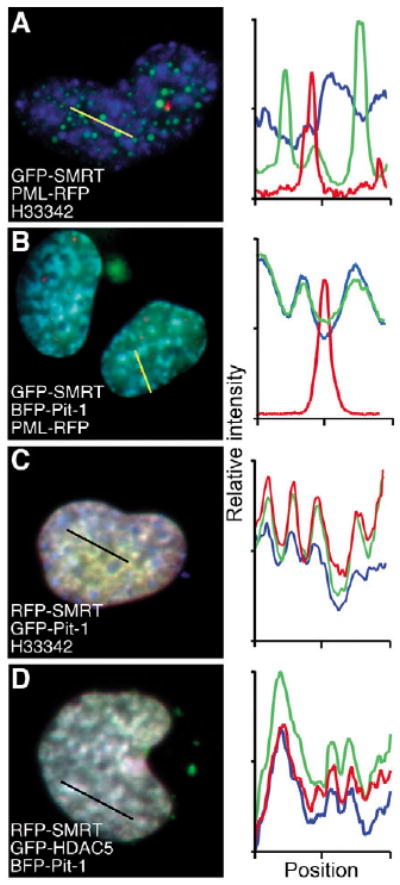
Pit-1 redistributes SMRT and HDAC to chromatin-containing compartments. WFM images of living GHFT1-5 cells were captured 24 hours after transfection with vectors encoding the indicated fusion proteins. The profile plots display the relative intensity of the fluorophores at the positions along the yellow line in the overlay images.
Pit-1 DNA-binding domain is crucial for subnuclear positioning of co-repressor complexes
We have previously showed that the Pit-1 DNA-binding activity is essential for its intranuclear targeting (Enwright et al., 2003). We next investigated the effect of a point mutation in the Pit-1 homeodomain that disrupts DNA binding on the ability of Pit-1 to reorganize the subnuclear distribution of the co-repressor. A naturally occurring homeodomain point mutation, changing residue 261 from tryptophan to cysteine (Pit-1W261C) disrupts binding to Pit-1 REs, preventing the activation of pituitary target genes and leading to the phenotype of the Snell Dwarf mouse strain (Li et al., 1990). When expressed alone, the BFP-Pit-1W261C was diffusely localized throughout the nucleus (data not shown) and, when co-expressed with YFP-SMRT, there was no reorganization of the SMRT foci. Instead, the BFP-Pit-1W261C became concentrated within the MAD bodies formed by SMRT (Fig. 8A and profile). This activity was not related to the FPs themselves because the co-expressed unfused BFP protein did not associate in the SMRT foci (Fig. 8B and profile). These qualitative imaging results indicate that the Pit-1 DNA-binding activity is necessary for its ability to reorganize the co-repressor protein in the cell nucleus. These results also suggest that the mutant Pit-1 protein was still capable of an association with the co-repressor.
Fig. 8.
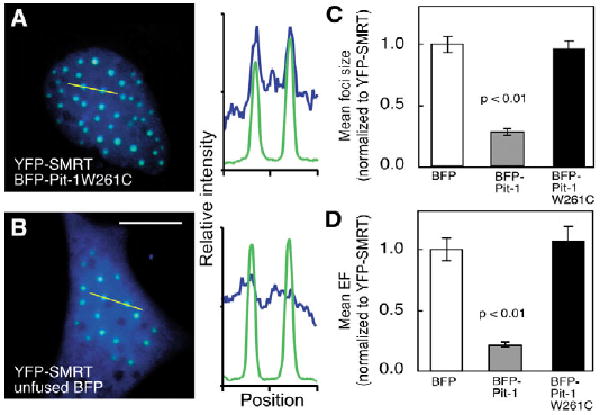
The Pit-1 DNA-binding domain is required to disperse SMRT. GHFT1-5 cells were co-transfected with expression vectors encoding YFP-SMRT and the indicated BFP fusion protein. (A,B) The profile plots display the relative intensity of the fluorophores at the positions along the yellow line in the overlay images. (C,D) Cells were selected using co-transfected mRFP and subcellular features were quantified using integrated image analysis. The focus size and enrichment factor were normalized for the relative level of YFP-SMRT expression in each cell. The data are shown as the means of each cell population with error bars denoting s.e.m.
To confirm and extend this observation, we used quantitative imaging to evaluate YFP-SMRT subnuclear organization in cell populations that co-expressed BFP-Pit-1W261C (79 cells) using the integrated image-analysis method and statistically compared these results with populations that expressed unfused BFP. These cell population data were normalized for the YFP-SMRT expression in each cell. The results (Fig. 8C,D) demonstrate that BFP-Pit-1 induced a statistically significant fourfold decrease in both focus size and EF compared with a similar population of cells that co-expressed the unfused BFP protein. In stark contrast to this, there was no significant change in either the YFP-SMRT focus size or EF in the population of cells that co-expressed BFP-Pit-1W261C (Fig. 8C,D). This rigorous analysis confirmed that DNA-binding activity was required for Pit-1 to redistribute co-repressor complexes within the nucleus. Taken together, these data suggest that chromatin interactions position the co-localized Pit-1/co-repressor complexes in widely dispersed intranuclear compartments.
Discussion
Many biochemical studies have established the related NCoR and SMRT co-repressor proteins as crucial regulators of transcription (Jepsen and Rosenfeld, 2002). However, the mechanisms that control co-repressor function in the complex environment of the intact cell nucleus remain unclear. Earlier studies demonstrated that the endogenous NCoR and SMRT were organized in at least two subnuclear compartments: the first, small focal bodies, are surrounded by the second, more diffuse and widely distributed nucleoplasmic co-repressor protein (Soderstrom et al., 1997; Wu et al., 2001). Here, we used immunocytochemistry to confirm this behavior of the endogenous NCoR protein and demonstrated that exogenous GFP-NCoR was co-localized with the endogenous NCoR (Fig. 1). This immunostaining also revealed that the size of the focal bodies was very sensitive to changes in relative protein expression levels. This indicated that the subnuclear distribution of co-repressor was highly regulated and suggested a mechanism whereby co-repressor protein exceeding the levels of binding partners in the nucleoplasmic compartment would be directed into focal bodies. If there was an equilibrium between these compartments, increasing the available binding partners in the nucleus should deplete the co-repressor in the focal bodies. Our study of co-repressor activity and subnuclear distribution, coupled with live-cell imaging of cell populations provides support for this mechanism, in which the balanced interactions between co-repressors and transcription factors tightly control the compartmentalization of the co-repressor.
Many proteins involved in transcriptional regulation and RNA-processing localize to specific compartments within the nucleus (for reviews, see Cook, 1999; Isogai and Tjian, 2003; Lamond and Spector, 2003; Spector, 2001). These self-organizing subnuclear compartments are maintained through protein-protein interactions, promoting coordinated multistep processes at specialized sites within the nucleus (Misteli, 2001a). For instance, extensive analysis has shown specific protein interactions are required for the formation of PML and Cajal bodies in distinct regions of the nucleus (Hebert and Matera, 2000; Maul et al., 2000). Although they are morphologically similar to these well-characterized intranuclear structures, the focal bodies formed by the co-repressor proteins were distinct from PML bodies, Daxx bodies and RNA splicing factor compartments (Wu et al., 2001). Furthermore, we have shown that the focal bodies formed by SMRT co-repressor and those formed by the coactivator glucocorticoid-receptor-interacting protein (GRIP1/TIF2) are also different (Voss et al., 2005). By contrast, the related co-repressor proteins SMRT and NCoR concentrate in the same intranuclear compartments (Fig. 2). Additionally, the Ataxin-1 protein, which functions as a transcriptional co-repressor and interacts physically with SMRT, was also found to co-localize with SMRT in the same subnuclear foci (Tsai et al., 2004). These co-repressor proteins function through their direct physical association with the HDACs, and earlier studies showed the recruitment of HDAC-1, HDAC-3 and HDAC-5 by SMRT and recruitment of HDAC-1 by NCoR (Downes et al., 2000; Soderstrom et al., 1997; Wu et al., 2001). We have demonstrated here that the co-producion of YFP-NCoR led to the recruitment of CFP-HDAC-5, but not the unfused CFP (Fig. 2), to the intranuclear focal bodies. These observations indicate that specific protein interactions target the co-repressors and their protein partners to specialized subnuclear compartments.
The proteins within these nuclear compartments are continuously exchanged with proteins in surrounding nuclear regions, providing a mechanism for dynamic regulation (Lamond and Spector, 2003; Misteli, 2001b). Using FRAP, we have demonstrated the exchange of FP-labeled co-repressor protein between focal bodies and the surrounding nucleoplasm (Fig. 2). Given that the size of the focal bodies is very sensitive to changes in co-repressor protein expression level, these results suggest that the relative concentrations of co-repressor complexes in different regions of the nucleus and the concentrations and distributions of other interacting proteins control the flux of co-repressor protein through this subnuclear compartment. In this regard, the co-repressors interact with many different classes of DNA-binding transcription factors (Bailey et al., 1999; Dhordain et al., 1997; Hong et al., 1997; Hu et al., 2001; Jimenez-Lara and Aranda, 1999; Kakizawa et al., 2001; Lavinsky et al., 1998; Lee et al., 2000) and these transcription factors compete for limiting quantities of co-repressor proteins (Shibata et al., 1997; Zhang et al., 1998). The activity of the pituitary-gland-specific transcription factor Pit-1 is regulated through its interactions with both coactivator and co-repressor proteins (Scully et al., 2000; Xu et al., 1998), and the precise balance of co-regulatory proteins is thought to play a key role in the regulation of gene expression (Glass and Rosenfeld, 2000; Sohn et al., 2003). In a previous study, we demonstrated that a GFP-Pit-1 fusion protein that had proper DNA-binding specificity was localized to distinct subnuclear compartments, where it interacted with other transcription factors to induce target gene expression (Day, 1998; Enwright et al., 2003).
Although best characterized for its role as an activator, Pit-1 also functions as an inhibitor of pituitary-hormone genes through an association with the nuclear co-repressor NCoR (Xu et al., 1998). We have showed here that tipping the balance in favor of the co-repressors resulted in the inhibition of Pit-1-dependent gene expression (Fig. 3). We confirmed that GFP-NCoR physically interacts with Pit-1 and functions to repress Pit-1-dependent transcriptional activity (Fig. 3). We found that SMRT also inhibited Pit-1-dependent transcription and retained this inhibitory activity when fused to GFP. These results are consistent with the overlapping activities of NCoR and SMRT that have been described for many DNA-binding factors (Gelmetti et al., 1998; Hu et al., 2001; Lavinsky et al., 1998; Yamamoto et al., 2001). Because both SMRT and NCoR are present in PRL-producing pituitary cells (Misiti et al., 1998), it seems likely that both function as important physiological regulators of Pit-1-dependent gene activity. Unlike NCoR, however, a strong interaction between SMRT and Pit-1 was not detected by co-IP experiments (Fig. 3). This could indicate different affinities of SMRT and NCoR for Pit-1 or could mean that in vitro binding conditions were not favorable for the interaction of SMRT with Pit-1. Together, the results support direct interactions of Pit-1 with the co-repressor protein, leading to the inhibition of Pit-1-dependent transcription.
We tested the hypothesis that Pit-1 could function to control co-repressor subnuclear positioning in living cells. We observed that, in mouse pituitary GHFT1-5 cells, endogenous NCoR and Pit-1 were co-localized within the nuclear compartment (Fig. 4). When GFP-NCoR was co-expressed with BFP-Pit-1, there was a striking reorganization of GFP-NCoR into the diffuse nuclear distribution of Pit-1 (Fig. 5). By contrast, the nuclear bodies formed by PML protein were unaffected by the co-produced Pit-1 protein. To quantify this, we used an integrated image analysis of populations of randomly selected cells from the transfected population (Voss et al., 2004; Voss et al., 2005). The co-expression of Pit-1 in the cell population resulted in statistically significant decreases in both YFP-SMRT focus size and the relative protein concentration in the foci compared with the surrounding nucleoplasm (Fig. 6). In cells in which the co-repressor concentration was high, focal bodies were still formed in the presence of Pit-1 but Pit-1 was never observed to localize to the co-repressor foci. This implies that most Pit-1/co-repressor interactions occur outside the spherical focal bodies. Interestingly, our linear regression models revealed that Pit-1 expression caused a graded dispersal of YFP-SMRT foci in the cell population instead of the ‘all or none’ dispersal implied by previous qualitative imaging studies of co-repressors and nuclear receptors (Tazawa et al., 2003; Wu et al., 2001). These are the first statistical data supporting the hypothesis that DNA-binding factors position co-repressor complexes in specific subnuclear domains, moving co-repressor protein out of spherical foci and into more widely distributed nuclear compartments.
The similar reorganization of HDAC-5 in cells that co-expressed SMRT and Pit-1 suggested that the co-repressor complex was reorganized by Pit-1 to the widely distributed compartments it occupied (Fig. 7). However, we could not distinguish a direct interaction between Pit-1 and the co-repressors from an indirect association of these proteins through common protein partners. Interestingly, several qualitative imaging studies have reported similar effects of DNA-binding nuclear receptors on co-repressor subnuclear organization. The non-ligand-bound retinoic-acid receptor α (RARα) physically interacts with SMRT and recruits SMRT into a diffuse nuclear pattern during repression of target gene promoters (Wu et al., 2001). In the presence of ligand, SMRT dissociates from the RARα, allowing coactivator complexes to bind the receptor and to stimulate target genes (Glass and Rosenfeld, 2000; Wu et al., 2001). Similar results were reported for receptor-interacting protein 140 (RIP140), another co-repressor protein that is redistributed from spherical foci to a diffuse pattern through physical interactions with the non-ligand-bound glucocorticoid receptor (GR). This dispersal of RIP140 also correlated with repression (Tazawa et al., 2003). In combination with these results, our imaging and functional studies indicate that transcriptional repression correlates with the dispersal of co-repressor complexes by DNA-binding factors in living cells.
Several lines of evidence suggest that the DNA-binding activity of Pit-1 is necessary for the recruitment of the co-repressor complexes. When expressed alone, Pit-1 distributed in a web-like pattern that was co-localized with H33342-stained euchromatin throughout the nucleus (Fig. 5A). In addition, the dispersed Pit-1/SMRT compartments are positioned in regions of H33342-stained euchromatin. Our earlier studies showed disruption of the Pit-1 DNA-binding activity prevented its co-localization with the chromatin stain (Enwright et al., 2003) (data not shown). This suggested that interactions with the chromatin control the position of Pit-1 in the nucleus. Here, we examined the naturally occurring point mutant Pit-1W261C, which is defective in DNA binding and failed to co-localize with the stained chromatin. Using quantitative image-analysis, we demonstrated that this Pit-1 point mutant failed to reorganize the focal bodies containing SMRT (Fig. 8). Instead, the DNA-binding-defective Pit-1 became co-localized with SMRT to the focal bodies. This indicated that, although the mutant Pit-1 retained its ability to interact with the co-repressor complex, DNA-binding activity was necessary to establish the final positioning of the Pit-1/co-repressor complexes in the cell nucleus. Similar results have been reported for qualitative imaging studies of the RIP140 co-repressor and a GR DNA-binding-domain mutant (Tazawa et al., 2003).
In summary, these observations provide striking evidence that DNA-binding factors alter the organization of their interacting co-repressor proteins and that this redistribution contributes to transcriptional regulation. The results indicated that dynamic protein interactions lead to the assembly of proteins in common subnuclear compartments and models that characterize these sites as aggregates of misfolded protein do not adequately account for this dynamic behavior. Our studies support the view that, in the presence of limiting amounts of DNA-binding transcription factors such as Pit-1, any excess of the co-repressor protein is organized in focal bodies. These foci are dynamic and exchange co-repressor complexes with the surrounding nucleoplasm. When Pit-1 is in excess, co-repressor protein is recruited to the nuclear sites occupied by Pit-1. The co-localization of these proteins was independent of the Pit-1 DNA-binding activity, but the dominant organizer activity of Pit-1 required the DNA-binding activity, suggesting either direct association with chromatin or through interactions with chromatin-associated factors. This dominant organizer activity overrides the co-repressor default targeting signal, resulting in dispersal of the co-repressor and HDAC to regions containing chromatin. We propose that active transcriptional regulation occurs in the widely distributed subnuclear compartments that contain high concentrations of Pit-1, SMRT/NCoR, HDAC and chromatin. Other nuclear proteins have subnuclear distributions that are similar to the patterns observed here for the co-repressor proteins, and may be similarly regulated.
Acknowledgments
We thank R. M. Evans (Salk Institute, San Diego, CA) for providing the SMRT-encoding cDNA, J. Torchia (University of Western Ontario) for providing the NCoR expression vector, S. Rhodes (Indiana University-Purdue University, Indianapolis, IN) for the Pit-1 antibody and F. Schaufele (University of California, San Francisco, CA) for the generation of the GFP-SMRT expression vector. We also thank A. Periasamy (W. M. Keck Center for Cellular Imaging, University of Virginia) for expert assistance. This study was supported by NIH grants RO1-DK-43701 (R.N.D.) and F32 DK 60315-01 (T.C.V.).
References
- Amirand C, Viari A, Ballini JP, Rezaei H, Beaujean N, Jullien D, Kas E, Debey P. Three distinct sub-nuclear populations of HMG-I protein of different properties revealed by co-localization image analysis. J Cell Sci. 1998;111:3551–3561. doi: 10.1242/jcs.111.23.3551. [DOI] [PubMed] [Google Scholar]
- Andrulis ED, Neiman AM, Zappulla DC, Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–595. doi: 10.1038/29100. [DOI] [PubMed] [Google Scholar]
- Ariumi Y, Ego T, Kaida A, Matsumoto M, Pandolfi PP, Shimotohno K. Distinct nuclear body components, PML and SMRT, regulate the trans-acting function of HTLV-1 Tax oncoprotein. Oncogene. 2003;22:1611–1619. doi: 10.1038/sj.onc.1206244. [DOI] [PubMed] [Google Scholar]
- Bailey P, Downes M, Lau P, Harris J, Chen SL, Hamamori Y, Sartorelli V, Muscat GE. The nuclear receptor corepressor N-CoR regulates differentiation: N-CoR directly interacts with MyoD. Mol Endocrinol. 1999;13:1155–1168. doi: 10.1210/mend.13.7.0305. [DOI] [PubMed] [Google Scholar]
- Belmont AS, Dietzel S, Nye AC, Strukov YG, Tumbar T. Large-scale chromatin structure and function. Curr Opin Cell Biol. 1999;11:307–311. doi: 10.1016/S0955-0674(99)80041-6. [DOI] [PubMed] [Google Scholar]
- Bradford AP, Wasylyk C, Wasylyk B, Gutierrez-Hartmann A. Interaction of Ets-1 and the POU-homeodomain protein GHF-1/Pit-1 reconstitutes pituitary-specific gene expression. Mol Cell Biol. 1997;17:1065–1074. doi: 10.1128/mcb.17.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- Brown KE, Baxter J, Graf D, Merkenschlager M, Fisher AG. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol Cell. 1999;3:207–217. doi: 10.1016/s1097-2765(00)80311-1. [DOI] [PubMed] [Google Scholar]
- Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- Cook PR. The organization of replication and transcription. Science. 1999;284:1790–1795. doi: 10.1126/science.284.5421.1790. [DOI] [PubMed] [Google Scholar]
- Day RN. Visualization of Pit-1 transcription factor interactions in the living cell nucleus by fluorescence resonance energy transfer microscopy. Mol Endocrinol. 1998;12:1410–1419. doi: 10.1210/mend.12.9.0168. [DOI] [PubMed] [Google Scholar]
- Day RN, Maurer RA. The distal enhancer region of the rat prolactin gene contains elements conferring response to multiple hormones. Mol Endocrinol. 1989;3:3–9. doi: 10.1210/mend-3-1-3. [DOI] [PubMed] [Google Scholar]
- Day RN, Koike S, Sakai M, Muramatsu M, Maurer RA. Both Pit-1 and the estrogen receptor are required for estrogen responsiveness of the rat prolactin gene. Mol Endocrinol. 1990;4:1964–1971. doi: 10.1210/mend-4-12-1964. [DOI] [PubMed] [Google Scholar]
- Day RN, Liu J, Sundmark V, Kawecki M, Berry D, Elsholtz HP. Selective inhibition of prolactin gene transcription by the ETS-2 repressor factor. J Biol Chem. 1998;273:31909–31915. doi: 10.1074/jbc.273.48.31909. [DOI] [PubMed] [Google Scholar]
- Day RN, Nordeen SK, Wan Y. Visualizing protein-protein interactions in the nucleus of the living cell. Mol Endocrinol. 1999;13:517–526. doi: 10.1210/mend.13.4.0259. [DOI] [PubMed] [Google Scholar]
- Day RN, Periasamy A, Schaufele F. Fluorescence resonance energy transfer microscopy of localized protein interactions in the living cell nucleus. Methods. 2001;25:4–18. doi: 10.1006/meth.2001.1211. [DOI] [PubMed] [Google Scholar]
- Dhordain P, Albagli O, Lin RJ, Ansieau S, Quief S, Leutz A, Kerckaert JP, Evans RM, Leprince D. Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc Natl Acad Sci USA. 1997;94:10762–10767. doi: 10.1073/pnas.94.20.10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes M, Ordentlich P, Kao HY, Alvarez JG, Evans RM. Identification of a nuclear domain with deacetylase activity. Proc Natl Acad Sci USA. 2000;97:10330–10335. doi: 10.1073/pnas.97.19.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsholtz HP, Lew AM, Albert PR, Sundmark VC. Inhibitory control of prolactin and Pit-1 gene promoters by dopamine. Dual signaling pathways required for D2 receptor-regulated expression of the prolactin gene. J Biol Chem. 1991;266:22919–22925. [PubMed] [Google Scholar]
- Enwright JF, 3rd, Kawecki-Crook MA, Voss TC, Schaufele F, Day RN. A PIT-1 homeodomain mutant blocks the intranuclear recruitment of the CCAAT/enhancer binding protein alpha required for prolactin gene transcription. Mol Endocrinol. 2003;17:209–222. doi: 10.1210/me.2001-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG, Bellini M, Wu Z, Murphy C. Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol Biol Cell. 1999;10:4385–4402. doi: 10.1091/mbc.10.12.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelmetti V, Zhang J, Fanelli M, Minucci S, Pelicci PG, Lazar MA. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol Cell Biol. 1998;18:7185–7191. doi: 10.1128/mcb.18.12.7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- Hebert MD, Matera AG. Self-association of coilin reveals a common theme in nuclear body localization. Mol Biol Cell. 2000;11:4159–4171. doi: 10.1091/mbc.11.12.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SH, David G, Wong CW, Dejean A, Privalsky ML. SMRT corepressor interacts with PLZF and with the PML-retinoic acid receptor alpha (RARalpha) and PLZF-RARalpha oncoproteins associated with acute promyelocytic leukemia. Proc Natl Acad Sci USA. 1997;94:9028–9033. doi: 10.1073/pnas.94.17.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass CK. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- Howard PW, Maurer RA. Thyrotropin releasing hormone stimulates transient phosphorylation of the tissue-specific transcription factor, Pit-1. J Biol Chem. 1994;269:28662–28669. [PubMed] [Google Scholar]
- Hu X, Li Y, Lazar MA. Determinants of CoRNR-dependent repression complex assembly on nuclear hormone receptors. Mol Cell Biol. 2001;21:1747–1758. doi: 10.1128/MCB.21.5.1747-1758.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingraham HA, Chen RP, Mangalam HJ, Elsholtz HP, Flynn SE, Lin CR, Simmons DM, Swanson L, Rosenfeld MG. A tissue-specific transcription factor containing a homeodomain specifies a pituitary phenotype. Cell. 1988;55:519–529. doi: 10.1016/0092-8674(88)90038-4. [DOI] [PubMed] [Google Scholar]
- Isogai Y, Tjian R. Targeting genes and transcription factors to segregated nuclear compartments. Curr Opin Cell Biol. 2003;15:296–303. doi: 10.1016/s0955-0674(03)00052-8. [DOI] [PubMed] [Google Scholar]
- Jepsen K, Rosenfeld MG. Biological roles and mechanistic actions of co-repressor complexes. J Cell Sci. 2002;115:689–698. doi: 10.1242/jcs.115.4.689. [DOI] [PubMed] [Google Scholar]
- Jimenez-Lara AM, Aranda A. The vitamin D receptor binds in a transcriptionally inactive form and without a defined polarity on a retinoic acid response element. FASEB J. 1999;13:1073–1081. doi: 10.1096/fasebj.13.9.1073. [DOI] [PubMed] [Google Scholar]
- Kakizawa T, Miyamoto T, Ichikawa K, Takeda T, Suzuki S, Mori J, Kumagai M, Yamashita K, Hashizume K. Silencing mediator for retinoid and thyroid hormone receptors interacts with octamer transcription factor-1 and acts as a transcriptional repressor. J Biol Chem. 2001;276:9720–9725. doi: 10.1074/jbc.M008531200. [DOI] [PubMed] [Google Scholar]
- Kim MK, Lesoon-Wood LA, Weintraub BD, Chung JH. A soluble transcription factor, Oct-1, is also found in the insoluble nuclear matrix and possesses silencing activity in its alanine-rich domain. Mol Cell Biol. 1996;16:4366–4377. doi: 10.1128/mcb.16.8.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- Lavinsky RM, Jepsen K, Heinzel T, Torchia J, Mullen TM, Schiff R, Del-Rio AL, Ricote M, Ngo S, Gemsch J, et al. Diverse signaling pathways modulate nuclear receptor recruitment of N-CoR and SMRT complexes. Proc Natl Acad Sci USA. 1998;95:2920–2925. doi: 10.1073/pnas.95.6.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Kim JH, Lee YC, Cheong J, Lee JW. Silencing mediator of retinoic acid and thyroid hormone receptors, as a novel transcriptional corepressor molecule of activating protein-1, nuclear factor-kappaB, and serum response factor. J Biol Chem. 2000;275:12470–12474. doi: 10.1074/jbc.275.17.12470. [DOI] [PubMed] [Google Scholar]
- Lew D, Brady H, Klausing K, Yaginuma K, Theill LE, Stauber C, Karin M, Mellon PL. GHF-1-promoter-targeted immortalization of a somatotropic progenitor cell results in dwarfism in transgenic mice. Genes Dev. 1993;7:683–693. doi: 10.1101/gad.7.4.683. [DOI] [PubMed] [Google Scholar]
- Li J, Wang J, Nawaz Z, Liu JM, Qin J, Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Crenshaw EBd, Rawson EJ, Simmons DM, Swanson LW, Rosenfeld MG. Dwarf locus mutants lacking three pituitary cell types result from mutations in the POU-domain gene Pit-1. Nature. 1990;347:528–533. doi: 10.1038/347528a0. [DOI] [PubMed] [Google Scholar]
- Maul GG, Negorev D, Bell P, Ishov AM. Properties and assembly mechanisms of ND10, PML bodies, or PODs. J Struct Biol. 2000;129:278–287. doi: 10.1006/jsbi.2000.4239. [DOI] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Lu J, Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller OJ, Schnedl W, Allen J, Erlanger BF. 5-Methylcytosine localised in mammalian constitutive heterochromatin. Nature. 1974;251:636–637. doi: 10.1038/251636a0. [DOI] [PubMed] [Google Scholar]
- Misiti S, Schomburg L, Yen PM, Chin WW. Expression and hormonal regulation of coactivator and corepressor genes. Endocrinology. 1998;139:2493–2500. doi: 10.1210/endo.139.5.5971. [DOI] [PubMed] [Google Scholar]
- Misteli T. The concept of self-organization in cellular architecture. J Cell Biol. 2001a;155:181–185. doi: 10.1083/jcb.200108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Protein dynamics: implications for nuclear architecture and gene expression. Science. 2001b;291:843–847. doi: 10.1126/science.291.5505.843. [DOI] [PubMed] [Google Scholar]
- Nagy L, Kao HY, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, Evans RM. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- Nelsen B, Tian G, Erman B, Gregoire J, Maki R, Graves B, Sen R. Regulation of lymphoid-specific immunoglobulin mu heavy chain gene enhancer by ETS-domain proteins. Science. 1993;261:82–86. doi: 10.1126/science.8316859. [DOI] [PubMed] [Google Scholar]
- Ordentlich P, Downes M, Xie W, Genin A, Spinner NB, Evans RM. Unique forms of human and mouse nuclear receptor corepressor SMRT. Proc Natl Acad Sci USA. 1999;96:2639–2644. doi: 10.1073/pnas.96.6.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- Patterson G, Day RN, Piston D. Fluorescent protein spectra. J Cell Sci. 2001;114:837–838. doi: 10.1242/jcs.114.5.837. [DOI] [PubMed] [Google Scholar]
- Pombo A, Cuello P, Schul W, Yoon JB, Roeder RG, Cook PR, Murphy S. Regional and temporal specialization in the nucleus: a transcriptionally-active nuclear domain rich in PTF, Oct1 and PIKA antigens associates with specific chromosomes early in the cell cycle. EMBO J. 1998;17:1768–1778. doi: 10.1093/emboj/17.6.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld MG, Glass CK. Coregulator codes of transcriptional regulation by nuclear receptors. J Biol Chem. 2001;276:36865–36868. doi: 10.1074/jbc.R100041200. [DOI] [PubMed] [Google Scholar]
- Schaufele F, Enwright JF, 3rd, Wang X, Teoh C, Srihari R, Erickson R, MacDougald OA, Day RN. CCAAT/enhancer binding protein alpha assembles essential cooperating factors in common subnuclear domains. Mol Endocrinol. 2001;15:1665–1676. doi: 10.1210/mend.15.10.0716. [DOI] [PubMed] [Google Scholar]
- Scully KM, Jacobson EM, Jepsen K, Lunyak V, Viadiu H, Carriere C, Rose DW, Hooshmand F, Aggarwal AK, Rosenfeld MG. Allosteric effects of Pit-1 DNA sites on long-term repression in cell type specification. Science. 2000;290:1127–1131. doi: 10.1126/science.290.5494.1127. [DOI] [PubMed] [Google Scholar]
- Shibata H, Nawaz Z, Tsai SY, O'Malley BW, Tsai MJ. Gene silencing by chicken ovalbumin upstream promoter-transcription factor I (COUP-TFI) is mediated by transcriptional corepressors, nuclear receptor-corepressor (N-CoR) and silencing mediator for retinoic acid receptor and thyroid hormone receptor (SMRT) Mol Endocrinol. 1997;11:714–724. doi: 10.1210/mend.11.6.0002. [DOI] [PubMed] [Google Scholar]
- Soderstrom M, Vo A, Heinzel T, Lavinsky RM, Yang WM, Seto E, Peterson DA, Rosenfeld MG, Glass CK. Differential effects of nuclear receptor corepressor (N-CoR) expression levels on retinoic acid receptor-mediated repression support the existence of dynamically regulated corepressor complexes. Mol Endocrinol. 1997;11:682–692. doi: 10.1210/mend.11.6.0018. [DOI] [PubMed] [Google Scholar]
- Sohn YC, Kim SW, Lee S, Kong YY, Na DS, Lee SK, Lee JW. Dynamic inhibition of nuclear receptor activation by corepressor binding. Mol Endocrinol. 2003;17:366–372. doi: 10.1210/me.2002-0150. [DOI] [PubMed] [Google Scholar]
- Spector DL. Nuclear domains. J Cell Sci. 2001;114:2891–2893. doi: 10.1242/jcs.114.16.2891. [DOI] [PubMed] [Google Scholar]
- Tazawa H, Osman W, Shoji Y, Treuter E, Gustafsson JA, Zilliacus J. Regulation of subnuclear localization is associated with a mechanism for nuclear receptor corepression by RIP140. Mol Cell Biol. 2003;23:4187–4198. doi: 10.1128/MCB.23.12.4187-4198.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CC, Kao HY, Mitzutani A, Banayo E, Rajan H, McKeown M, Evans RM. Ataxin 1, a SCA1 neurodegenerative disorder protein, is functionally linked to the silencing mediator of retinoid and thyroid hormone receptors. Proc Natl Acad Sci USA. 2004;101:4047–4052. doi: 10.1073/pnas.0400615101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Roessel P, Brand AH. Imaging into the future: visualizing gene expression and protein interactions with fluorescent proteins. Nat Cell Biol. 2002;4:E15–E20. doi: 10.1038/ncb0102-e15. [DOI] [PubMed] [Google Scholar]
- van Wijnen AJ, Bidwell JP, Fey EG, Penman S, Lian JB, Stein JL, Stein GS. Nuclear matrix association of multiple sequence-specific DNA binding activities related to SP-1, ATF, CCAAT, C/EBP, OCT-1, and AP-1. Biochemistry. 1993;32:8397–8402. doi: 10.1021/bi00084a003. [DOI] [PubMed] [Google Scholar]
- Voss TC, Demarco IA, Booker CF, Day RN. A computer-assisted image analysis protocol that quantitatively measures subnuclear protein organization in cell populations. BioTechniques. 2004;36:240–247. doi: 10.2144/04362BI01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss TC, Demarco IA, Booker CF, Day RN. Quantitative methods analyze subnuclear protein organization in cell populations with varying degrees of protein expression. J Biomed Opt. 2005;10:24011. doi: 10.1117/1.1891085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Green S, Greene G, Krust A, Bornert JM, Jeltsch JM, Staub A, Jensen E, Scrace G, Waterfield M, et al. Cloning of the human estrogen receptor cDNA. Proc Natl Acad Sci USA. 1985;82:7889–7893. doi: 10.1073/pnas.82.23.7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Li H, Park EJ, Chen JD. SMRTe inhibits MEF2c transcriptional activation by targeting HDAC4 and 5 to nuclear domains. J Biol Chem. 2001;276:24177–24185. doi: 10.1074/jbc.M100412200. [DOI] [PubMed] [Google Scholar]
- Xu L, Lavinsky RM, Dasen JS, Flynn SE, McInerney EM, Mullen TM, Heinzel T, Szeto D, Korzus E, Kurokawa R, et al. Signal-specific co-activator domain requirements for Pit-1 activation. Nature. 1998;395:301–306. doi: 10.1038/26270. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Wada O, Suzawa M, Yogiashi Y, Yano T, Kato S, Yanagisawa J. The tamoxifen-responsive estrogen receptor alpha mutant D351Y shows reduced tamoxifen dependent interaction with corepressor complexes. J Biol Chem. 2001;276:42684–42691. doi: 10.1074/jbc.M107844200. [DOI] [PubMed] [Google Scholar]
- Zanger K, Cohen LE, Hashimoto K, Radovick S, Wondisford FE. A novel mechanism for cyclic adenosine 3′,5′-monophosphate regulation of gene expression by CREB-binding protein. Mol Endocrinol. 1999;13:268–275. doi: 10.1210/mend.13.2.0245. [DOI] [PubMed] [Google Scholar]
- Zeng C, McNeil S, Pockwinse S, Nickerson J, Shopland L, Lawrence JB, Penman S, Hiebert S, Lian JB, van Wijnen AJ, et al. Intranuclear targeting of AML/CBFalpha regulatory factors to nuclear matrix-associated transcriptional domains. Proc Natl Acad Sci USA. 1998;95:1585–1589. doi: 10.1073/pnas.95.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Jeyakumar M, Petukhov S, Bagchi MK. A nuclear receptor corepressor modulates transcriptional activity of antagonist-occupied steroid hormone receptor. Mol Endocrinol. 1998;12:513–524. doi: 10.1210/mend.12.4.0089. [DOI] [PubMed] [Google Scholar]
- Zhong S, Salomoni P, Pandolfi PP. The transcriptional role of PML and the nuclear body. Nat Cell Biol. 2000;2:E85–E90. doi: 10.1038/35010583. [DOI] [PubMed] [Google Scholar]


